Abstract
Pontin (also known as RUVBL1 and RVB1) and Reptin (also called RUVBL2 and RVB2) are related members of the large AAA+ (adenosine triphosphatase associated with diverse cellular activities) superfamily of conserved proteins. Various cellular functions depend on Pontin and Reptin, mostly because of their functions in the assembly of protein complexes that play a role in the regulation of cellular energetic metabolism, transcription, chromatin remodeling, and the DNA damage response. Little is known, though, about the interconnections between these multiple functions, how the relevant signaling pathways are regulated, whether the interconnections are affected in human disease, and whether components of these pathways are suitable targets for therapeutic intervention. The First International Workshop on Pontin (RUVBL1) and Reptin (RUVBL2), held between 16 and 19 October 2012, discussed the nature of the oligomeric organization of these proteins, their structures, their roles as partners in various protein complexes, and their involvement in cellular regulation, signaling, and pathophysiology, as well as their potential for therapeutic targeting. A major outcome of the meeting was a general consensus that most functions of Pontin and Reptin are related to their roles as chaperones or adaptor proteins that are important for the assembly and function of large signaling protein complexes.
Introduction
Pontin (also known as RUVBL1, RVB1, Tip49a, ECP-54, Tih1, p50, and Tap54β) and Reptin (RUVBL2, RVB2, Tip49b, ECP-51, Tih2, p47, and Tap54α) came to attention in the late 1990s with their cloning and identification as putative DNA helicases with homology to bacterial RuvB (1, 2). They belong to the AAA+ [adenosine triphosphatase (ATPase) associated with various cellular activities] superfamily of proteins, members of which are characterized by the presence of conserved Walker A and B sequences that are involved in ATP binding and hydrolysis, respectively. It subsequently became clear that these proteins play diverse roles in many essential cellular processes. Because Pontin and Reptin help assemble complexes containing members of the phosphatidylinositol-3 kinase–related kinase (PIKK) family, these proteins are involved in many signaling pathways, including those regulating nutrient sensing, RNA metabolism, and DNA damage repair. How can Pontin and Reptin be involved in the assembly of protein complexes in the cytoplasm as well as the remodeling of chromatin in the nucleus? Although they are most often present in the same complexes, Pontin and Reptin can have opposing functions, notably in the regulation of transcription. Controversies have arisen regarding the nature of their oligomeric organization, which is likely the key to understanding the functions of these proteins. Some of these issues have been discussed in several recent reviews (3–6). The First International Workshop on Pontin (RUVBL1) and Reptin (RUVBL2), held near Bordeaux France, brought together ~60 scientists from 14 countries. At the meeting, these researchers discussed many aspects of Pontin and Reptin structure and function. The meeting was organized by Jean Rosenbaum (Université de Bordeaux, Bordeaux, France), Otmar Huber (Jena University Hospital, Jena, Germany), and Ted Hupp (University of Edinburgh, Edinburgh, UK).
One important objective of the meeting was to adopt a common terminology for referring to Pontin and Reptin, because each is known by up to 10 different names. Though this proved to be a difficult task, a consensus was reached to restrict each to three names: the HUGO terms RUVBL1 and RUVBL2; Pontin and Reptin, originally coined by researchers in the Drosophila melanogaster community and favored by many; and RVB1 and RVB2, the names preferred by those using yeast as a model system. For clarity, only Pontin and Reptin will be used in this Meeting Report.
Anindya Dutta (University of Virginia, Charlottesville, Virginia, USA) offered an overview of the field, pointing out that many groups have described Pontin and Reptin based on their association with other proteins of interest. Pontin and Reptin have now been implicated in functions related to chromatin remodeling complexes, like INO80 or NuA4; stimulation or repression of transcription factors, such as Myc or β-catenin; and assembly of ribonucleoprotein complexes, like small nucleolar ribonucleoproteins (snoRNPs) and telomerase. Consistent with their role as assembly factors or chaperones, Pontin and Reptin are associated with heat shock protein 90 as part of the R2TP complex. Pontin and Reptin have also been implicated in cell transformation, the DNA damage response, apoptosis, and mitosis. After this introduction, Dutta summarized work from his own laboratory demonstrating that the ATPase activities of Pontin and Reptin were equally and independently essential for viability in the budding yeast Saccharomyces cerevisiae. Pontin and Reptin were an integral part of the yeast INO80 complex and are required for its chromatin remodeling activity, most likely because they are required to incorporate actin-related protein 5 (ARP5) into the complex (7). Dutta reported that human Pontin and Reptin were also associated with one another and protected each other from degradation. Further, Pontin and Reptin are part of the multisubunit NuA4 remodeling complex, which plays a key role in both transcription and DNA repair.
Key subunits of NuA4 include Reptin and Pontin, the acetyltransferase TIP60, and the motor ATPase p400. The Pontin and Reptin subunits of NuA4 are essential for the acetyltransferase activity of the TIP60 subunit during DNA repair (8) and also contribute to the assembly and structural organization of the NuA4 complex (9). Knocking down Pontin and Reptin in cultured human cells decreased the acetyltransferase activity of the NuA4 complex, leading to increased longevity and abundance of the phosphorylated form of the histone H2AX after DNA damage. Dutta concluded by enumerating important unanswered questions: Do Pontin and Reptin always work together, or can they function independently in different complexes? Are their ATPase activities required for all their functions? Do they work as hexamers, dodecamers, or simply as dimers?
Biochemistry and Structural Biology
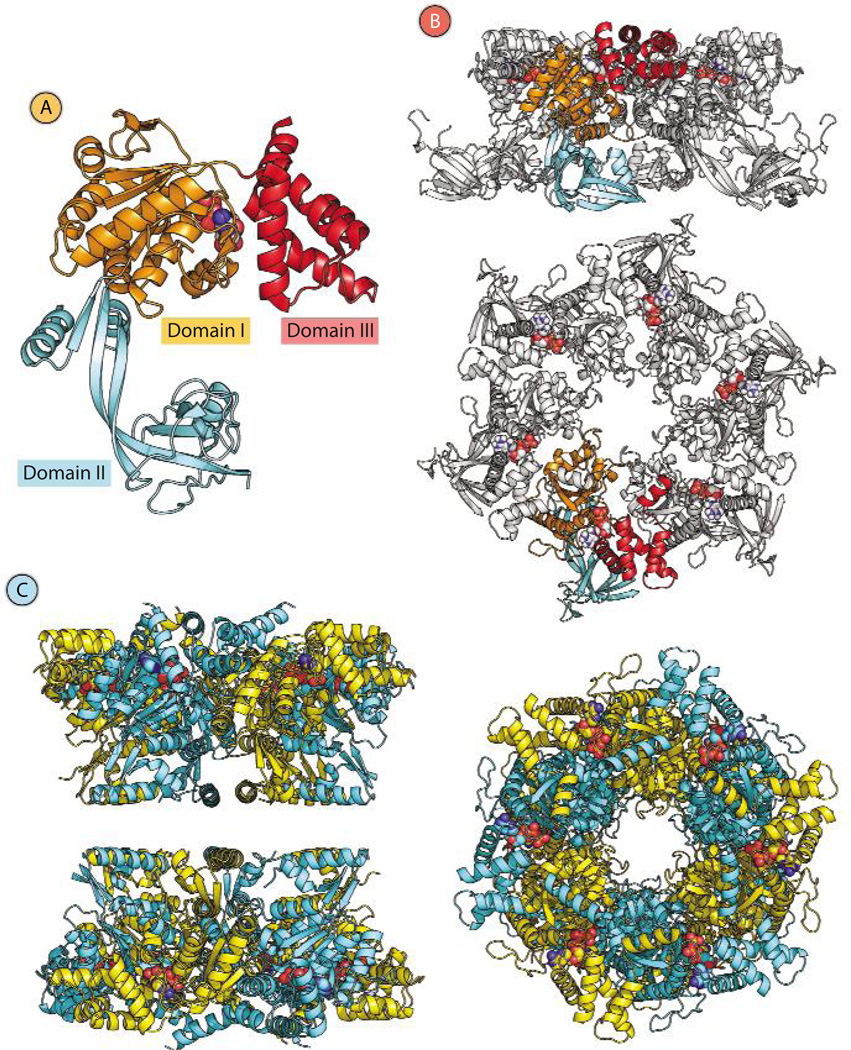
After several years of conflicting results, structural data on human Pontin and Reptin complexes are now converging. Pedro Matias (Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa, Oeiras, Portugal) presented x-ray crystallographic three-dimensional structures of Pontin in complex with Reptin (Fig. 1). Pontin forms hexamers, and each monomer is composed of three domains, with domains I and III cooperating in ATP binding and domain I mediating ATP hydrolysis. The role of domain II is still poorly understood but likely involves interactions with other proteins, DNA, or RNA. The ATPase and helicase activities of Pontin are weak, probably due to a tight-binding ATP pocket, which requires conformational changes to become active (10). Crystals of the Pontin-Reptin complex, obtained only after truncation of the central (second) domain of each protein, revealed a dodecameric structure formed by two heterohexameric rings that contact one another through the truncated second domains. Surprisingly, Pontin and Reptin variants with a truncated domain II displayed greater ATPase and helicase activities than the full-length proteins, supporting the hypothesis that these activities are regulated by domain II, presumably through interactions with other binding partners (11).
Fig. 1.
Structures of the Pontin monomer, the Pontin hexamer, and the Pontin-Reptin dodecamer. (A) Ribbon diagram of the Pontin monomer [Protein Data Bank identification number (PDB ID) 2c9o] showing its domain structure. Domains I and III are involved in binding and hydrolysis of ATP, whereas the function of domain II likely involves interactions with protein partners, DNA, or RNA. A molecule of adenosine diphosphate (ADP) is shown bound to Domain I. (B) Ribbon diagrams of the Pontin hexamer (PDB ID 2c9o). The “side” (upper diagram) and “top” (lower diagram) views are related by a 90° rotation about the horizontal axis. One monomer is shaded according to the domain structure as represented in (A). (C) Ribbon diagrams of the Pontin-Reptin dodecamer with truncated domain II (PDB ID 2xsz). Pontin monomers are shown in yellow; Reptin monomers are shown in light blue. The two views are 90° apart about a horizontal rotation axis. The ADP ligands in panels A and B and the ATP ligands in panel C are represented as spheres with gray for carbon, blue for nitrogen, red for oxygen, and orange for phosphorus.
In parallel, Oscar Llorca’s lab (Centro de Investigaciones Biologicas, Consejo Superior de Investigaciones Cientificas, Madrid, Spain) used cryogenic electron microscopy to obtain a structure of dodecameric complexes of full-length human Pontin and Reptin that proved very similar to that from Matias’s lab (12). In addition, Llorca’s team demonstrated the coexistence of different conformations of the dodecamer within the same preparation, which might affect the ATPase activity. Key residues likely to be required for conformational changes were identified by Mikhail Grigoriev (Université Paul Sabatier, Toulouse, France), Mikhail Petukhov (Saint Petersburg State Polytechnical University, Saint Petersburg, Russia), and their collaborators with the use of molecular dynamics simulations (13). Further analysis of alternative conformations of Pontin and Reptin should contribute to a better understanding of their extraordinarily diverse biological functions.
Nuclear Functions: Transcription and Chromatin Remodeling
Kai Albring and Otmar Huber (Jena University Hospital, Jena, Germany) demonstrated multiple roles of Pontin and Reptin in transcriptional regulation. Albring and Huber previously reported that within the nucleolus, Pontin associates with RNA polymerase I and forms a complex at regulatory sites of ribosomal RNA transcription with c-Myc (14). They now report that the estrogen receptor forms complexes with Pontin or Reptin, or both, at regulatory cis elements in a gene-specific manner. The interaction of Reptin with the estrogen receptor provided Mathieu Dalvai (Laboratoire de Biologie Moléculaire Eucaryote, Université Paul Sabatier, Toulouse, France) with a means to investigate how Reptin is involved in the positive regulation of gene expression through the control of chromatin structure. He showed that the histone variant H2A.Z is present at the transcription start site and at downstream enhancer sequences of the cyclin D1 gene (CCND1 when the gene is poorly transcribed (15). Reptin promoted the release of H2A.Z, which, in turn, reduced the frequency of enhancer-promoter contacts, thereby releasing a repressive intragenic loop and enabling the estrogen receptor to bind the CCND1 promoter.
Sung Hee Baek’s group (Seoul National University, Seoul, South Korea) studied the role of Pontin and Reptin in the regulation of transcription in the context of cancer. Baek’s lab showed how posttranslational modifications of Pontin and Reptin affect their transcriptional activities and proposed a model in which these proteins can have roles independent of one another in the regulation of gene expression. For example, Baek’s team showed that a specific TIP60-Pontin complex (which is distinct from the previously described NuA4 complex) promotes expression of the metastasis suppressor KAI1 in nonmetastatic prostate cancer cells, whereas a β-catenin–Reptin complex represses expression of KAI1 in metastatic cells. Sumoylation of Reptin further stimulates the repressive potential of Reptin by promoting its nuclear localization and increasing its interaction with the histone deacetylase HDAC1 (16, 17), whereas sumoylated Pontin transcriptionally activates androgen receptor–responsive genes and, consequently, increases the proliferation of prostate cancer cells (18). The group also reported that Reptin and Pontin are modified by the lysine methyltransferase G9a during hypoxia. Lysine-methylated Reptin participates in the repression of genes involved in hypoxic signaling and inhibits tumor growth in vivo (19), whereas lysine-methylated Pontin increases the transcriptional activity of the α subunit of hypoxia-inducible factor 1 (HIF1α) and enhances cell proliferation. Genome-wide analysis revealed that Pontin-and Reptin-dependent target genes do not overlap in the cellular response to hypoxia (20).
Esther Marza (University of Bordeaux, Bordeaux, France) studied the regulation of transcription in the context of endoplasmic reticulum (ER) stress. In an in vivo genome-wide RNA interference screen, Marza’s group showed that Caenorhabditis elegans Reptin functionally interacts with the AAA+ family member cell-division cycle protein 48 (CDC48) during ER stress. Marza’s team suggests that CDC48-dependent degradation of Reptin is required to release the inhibition of transcription of ER stress target genes. This mechanism was conserved in mammalian cells.
As mentioned previously, Reptin and Pontin are components of the mammalian NuA4 complex, which plays a key role in regulating DNA repair. Brendan Price (Dana-Farber Cancer Institute, Boston, Massachusetts, USA) demonstrated that the NuA4 remodeling complex exchanged H2A.Z for H2A onto nucleosomes at sites of DNA damage before double-strand break repair. Exchange of H2A.Z required two subunits of NuA4, TIP60 and p400, and was essential for correct processing and repair of DNA damage (21). Furthermore, both the Pontin and Reptin subunits of NuA4 were critical for H2A.Z exchange and played an unexpected role in regulating the activity of the p400 and TIP60 subunits of NuA4, similar to the observed regulation of NuA4 by Reptin and Pontin reported by the Dutta group.
In addition to functioning as a component of NuA4, several isoforms of TIP60 exist that function independently of NuA4 but whose function remains poorly understood. John Lough (Medical College of Wisconsin, Milwaukee, Wisconsin, USA) showed that adult cardiomyocytes exclusively express the TIP60®isoprotein (22). He then showed that upon stress imposed by either overexpression of c-Myc or aortic banding, cell cycle markers increased in cardiomyocytes of TIP60+/− adult mice, consistent with the suggested role for TIP60 in cell cycle regulation (23).
Finally, Wolfram Antonin (Friedrich Miescher Laboratory of the Max Planck Society, Tübingen, Germany) identified yet another function for Pontin and Reptin. He showed that both are required for chromatin decondensation at the end of mitosis in both Xenopus laevis egg extracts and in human HeLa cells. These data indicate that chromatin decondensation is an ATP-dependent process requiring specific cellular machinery rather than the mere inactivation of condensation factors.
Protein-Protein Complex Assembly and Molecular Chaperoning
Walid A. Houry (University of Toronto, Toronto, Canada) presented an overview of the R2TP complex that his group identified (24). This complex consists of Pontin, Reptin, and two heat shock protein 90 (Hsp90) partners: tetratricopeptide repeat domain–containing protein associated with Hsp90 [Tah1 in yeast; known as RNA polymerase II associated protein 3 (RPAP3) or Spagh in mammalian cells] and protein interacting with Hsp90 [Pih1 in yeast; also called nucleolar protein 17, but PIH1 domain–containing 1 (PIH1D1) in mammalian cells]. Makio Saeki (Osaka University, Osaka, Japan) identified the presence of three isoforms of RPAP3 in mammalian cells. Saeki showed that only isoform 1 is part of the R2TP complex and stabilizes PIH1D1 (25). Houry also presented the recently solved nuclear magnetic resonance structure of Tah1 (26). His group, in collaboration with Joaquin Ortega’s group (McMaster University, Hamilton, Canada), showed that yeast Pontin and Reptin form a heterohexameric complex (27, 28). A pathway for the assembly of the box C/D class of snoRNPs, as mediated by Hsp90-R2TP, was proposed. In parallel, Tom Meier (Albert Einstein College of Medicine, New York, USA) provided important clues about the molecular role of Pontin and Reptin in the biogenesis of the H/ACA class of RNPs that includes telomerase, showing that Pontin and Reptin function in separating the pseudouridine synthase NAP57 (dyskerin) from the assembly factor SHQ1 (29).
Bérengère Pradet-Balade and colleagues (Institut de Génétique Moléculaire, Montpellier, France) have also demonstrated a role for the R2TP complex in snoRNP assembly (30) and on the cytoplasmic assembly of RNA polymerase II (31). They now show that the pathway is conserved in Drosophila, establishing it as a powerful system to investigate new R2TP clients.
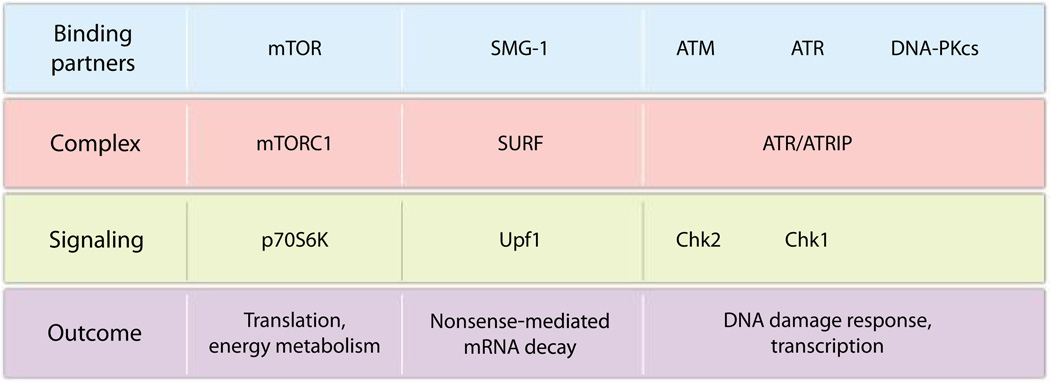
Shigeo Ohno (Yokohama City University School of Medicine, Yokohama, Japan) showed that Pontin and Reptin, together with the other R2TP components, interact with many PIKK family members, including ataxia telangiectasia mutated (ATM), ataxia telangiectasia Rad3-related (ATR), DNA-dependent protein kinase catalytic subunit (DNA-PKcs), suppressor with morphogenetic effect on genitalia-1 (SMG-1), transformation/transcription domain–associated protein, and the mechanistic target of rapamycin (mTOR) (32). Depletion of Pontin or Reptin in cultured human cells destabilizes PIKKs, leading to dramatic phenotypic effects— notably, a defect in SMG-1–dependent non-sense-mediated mRNA decay. Data published after this meeting show that Pontin and Reptin are also required for the association of mTOR with lysosomes (33). The central position of PIKKs in several major signaling pathways establishes Pontin and Reptin as important actors in cell signaling (Fig. 2).
Fig. 2.
Pontin and Reptin as major actors in cell signaling. Pontin and Reptin are required for the stabilization of every PIKK family member and for the assembly of supramolecular complexes involving PIKKs. Their functional role has been established by showing decreased activation of PIKK effectors upon Pontin or Reptin silencing, with consequences on biological outcomes. Pontin and Reptin help assemble the mTOR complex 1 (mTORC1), thus modulating signaling through the p70 ribosomal S6 kinase (p70S6K) and thereby affecting cellular processes that include protein translation and energy metabolism. Similarly, Pontin and Reptin stabilize SMG-1 that will be incorporated into the SMG-1, Upf1, eRF1, and eRF3 (SURF) complex, enabling SMG-1 to phosphorylate Upf1 and leading to nonsense-mediated decay of mRNAs. Pontin and Reptin also stabilize ATM, ATR, and DNA-PKcs and help assemble the ATR-ATR interacting protein (ATR-ATRIP) complex, which promotes DNA damage response signaling.
Although separate from their chromatin modification and transcriptional regulatory functions, it is clear that the chaperone functions of Pontin and Reptin also probably affect nuclear processes due to their role in the assembly of nuclear complexes, such as INO80 and NuA4-TIP60, as highlighted by Anindya Dutta. Whether this only happens when Pontin and Reptin are part of the R2TP complex and whether R2TP also has clients in addition to those already identified remain open questions.
Pathophysiology and Therapeutic Targeting
Jean Rosenbaum’s team (University of Bordeaux, Bordeaux, France) demonstrated that the genes encoding Pontin and Reptin were overexpressed in human liver cancer, where they were required for growth and viability of tumor cells both in vitro and in vivo (34–36). Rosenbaum’s group now shows that the ATPase activity of Reptin is required for sustaining tumor cell growth, suggesting that Reptin could be a viable drug target for cancer therapy (37). In collaboration with Michel Laguerre’s group (Institut Européen de Chimie et de Biologie, Pessac, France), Patrick Lestienne, from the Rosenbaum group, presented small molecule inhibitors of Pontin’s ATPase activity identified through in silico modeling and in vitro testing (38).
Pontin and Reptin are required for the assembly of telomerase (39, 40). An alternative mechanism (ALT) to maintain telomeres instead of telomerase activity, already long known to be used in some tumor cells, has recently been shown to occur in normal mammalian somatic cells also (41). Klaus Holzmann (Medizinische Universität Wien, Vienna, Austria) reported that Pontin, but not Reptin, transcripts were reduced in ALT as compared with non-ALT tumor cell lines. Although Pontin was generally over-expressed in colorectal cancer (CRC) (42), preliminary data from human specimens of CRC with ALT showed reduced expression of Pontin in tumors compared with nontumor tissues. This change may influence the activity of Pontin- and Reptin-containing complexes in several contexts—notably in the DNA damage response.
Oxana Bereshchenko (University of Perugia, Italy) reported on the function of Pontin in an intact mammalian organism. Deletion of Pontin in mice resulted in embryonic lethality due to a loss of embryonic stem (ES) cell function. The defect was not characterized further, but no Pontin knock-out colonies were detected in ES cell outgrowth cultures in vitro. Conditional ablation of Pontin in hematopoietic tissues led to bone marrow failure characterized by the apoptotic loss of hematopoietic stem cells in adult mice. Thus, Pontin is critical for the function of both embryonic stem cells and adult hematopoietic stem cells (43). Further in vivo studies will be important to better define the physiological and pathophysiological functions of Pontin and Reptin.
Ted Hupp (University of Edinburgh, Edinburgh, UK) previously demonstrated an interaction between anterior gradient-2 (AGR2) and Reptin (44). AGR2 is a component of a pro-oncogenic signaling pathway that mediates cellular transformation, ER homeostasis, and p53 inhibition. Hupp demonstrated an interaction between AGR2, Reptin, and p53 that is mediated by the p53 tetramerization domain and stimulated by AGR2. This interaction defines a previously unidentified pathway for suppressing the activity of p53.
In contrast to Pontin and Reptin, the two RuvB-like proteins found in humans and yeast, the malaria parasite Plasmodium falciparum contains three RuvB-related proteins. A phylogenetic analysis presented by Moaz Ahmad (International Centre for Genetic Engineering and Biotechnology, New Delhi, India) revealed that PfRuvB1 and PfRuvB2 are most similar to Pontin, whereas PfRuvB3 is more closely related to Reptin (45). Further studies are required to explore the roles of these proteins and why this parasite requires three such proteins.
Conclusions
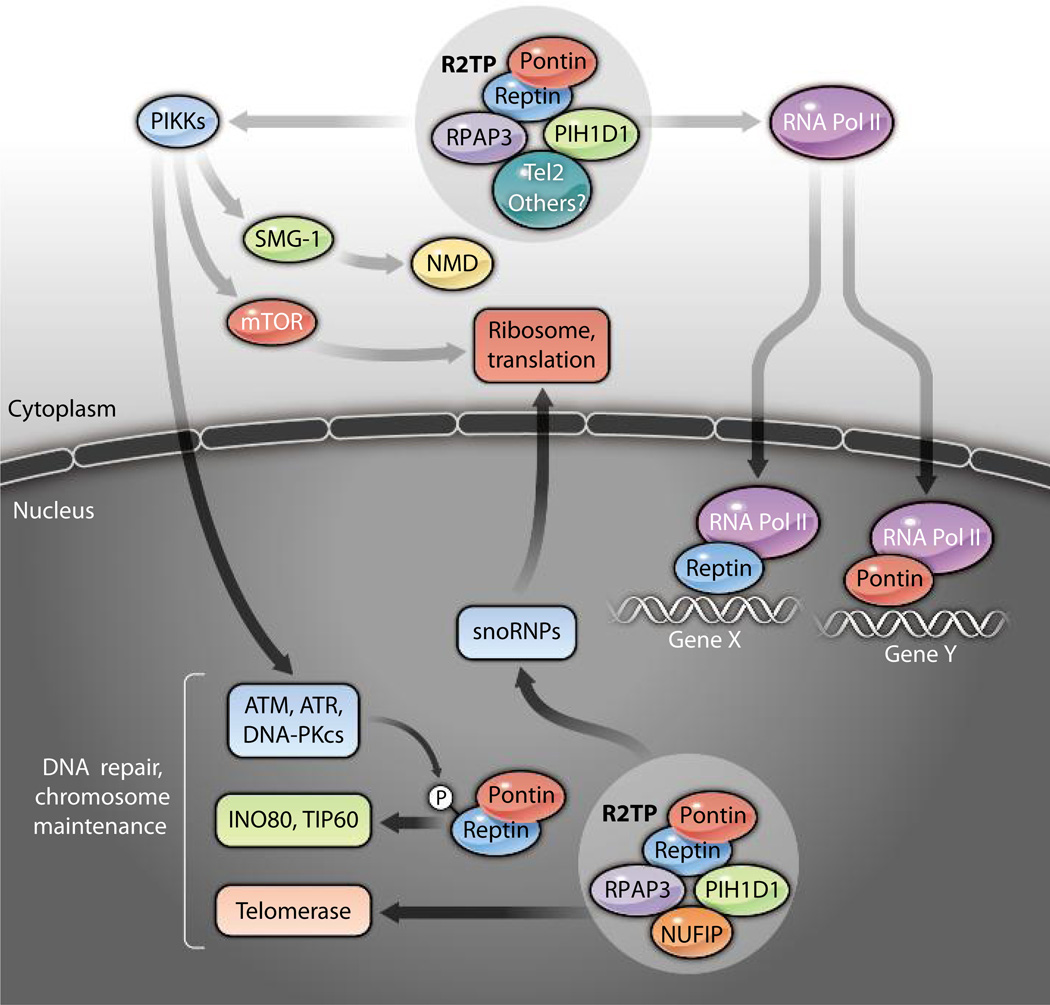
This First International Workshop on Pontin (RUVBL1) and Reptin (RUVBL2) has met its objectives, allowing researchers working independently on these proteins to meet and exchange ideas across diverse disciplines in the life sciences. The emerging view is that the diverse and largely overlapping functions of these two closely related proteins result from a highly diverse set of protein-protein interactions that are likely controlled, in part, by the oligomeric nature of Pontin and Reptin and the possibility that these proteins act as molecular scaffolds that regulate the assembly and disassembly of protein complexes (Fig. 3). The diverse nature of these protein-protein interactions and their rewiring in pathophysiological states may impact the development of diseases such as cancer. Still, many issues remain unsettled, leaving ample room for discussions in the next meeting to be held in Portugal in 2014.
Fig. 3.
Major functions of human Pontin and Reptin. In the cytoplasm, Pontin (red ovals) and Reptin (blue ovals) associate with RPAP3 and PIH1D1 to form the R2TP complex. This complex associates with the adaptor Tel2 and helps stabilize all members of the PIKK family, such as SMG-1 [which is involved in nonsense-mediated mRNA decay (NMD)]; mTOR (which regulates translation); and ATM, ATR, and DNA-PKcs (which are involved in DNA damage repair). Likely cooperating with another unidentified adaptor, the R2TP complex assembles RNA polymerase II (RNA Pol II) in the cytosol. To participate in the biogenesis of snoRNPs and telomerase, the nuclear R2TP complex associates with the adaptor nuclear fragile X mental retardation–interacting protein 1 (NUFIP) instead of Tel2 (30). In the nucleus, Pontin and Reptin are also involved in the assembly of INO80 and NuA4-TIP60 complexes, but it is unknown whether additional partners are required. A large-scale study has shown that Reptin is phosphorylated (P) on an ATM/ATR consensus site after DNA damage (46); thus far, the functional consequences are unknown. Pontin and Reptin also regulate gene transcription in association with RNA Pol II. Transcriptional regulation is the only context in which Pontin and Reptin appear to function separately.
Acknowledgments
Funding: J.R. was supported by the Equipe Labélisée Ligue Contre le Cancer 2011 and the National Cancer Institute (grant PLBIO10-155); S.H.B. was supported by the Creative Research Initiatives Program (Research Center for Chromatin Dynamics, grant 2009-0081563); A.D. was supported by the NIH (grant R01 GM084465); W.A.H. was supported by the Canadian Institutes of Health Research (grant MOP-93778); O.H. was supported by the Deutsche Forschungsgemeinschaft (grants SFB366-C13 and HU881/6-1); T.R.H. was supported by the Medical Research Council UK, the Biotechnology and Biological Sciences Research Council, and Cancer Research UK; and P.M.M. was supported by the European Synchrotron Research Facility (Grenoble, France), Bayer (Berlin, Germany), and SPINE2-COMPLEXES project LSHG-CT-2006-031220 (principal investigator: M. A. Carrondo).
References and Notes
- 1.Kanemaki M, Makino Y, Yoshida T, Kishimoto T, Koga A, Yamamoto K, Yamamoto M, Moncollin V, Egly JM, Muramatsu M, Tamura T. Molecular cloning of a rat 49-kDa TBP-interacting protein (TIP49) that is highly homologous to the bacterial RuvB. Biochem Biophys Res Commun. 1997;235:64–68. doi: 10.1006/bbrc.1997.6729. [DOI] [PubMed] [Google Scholar]
- 2.Kanemaki M, Kurokawa Y, Matsu-ura T, Makino Y, Masani A, Okazaki K, Morishita T, Tamura TA. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J Biol Chem. 1999;274:22437–22444. doi: 10.1074/jbc.274.32.22437. [DOI] [PubMed] [Google Scholar]
- 3.Huber O, Menard L, Haurie V, Nicou A, Taras D, Rosenbaum J. Pontin and reptin, two related ATPases with multiple roles in cancer. Cancer Res. 2008;68:6873–6876. doi: 10.1158/0008-5472.CAN-08-0547. [DOI] [PubMed] [Google Scholar]
- 4.Jha S, Dutta A. RVB1/RVB2: running rings around molecular biology. Mol Cell. 2009;34:521–533. doi: 10.1016/j.molcel.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huen J, Kakihara Y, Ugwu F, Cheung KL, Ortega J, Houry WA. Rvb1-Rvb2: essential ATP-dependent helicases for critical complexes. Biochem Cell Biol. 2010;88:29–40. doi: 10.1139/o09-122. [DOI] [PubMed] [Google Scholar]
- 6.Grigoletto A, Lestienne P, Rosenbaum J. The multifaceted proteins Reptin and Pontin as major players in cancer. Biochim Biophys Acta. 2011;31:91–103. doi: 10.1016/j.bbcan.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Jonsson ZO, Jha S, Wohlschlegel JA, Dutta A. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol Cell. 2004;16:465–477. doi: 10.1016/j.molcel.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 8.Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol. 2008;28:2690–2700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jha S, Gupta A, Dar A, Dutta A. RVBs are required for assembling a functional TIP60 complex. Mol Cell Biol. doi: 10.1128/MCB.01567-12. Published 2013 as doi: 10.1128/MCB.01567-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matias PM, Gorynia S, Donner P, Carrondo MA. Crystal structure of the human AAA+ protein RuvBL1. J Biol Chem. 2006;281:38918–38929. doi: 10.1074/jbc.M605625200. [DOI] [PubMed] [Google Scholar]
- 11.Gorynia S, Bandeiras TM, Pinho FG, McVey CE, Vonrhein C, Round A, Svergun DI, Donner P, Matias PM, Carrondo MA. Structural and functional insights into a dodecameric molecular machine - The RuvBL1/RuvBL2 complex. J Struct Biol. 2011;176:279–291. doi: 10.1016/j.jsb.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Perrote A, Munoz-Hernandez H, Gil D, Llorca O. Conformational transitions regulate the exposure of a DNA-binding domain in the RuvBL1-RuvBL2 complex. Nucleic Acids Res. 2012;40:11086–11099. doi: 10.1093/nar/gks871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petukhov M, Dagkessamanskaja A, Bommer M, Barrett T, Tsaneva I, Yakimov A, Queval R, Shvetsov A, Khodorkovskiy M, Kas E, Grigoriev M. Large-Scale Conformational Flexibility Determines the Properties of AAA+ TIP49 ATPases. Structure. 2012;20:1321–1331. doi: 10.1016/j.str.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Cvackova Z, Albring KF, Koberna K, Ligasova A, Huber O, Raska I, Stanek D. Pontin is localized in nucleolar fibrillar centers. Chromosoma. 2008;117:487–497. doi: 10.1007/s00412-008-0170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalvai M, Bellucci L, Fleury L, Lavigne AC, Moutahir F, Bystricky K. H2A.Z-dependent crosstalk between enhancer and promoter regulates Cyclin D1 expression. Oncogene. 2012 doi: 10.1038/onc.2012.442. [DOI] [PubMed] [Google Scholar]
- 16.Kim JH, Choi HJ, Kim B, Kim MH, Lee JM, Kim IS, Lee MH, Choi SJ, Kim KI, Kim SI, Chung CH, Baek SH. Roles of sumoylation of a reptin chromatin-remodelling complex in cancer metastasis. Nat Cell Biol. 2006;8:631–639. doi: 10.1038/ncb1415. [DOI] [PubMed] [Google Scholar]
- 17.Kim JH, Kim B, Cai L, Choi HJ, Ohgi KA, Tran C, Chen C, Chung CH, Huber O, Rose DW, Sawyers CL, Rosenfeld MG, Baek SH. Transcriptional regulation of a metastasis suppressor gene by Tip60 and beta-catenin complexes. Nature. 2005;434:921–926. doi: 10.1038/nature03452. [DOI] [PubMed] [Google Scholar]
- 18.Kim JH, Lee JM, Nam HJ, Choi HJ, Yang JW, Lee JS, Kim MH, Kim SI, Chung CH, Kim KI, Baek SH. SUMOylation of pontin chromatin-remodeling complex reveals a signal integration code in prostate cancer cells. Proc Natl Acad Sci U S A. 2007;104:20793–20798. doi: 10.1073/pnas.0710343105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JS, Kim Y, Kim IS, Kim B, Choi HJ, Lee JM, Shin HJ, Kim JH, Kim JY, Seo SB, Lee H, Binda O, Gozani O, Semenza GL, Kim M, Kim KI, Hwang D, Baek SH. Negative regulation of hypoxic responses via induced Reptin methylation. Mol Cell. 2010;39:71–85. doi: 10.1016/j.molcel.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JS, Kim Y, Bhin J, Shin HJ, Nam HJ, Lee SH, Yoon JB, Binda O, Gozani O, Hwang D, Baek SH. Hypoxia-induced methylation of a pontin chromatin remodeling factor. Proc Natl Acad Sci U S A. 2011;108:13510–13515. doi: 10.1073/pnas.1106106108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z Controls a Critical Chromatin Remodeling Step Required for DNA Double-Strand Break Repair. Mol Cell. 2012;48:723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher JB, Kim MS, Blinka S, Ge ZD, Wan T, Duris C, Christian D, Twaroski K, North P, Auchampach J, Lough J. Stress-induced cell-cycle activation in Tip60 haploinsufficient adult cardiomyocytes. PLoS One. 2012;7:e31569. doi: 10.1371/journal.pone.0031569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, Confalonieri S, Cesaroni M, Marchesi F, Gasco M, Scanziani E, Capra M, Mai S, Nuciforo P, Crook T, Lough J, Amati B. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–1067. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- 24.Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, Boone C, Emili A, Houry WA. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida M, Saeki M, Egusa H, Irie Y, Kamano Y, Uraguchi S, Sotozono M, Niwa H, Kamisaki Y. RPAP3 splicing variant isoform 1 interacts with PIH1D1 to compose R2TP complex for cell survival. Biochemical and Biophysical Research Communications. 2012 doi: 10.1016/j.bbrc.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 26.Jimenez B, Ugwu F, Zhao R, Orti L, Makhnevych T, Pineda-Lucena A, Houry WA. Structure of minimal tetratricopeptide repeat domain protein Tah1 reveals mechanism of its interaction with Pih1 and Hsp90. J Biol Chem. 2012;287:5698–5709. doi: 10.1074/jbc.M111.287458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung KL, Huen J, Kakihara Y, Houry WA, Ortega J. Alternative oligomeric states of the yeast Rvb1/Rvb2 complex induced by histidine tags. J Mol Biol. 2010;404:478–492. doi: 10.1016/j.jmb.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gribun A, Cheung KL, Huen J, Ortega J, Houry WA. Yeast Rvb1 and Rvb2 are ATP-dependent DNA helicases that form a heterohexameric complex. J Mol Biol. 2008;376:1320–1333. doi: 10.1016/j.jmb.2007.12.049. [DOI] [PubMed] [Google Scholar]
- 29.Machado-Pinilla R, Liger D, Leulliot N, Meier UT. Mechanism of the AAA+ ATPases pontin and reptin in the biogenesis of H/ACA RNPs. RNA. 2012;18:1833–1845. doi: 10.1261/rna.034942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boulon S, Marmier-Gourrier N, Pradet-Balade B, Wurth L, Verheggen C, Jady BE, Rothe B, Pescia C, Robert MC, Kiss T, Bardoni B, Krol A, Branlant C, Allmang C, Bertrand E, Charpentier B. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J Cell Biol. 2008;180:579–595. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, Georgieva M, Azzag K, Robert MC, Ahmad Y, Neel H, Lamond AI, Bertrand E. HSP90 and Its R2TP/Prefoldin-like Cochaperone Are Involved in the Cytoplasmic Assembly of RNA Polymerase II. Mol Cell. 2010;39:912–924. doi: 10.1016/j.molcel.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izumi N, Yamashita A, Iwamatsu A, Kurata R, Nakamura H, Saari B, Hirano H, Anderson P, Ohno S. AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay. Sci Signal. 2010;3:ra27. doi: 10.1126/scisignal.2000468. [DOI] [PubMed] [Google Scholar]
- 33.Kim SG, Hoffman GR, Poulogiannis G, Buel GR, Jang YJ, Lee KW, Kim BY, Erikson RL, Cantley LC, Choo AY, Blenis J. Metabolic Stress Controls mTORC1 Lysosomal Localization and Dimerization by Regulating the TTT-RUVBL1/2 Complex. Mol Cell. doi: 10.1016/j.molcel.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haurie V, Menard L, Nicou A, Touriol C, Metzler P, Fernandez J, Taras D, Lestienne P, Balabaud C, Bioulac-Sage P, Prats H, Zucman-Rossi J, Rosenbaum J. Adenosine triphosphatase pontin is overexpressed in hepatocellular carcinoma and coregulated with reptin through a new posttranslational mechanism. Hepatology. 2009;50:1871–1883. doi: 10.1002/hep.23215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menard L, Taras D, Grigoletto A, Haurie V, Nicou A, Dugot-Senant N, Costet P, Rousseau B, Rosenbaum J. In vivo silencing of Reptin blocks the progression of human hepatocellular carcinoma in xenografts and is associated with replicative senescence. J Hepatol. 2010;52:681–689. doi: 10.1016/j.jhep.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 36.Rousseau B, Menard L, Haurie V, Taras D, Blanc J, Moreau-Gaudry F, Metzler P, Hugues M, Boyault S, Lemiere S, Canron X, Costet P, Cole M, Balabaud C, Bioulac-Sage P, Zucman-Rossi J, Rosenbaum J. Overexpression and role of the ATPase and putative DNA helicase RuvB-like 2 in human hepatocellular carcinoma. Hepatology. 2007;46:1108–1118. doi: 10.1002/hep.21770. [DOI] [PubMed] [Google Scholar]
- 37.Grigoletto A, Neaud V, Allain-Courtois N, Lestienne P, Rosenbaum J. The ATPase activity of Reptin is required for its effects on tumor cell growth and viability in hepatocellular carcinoma. Mol Cancer Res. doi: 10.1158/1541-7786.MCR-12-0455. Published 2012 as DOI:10.1158/1541-7786.MCR-12-0455. [DOI] [PubMed] [Google Scholar]
- 38.Elkaim J, Castroviejo M, Bennani D, Taouji S, Allain N, Laguerre M, Rosenbaum J, Dessolin J, Lestienne P. First identification of small molecule inhibitors of Pontin by combining virtual screening and enzymatic assay. Biochem J. 2012;443:449–459. doi: 10.1042/BJ20111779. [DOI] [PubMed] [Google Scholar]
- 39.Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baek SH. When ATPases pontin and reptin met telomerase. Dev Cell. 2008;14:459–461. doi: 10.1016/j.devcel.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 41.Neumann AA, Watson CM, Noble JR, Pickett HA, Tam PP, Reddel RR. Alternative lengthening of telomeres in normal mammalian somatic cells. Genes Dev. 2013;27:18–23. doi: 10.1101/gad.205062.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lauscher JC, Loddenkemper C, Kosel L, Grone J, Buhr HJ, Huber O. Increased pontin expression in human colorectal cancer tissue. Hum Pathol. 2007;38:978–985. doi: 10.1016/j.humpath.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 43.Bereshchenko O, Mancini E, Luciani L, Gambardella A, Riccardi C, Nerlov C. Pontin is essential for murine hematopoietic stem cell survival. Haematologica. 2012;97:1291–1294. doi: 10.3324/haematol.2011.060251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maslon MM, Hrstka R, Vojtesek B, Hupp TR. A Divergent Substrate-Binding Loop within the Pro-oncogenic Protein Anterior Gradient-2 forms a Docking Site for Reptin. J Mol Biol. 2010;404:418–438. doi: 10.1016/j.jmb.2010.09.035. [DOI] [PubMed] [Google Scholar]
- 45.Ahmad M, Singh S, Afrin F, Tuteja R. Novel RuvB nuclear ATPase is specific to intraerythrocytic mitosis during schizogony of Plasmodium falciparum. Mol Biochem Parasitol. 2012;185:58–65. doi: 10.1016/j.molbiopara.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]