Abstract
Nonmuscle myosin-II is an actin-based motor that converts chemical energy into force and movement, and thus functions as a key regulator of the eukaryotic cytoskeleton. Although it is established that phosphorylation on the regulatory light chain increases the actin-activated MgATPase activity of the motor and promotes myosin-II filament assembly, studies have begun to characterize alternative mechanisms that regulate filament assembly and disassembly. These investigations have revealed that all three nonmuscle myosin-II isoforms are subject to additional regulatory controls, which impact diverse cellular processes. In this review, we discuss current knowledge on mechanisms that regulate the oligomerization state of nonmuscle myosin-II filaments by targeting the myosin heavy chain.
Keywords: nonmuscle myosin-II, NMHC-IIA, NMHC-IIB, NMHC-IIC, phosphorylation, S100A4, Lgl, filament assembly
Introduction
Myosins constitute a large and diverse superfamily of motor proteins that bind actin filaments to produce force and tension. The overall domain organization of the family is characterized by a motor domain, which binds to F-actin, a neck domain composed of variable numbers of IQ motifs that bind light chains, and a tail domain of variable length and function.1 The founding member of the myosin superfamily is the class II or conventional myosin, whose actin-activated ATPase activity was described over 70 y ago.2 Class II myosins are the primary contractile proteins in smooth, skeletal and cardiac muscles, and also perform similar functions in eukaryotic nonmuscle cells. The myosin-II molecule is a hexamer composed of two myosin-II heavy chains (MHC-II), two essential light chains (ELCs) that stabilize the neck domain and two regulatory light chains (RLCs) that stabilize the neck and regulate motor activity.1 This hexameric molecule is generally referred to as the myosin-II monomer. With respect to the domain structure of the myosin-II molecule, each individual heavy chain consists of (1) a globular N-terminal motor or head domain (~800 amino acids) that contains the ATP and actin binding sites; (2) a neck domain (~50 amino acids) composed of two IQ motifs that bind one ELC and one RLC; (3) a coiled coil or rod domain (~1100 amino acids) composed of heptad repeats; and (4) a short C-terminal segment, termed the tailpiece of ~34–47 amino acids in length (Fig. 1). The rod domain mediates the association of the two myosin-II heavy chains through the formation of a long, parallel α-helical coiled coil and it is this interaction that underlies the hexameric organization of the myosin monomer. In addition to directing the assembly of the hexameric myosin monomer, intermolecular interactions between the coiled coils of myosin-II molecules are crucial for the formation of myosin-II filaments. The assembly of myosin-II bipolar filaments occurs via both parallel and anti-parallel intermolecular interactions between myosin-II monomers. For nonmuscle myosin-II, these filaments are composed of ~28 hexameric units (i.e., monomers), and are considerably smaller than those observed in skeletal and smooth muscle.3,4
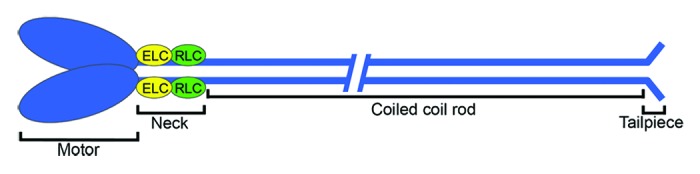
Figure 1. Domain organization of myosin-II. Each myosin-II heavy chain is composed of an N-terminal motor domain, a neck domain that binds one essential light chain (yellow) and one regulatory light chain (green), a coiled coil rod domain and a C-terminal tailpiece. The head domain contains the ATP binding site and the actin binding domain. The two myosin-II heavy chains dimerize via interactions between the rod domains, which form an α-helical coiled coil. This hexameric assembly is referred to as the myosin-II monomer.
In vertebrates, there are three nonmuscle myosin heavy chain genes (Myh9, Myh10 and Myh14) that encode NMHC-IIA, NMHC-IIB and NMHC-IIC, respectively.5-9 The three NMHC-II isoforms are highly conserved with 79% amino acid identity between NMHC-IIA and NMHC-IIB, 65% identity between NMHC-IIA and NMHC-IIC, and 68% identity between NMHC-IIB and NMHC-IIC, with the majority of the amino acid differences occurring within the C-terminal tailpiece. Despite the overall sequence similarity between the heavy chains, the three NMHC-II isoforms have different actin-activated MgATPase activities and duty ratios (the fraction of time that the myosin motor is bound to an actin filament),10-13 exhibit distinct patterns of tissue and cell expression,9,14 and have non-redundant as well as overlapping functional roles in vivo (for a review see refs. 15, 16). In addition, the myosin-II isoforms can exhibit distinct subcellular distributions, which are mediated by the C-terminal ~180 amino acids of the myosin-II heavy chain that includes the C-terminal end of the coiled coil and the tailpiece.17,18 These observations highlight the importance of the myosin-II tail in modulating the spatiotemporal localization of myosin-II molecules and further suggest that regulation via the tail could modulate myosin-II localization and/or activity. In this review, we discuss the regulation of myosin-II filament assembly with a particular emphasis on mechanisms that target the heavy chains of nonmuscle myosin-II.
Regulation of Filament Assembly by RLC Phosphorylation
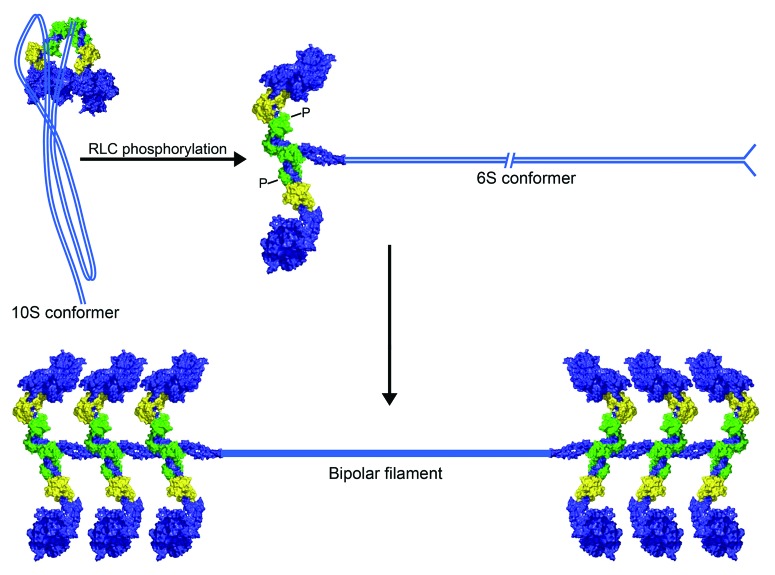
In the fully dephosphorylated state, the myosin-II molecule adopts a compact, folded structure referred to as 10S myosin based on its sedimentation coefficient, which differs from that of the extended myosin-II monomer (6S myosin).19,20 The inactive monomer is characterized by asymmetric head-head interactions in the two-headed myosin molecule that block the interaction of one head with F-actin and of the second head with nucleotide (Fig. 2).21,22 Electron microscopy studies indicate that folding of the coiled coil rod domain into three segments, which permits head-tail interactions, stabilizes the compact monomer conformation and prevents filament assembly.19,23-27 While under certain in vitro conditions nonphosphorylated myosin-II can polymerize, these filaments are highly susceptible to MgATP, which disassembles them into folded monomers.19,23,28 Phosphorylation on Ser19 of the regulatory light chain (RLC) by myosin light chain kinase or Rho kinase relieves the autoinhibited state and promotes the assembly competent, extended 6S conformation.29,30 In addition, RLC phosphorylation decreases the critical monomer concentration required for filament assembly and protects against MgATP-induced filament disassembly, thus providing a mechanism for filament stabilization in vivo, where intracellular ATP concentrations are high.28,31 Lastly, there is increasing evidence that interconversion between these two conformers regulates the assembly of smooth muscle and nonmuscle myosin-II filaments in vivo.32-34 For example, a myosin-IIA mutant that is incapable of forming a stable 10S monomer shows poor localization to the edge of spreading cells as compared with wild-type myosin-IIA and over-assembles in the central cell body.32 These data suggest that the 10S conformer contributes to both the proper localization and assembly of myosin-IIA.
Figure 2. Conformation of 10S and 6S myosin-II. The myosin-II motor and neck domains are shown in blue and the associated essential and regulatory light chains (ELCs and RLCs) are shown in yellow and green, respectively. In the absence of RLC phosphorylation, head-tail interactions stabilize the compact, assembly-incompetent 10S conformation. Phosphorylation on Ser19 of the RLC promotes unfolding of the tail to the assembly competent, extended 6S conformation. The conversion to the extended conformation also triggers the MgATPase activity of the motor and myosin-II assembly into bipolar filaments. Filament assembly requires both parallel and anti-parallel interactions between myosin-II monomers.
Heavy Chain Requirements for Filament Assembly
While RLC phosphorylation regulates the conformation of the myosin-II monomer, it is the α-helical coiled coil rod domain and C-terminal tailpiece that mediate the assembly of myosin-II into filaments. The myosin-II rod domain is characterized by a 28 amino acid repeat along the length of the α-helical coiled coil that produces a pattern of alternating positive and negative charges that mediate the axial staggering between myosin-II molecules within a filament.35,36 In skeletal myosin-II, a 29-residue sequence near the C-terminus of the coiled coil was identified as a critical region for myosin-II filament assembly.37 This assembly competence domain or ACD is part of a longer conserved region present in all type II myosins (termed the extended ACD) (Fig. 3, ACD1).38 The extended ACD lacks the 28-residue periodicity of positive and negative charge that is characteristic of most of the myosin-II coiled coil, and instead exhibits a unique electrostatic profile that is composed of short alternating segments of negative and positive charge (Fig. 3). The extended ACD also contains a high proportion of solvent exposed large apolar residues.38 Deletion studies have confirmed the requirement of the extended ACD for both the in vitro and in vivo assembly of nonmuscle myosin-II filaments.17,39-41 In addition, studies on myosin-IIB have identified a second ACD (ACD2), which is poorly conserved among sarcomeric myosin-II molecules, that is upstream of the extended ACD (ACD1) (Fig. 3).39 Electrostatic interactions between these two regions of the myosin-II heavy chain are thought to regulate both parallel and anti-parallel interactions between myosin-II monomers during filament assembly, although this has yet to be formally tested.
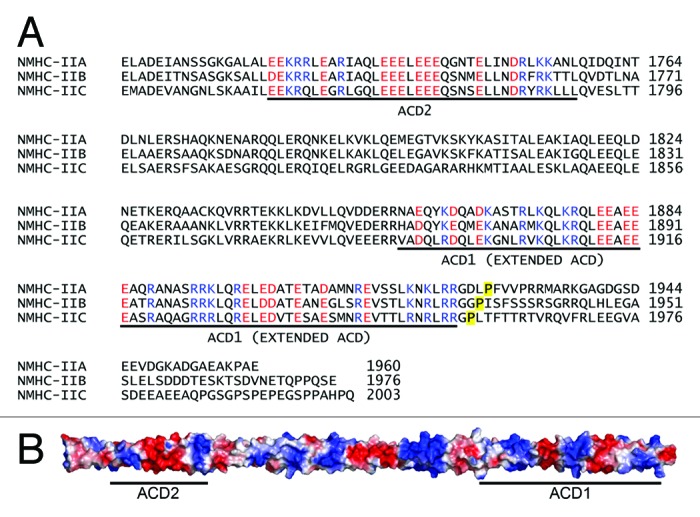
Figure 3. Assembly competence domains in nonmuscle myosin-II. (A) Alignment of the human nonmuscle myosin-II heavy chains (Myh9: NP_002464; Myh10: NP_005955.3; and Myh14: NP_001070654). The positively and negatively charged residues in ACD1 (extended ACD) and ACD2 are highlighted blue and red, respectively. The beginning of the C–terminal tailpiece is denoted by the yellow highlighted proline. (B) Structural model of the NMHC-IIA coiled coil (residues Glu1705-Leu1926). The protein surface is colored according to the electrostatic potential. Positively charged regions are blue, negatively charged regions are red, and neutral regions are white. The regions corresponding to ACD1 and ACD2 are denoted with a line.
In addition to ACD1 and ACD2, the C-terminal tailpiece, which is ~34–47 amino acids in length depending on the NMHC-II isoform, is also critical for filament assembly. In vitro studies with myosin-II constructs comprising two-thirds of the coiled coil domain, which contain all known heavy chain assembly determinants, and in vivo studies with GFP-tagged full-length NMHC-IIA, demonstrated that deletion of the 35 amino acid tailpiece from myosin-IIA enhances myosin-II assembly.32,41 Similar to myosin-IIA, deletion of the myosin-IIB tailpiece increases assembly, whereas removal of the tailpiece reduces myosin-IIC assembly.41 These investigations, as well as tailpiece swapping studies, have established the C-terminal tailpiece as an important mediator of isoform-specific nonmuscle myosin-II filament assembly.41,42 In addition, studies on myosin-IIC have shown that in conjunction with the C-terminal region of the extended ACD (ACD1), the tailpiece directly binds the coiled coil to mediate the association between myosin-IIC molecules.43 Whether the tailpiece has a similar function in myosin-IIA and myosin-IIB will require further investigation.
Regulation of Filament Assembly by Heavy Chain Phosphorylation
Although heavy chain phosphorylation was observed more than 30 y ago in vertebrates,44-46 RLC phosphorylation has been considered the primary mechanism for controlling myosin-II filament assembly in vivo. In Dictyostelium it is well established that heavy chain phosphorylation induces an assembly-incompetent, bent monomeric conformation that inhibits filament oligomerization.47,48 In contrast, the structural and biological consequences of heavy chain phosphorylation in vertebrates are still being defined. Nonetheless biochemical and cellular studies have suggested that heavy chain phosphorylation provides another physiologically relevant regulatory mechanism for myosin-II assembly.
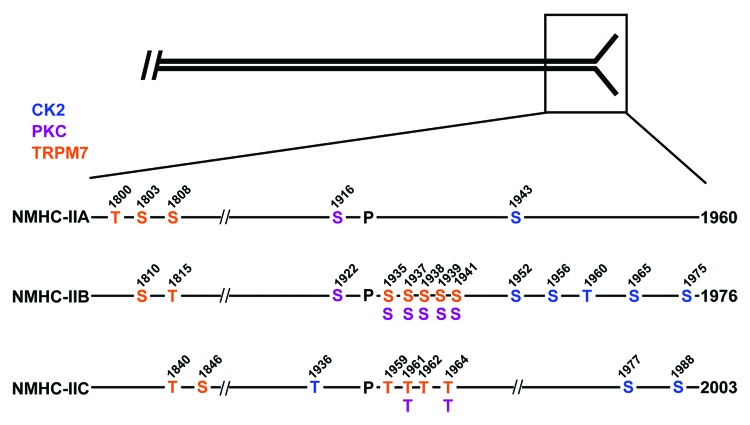
The majority of myosin-II heavy chain phosphorylation sites are located at the C-terminal end of the heavy chain, in the coiled coil and tailpiece regions (Fig. 4). Multiple kinases phosphorylate these sites, including the transient receptor potential melastatin 7 (TRPM7),49,50 members of the protein kinase C (PKC) family,41,51,52 and casein kinase 2 (CK2) (Fig. 4).41,51,52 A common feature of these phosphorylation events is that phosphorylation on the tailpiece, as well as the coiled coil domain, inhibits filament formation in vitro. Consistent with these data, receptor agonist-mediated myosin-II heavy chain phosphorylation is associated with myosin-II disassembly and cytoskeletal reorganization in vivo.53-57
Figure 4. Myosin-II filament assembly is regulated by phosphorylation on the heavy chain. NMHC-IIA, NMHC-IIB and NMHC-IIC are phosphorylated on the coiled coil and C-terminal tailpiece by the transient receptor potential melastatin 7 (TRPM7, orange residues), protein kinase C (PKC, purple residues), and casein kinase 2 (CK2, blue residues). The proline indicates the start of the C-terminal tailpiece. Phosphorylation on either the coiled coil domain or the tailpiece inhibits myosin-II filament formation in vitro.
TRPM7 and TRPM6, which are bifunctional proteins composed of a transient receptor potential (TRP) cation channel fused to a C-terminal intracellular α-kinase domain, phosphorylate all three NMHC-II isoforms on the coiled coil domain. While TRPM7 and TRPM6 phosphorylation of myosin-IIA is restricted to the coiled coil, these kinases also phosphorylate myosin-IIB and myosin-IIC on several sites in the tailpiece.49 Although the biochemical effects of TRPM7-mediated phosphorylation on the tailpiece have yet to be investigated; phosphorylation on Thr1800, Ser1803 and Ser1808 of the myosin-IIA coiled coil reduces filament assembly in vitro and diminishes the incorporation of myosin-IIA into the actin cytoskeleton in vivo.50 Cellular studies have shown that TRPM7 regulates cell polarization and migration by mediating the production of Ca2+ flickers that promote Ca2+- and kinase-dependent cytoskeletal rearrangements, including the reorganization of myosin-II assemblies.55,58
Phosphorylation of all three nonmuscle myosin-II isoforms by protein kinase C inhibits myosin-II filament assembly in vitro. PKC phosphorylates myosin-IIA on a single site (Ser1916) near the C-terminal end of the coiled coil.59 In addition, it phosphorylates multiple serines in the myosin-IIB coiled-coil and tailpiece,51 and two threonines in the myosin-IIC tailpiece.41 Biochemical studies with myosin-IIB rod domain constructs containing four Asp substitutions in the tailpiece suggest that PKC phosphorylation increases the critical concentration for myosin-IIB filament assembly.60 Additional studies suggest that PKC-mediated phosphorylation on the myosin-IIC tailpiece inhibits filament assembly by disrupting intermolecular interactions between the tailpiece and coiled coil.61
Cellular studies have shown that phorbol esters induce NMHC-IIA Ser1916 phosphorylation in platelets and T-lymphocytes,62,63 and Ser1916 phosphorylation is upregulated during the TGF-β-induced epithelial to mesenchymal transition of mouse mammary epithelial cells.64 In mast cells, myosin-IIA Ser1916 phosphorylation is mediated by protein kinase CβII and is associated with the onset of degranulation.56 For myosin-IIB, aPKCζ mediates phosphorylation on Ser1937 via a signaling pathway that involves the EGF receptor and p21-activated kinase.65 These studies demonstrate PKC-mediated regulation of myosin-II activity under a number of physiological and pathological stimuli. Future studies will be required to identify the PKC isoforms that mediate myosin-II phosphorylation during these different cellular processes and to determine whether phosphorylation on all identified PKC sites or a subset of sites is sufficient for the regulation of assembly.
Phosphorylation of the myosin-IIA heavy chain on S1943 is one of the most widely observed posttranslational modifications detected for this isoform in vivo. EGF stimulation induces transient S1943 myosin-IIA phosphorylation in multiple cell types,43,57,66 and is associated with reduced myosin-IIA assembly.57 In addition, S1943 phosphorylation is upregulated during integrin engagement on fibronectin.61 Myosin-IIA phosphorylation on Ser1943 also regulates the association of individual myosin IIA molecules with lytic granules in Natural killer cells,67 and a reduction in S1943 phosphorylation mediates myosin-IIA filament assembly during durotaxis (the crawling from soft to stiff matrices) in mesenchymal stem cells.68 Consistent with in vitro biochemical studies demonstrating that S1943 phosphorylation inhibits myosin-IIA filament assembly,52 FRAP studies showed that a myosin-IIA S1943D phosphomimetic exhibits increased filament turnover and reduced assembly compared with wild-type myosin-IIA.68 Breast tumor cells expressing myosin-IIA S1943E or S1943D phosphomimetic mutants exhibit increased migration and EGF-stimulated lamellipod extension as compared with cells expressing wild-type myosin-IIA.57 Conversely, a study with a non-phosphorylatable mutant of myosin-IIA (S1943A) demonstrates that inhibition of tailpiece phosphorylation results in myosin-IIA over-assembly at the lamellar margin of spreading cells and inhibits cell migration.57,64 The involvement of S1943 phosphorylation in the temporal and spatial control of myosin-IIA assembly during cell migration and the observation that S1943 phosphorylation is induced during the epithelial to mesenchymal transition64 and is elevated in human glioblastoma samples,66 suggests that S1943 phosphorylation may be an important regulatory control for the acquisition of an invasive phenotype.
In vitro, casein kinase 2 (CK2) directly phosphorylates S1943 on myosin-IIA. In addition to phosphorylating myosin-IIA, CK2 phosphorylates myosin-IIB on four serines in the tailpiece,51 and myosin-IIC on a single threonine in the coiled coil and on two serines in the tailpiece.41 CK2 phosphorylation of myosin-IIB inhibits filament assembly in vitro,51 and studies with Asp substitutions at the CK2 sites indicate that phosphorylation increases the critical concentration for assembly.60 In addition, phosphomimetic substitutions of the CK2 sites in the NMHC-IIC tailpiece inhibit myosin-IIC filament assembly and induce a diffuse cellular distribution consistent with filament disassembly.41 Although the CK2 site on myosin-IIA (S1943) is highly phosphorylated in breast tumor cells during cell spreading and integrin-fibronectin engagement, neither pharmacological nor siRNA-mediated inhibition of the α and α′ CK2 subunits significantly reduces S1943 phosphorylation.61 While there is no information on the contribution of CK2 to the in vivo phosphorylation of myosin-IIB and myosin-IIC, these observations raise the question as to whether CK2 is the in vivo heavy chain kinase for myosin-IIA.
Regulation of Filament Assembly via Interactions with Regulatory Proteins
In addition to light and heavy chain phosphorylation-mediated regulation of myosin-II filament assembly, there is accumulating evidence that interactions with a range of regulatory proteins can modulate the subcellular localization and organization of myosin-II assemblies. Several proteins have been reported to bind and regulate nonmuscle myosin-II filament assembly, including lethal giant larvae,69 myosin binding protein H,42 S100A470,71 and S100P.72 Interestingly, the myosin-II binding proteins identified to date all promote the unassembled form of nonmuscle myosin-II. These observations are intriguing as recent studies suggest that activated (e.g., RLC Ser19-phosphorylated), but unassembled myosin-II plays a role in initiating focal complex formation and lamellipodial protrusion.73 Myosin-II regulatory proteins may provide a mechanism for maintaining a monomeric pool of activated myosin-II. This would allow for the tight spatiotemporal control of myosin-II assembly, and as a consequence, myosin-II-mediated contractility.
Lethal(2)giant larvae (Lgl)
Lgl, a tumor suppressor first identified in Drosophila, regulates apical-basal polarization and the asymmetric division of Drosophila progenitor cells.74-77 Lgl mutant progenitors fail to differentiate and continue to proliferate, resulting in hyperplasia and dysplasia.78,79 Mammals express two Lgl homologs, Lgl1 and Lgl2, whose loss is associated with the disruption of cell polarity and epithelial architecture, and uncontrolled proliferation.80,81 The overall organization of the Lgl proteins is conserved in eukaryotes. The protein is composed of an N-terminal domain containing two WD40 β-propellers and an Lgl-specific C-terminal domain of unknown function.69,82
Biochemical studies have shown that Lgl is a component of a multi-protein complex that is associated with the membrane cytoskeleton.83,84 Lgl interacts with nonmuscle myosin-II, and aPKCζ-mediated phosphorylation of Lgl disrupts the Lgl/myosin-II interaction and induces Lgl dissociation from the membrane cytoskeleton.84-87 Phosphorylation by aPKC in the Lgl domain promotes an autoinhibitory intramolecular interaction between the N-terminal and Lgl domains that prevents the interaction of Lgl with its binding partners such as myosin-II.87 Lgl1 has been reported to associate with both myosin-IIA and myosin-IIB.88,89 Consistent with Lgl binding to the coiled coil region between ACD1 and ACD2, Lg1 blocks the assembly of myosin-IIA filaments in vitro and regulates myosin-IIA localization in vivo.89,90
S100A4
S100A4 (also known as FSP1, metastasin (mts1), pEL98, CAPL, p9Ka and calvasculin) is a member of the S100 family of small EF-hand proteins,91,92 and is a well characterized metastasis factor.93 The protein is a symmetric homodimer whose monomers contain four α-helices. Two α-helices from each S100A4 monomer (1, 4, 1′, and 4′) form a tight X-type four-helix bundle that constitutes the dimer interface.94 In addition, there are two EF-hand Ca2+-binding loops per S100A4 monomer: an N-terminal pseudo EF-hand (EF1) consisting of 14 residues and a C-terminal canonical EF-hand (EF2) consisting of 12 residues.91,92 Calcium binding to the C-terminal EF-hand induces a large conformational rearrangement in helix 3, resulting in the exposure of a large hydrophobic pocket that forms the binding site for target proteins (Fig. 5).95-97 One important consequence of this structural rearrangement, which is unique to the S100 family, is that S100A4 target binding is Ca2+-dependent.71 Given the two binding sites present in a S100 dimer, family members typically bind two target molecules.
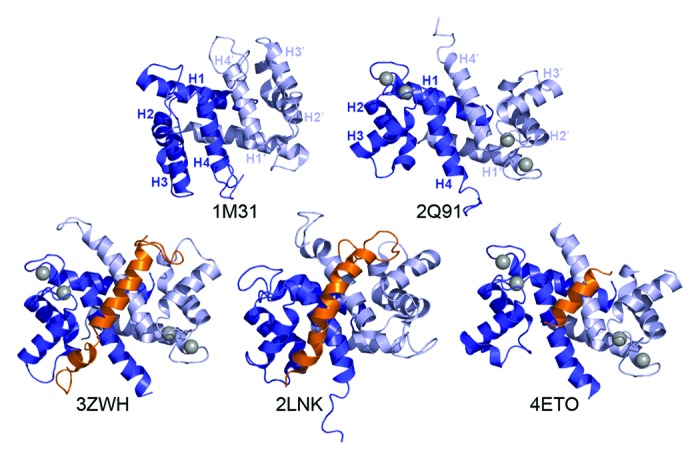
Figure 5. S100A4 structures. Ribbon diagrams of apo-S100A4 (1M31), Ca2+-S100A4 (2Q91) and three S100A4/myosin-IIA peptide complex structures; S100A4/MIIA1893–1935 (3ZWH), S100A4/MIIA1897–1935 (2LNK) and S100A4/MIIA1908–1923 (4ETO). The S100A4 monomers are shown in dark and light blue, and the myosin-IIA peptide in orange. The Ca2+ ions are denoted as gray spheres. The dimer interface is composed of helices 1, 4, 1′, and 4′. Ca2+-binding results in reorientation of helix 3 to expose a hydrophobic cleft that mediates interactions with the myosin-IIA target peptide.
S100A4 expression is associated with enhanced cell migration in multiple cell types and detailed phenotypic studies have demonstrated that S100A4 enhances chemotactic migration by maintaining cell polarization and inhibiting cell turning.98-104 Consistent with a role in cell migration, proteomic and localization studies have shown that S100A4 is enriched in the pseudopodia of crawling cells and the active Ca2+-bound form of S100A4 localizes to the leading edge of migrating cells.93,105,106 Notably, the equilibrium dissociation constants (KD) for the Ca2+:S100A4 interaction are relatively weak in the absence of a protein target (EF1 = 54 µM; EF2 = 3.3 µM); but increase approximately 10-fold in the presence of target (EF1 = 3.6 µM; EF2 = 0.26 µM).95 The localized calcium flickers observed at the leading edge of migrating cells provide a possible mechanism for restricted S100A4 activation.58,107,108 Moreover, target-mediated enhancement of Ca2+-binding to S100A4 would allow for high intracellular S100A4 concentrations (e.g., 3–5 μM104) without the disruption of intracellular of Ca2 + dynamics.
Biochemical studies have shown that S100A4 binds with low nanomolar affinity to isolated peptides derived from the myosin-IIA heavy chain and preferentially depolymerizes myosin-IIA filaments.109-113 S100A4 is also reported to bind with high affinity to peptides from the myosin-IIC heavy chain, but the ability of S100A4 to disassemble myosin-IIC filaments has not been investigated.113 Consistent with the in vitro regulation of myosin-IIA assembly by S100A4, S100A4−/− macrophages over-assemble myosin-IIA filaments, which results in lamellipodial instability and reduced persistence during chemotaxis.104 In addition, studies in mammary adenocarcinoma cells demonstrate that the S100A4/myosin-IIA interaction mediates the cellular location of pseudopodial protrusions during chemotaxis.103
The recent X-ray and NMR structures of the S100A4/myosin-IIA complex revealed that a single NMHC-IIA polypeptide wraps around the S100A4 dimer to form contacts with the canonical target binding pocket in each S100A4 monomer (Fig. 5).112-114 These structural studies demonstrate that the S100A4 binding site encompasses NMHC-IIA residues 1893–1943, and is composed of the C-terminal region of ACD1 and the N-terminal region of the tailpiece. A particular attribute of the S100A4/NMHC-IIA interaction is that myosin-IIA heavy chain residues at the a and d positions of the coiled coil heptad repeat, which mediate interactions between the two strands of the coiled coil, also mediate S100A4 binding. These findings suggest a mechanism in which S100A4 binding induces local unwinding of the myosin-IIA coiled coil that disrupts intermolecular interactions between adjacent myosin-IIA molecules and promotes filament disassembly. Although S100A4 binds with high affinity (e.g., nanomolar) to single myosin-IIA peptides, binding to dimeric rod domain constructs and full-length myosin-IIA appears to be weaker.71,109 These data suggest that some of the binding energy associated with the formation of the S100A4/myosin-IIA complex is used to unzip the myosin-IIA coiled coil, which would result in a lower apparent dissociation constant. A complete mechanistic characterization of S100A4-mediated myosin-IIA disassembly will require future investigations on the energetic coupling of S100A4 binding and local myosin-IIA coiled coil unzipping.
Although the S100A4 binding site on the myosin-IIA heavy chain encompasses the Ser1916 phosphorylation site, structures of the S100A4/myosin-IIA complex demonstrate that the Ser1916 side chain points away from the S100A4/myosin-IIA binding interface; consistent with biochemical studies showing that phosphorylation on this site does not affect S100A4 binding.52 In contrast, phosphorylation on S1943, which is located outside the S100A4 binding site in the tailpiece, inhibits S100A4 binding. These observations suggest a possible mechanism in which incorporation of a negatively charged phosphate group on the tailpiece facilitates a conformational rearrangement that supports intramolecular interactions between the tailpiece and coiled coil domain to prevent S100A4 binding. Given that both S100A4 and heavy chain phosphorylation promote myosin-IIA filament disassembly, and that S1943 heavy chain phosphorylation also inhibits S100A4 binding, this raises the interesting question as to how these two regulatory pathways might function in vivo. One possibility is that these are independent mechanisms, which operate in distinct subcellular compartments. Alternatively, these two pathways could function in a synergistic manner. For example, S100A4 binding could increase the substrate specificity of the heavy chain kinase for the myosin-IIA heavy chain. Phosphorylation would then mediate S100A4 dissociation from the heavy chain, and permit recycling of S100A4 for additional rounds of myosin-IIA depolymerization. Continued investigation into the molecular details of myosin-II filament disassembly by covalent modification and/or interactions with regulatory proteins should reveal how these regulatory mechanisms are integrated to control overall cell physiology.
Conclusions
Biochemical and cellular studies have established that regulation via the myosin-II heavy chain provides a mechanism for modulating the assembly of myosin-II filaments. The identification of missense mutations and small in frame deletions in the myosin-IIA heavy chain coiled-coil and C-terminal tailpiece (e.g., E1841K and R1933stop), which result in an autosomal dominant disorder manifested by leukocyte inclusions, thrombocytopenia and giant platelets,115,116 underscores the physiological need for regulated myosin-II assembly. Although we have a basic understanding of the contribution of heavy chain phosphorylation to the control of filament assembly, the structural consequences of these phosphorylation events and how they affect intramolecular interactions in the myosin-II monomer and intermolecular interactions within a myosin-II filament needs further investigation. In addition, biochemical evaluation of myosin-II heavy chain phosphorylation has primarily used assembly-competent myosin-II rod domain constructs that lack the motor domain and associated light chains. As a consequence, we do not have information on how heavy chain phosphorylation regulates myosin-II filament assembly in the context of RLC phosphorylation. Likewise, while several myosin-II heavy chain kinases have been identified from in vitro studies, our knowledge of the kinases that regulate myosin-II phosphorylation in vivo is still incomplete. Lastly, there have been no investigations into the vertebrate nonmuscle myosin-II heavy chain phosphatases. A complete understanding of the role of myosin-II heavy chain phosphorylation in cellular physiology will require the identification and characterization of these molecules.
The identification of myosin-II regulatory proteins has opened an exciting new area of myosin-II regulation. While it is clear that regulatory proteins modulate myosin-IIA oligomerization, proteins that preferentially regulate myosin-IIB and myosin-IIC filament assembly have yet to be described. In humans, there are 21 S100 family members that exhibit both cell and tissue-specific expression.92,117,118 The recent report of a second S100 family member, S100P,72 as a myosin-IIA regulator raises the possibility that other S100 proteins may selectively modulate the assembly of myosin-IIB and myosin-IIC. A major goal for the future is the discovery new myosin-II regulatory proteins as well as the biochemical, structural and cellular characterization of how these proteins regulate myosin-II function and cell physiology.
Acknowledgments
The authors regret the omission of some studies due to space limitations. We thank KA Taylor (Florida State University) for providing the refined model of the 10S myosin head, and JM Backer and SC Almo (Albert Einstein College of Medicine) for helpful suggestions and reading the manuscript. This work was supported by National Institutes of Health grant PO1 CA100324.
Submitted
07/14/2013
Revised
08/12/2013
Accepted
08/12/2013
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
References
- 1.Sellers JR. Myosins: a diverse superfamily. Biochim Biophys Acta. 2000;1496:3–22. doi: 10.1016/S0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- 2.Engelhardt WA, Ljubimowa MN. Myosine and Adenosinetriphosphatase. Nature. 1939;144:668–9. doi: 10.1038/144668b0. [DOI] [Google Scholar]
- 3.Niederman R, Pollard TD. Human platelet myosin. II. In vitro assembly and structure of myosin filaments. J Cell Biol. 1975;67:72–92. doi: 10.1083/jcb.67.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verkhovsky AB, Svitkina TM, Borisy GG. Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J Cell Biol. 1995;131:989–1002. doi: 10.1083/jcb.131.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saez CG, Myers JC, Shows TB, Leinwand LA. Human nonmuscle myosin heavy chain mRNA: generation of diversity through alternative polyadenylylation. Proc Natl Acad Sci U S A. 1990;87:1164–8. doi: 10.1073/pnas.87.3.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simons M, Wang M, McBride OW, Kawamoto S, Yamakawa K, Gdula D, Adelstein RS, Weir L. Human nonmuscle myosin heavy chains are encoded by two genes located on different chromosomes. Circ Res. 1991;69:530–9. doi: 10.1161/01.RES.69.2.530. [DOI] [PubMed] [Google Scholar]
- 7.Phillips CL, Yamakawa K, Adelstein RS. Cloning of the cDNA encoding human nonmuscle myosin heavy chain-B and analysis of human tissues with isoform-specific antibodies. J Muscle Res Cell Motil. 1995;16:379–89. doi: 10.1007/BF00114503. [DOI] [PubMed] [Google Scholar]
- 8.Berg JS, Powell BC, Cheney RE. A millennial myosin census. Mol Biol Cell. 2001;12:780–94. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golomb E, Ma X, Jana SS, Preston YA, Kawamoto S, Shoham NG, Goldin E, Conti MA, Sellers JR, Adelstein RS. Identification and characterization of nonmuscle myosin II-C, a new member of the myosin II family. J Biol Chem. 2004;279:2800–8. doi: 10.1074/jbc.M309981200. [DOI] [PubMed] [Google Scholar]
- 10.Kovács M, Wang F, Hu A, Zhang Y, Sellers JR. Functional divergence of human cytoplasmic myosin II: kinetic characterization of the non-muscle IIA isoform. J Biol Chem. 2003;278:38132–40. doi: 10.1074/jbc.M305453200. [DOI] [PubMed] [Google Scholar]
- 11.Rosenfeld SS, Xing J, Chen LQ, Sweeney HL. Myosin IIb is unconventionally conventional. J Biol Chem. 2003;278:27449–55. doi: 10.1074/jbc.M302555200. [DOI] [PubMed] [Google Scholar]
- 12.Wang F, Kovacs M, Hu A, Limouze J, Harvey EV, Sellers JR. Kinetic mechanism of non-muscle myosin IIB: functional adaptations for tension generation and maintenance. J Biol Chem. 2003;278:27439–48. doi: 10.1074/jbc.M302510200. [DOI] [PubMed] [Google Scholar]
- 13.Heissler SM, Manstein DJ. Comparative kinetic and functional characterization of the motor domains of human nonmuscle myosin-2C isoforms. J Biol Chem. 2011;286:21191–202. doi: 10.1074/jbc.M110.212290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawamoto S, Adelstein RS. Chicken nonmuscle myosin heavy chains: differential expression of two mRNAs and evidence for two different polypeptides. J Cell Biol. 1991;112:915–24. doi: 10.1083/jcb.112.5.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–90. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heissler SM, Manstein DJ. Nonmuscle myosin-2: mix and match. Cell Mol Life Sci. 2013;70:1–21. doi: 10.1007/s00018-012-1002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandquist JC, Means AR. The C-terminal tail region of nonmuscle myosin II directs isoform-specific distribution in migrating cells. Mol Biol Cell. 2008;19:5156–67. doi: 10.1091/mbc.E08-05-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vicente-Manzanares M, Koach MA, Whitmore L, Lamers ML, Horwitz AF. Segregation and activation of myosin IIB creates a rear in migrating cells. J Cell Biol. 2008;183:543–54. doi: 10.1083/jcb.200806030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trybus KM, Huiatt TW, Lowey S. A bent monomeric conformation of myosin from smooth muscle. Proc Natl Acad Sci U S A. 1982;79:6151–5. doi: 10.1073/pnas.79.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kendrick-Jones J, Smith RC, Craig R, Citi S. Polymerization of vertebrate non-muscle and smooth muscle myosins. J Mol Biol. 1987;198:241–52. doi: 10.1016/0022-2836(87)90310-X. [DOI] [PubMed] [Google Scholar]
- 21.Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci U S A. 2001;98:4361–6. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Wendt T, Taylor D, Taylor K. Refined model of the 10S conformation of smooth muscle myosin by cryo-electron microscopy 3D image reconstruction. J Mol Biol. 2003;329:963–72. doi: 10.1016/S0022-2836(03)00516-3. [DOI] [PubMed] [Google Scholar]
- 23.Onishi H, Wakabayashi T. Electron microscopic studies of myosin molecules from chicken gizzard muscle I: the formation of the intramolecular loop in the myosin tail. J Biochem. 1982;92:871–9. doi: 10.1093/oxfordjournals.jbchem.a134001. [DOI] [PubMed] [Google Scholar]
- 24.Burgess SA, Yu S, Walker ML, Hawkins RJ, Chalovich JM, Knight PJ. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J Mol Biol. 2007;372:1165–78. doi: 10.1016/j.jmb.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 25.Jung HS, Komatsu S, Ikebe M, Craig R. Head-head and head-tail interaction: a general mechanism for switching off myosin II activity in cells. Mol Biol Cell. 2008;19:3234–42. doi: 10.1091/mbc.E08-02-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung HS, Burgess SA, Billington N, Colegrave M, Patel H, Chalovich JM, Chantler PD, Knight PJ. Conservation of the regulated structure of folded myosin 2 in species separated by at least 600 million years of independent evolution. Proc Natl Acad Sci U S A. 2008;105:6022–6. doi: 10.1073/pnas.0707846105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung HS, Billington N, Thirumurugan K, Salzameda B, Cremo CR, Chalovich JM, Chantler PD, Knight PJ. Role of the tail in the regulated state of myosin 2. J Mol Biol. 2011;408:863–78. doi: 10.1016/j.jmb.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kendrick-Jones J, Smith RC, Craig R, Citi S. Polymerization of vertebrate non-muscle and smooth muscle myosins. J Mol Biol. 1987;198:241–52. doi: 10.1016/0022-2836(87)90310-X. [DOI] [PubMed] [Google Scholar]
- 29.Craig R, Smith R, Kendrick-Jones J. Light-chain phosphorylation controls the conformation of vertebrate non-muscle and smooth muscle myosin molecules. Nature. 1983;302:436–9. doi: 10.1038/302436a0. [DOI] [PubMed] [Google Scholar]
- 30.Trybus KM, Lowey S. Conformational states of smooth muscle myosin. Effects of light chain phosphorylation and ionic strength. J Biol Chem. 1984;259:8564–71. [PubMed] [Google Scholar]
- 31.Trybus KM, Lowey S. Assembly of smooth muscle myosin minifilaments: effects of phosphorylation and nucleotide binding. J Cell Biol. 1987;105:3007–19. doi: 10.1083/jcb.105.6.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breckenridge MT, Dulyaninova NG, Egelhoff TT. Multiple regulatory steps control mammalian nonmuscle myosin II assembly in live cells. Mol Biol Cell. 2009;20:338–47. doi: 10.1091/mbc.E08-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milton DL, Schneck AN, Ziech DA, Ba M, Facemyer KC, Halayko AJ, Baker JE, Gerthoffer WT, Cremo CR. Direct evidence for functional smooth muscle myosin II in the 10S self-inhibited monomeric conformation in airway smooth muscle cells. Proc Natl Acad Sci U S A. 2011;108:1421–6. doi: 10.1073/pnas.1011784108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiboku T, Katoh T, Nakamura A, Kitamura A, Kinjo M, Murakami Y, Takahashi M. Nonmuscle myosin II folds into a 10S form via two portions of tail for dynamic subcellular localization. Genes Cells. 2013;18:90–109. doi: 10.1111/gtc.12021. [DOI] [PubMed] [Google Scholar]
- 35.McLachlan AD, Karn J. Periodic charge distributions in the myosin rod amino acid sequence match cross-bridge spacings in muscle. Nature. 1982;299:226–31. doi: 10.1038/299226a0. [DOI] [PubMed] [Google Scholar]
- 36.Atkinson SJ, Stewart M. Molecular interactions in myosin assembly. Role of the 28-residue charge repeat in the rod. J Mol Biol. 1992;226:7–13. doi: 10.1016/0022-2836(92)90118-4. [DOI] [PubMed] [Google Scholar]
- 37.Sohn RL, Vikstrom KL, Strauss M, Cohen C, Szent-Gyorgyi AG, Leinwand LA. A 29 residue region of the sarcomeric myosin rod is necessary for filament formation. J Mol Biol. 1997;266:317–30. doi: 10.1006/jmbi.1996.0790. [DOI] [PubMed] [Google Scholar]
- 38.Cohen C, Parry DA. A conserved C-terminal assembly region in paramyosin and myosin rods. J Struct Biol. 1998;122:180–7. doi: 10.1006/jsbi.1998.3983. [DOI] [PubMed] [Google Scholar]
- 39.Nakasawa T, Takahashi M, Matsuzawa F, Aikawa S, Togashi Y, Saitoh T, Yamagishi A, Yazawa M. Critical regions for assembly of vertebrate nonmuscle myosin II. Biochemistry. 2005;44:174–83. doi: 10.1021/bi048807h. [DOI] [PubMed] [Google Scholar]
- 40.Rosenberg M, Straussman R, Ben-Ya’acov A, Ronen D, Ravid S. MHC-IIB filament assembly and cellular localization are governed by the rod net charge. PLoS One. 2008;3:e1496. doi: 10.1371/journal.pone.0001496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ronen D, Ravid S. Myosin II tailpiece determines its paracrystal structure, filament assembly properties, and cellular localization. J Biol Chem. 2009;284:24948–57. doi: 10.1074/jbc.M109.023754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hosono Y, Usukura J, Yamaguchi T, Yanagisawa K, Suzuki M, Takahashi T. MYBPH inhibits NM IIA assembly via direct interaction with NMHC IIA and reduces cell motility. Biochem Biophys Res Commun. 2012;428:173–8. doi: 10.1016/j.bbrc.2012.10.036. [DOI] [PubMed] [Google Scholar]
- 43.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–48. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 44.Trotter JA. Living macrophages phosphorylate the 20,000 Dalton light chains and heavy chains of myosin. Biochem Biophys Res Commun. 1982;106:1071–7. doi: 10.1016/0006-291X(82)91820-4. [DOI] [PubMed] [Google Scholar]
- 45.Trotter JA, Nixon CS, Johnson MA. The heavy chain of macrophage myosin is phosphorylated at the tip of the tail. J Biol Chem. 1985;260:14374–8. [PubMed] [Google Scholar]
- 46.Barylko B, Tooth P, Kendrick-Jones J. Proteolytic fragmentation of brain myosin and localisation of the heavy-chain phosphorylation site. Eur J Biochem. 1986;158:271–82. doi: 10.1111/j.1432-1033.1986.tb09747.x. [DOI] [PubMed] [Google Scholar]
- 47.Egelhoff TT, Lee RJ, Spudich JA. Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell. 1993;75:363–71. doi: 10.1016/0092-8674(93)80077-R. [DOI] [PubMed] [Google Scholar]
- 48.Pasternak C, Flicker PF, Ravid S, Spudich JA. Intermolecular versus intramolecular interactions of Dictyostelium myosin: possible regulation by heavy chain phosphorylation. J Cell Biol. 1989;109:203–10. doi: 10.1083/jcb.109.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clark K, Middelbeek J, Dorovkov MV, Figdor CG, Ryazanov AG, Lasonder E, van Leeuwen FN. The alpha-kinases TRPM6 and TRPM7, but not eEF-2 kinase, phosphorylate the assembly domain of myosin IIA, IIB and IIC. FEBS Lett. 2008;582:2993–7. doi: 10.1016/j.febslet.2008.07.043. [DOI] [PubMed] [Google Scholar]
- 50.Clark K, Middelbeek J, Lasonder E, Dulyaninova NG, Morrice NA, Ryazanov AG, Bresnick AR, Figdor CG, van Leeuwen FN. TRPM7 regulates myosin IIA filament stability and protein localization by heavy chain phosphorylation. J Mol Biol. 2008;378:790–803. doi: 10.1016/j.jmb.2008.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murakami N, Chauhan VP, Elzinga M. Two nonmuscle myosin II heavy chain isoforms expressed in rabbit brains: filament forming properties, the effects of phosphorylation by protein kinase C and casein kinase II, and location of the phosphorylation sites. Biochemistry. 1998;37:1989–2003. doi: 10.1021/bi971959a. [DOI] [PubMed] [Google Scholar]
- 52.Dulyaninova NG, Malashkevich VN, Almo SC, Bresnick AR. Regulation of myosin-IIA assembly and Mts1 binding by heavy chain phosphorylation. Biochemistry. 2005;44:6867–76. doi: 10.1021/bi0500776. [DOI] [PubMed] [Google Scholar]
- 53.van Leeuwen FN, van Delft S, Kain HE, van der Kammen RA, Collard JG. Rac regulates phosphorylation of the myosin-II heavy chain, actinomyosin disassembly and cell spreading. Nat Cell Biol. 1999;1:242–8. doi: 10.1038/12068. [DOI] [PubMed] [Google Scholar]
- 54.Straussman R, Even L, Ravid S. Myosin II heavy chain isoforms are phosphorylated in an EGF-dependent manner: involvement of protein kinase C. J Cell Sci. 2001;114:3047–57. doi: 10.1242/jcs.114.16.3047. [DOI] [PubMed] [Google Scholar]
- 55.Clark K, Langeslag M, van Leeuwen B, Ran L, Ryazanov AG, Figdor CG, Moolenaar WH, Jalink K, van Leeuwen FN. TRPM7, a novel regulator of actomyosin contractility and cell adhesion. EMBO J. 2006;25:290–301. doi: 10.1038/sj.emboj.7600931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ludowyke RI, Elgundi Z, Kranenburg T, Stehn JR, Schmitz-Peiffer C, Hughes WE, Biden TJ. Phosphorylation of nonmuscle myosin heavy chain IIA on Ser1917 is mediated by protein kinase C beta II and coincides with the onset of stimulated degranulation of RBL-2H3 mast cells. J Immunol. 2006;177:1492–9. doi: 10.4049/jimmunol.177.3.1492. [DOI] [PubMed] [Google Scholar]
- 57.Dulyaninova NG, House RP, Betapudi V, Bresnick AR. Myosin-IIA heavy-chain phosphorylation regulates the motility of MDA-MB-231 carcinoma cells. Mol Biol Cell. 2007;18:3144–55. doi: 10.1091/mbc.E06-11-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei C, Wang X, Chen M, Ouyang K, Song LS, Cheng H. Calcium flickers steer cell migration. Nature. 2009;457:901–5. doi: 10.1038/nature07577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Conti MA, Sellers JR, Adelstein RS, Elzinga M. Identification of the serine residue phosphorylated by protein kinase C in vertebrate nonmuscle myosin heavy chains. Biochemistry. 1991;30:966–70. doi: 10.1021/bi00218a012. [DOI] [PubMed] [Google Scholar]
- 60.Murakami N, Kotula L, Hwang YW. Two distinct mechanisms for regulation of nonmuscle myosin assembly via the heavy chain: phosphorylation for MIIB and mts 1 binding for MIIA. Biochemistry. 2000;39:11441–51. doi: 10.1021/bi000347e. [DOI] [PubMed] [Google Scholar]
- 61.Betapudi V, Gokulrangan G, Chance MR, Egelhoff TT. A proteomic study of myosin II motor proteins during tumor cell migration. J Mol Biol. 2011;407:673–86. doi: 10.1016/j.jmb.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kawamoto S, Bengur AR, Sellers JR, Adelstein RS. In situ phosphorylation of human platelet myosin heavy and light chains by protein kinase C. J Biol Chem. 1989;264:2258–65. [PubMed] [Google Scholar]
- 63.Moussavi RS, Kelley CA, Adelstein RS. Phosphorylation of vertebrate nonmuscle and smooth muscle myosin heavy chains and light chains. Mol Cell Biochem. 1993;127-128:219–27. doi: 10.1007/BF01076773. [DOI] [PubMed] [Google Scholar]
- 64.Beach JR, Hussey GS, Miller TE, Chaudhury A, Patel P, Monslow J, Zheng Q, Keri RA, Reizes O, Bresnick AR, et al. Myosin II isoform switching mediates invasiveness after TGF-β-induced epithelial-mesenchymal transition. Proc Natl Acad Sci U S A. 2011;108:17991–6. doi: 10.1073/pnas.1106499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Even-Faitelson L, Ravid S. PAK1 and aPKCzeta regulate myosin II-B phosphorylation: a novel signaling pathway regulating filament assembly. Mol Biol Cell. 2006;17:2869–81. doi: 10.1091/mbc.E05-11-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ivkovic S, Beadle C, Noticewala S, Massey SC, Swanson KR, Toro LN, Bresnick AR, Canoll P, Rosenfeld SS. Direct inhibition of myosin II effectively blocks glioma invasion in the presence of multiple motogens. Mol Biol Cell. 2012;23:533–42. doi: 10.1091/mbc.E11-01-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sanborn KB, Mace EM, Rak GD, Difeo A, Martignetti JA, Pecci A, Bussel JB, Favier R, Orange JS. Phosphorylation of the myosin IIA tailpiece regulates single myosin IIA molecule association with lytic granules to promote NK-cell cytotoxicity. Blood. 2011;118:5862–71. doi: 10.1182/blood-2011-03-344846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Raab M, Swift J, Dingal PC, Shah P, Shin JW, Discher DE. Crawling from soft to stiff matrix polarizes the cytoskeleton and phosphoregulates myosin-II heavy chain. J Cell Biol. 2012;199:669–83. doi: 10.1083/jcb.201205056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vasioukhin V. Lethal giant puzzle of Lgl. Dev Neurosci. 2006;28:13–24. doi: 10.1159/000090749. [DOI] [PubMed] [Google Scholar]
- 70.Kriajevska MV, Cardenas MN, Grigorian MS, Ambartsumian NS, Georgiev GP, Lukanidin EM. Non-muscle myosin heavy chain as a possible target for protein encoded by metastasis-related mts-1 gene. J Biol Chem. 1994;269:19679–82. [PubMed] [Google Scholar]
- 71.Ford HL, Silver DL, Kachar B, Sellers JR, Zain SB. Effect of Mts1 on the structure and activity of nonmuscle myosin II. Biochemistry. 1997;36:16321–7. doi: 10.1021/bi971182l. [DOI] [PubMed] [Google Scholar]
- 72.Du M, Wang G, Ismail TM, Gross S, Fernig DG, Barraclough R, Rudland PS. S100P dissociates myosin IIA filaments and focal adhesion sites to reduce cell adhesion and enhance cell migration. J Biol Chem. 2012;287:15330–44. doi: 10.1074/jbc.M112.349787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shutova M, Yang C, Vasiliev JM, Svitkina T. Functions of nonmuscle myosin II in assembly of the cellular contractile system. PLoS One. 2012;7:e40814. doi: 10.1371/journal.pone.0040814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gateff E, Schneiderman HA. Neoplasms in mutant and cultured wild-tupe tissues of Drosophila. Natl Cancer Inst Monogr. 1969;31:365–97. [PubMed] [Google Scholar]
- 75.Mechler BM, McGinnis W, Gehring WJ. Molecular cloning of lethal(2)giant larvae, a recessive oncogene of Drosophila melanogaster. EMBO J. 1985;4:1551–7. doi: 10.1002/j.1460-2075.1985.tb03816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ohshiro T, Yagami T, Zhang C, Matsuzaki F. Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature. 2000;408:593–6. doi: 10.1038/35046087. [DOI] [PubMed] [Google Scholar]
- 77.Peng CY, Manning L, Albertson R, Doe CQ. The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature. 2000;408:596–600. doi: 10.1038/35046094. [DOI] [PubMed] [Google Scholar]
- 78.Bilder D, Li M, Perrimon N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 2000;289:113–6. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- 79.Bilder D. Epithelial polarity and proliferation control: links from the Drosophila neoplastic tumor suppressors. Genes Dev. 2004;18:1909–25. doi: 10.1101/gad.1211604. [DOI] [PubMed] [Google Scholar]
- 80.Klezovitch O, Fernandez TE, Tapscott SJ, Vasioukhin V. Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 2004;18:559–71. doi: 10.1101/gad.1178004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Russ A, Louderbough JM, Zarnescu D, Schroeder JA. Hugl1 and Hugl2 in mammary epithelial cells: polarity, proliferation, and differentiation. PLoS One. 2012;7:e47734. doi: 10.1371/journal.pone.0047734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hattendorf DA, Andreeva A, Gangar A, Brennwald PJ, Weis WI. Structure of the yeast polarity protein Sro7 reveals a SNARE regulatory mechanism. Nature. 2007;446:567–71. doi: 10.1038/nature05635. [DOI] [PubMed] [Google Scholar]
- 83.Strand D, Raska I, Mechler BM. The Drosophila lethal(2)giant larvae tumor suppressor protein is a component of the cytoskeleton. J Cell Biol. 1994;127:1345–60. doi: 10.1083/jcb.127.5.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kalmes A, Merdes G, Neumann B, Strand D, Mechler BM. A serine-kinase associated with the p127-l(2)gl tumour suppressor of Drosophila may regulate the binding of p127 to nonmuscle myosin II heavy chain and the attachment of p127 to the plasma membrane. J Cell Sci. 1996;109:1359–68. doi: 10.1242/jcs.109.6.1359. [DOI] [PubMed] [Google Scholar]
- 85.Strand D, Jakobs R, Merdes G, Neumann B, Kalmes A, Heid HW, Husmann I, Mechler BM. The Drosophila lethal(2)giant larvae tumor suppressor protein forms homo-oligomers and is associated with nonmuscle myosin II heavy chain. J Cell Biol. 1994;127:1361–73. doi: 10.1083/jcb.127.5.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Plant PJ, Fawcett JP, Lin DC, Holdorf AD, Binns K, Kulkarni S, Pawson T. A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat Cell Biol. 2003;5:301–8. doi: 10.1038/ncb948. [DOI] [PubMed] [Google Scholar]
- 87.Betschinger J, Eisenhaber F, Knoblich JA. Phosphorylation-induced autoinhibition regulates the cytoskeletal protein Lethal (2) giant larvae. Curr Biol. 2005;15:276–82. doi: 10.1016/j.cub.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 88.Solinet S, Akpovi CD, Garcia CJ, Barry A, Vitale ML. Myosin IIB deficiency in embryonic fibroblasts affects regulators and core members of the par polarity complex. Histochem Cell Biol. 2011;136:245–66. doi: 10.1007/s00418-011-0840-0. [DOI] [PubMed] [Google Scholar]
- 89.Dahan I, Yearim A, Touboul Y, Ravid S. The tumor suppressor Lgl1 regulates NMII-A cellular distribution and focal adhesion morphology to optimize cell migration. Mol Biol Cell. 2012;23:591–601. doi: 10.1091/mbc.E11-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.De Lorenzo C, Mechler BM, Bryant PJ. What is Drosophila telling us about cancer? Cancer Metastasis Rev. 1999;18:295–311. doi: 10.1023/A:1006381526008. [DOI] [PubMed] [Google Scholar]
- 91.Zimmer DB, Cornwall EH, Landar A, Song W. The S100 protein family: history, function, and expression. Brain Res Bull. 1995;37:417–29. doi: 10.1016/0361-9230(95)00040-2. [DOI] [PubMed] [Google Scholar]
- 92.Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature) Biochem Biophys Res Commun. 2004;322:1111–22. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 93.House RP, Garrett SC, Bresnick AR. Moving Aggressively: S100A4 and Tumor Invasion. In: Fatatis A, ed. Signaling Pathways and Molecular Mediators in Metastasis: Springer Netherlands, 2012:91-113. [Google Scholar]
- 94.Vallely KM, Rustandi RR, Ellis KC, Varlamova O, Bresnick AR, Weber DJ. Solution structure of human Mts1 (S100A4) as determined by NMR spectroscopy. Biochemistry. 2002;41:12670–80. doi: 10.1021/bi020365r. [DOI] [PubMed] [Google Scholar]
- 95.Malashkevich VN, Varney KM, Garrett SC, Wilder PT, Knight D, Charpentier TH, Ramagopal UA, Almo SC, Weber DJ, Bresnick AR. Structure of Ca2+-bound S100A4 and its interaction with peptides derived from nonmuscle myosin-IIA. Biochemistry. 2008;47:5111–26. doi: 10.1021/bi702537s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gingras AR, Basran J, Prescott A, Kriajevska M, Bagshaw CR, Barsukov IL. Crystal structure of the Ca(2+)-form and Ca(2+)-binding kinetics of metastasis-associated protein, S100A4. FEBS Lett. 2008;582:1651–6. doi: 10.1016/j.febslet.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 97.Pathuri P, Vogeley L, Luecke H. Crystal structure of metastasis-associated protein S100A4 in the active calcium-bound form. J Mol Biol. 2008;383:62–77. doi: 10.1016/j.jmb.2008.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Takenaga K, Nakamura Y, Endo H, Sakiyama S. Involvement of S100-related calcium-binding protein pEL98 (or mts1) in cell motility and tumor cell invasion. Jpn J Cancer Res. 1994;85:831–9. doi: 10.1111/j.1349-7006.1994.tb02955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takenaga K, Nakamura Y, Sakiyama S. Expression of antisense RNA to S100A4 gene encoding an S100-related calcium-binding protein suppresses metastatic potential of high-metastatic Lewis lung carcinoma cells. Oncogene. 1997;14:331–7. doi: 10.1038/sj.onc.1200820. [DOI] [PubMed] [Google Scholar]
- 100.Bjørnland K, Winberg JO, Odegaard OT, Hovig E, Loennechen T, Aasen AO, Fodstad O, Maelandsmo GM. S100A4 involvement in metastasis: deregulation of matrix metalloproteinases and tissue inhibitors of matrix metalloproteinases in osteosarcoma cells transfected with an anti-S100A4 ribozyme. Cancer Res. 1999;59:4702–8. [PubMed] [Google Scholar]
- 101.Grum-Schwensen B, Klingelhofer J, Berg CH, El-Naaman C, Grigorian M, Lukanidin E, Ambartsumian N. Suppression of tumor development and metastasis formation in mice lacking the S100A4(mts1) gene. Cancer Res. 2005;65:3772–80. doi: 10.1158/0008-5472.CAN-04-4510. [DOI] [PubMed] [Google Scholar]
- 102.Stein U, Arlt F, Walther W, Smith J, Waldman T, Harris ED, Mertins SD, Heizmann CW, Allard D, Birchmeier W, et al. The metastasis-associated gene S100A4 is a novel target of beta-catenin/T-cell factor signaling in colon cancer. Gastroenterology. 2006;131:1486–500. doi: 10.1053/j.gastro.2006.08.041. [DOI] [PubMed] [Google Scholar]
- 103.Li ZH, Bresnick AR. The S100A4 metastasis factor regulates cellular motility via a direct interaction with myosin-IIA. Cancer Res. 2006;66:5173–80. doi: 10.1158/0008-5472.CAN-05-3087. [DOI] [PubMed] [Google Scholar]
- 104.Li ZH, Dulyaninova NG, House RP, Almo SC, Bresnick AR. S100A4 regulates macrophage chemotaxis. Mol Biol Cell. 2010;21:2598–610. doi: 10.1091/mbc.E09-07-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim EJ, Helfman DM. Characterization of the metastasis-associated protein, S100A4. Roles of calcium binding and dimerization in cellular localization and interaction with myosin. J Biol Chem. 2003;278:30063–73. doi: 10.1074/jbc.M304909200. [DOI] [PubMed] [Google Scholar]
- 106.Wang Y, Ding SJ, Wang W, Jacobs JM, Qian WJ, Moore RJ, Yang F, Camp DG, 2nd, Smith RD, Klemke RL. Profiling signaling polarity in chemotactic cells. Proc Natl Acad Sci U S A. 2007;104:8328–33. doi: 10.1073/pnas.0701103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Evans JH, Falke JJ. Ca2+ influx is an essential component of the positive-feedback loop that maintains leading-edge structure and activity in macrophages. Proc Natl Acad Sci U S A. 2007;104:16176–81. doi: 10.1073/pnas.0707719104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wei C, Wang X, Zheng M, Cheng H. Calcium gradients underlying cell migration. Curr Opin Cell Biol. 2012;24:254–61. doi: 10.1016/j.ceb.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 109.Li Z-H, Spektor A, Varlamova O, Bresnick AR. Mts1 regulates the assembly of nonmuscle myosin-IIA. Biochemistry. 2003;42:14258–66. doi: 10.1021/bi0354379. [DOI] [PubMed] [Google Scholar]
- 110.Mitsuhashi M, Sakata H, Kinjo M, Yazawa M, Takahashi M. Dynamic assembly properties of nonmuscle myosin II isoforms revealed by combination of fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy. J Biochem. 2011;149:253–63. doi: 10.1093/jb/mvq134. [DOI] [PubMed] [Google Scholar]
- 111.Badyal SK, Basran J, Bhanji N, Kim JH, Chavda AP, Jung HS, Craig R, Elliott PR, Irvine AF, Barsukov IL, et al. Mechanism of the Ca²+-dependent interaction between S100A4 and tail fragments of nonmuscle myosin heavy chain IIA. J Mol Biol. 2011;405:1004–26. doi: 10.1016/j.jmb.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Elliott PR, Irvine AF, Jung HS, Tozawa K, Pastok MW, Picone R, Badyal SK, Basran J, Rudland PS, Barraclough R, et al. Asymmetric mode of Ca²⁺-S100A4 interaction with nonmuscle myosin IIA generates nanomolar affinity required for filament remodeling. Structure. 2012;20:654–66. doi: 10.1016/j.str.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kiss B, Duelli A, Radnai L, Kékesi KA, Katona G, Nyitray L. Crystal structure of the S100A4-nonmuscle myosin IIA tail fragment complex reveals an asymmetric target binding mechanism. Proc Natl Acad Sci U S A. 2012;109:6048–53. doi: 10.1073/pnas.1114732109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ramagopal UA, Dulyaninova NG, Varney KM, Wilder PT, Nallamsetty S, Brenowitz M, et al. Structure of the S100A4/myosin-IIA complex. 2013. [DOI] [PMC free article] [PubMed]
- 115.Seri M, Cusano R, Gangarossa S, Caridi G, Bordo D, Lo Nigro C, Ghiggeri GM, Ravazzolo R, Savino M, Del Vecchio M, et al. The May-Heggllin/Fechtner Syndrome Consortium Mutations in MYH9 result in the May-Hegglin anomaly, and Fechtner and Sebastian syndromes. Nat Genet. 2000;26:103–5. doi: 10.1038/79063. [DOI] [PubMed] [Google Scholar]
- 116.Heath KE, Campos-Barros A, Toren A, Rozenfeld-Granot G, Carlsson LE, Savige J, Denison JC, Gregory MC, White JG, Barker DF, et al. Nonmuscle myosin heavy chain IIA mutations define a spectrum of autosomal dominant macrothrombocytopenias: May-Hegglin anomaly and Fechtner, Sebastian, Epstein, and Alport-like syndromes. Am J Hum Genet. 2001;69:1033–45. doi: 10.1086/324267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ravasi T, Hsu K, Goyette J, Schroder K, Yang Z, Rahimi F, Miranda LP, Alewood PF, Hume DA, Geczy C. Probing the S100 protein family through genomic and functional analysis. Genomics. 2004;84:10–22. doi: 10.1016/j.ygeno.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 118.Donato R. Intracellular and extracellular roles of S100 proteins. Microsc Res Tech. 2003;60:540–51. doi: 10.1002/jemt.10296. [DOI] [PubMed] [Google Scholar]