Abstract
A baffling aspect of metazoan proteostasis is the lack of an Hsp104 ortholog that rapidly disaggregates and reactivates misfolded polypeptides trapped in stress induced disordered aggregates, preamyloid oligomers, or amyloid fibrils. By contrast, in bacteria, protozoa, chromista, fungi, and plants, Hsp104 orthologs are highly conserved and confer huge selective advantages in stress tolerance. Moreover, in fungi, the amyloid remodeling activity of Hsp104 has enabled deployment of prions for various beneficial modalities. Thus, a longstanding conundrum has remained unanswered: how do metazoan cells renature aggregated proteins or resolve amyloid fibrils without Hsp104? Here, we highlight recent advances that unveil the metazoan protein-disaggregase machinery, comprising Hsp110, Hsp70, and Hsp40, which synergize to dissolve disordered aggregates, but are unable to rapidly solubilize stable amyloid fibrils. However, Hsp110, Hsp70, and Hsp40 exploit the slow monomer exchange dynamics of amyloid, and can slowly depolymerize amyloid fibrils from their ends in a manner that is stimulated by small heat shock proteins. Upregulation of this system could have key therapeutic applications in various protein-misfolding disorders. Intriguingly, yeast Hsp104 can interface with metazoan Hsp110, Hsp70, and Hsp40 to rapidly eliminate disease associated amyloid. Thus, metazoan proteostasis is receptive to augmentation with exogenous disaggregases, which opens a number of therapeutic opportunities.
Keywords: Hsp104, Hsp110, Hsp70, metazoa, protein disaggregation
Introduction
Controlling the “quality” of proteins beyond translation is key in assuring correct cellular functioning and preserving organismal health.1 For a nascent polypeptide chain, folding into the correct native structure and maintaining the correct native form is challenging within the crowded environment of a cell.2-4 Thus, proteins occasionally assume defective, nonfunctional conformations. To circumvent this fundamental problem of cell biology, nature has devised several mechanisms to prevent or resolve protein misfolding in the cell. For instance, molecular chaperones block protein aggregation and actively help proteins reach their native conformations.5 If a protein has been damaged beyond repair, it can be cleared by specialized degradation systems.6-8 Alas, protein misfolding and aggregation can overcome these systems, especially upon environmental stress, which can even elicit aging and disease.9 Indeed, protein misfolding underpins several devastating neurodegenerative diseases, including Alzheimer disease, Parkinson disease, Creutzfeldt-Jakob disease, and Huntington's disease.10,11
In the 1990s, Susan Lindquist and coworkers unequivocally defined a new branch in protein quality control: protein disaggregation coupled to protein reactivation.12-14 Although it had been speculated that protein disaggregation and reactivation might occur,15 it had never been convincingly demonstrated. Lindquist and colleagues discovered a new heat shock protein (Hsp) in Saccharomyces cerevisiae, Hsp104, which was found to have a key role in allowing cells to survive severe stress after heat treatment (thermotolerance).14 In subsequent investigations, Hsp104 was found to solubilize large protein aggregates resulting from severe heat stress and recover enzymatically active proteins from these aggregates.12 Accordingly, yeast cells lacking Hsp104 were no longer able to rapidly solubilize and reactivate proteins from an aggregated state following thermal stress.13,16
Since then, we have learned much about the way Hsp104 functions. Hsp104 is a ring-shaped homohexamer with two AAA+ nucleotide-binding domains (NBDs) per subunit that couple ATP binding and hydrolysis to protein disaggregation.17-22 Hsp104 is thought to drive protein disaggregation by threading substrates through its central channel to solution.23-26 Hsp104 disaggregates a diverse array of structures, ranging from stable amyloid to less stable disordered aggregates.12,13,27-33 Hsp104 hexamers adapt different mechanisms of intersubunit collaboration to disaggregate stress-induced aggregates vs. amyloid.27,34 Hsp104 acts alone or in concert with other molecular chaperones to rescue aggregated polypeptides.12,20,28,32,34-36 In particular, Hsp70, Hsp40, and small heat shock proteins (Hsp26 and Hsp42) can synergize with Hsp104 to promote the reactivation of protein aggregates.35,37,38 Hsp104 is also essential for the formation and propagation of several yeast prions; protein-based genetic elements comprised of amyloid fibers that can confer advantageous self-perpetuating changes in protein structure and function.32,39-46
Hsp104 is highly conserved in eubacteria and eukaryotes. Inexplicably, however, Hsp104 has no exact homolog or ortholog in metazoa.47 NBD2, but not other parts of Hsp104, appears to be partially conserved in the four ER-resident AAA+ proteins: torsin A, B, 2A, and 3A, as well as the mitochondrial AAA+ protein, SKD3.48-50 This deficiency of Hsp104 in animals is puzzling, as a protein that reverses protein aggregation and restores protein function would be pivotal in combating aberrant protein aggregation.17,47 The reason underlying the loss of Hsp104 is unknown, and is even more baffling because Hsp104 is well tolerated and even neuroprotective in animal systems.30,51-55 For example, Hsp104 rescues α-synuclein aggregation and dopaminergic neurodegeneration in a rat model of Parkinson disease.30 Whether mammals boast an equivalent protein disaggregase has endured as a persistent unanswered question.
Hsp110, Hsp70 and Hsp40 as a novel protein disaggregase system
We have recently shed some light on this issue and have identified the mammalian disaggregase system via biochemical fractionation of mammalian cytosol and reconstitution with pure components.56 The mammalian disaggregase system is comprised of an Hsp110 (Apg-2), an Hsp70 (Hsc70 or Hsp70), and an Hsp40 (Hdj1 or Hdj2).56 The combination of these three proteins was found to establish an active disaggregase system in the mammalian cytosol prepared from rat liver or sHeLa cells.56 Hsp110, Hsp70, and Hsp40 were able to refold proteins from large chemically or thermally denatured protein aggregates.56 Using pure proteins, we established that Hsp70 and Hsp40 alone are not sufficient for robust disaggregase activity, but must be supplemented with Hsp110.56 Hsp110 homologs are found in all eukaryotes and contribute toward thermotolerance in mammalian cells.57,58 Hsp110 can serve as a nucleotide exchange factor (NEF) for Hsp70 but also displays chaperone activity.59-64 We established that Hsp110-Hsp70-Hsp40 disaggregase activity was most effective against disordered, amorphous aggregates.56 Indeed, Hsp110, Hsp70 and Hsp40 were unable to rapidly disaggregate Sup35 prions or amyloid forms of α-synuclein.56 Disaggregase activity was conserved to the yeast homologs.56 Thus, Sse1 (Hsp110), Ssa1 (Hsp70), and Sis1 or Ydj1 (Hsp40) could synergize to rescue proteins from large disordered aggregates.56 This activity was slow in comparison to Hsp104- catalyzed protein disaggregation, which might help explain why minimal disaggregase activity is observed in yeast lacking Hsp104 immediately after heat shock.13 In yeast, Sse1 contributes to prion propagation65-67 and might also be involved in the dissolution of “Q-bodies” or “stress foci”: punctate cytoplasmic structures where misfolded proteins are collected prior to maturation into larger inclusions.68-70 Using a series of Sse1 mutants,61 we determined that Sse1 must engage both substrate and Hsp70, promote nucleotide exchange on Hsp70, and bind and hydrolyze ATP itself to promote disaggregation of disordered aggregates. Thus, simply providing Hsp70 with a NEF, such as Fes1 or Snl1ΔN, in place of Sse1 was insufficient to promote protein disaggregation.56 Likewise, using a series of Ssa1 mutants, we determined that Hsp70 must engage substrate and Hsp110, and hydrolyze ATP for protein disaggregation.56 Hsp40 must harbor a functional J domain to promote protein disaggregation, but the J domain alone is insufficient.56 Optimal disaggregase activity was achieved when the Hsp40 could stimulate Hsp110 and Hsp70 ATPase activity.56 Finally, while Hsp110, Hsp70, and Hsp40 were unable to rapidly resolve amyloid conformers directly, they enhanced disaggregation of Sup35 prions and α-synuclein amyloid fibrils by Hsp104.56
About a year later, a subsequent study confirmed the metazoan disaggregation activity exerted by Hsp110 (Apg-1, Apg-2, or Hsp105), Hsp70 (Hsc70 or Hsp70), and Hsp40 (Hdj1 or DNAJA2) in vitro.71 Curiously, under the in vitro conditions employed the ATPase activity of Hsp110 was not required to promote protein disaggregation.71 Hsp110 appeared to contribute primarily by acting as a nucleotide exchange factor (NEF) for Hsp70. Mild aggregation conditions were even established where Hsc70 and the alternative Hsp40, DNAJA2, could disaggregate substrates if provided with the Hsp70 NEFs Bag-1 or Snl1ΔN instead of Hsp110.71 Under these circumstances, stimulation of Hsp70 nucleotide exchange was sufficient for disaggregation.71
Using C. elegans as a model system, knockdown of Hsp110 in briefly heatshocked C. elegans resulted in persistent luciferase-YFP aggregates and a drastically reduced lifespan.71 The persistence of protein aggregates and lifespan reduction could reflect a requirement for Hsp110 in the solubilization of protein aggregates in vivo.71
Alternatively, it might point to a role for Hsp110 in the inhibition of ongoing aggregation after the transient heat shock. Unfortunately, the experiments performed could not differentiate between these two possibilities, as the expression of luciferase-YFP was not shut down after the transient heat shock. Thus, it is unclear whether the persistence of luciferase-YFP aggregates reflects a failure to disaggregate pre-existing luciferase-YFP or whether newly synthesized luciferase-YFP continued to aggregate after the transient heat shock (perhaps due to seeding by preformed aggregates).
Originally, to convincingly establish the disaggregase activity of Hsp104 in vivo, it was necessary to stringently shut down protein synthesis using cycloheximide immediately after the heat shock.13 In this way, one could be absolutely certain that any protein reactivation that occurred was due to recovery of previously aggregated protein and not due to the accumulation of newly synthesized material.13 A similar strategy has been employed to demonstrate that Hsp104 can solubilize amyloid in vivo.29 Thus, in the study by Rampelt et al. one cannot be certain what proportion of the soluble luciferase- YFP observed in wild-type C. elegans after a 12h or 24h recovery from heat shock represents newly synthesized protein or bona fide resolubilized protein.71 A similar issue arises in more recent experiments that also aimed to demonstrate in vivo disaggregase activity of Hsp110 using Drosophila S2 cells, but again the critical cycloheximide control was also omitted.72 As such, although the foregoing experiments provide compelling indications,71,72 we still await an unequivocal in vivo demonstration of Hsp110, Hsp70, and Hsp40 disaggregase activity.
More recently, Goloubinoff and colleagues corroborated that the cytosol and the endoplasmic reticulum of mammalian cells contain Hsp110 and Hsp70 machineries that can unfold and solubilize stably misfolded and aggregated protein.62 Outstandingly, Hsp110 (Hsp105) was found to be an ATP-dependent unfoldase that can prevent aggregation, catalyze the unfolding of misfolded polypeptides, and favor their conversion into native protein on its own.62 Thus, Hsp110 can act as a bona fide chaperone with unfolding activity, and does not simply serve as a NEF for Hsp70.62 Furthermore, titration of the ATP- and Hsp40-dependent refolding activity in the presence of various amounts of Hsp110 and Hsp70 showed optimal disaggregation activity at a 1:1 ratio.62
Intriguingly, even without ATP, Hsp110 promotes the release of a prebound substrate from Hsp70, and Hsp70 could activate the release of a prebound substrate from Hsp110.62 Together with our study,56 these findings conflict with the notion that the only function for Hsp110 in protein disaggregation is as a NEF for Hsp70.62 Indeed, it should also be noted that several key in vivo functions of Sse1 require its ATPase activity.73,74
Collectively, these results from three different groups independently corroborate the existence of a disaggregase system, consisting of Hsp110, Hsp70, and Hsp40, that couple protein disaggregation to protein renaturation in metazoa (Fig. 1). Discrepancies in the details of the system regarding the requirement for Hsp110 ATPase activity in disaggregation and the relative level of activity of the system are likely the result of different experimental conditions. For instance, we used equal concentrations of Hsp70 and Hsp110,56 while Rampelt and colleagues used a 1:10 ratio of Hsp110 to Hsp70.71 Analogously to our work, Goloubinoff and coworkers identified an optimal 1:1 ratio for the concentrations of Hsp110 and Hsp70, which might even suggest an Hsp110:Hsp70 heterodimer that co-operatively drives disaggregation via synergistic entropic pulling.56,62,75-77 Indeed, Sse1 has been co-crystallized in 1:1 complex with the Hsc70 nucleotide-binding domain, suggesting that a 1:1 complex could be critical.78
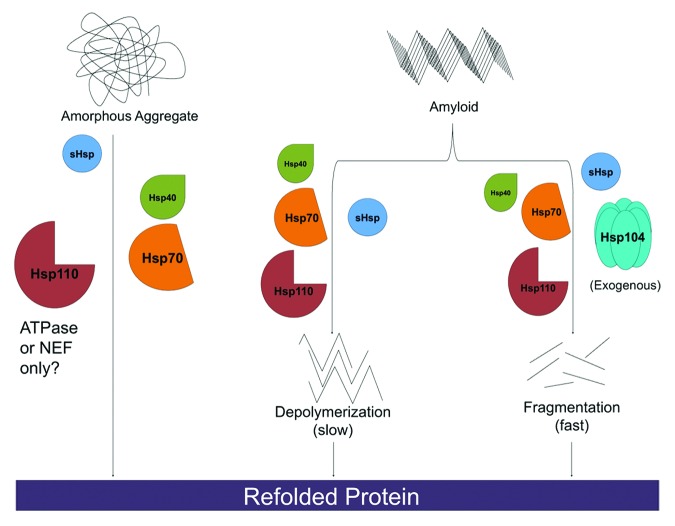
Figure 1. Hsp110, Hsp70, Hsp40 and sHsps are a disaggregation system in metazoan cells. Heat shock proteins Hsp110, Hsp70, and Hsp40 are capable of dissolving disordered aggregates. For labile aggregates, Hsp110 may only need to operate as a nucleotide exchange factor (NEF) for Hsp70, whereas for more stable aggregates it may need to serve as a NEF for Hsp70, engage substrate, and bind and hydrolyze ATP. Hsp110, Hsp70, and Hsp40 can also slowly depolymerize ordered amyloid substrates from their ends. Rapid amyloid dissolution can be achieved by supplementing Hsp110, Hsp70 and Hsp40 with exogenous Hsp104. Here, fibrils can be fragmented and monomers extracted from anywhere in the fibril (not just the ends), which leads to more rapid dissolution. sHsps can stimulate all of these protein disaggregation reactions, but are not absolutely required.
Moreover, Ssa1 and Sse1 display high affinity for different peptides,79 indicating that they might interact and exert force on different regions of the polypeptide to cooperatively drive disaggregation. In each of these studies,56,62,71 different conditions were used to generate the protein aggregates studied. For instance, in our study, more severe chemical or thermal denaturation was used to generate aggregates.56 Hence, it is highly probable that different aggregated conformers were studied in each case. We suggest that proteins can adopt a wide variety of conformations in the aggregated state, some of which are more labile and do not require the full chaperone repertoire of Hsp110 for disaggregation (NEF activity is sufficient), whereas others are more recalcitrant and require the full complement of Hsp110 activities encompassing: substrate binding, Hsp70 binding, promotion of nucleotide exchange on Hsp70, and ATP binding and hydrolysis. Additional studies are required to explore this hypothesis further. However, the Hsp70 NEF, Snl1ΔN, could not substitute for Hsp110 under our more stringent aggregation conditions,56 whereas it could under much milder aggregation conditions.71 An interesting parallel may be drawn with Hsp104, which employs distinct mechanisms to dissolve labile aggregates vs. stable amyloid.27 Hsp104 subunits within the hexamer can function independently to resolve disordered aggregates.27 Thus, even a single subunit within the hexamer can drive disaggregation of disordered aggregates.27 By contrast, multiple Hsp104 subunits must work together within the hexamer in a co-operative manner to drive amyloid dissolution.27 Some very stable amyloid conformations even require direct co-operation between two Hsp104 hexamers.27,56 By analogy, the Hsp110-Hsp70-Hsp40 system might also exhibit mechanistic plasticity in protein disaggregation. We suggest that more stable aggregated structures might necessitate the full repertoire of Hsp110 modalities, whereas the NEF activity might suffice for more facile conformers.
Amyloid Disaggregation in Metazoa
In addition to amorphous aggregates, misfolded proteins in the cell can form amyloids and prions.10,80,81 Amyloids are self-templating protein conformers.10,80,81 They form long, stable fibers by self-replicating their “cross-β” conformation at their growing ends and by converting other copies of the same protein to the “cross-β” amyloid form.10,80,81 When amyloid fibers become infectious, they are termed prions.10,80,81 Initially, we found Hsp110, Hsp70, and Hsp40 were unable to rapidly remodel amyloid in the absence of Hsp104.56 However, in a later study, we found Hsp110, Hsp70, and Hsp40 especially in conjunction with small heat shock proteins (sHsps), can very slowly depolymerize amyloid fibers (Sup35 prions or α-synuclein fibrils) from their ends,35 providing a pathway for amyloid disaggregation in metazoans in the absence of Hsp104. Amyloid depolymerization is a lengthy process that occurs on a similar timeframe to molecular recycling within amyloid fibers (days).35,82,83 The disaggregase system involving Hsp110, Hsp70, Hsp40, and sHsps might exploit this process to slowly eliminate amyloid by accelerating monomer dissociation or by capturing released monomers or by sealing off fibril ends once a monomer has been released thereby preventing monomer reassociation.35 While newly released monomers could hypothetically collect into toxic oligomers, the chaperone system would likely prevent any toxic oligomer formation. Remarkably, we found this activity to be conserved in humans.35 Thus, Hsp110 (Apg-2), Hsp70 (Hsc70), Hsp40 (Hdj1) and a small heat shock protein (HspB5) slowly depolymerized α-synuclein fibrils, which are connected to Parkinson disease.19 These data suggest that in metazoa, which lack an Hsp104 homolog, Hsp110, Hsp70, and Hsp40 can slowly eliminate amyloid forms by specifically hijacking their intrinsic monomer recycling process.35,82,83
Treating Neurodegenerative Disease: Can We Give Hsp110 a Boost?
The Hsp110-Hsp70-Hsp40 disaggregase system might prove to be an advantageous therapeutic target against the numerous neurological disorders connected to protein misfolding and aggregation.10 Indeed, co-expression of Hsp110 and Hsp40 in Drosophila melanogaster suppresses the cytotoxicity of polyglutamine aggregation.84 Importantly, the ATPase activity of Hsp110 was critical for this rescue.84 A feasible explanation for this effect invokes Hsp110 and Hsp40 interfacing with members of the Hsp70 family to disassemble polyglutamine aggregates and thus reduce the cellular toxicity of protein aggregation. Polyglutamine aggregation is connected to several neurodegenerative diseases, including Huntington’s disease and Spinocerebellar Ataxias.85 Similarly, Hsp110 was found to completely reverse a vesicle transport defect produced by a mutant (G85R) of Superoxide Dismutase 1 (SOD1) associated with amyotrophic lateral sclerosis (ALS) in the isolated axoplasm from the giant axon of the squid Loligo pealei.86 Hsp110 appears to either directly bind to the mutant SOD1, or to associate with the mutant protein via Hsp70, and occlude binding surfaces that would otherwise interact with endogenous proteins leading to a gain of toxic function. Further studies are warranted to determine whether Hsp110, Hsp70, and Hsp40 can also disaggregate misfolded SOD1 conformers connected to ALS or polyglutamine fibrils and oligomers connected to Huntington’s disease and Spinocerebellar Ataxias. Treatment of several neurodegenerative disorders could entail the activation of the Hsp110-Hsp70-Hsp40 disaggregase system. As Hsp110, Hsp70, and Hsp40 enhanced amyloid remodeling by Hsp104,56 one alternative possibility to achieve such activation would be to supplement metazoan cells with Hsp104.47 For instance, Hsp104 prevented the aggregation and toxicity of polyglutamine in C. elegans.54 In mouse and rat, Hsp104 expression resulted in extension the animal’s lifespan and rescue of striatal dysfunction respectively.53,55 We have recently introduced Hsp104 into Drosophila models of Spinocerebellar Ataxia Type-3.51 Notably, Hsp104 suppressed toxicity of a C-terminal ataxin-3 fragment when expressed even after the onset of polyglutamine-induced degeneration.51 This constitutes the first disaggregase treatment that halts disease progression after the start of pathogenic degeneration.51 Notably, induction of Hsp70 after polyglutamine-mediated degeneration had already initiated was unable to significantly mitigate disease progression.51 It is possible that simultaneous induction of not only Hsp70, but also Hsp110 and Hsp40 is necessary to achieve toxicity suppression after degeneration and aggregation have initiated.84
Another possibility to enhance clearance of harmful amyloids would be to boost sHsp levels or activity to facilitate the action of endogenous human Hsp110, Hsp70, and Hsp40. Thus, small molecules that induce the expression of these proteins without compromising other components of the stress response could be critical.11,87,88 For example, the ability to stimulate the dissolution of α-synuclein fibers in patients with Parkinson disease might provide an unprecedented therapeutic leap in the treatment of this disease. Although released monomers could theoretically reassemble into toxic oligomers, the proteostasis network would likely prevent this situation. Lastly, direct pharmacological activation of Hsp110 or Hsp70 is another attractive possibility.89 While pharmaceutical discovery efforts generally focus on protein inhibition, protein activation is an emerging field.90
In conclusion, the metazoan disaggregase machinery is comprised of the heat shock proteins Hsp110, Hsp70, and Hsp40, which dissolve disordered aggregates.56,62,71 Hsp110, Hsp70, and Hsp40 exploit the exchange dynamics of amyloid, and can slowly depolymerize amyloid fibrils from their ends.35 This amyloid depolymerase activity as well as the disaggregation of disordered aggregates is stimulated by sHsps.35,71 Augmentation of this disaggregase network could have key applications in various neurological disorders linked to protein misfolding. Fascinatingly, this metazoan disaggregation network is amenable to augmentation with exogenous disaggregases, which opens several exciting avenues for potential treatments.47
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Laura Castellano, Jamie DeNizio, Dr Meredith Jackrel, and Elizabeth Sweeny for helpful comments on the manuscript. MPT is supported by a PENN-PORT postdoctoral fellowship (K12GM081259). JS is supported by grants: NIH Director's New Innovator Award DP2OD002177, R21HD074510, R01GM099836, a Muscular Dystrophy Association Research Award (MDA277268), Packard Center for ALS Research at Johns Hopkins University, Target ALS, and an Ellison Medical Foundation New Scholar in Aging Award.
References
- 1.Lindquist SL, Kelly JW. Chemical and biological approaches for adapting proteostasis to ameliorate protein misfolding and aggregation diseases: progress and prognosis. Cold Spring Harb Perspect Biol. 2011;3:3. doi: 10.1101/cshperspect.a004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarkar M, Smith AE, Pielak GJ. Impact of reconstituted cytosol on protein stability. Proc Natl Acad Sci U S A. 2013;110:19342–7. doi: 10.1073/pnas.1312678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uversky VN, M Cooper E, Bower KS, Li J, Fink AL. Accelerated alpha-synuclein fibrillation in crowded milieu. FEBS Lett. 2002;515:99–103. doi: 10.1016/S0014-5793(02)02446-8. [DOI] [PubMed] [Google Scholar]
- 4.van den Berg B, Ellis RJ, Dobson CM. Effects of macromolecular crowding on protein folding and aggregation. EMBO J. 1999;18:6927–33. doi: 10.1093/emboj/18.24.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–55. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 6.Wong E, Cuervo AM. Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb Perspect Biol. 2010;2:a006734. doi: 10.1101/cshperspect.a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maday S, Holzbaur EL. Autophagosome assembly and cargo capture in the distal axon. Autophagy. 2012;8:858–60. doi: 10.4161/auto.20055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maday S, Wallace KE, Holzbaur EL. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol. 2012;196:407–17. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coelho M, Dereli A, Haese A, Kühn S, Malinovska L, DeSantis ME, Shorter J, Alberti S, Gross T, Tolić-Nørrelykke IM. Fission yeast does not age under favorable conditions, but does so after stress. Curr Biol. 2013;23:1844–52. doi: 10.1016/j.cub.2013.07.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cushman M, Johnson BS, King OD, Gitler AD, Shorter J. Prion-like disorders: blurring the divide between transmissibility and infectivity. J Cell Sci. 2010;123:1191–201. doi: 10.1242/jcs.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackrel ME, Shorter J. Shock and awe: unleashing the heat shock response to treat Huntington disease. J Clin Invest. 2011;121:2972–5. doi: 10.1172/JCI59190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/S0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 13.Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–8. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez Y, Lindquist SL. HSP104 required for induced thermotolerance. Science. 1990;248:1112–5. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 15.Pelham HR. Speculations on the functions of the major heat shock and glucose-regulated proteins. Cell. 1986;46:959–61. doi: 10.1016/0092-8674(86)90693-8. [DOI] [PubMed] [Google Scholar]
- 16.Vogel JL, Parsell DA, Lindquist S. Heat-shock proteins Hsp104 and Hsp70 reactivate mRNA splicing after heat inactivation. Curr Biol. 1995;5:306–17. doi: 10.1016/S0960-9822(95)00061-3. [DOI] [PubMed] [Google Scholar]
- 17.Vashist S, Cushman M, Shorter J. Applying Hsp104 to protein-misfolding disorders. Biochem Cell Biol. 2010;88:1–13. doi: 10.1139/O09-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeSantis ME, Shorter J. The elusive middle domain of Hsp104 and ClpB: location and function. Biochim Biophys Acta. 2012;1823:29–39. doi: 10.1016/j.bbamcr.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013;14:630–42. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desantis ME, Sweeny EA, Snead D, Leung EH, Go MS, Gupta K, Wendler P, Shorter J. Conserved distal loop residues in the Hsp104 and ClpB middle domain contact nucleotide-binding domain 2 and enable Hsp70-dependent protein disaggregation. J Biol Chem. 2014;289:848–67. doi: 10.1074/jbc.M113.520759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doyle SM, Genest O, Wickner S. Protein rescue from aggregates by powerful molecular chaperone machines. Nat Rev Mol Cell Biol. 2013;14:617–29. doi: 10.1038/nrm3660. [DOI] [PubMed] [Google Scholar]
- 22.Wendler P, Shorter J, Plisson C, Cashikar AG, Lindquist S, Saibil HR. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell. 2007;131:1366–77. doi: 10.1016/j.cell.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shorter J, Lindquist S. Navigating the ClpB channel to solution. Nat Struct Mol Biol. 2005;12:4–6. doi: 10.1038/nsmb0105-4. [DOI] [PubMed] [Google Scholar]
- 24.Wendler P, Shorter J, Snead D, Plisson C, Clare DK, Lindquist S, Saibil HR. Motor mechanism for protein threading through Hsp104. Mol Cell. 2009;34:81–92. doi: 10.1016/j.molcel.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lum R, Niggemann M, Glover JR. Peptide and protein binding in the axial channel of Hsp104. Insights into the mechanism of protein unfolding. J Biol Chem. 2008;283:30139–50. doi: 10.1074/jbc.M804849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lum R, Tkach JM, Vierling E, Glover JR. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem. 2004;279:29139–46. doi: 10.1074/jbc.M403777200. [DOI] [PubMed] [Google Scholar]
- 27.DeSantis ME, Leung EH, Sweeny EA, Jackrel ME, Cushman-Nick M, Neuhaus-Follini A, Vashist S, Sochor MA, Knight MN, Shorter J. Operational plasticity enables hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell. 2012;151:778–93. doi: 10.1016/j.cell.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeSantis ME, Shorter J. Hsp104 drives “protein-only” positive selection of Sup35 prion strains encoding strong [PSI(+)] Chem Biol. 2012;19:1400–10. doi: 10.1016/j.chembiol.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DiSalvo S, Derdowski A, Pezza JA, Serio TR. Dominant prion mutants induce curing through pathways that promote chaperone-mediated disaggregation. Nat Struct Mol Biol. 2011;18:486–92. doi: 10.1038/nsmb.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo Bianco C, Shorter J, Régulier E, Lashuel H, Iwatsubo T, Lindquist S, Aebischer P. Hsp104 antagonizes alpha-synuclein aggregation and reduces dopaminergic degeneration in a rat model of Parkinson disease. J Clin Invest. 2008;118:3087–97. doi: 10.1172/JCI35781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–7. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 32.Sweeny EA, Shorter J. Prion proteostasis: Hsp104 meets its supporting cast. Prion. 2008;2:135–40. doi: 10.4161/pri.2.4.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell. 2006;23:425–38. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doyle SM, Shorter J, Zolkiewski M, Hoskins JR, Lindquist S, Wickner S. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol. 2007;14:114–22. doi: 10.1038/nsmb1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duennwald ML, Echeverria A, Shorter J. Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol. 2012;10:e1001346. doi: 10.1371/journal.pbio.1001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008;27:2712–24. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cashikar AG, Duennwald M, Lindquist SL. A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J Biol Chem. 2005;280:23869–75. doi: 10.1074/jbc.M502854200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haslbeck M, Miess A, Stromer T, Walter S, Buchner J. Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J Biol Chem. 2005;280:23861–8. doi: 10.1074/jbc.M502697200. [DOI] [PubMed] [Google Scholar]
- 39.Newby GA, Lindquist S. Blessings in disguise: biological benefits of prion-like mechanisms. Trends Cell Biol. 2013;23:251–9. doi: 10.1016/j.tcb.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 40.True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–7. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- 41.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–83. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 42.Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482:363–8. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–58. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–4. doi: 10.1126/science.7754373. [psi+] [DOI] [PubMed] [Google Scholar]
- 45.Liebman SW, Chernoff YO. Prions in yeast. Genetics. 2012;191:1041–72. doi: 10.1534/genetics.111.137760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–34. [PMC free article] [PubMed] [Google Scholar]
- 47.Shorter J. Hsp104: a weapon to combat diverse neurodegenerative disorders. Neurosignals. 2008;16:63–74. doi: 10.1159/000109760. [DOI] [PubMed] [Google Scholar]
- 48.Murdock DG, Boone BE, Esposito LA, Wallace DC. Up-regulation of nuclear and mitochondrial genes in the skeletal muscle of mice lacking the heart/muscle isoform of the adenine nucleotide translocator. J Biol Chem. 1999;274:14429–33. doi: 10.1074/jbc.274.20.14429. [DOI] [PubMed] [Google Scholar]
- 49.Ozelius LJ, Page CE, Klein C, Hewett JW, Mineta M, Leung J, Shalish C, Bressman SB, de Leon D, Brin MF, et al. The TOR1A (DYT1) gene family and its role in early onset torsion dystonia. Genomics. 1999;62:377–84. doi: 10.1006/geno.1999.6039. [DOI] [PubMed] [Google Scholar]
- 50.Périer F, Radeke CM, Raab-Graham KF, Vandenberg CA. Expression of a putative ATPase suppresses the growth defect of a yeast potassium transport mutant: identification of a mammalian member of the Clp/HSP104 family. Gene. 1995;152:157–63. doi: 10.1016/0378-1119(94)00697-Q. [DOI] [PubMed] [Google Scholar]
- 51.Cushman-Nick M, Bonini NM, Shorter J. Hsp104 suppresses polyglutamine-induced degeneration post onset in a drosophila MJD/SCA3 model. PLoS Genet. 2013;9:e1003781. doi: 10.1371/journal.pgen.1003781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dandoy-Dron F, Bogdanova A, Beringue V, Bailly Y, Tovey MG, Laude H, Dron M. Infection by ME7 prion is not modified in transgenic mice expressing the yeast chaperone Hsp104 in neurons. Neurosci Lett. 2006;405:181–5. doi: 10.1016/j.neulet.2006.05.066. [DOI] [PubMed] [Google Scholar]
- 53.Perrin V, Régulier E, Abbas-Terki T, Hassig R, Brouillet E, Aebischer P, Luthi-Carter R, Déglon N. Neuroprotection by Hsp104 and Hsp27 in lentiviral-based rat models of Huntington’s disease. Mol Ther. 2007;15:903–11. doi: 10.1038/mt.sj.6300141. [DOI] [PubMed] [Google Scholar]
- 54.Satyal SH, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer JM, Morimoto RI. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2000;97:5750–5. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vacher C, Garcia-Oroz L, Rubinsztein DC. Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington’s disease. Hum Mol Genet. 2005;14:3425–33. doi: 10.1093/hmg/ddi372. [DOI] [PubMed] [Google Scholar]
- 56.Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS One. 2011;6:e26319. doi: 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Easton DP, Kaneko Y, Subjeck JR. The hsp110 and Grp1 70 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones. 2000;5:276–90. doi: 10.1379/1466-1268(2000)005<0276:THAGSP>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oh HJ, Chen X, Subjeck JR. Hsp110 protects heat-denatured proteins and confers cellular thermoresistance. J Biol Chem. 1997;272:31636–40. doi: 10.1074/jbc.272.50.31636. [DOI] [PubMed] [Google Scholar]
- 59.Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25:2510–8. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–28. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Polier S, Dragovic Z, Hartl FU, Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008;133:1068–79. doi: 10.1016/j.cell.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 62.Mattoo RU, Sharma SK, Priya S, Finka A, Goloubinoff P. Hsp110 is a bona fide chaperone using ATP to unfold stable misfolded polypeptides and reciprocally collaborate with Hsp70 to solubilize protein aggregates. J Biol Chem. 2013;288:21399–411. doi: 10.1074/jbc.M113.479253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Polier S, Hartl FU, Bracher A. Interaction of the Hsp110 molecular chaperones from S. cerevisiae with substrate protein. J Mol Biol. 2010;401:696–707. doi: 10.1016/j.jmb.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 64.Shaner L, Sousa R, Morano KA. Characterization of Hsp70 binding and nucleotide exchange by the yeast Hsp110 chaperone Sse1. Biochemistry. 2006;45:15075–84. doi: 10.1021/bi061279k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fan Q, Park KW, Du Z, Morano KA, Li L. The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics. 2007;177:1583–93. doi: 10.1534/genetics.107.077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moran C, Kinsella GK, Zhang ZR, Perrett S, Jones GW. Mutational analysis of Sse1 (Hsp110) suggests an integral role for this chaperone in yeast prion propagation in vivo. G3 (Bethesda) 2013;3:1409–18. doi: 10.1534/g3.113.007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sadlish H, Rampelt H, Shorter J, Wegrzyn RD, Andréasson C, Lindquist S, Bukau B. Hsp110 chaperones regulate prion formation and propagation in S. cerevisiae by two discrete activities. PLoS One. 2008;3:e1763. doi: 10.1371/journal.pone.0001763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Escusa-Toret S, Vonk WI, Frydman J. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat Cell Biol. 2013;15:1231–43. doi: 10.1038/ncb2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spokoini R, Moldavski O, Nahmias Y, England JL, Schuldiner M, Kaganovich D. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep. 2012;2:738–47. doi: 10.1016/j.celrep.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 70.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–95. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rampelt H, Kirstein-Miles J, Nillegoda NB, Chi K, Scholz SR, Morimoto RI, Bukau B. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 2012;31:4221–35. doi: 10.1038/emboj.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cherkasov V, Hofmann S, Druffel-Augustin S, Mogk A, Tyedmers J, Stoecklin G, Bukau B. Coordination of Translational Control and Protein Homeostasis during Severe Heat Stress. Curr Biol. 2013;23:2452–62. doi: 10.1016/j.cub.2013.09.058. [DOI] [PubMed] [Google Scholar]
- 73.Goeckeler JL, Stephens A, Lee P, Caplan AJ, Brodsky JL. Overexpression of yeast Hsp110 homolog Sse1p suppresses ydj1-151 thermosensitivity and restores Hsp90-dependent activity. Mol Biol Cell. 2002;13:2760–70. doi: 10.1091/mbc.02-04-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hrizo SL, Gusarova V, Habiel DM, Goeckeler JL, Fisher EA, Brodsky JL. The Hsp110 molecular chaperone stabilizes apolipoprotein B from endoplasmic reticulum-associated degradation (ERAD) J Biol Chem. 2007;282:32665–75. doi: 10.1074/jbc.M705216200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ben-Zvi A, De Los Rios P, Dietler G, Goloubinoff P. Active solubilization and refolding of stable protein aggregates by cooperative unfolding action of individual hsp70 chaperones. J Biol Chem. 2004;279:37298–303. doi: 10.1074/jbc.M405627200. [DOI] [PubMed] [Google Scholar]
- 76.De Los Rios P, Ben-Zvi A, Slutsky O, Azem A, Goloubinoff P. Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proc Natl Acad Sci U S A. 2006;103:6166–71. doi: 10.1073/pnas.0510496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goloubinoff P, De Los Rios P. The mechanism of Hsp70 chaperones: (entropic) pulling the models together. Trends Biochem Sci. 2007;32:372–80. doi: 10.1016/j.tibs.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 78.Schuermann JP, Jiang J, Cuellar J, Llorca O, Wang L, Gimenez LE, Jin S, Taylor AB, Demeler B, Morano KA, et al. Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol Cell. 2008;31:232–43. doi: 10.1016/j.molcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goeckeler JL, Petruso AP, Aguirre J, Clement CC, Chiosis G, Brodsky JL. The yeast Hsp110, Sse1p, exhibits high-affinity peptide binding. FEBS Lett. 2008;582:2393–6. doi: 10.1016/j.febslet.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shorter J. Emergence and natural selection of drug-resistant prions. Mol Biosyst. 2010;6:1115–30. doi: 10.1039/c004550k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carulla N, Caddy GL, Hall DR, Zurdo J, Gairí M, Feliz M, Giralt E, Robinson CV, Dobson CM. Molecular recycling within amyloid fibrils. Nature. 2005;436:554–8. doi: 10.1038/nature03986. [DOI] [PubMed] [Google Scholar]
- 83.Carulla N, Zhou M, Giralt E, Robinson CV, Dobson CM. Structure and intermolecular dynamics of aggregates populated during amyloid fibril formation studied by hydrogen/deuterium exchange. Acc Chem Res. 2010;43:1072–9. doi: 10.1021/ar9002784. [DOI] [PubMed] [Google Scholar]
- 84.Kuo Y, Ren S, Lao U, Edgar BA, Wang T. Suppression of polyglutamine protein toxicity by co-expression of a heat-shock protein 40 and a heat-shock protein 110. Cell Death Dis. 2013;4:e833. doi: 10.1038/cddis.2013.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Blum ES, Schwendeman AR, Shaham S. PolyQ disease: misfiring of a developmental cell death program? Trends Cell Biol. 2013;23:168–74. doi: 10.1016/j.tcb.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Song Y, Nagy M, Ni W, Tyagi NK, Fenton WA, López-Giráldez F, Overton JD, Horwich AL, Brady ST. Molecular chaperone Hsp110 rescues a vesicle transport defect produced by an ALS-associated mutant SOD1 protein in squid axoplasm. Proc Natl Acad Sci U S A. 2013;110:5428–33. doi: 10.1073/pnas.1303279110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Calamini B, Silva MC, Madoux F, Hutt DM, Khanna S, Chalfant MA, Saldanha SA, Hodder P, Tait BD, Garza D, et al. Small-molecule proteostasis regulators for protein conformational diseases. Nat Chem Biol. 2012;8:185–96. doi: 10.1038/nchembio.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Labbadia J, Cunliffe H, Weiss A, Katsyuba E, Sathasivam K, Seredenina T, Woodman B, Moussaoui S, Frentzel S, Luthi-Carter R, et al. Altered chromatin architecture underlies progressive impairment of the heat shock response in mouse models of Huntington disease. J Clin Invest. 2011;121:3306–19. doi: 10.1172/JCI57413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang AM, Miyata Y, Klinedinst S, Peng HM, Chua JP, Komiyama T, Li X, Morishima Y, Merry DE, Pratt WB, et al. Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat Chem Biol. 2013;9:112–8. doi: 10.1038/nchembio.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zorn JA, Wells JA. Turning enzymes ON with small molecules. Nat Chem Biol. 2010;6:179–88. doi: 10.1038/nchembio.318. [DOI] [PubMed] [Google Scholar]