Abstract
A bifunctional oligonucleotide integrates in situ synthesis of a fluorogenic silver cluster with recognition of a target DNA sequence. With the template C3AC3AC3GC3A, a complex forms with 10 silver atoms that possesses electronic transitions in the near-infrared and that is detected at nM concentrations using diode laser excitation. Pendant to this cluster encoding region, the recognition component binds a target DNA strand through hybridization, and decoupling of these two regions of the composite sensor renders a modular sensor for specific oligonucleotides. A target is detected using a quencher strand that bridges the cluster template and recognition components and disturbs cluster binding, as indicated by static quenching. Competitive displacement of the quencher by the target strand restores the favored cluster environment, and our key finding is that this exchange enhances emission through a proportional increase in the number of emissive clusters. DNA detection is also accomplished in serum-containing buffers, by taking advantage of the high brightness of this fluorophore and the inherently low endogenous background in the near-infrared spectral region. Cluster stability in this biological environment is enhanced by supplementing the solutions with Ag+.
Keywords: Silver Cluster, DNA Template, Fluorescence, Biosensing
As sensors, nucleic acids offer robust platforms for detecting a diversity of analytes.1,2 DNA and RNA are prominent targets for analysis, as stringent detection is imposed by base pairing and stacking between the target and sensor.3 This selectivity is exemplified by molecular beacons that balance intramolecular association within the stem against intermolecular hybridization of the desired sequence within the loop, thereby allowing discrimination of sequences that differ by one nucleotide.4–6 A wider range of analytes is accessible through the multifarious structures available to nucleic acids.1,7,8 Folding of secondary structural elements such as stems, loops, and bulges into a composite aptamer structure produces binding pockets that allow fine distinctions between ligands, as illustrated by the 10,000 fold difference in affinity for theophylline and its methylated variant caffeine.9–11 Through the process of systematic evolution, aptamer sequences have been developed for a diverse range of analytes, from metal ions to proteins to cells.1,10,12
When target recognition is coupled with signal transduction, the diagnostic capabilities of nucleic acid sensors are experimentally realized. A diverse range of techniques are fundamentally linked by the ability to conveniently and precisely modify the sensing nucleic acid strand.1,7 For surface-based techniques, oligonucleotides are terminally attached to project sensor binding sites into the bulk solution, and target analytes can trigger electrochemical changes that achieve sensitive analyte detection in a range of matrices.2 Nanomaterials provide alternative sensing platforms that exhibit high surface areas with enhanced mass transport, high receptor loading with synergistic amplification of the target response, and translation of the surface binding event to a spectroscopic response.13 Homogeneous assays based on fluorescence rely on the positioning of exogenous chromophores that transduce ligand binding by intensity, lifetime, and anisotropy changes.14,15 Such assays are rapid and cost-effective because sensitivity minimizes sample demand while selectivity minimizes purification and enables multiplexed analysis.
A new perspective on nanomaterial biosensors is provided by DNA-based silver clusters, in which a DNA matrix encapsulates a small (~10 atoms) aggregate of silver atoms.16–18 The base sequence of the oligonucleotide host encodes the formation of specific clusters that have distinct spectra covering the blue-green to near-infrared region.19,20 These metallic fluorophores efficiently excite to and relax from higher-lying electronic states, and they feature short-lived dark states that both enhance the overall emission rate and that can be optically addressed.21,22 Combining these spectroscopic features with the functionality of the host DNA matrix, these hybrid nanomaterials are promising biosensors. Emission quenching represents one method of target recognition, as illustrated by the Hg2+ suppression of red emission from a dC12-based cluster.23 Likewise, thiols quench cluster emission, and their association can be specifically reversed by Cu2+ via oxidation and concomitant disulfide bond formation.24 The range of analytes has been extended to proteins, with nM detection of thrombin using an aptamer combined with the DNA sequence for a red emitting cluster.25 Another approach uses DNA context to influence the cluster environment and hence emission properties. The potential impact of DNA-encapsulated silver clusters as sensors is exemplified by a 500-fold increase in emission intensity that accompanies the conversion of nonemissive to emissive clusters based on guanine proximity, exceeding responses from related sensors utilizing organic chromophores.26,27 This context-dependent emission has been used to create NanoCluster Beacons that spectroscopically signal target recognition with high discrimination. Spectroscopic signatures of base pairing and mispairing arise from perturbed base pairing in regions adjacent to the cluster template.28 All of these assays feature fluorescent reporters that are conveniently synthesized simply by varying the nucleobase sequence and that enable sensitive, in situ detection of the analyte using fluorescence spectroscopy.
This work focuses on a bifunctional oligonucleotide that both stabilizes a specific silver cluster and binds a target DNA strand (Fig. 1). Using C3AC3AC3GC3A, a 10-atom silver conjugate forms, whose fluorescence brightness and lifetime make it a viable probe for near-infrared detection.29,30 This cluster favors the 3' region of its DNA template, so the strand recognition region was appended to this terminus with the goal of spectroscopically signaling target binding. Decoupling of the target and recognition components of this bifunctional oligonucleotide is indicated by the spectral similarities of the clusters bound to the base template C3AC3AC3GC3A vs C3AC3AC3GC3A-duplex constructs, irrespective of the base composition and length of the pendant duplex. Central to this detection scheme is the quencher sequence that overlaps the target and recognition components within the composite oligonucleotide. This strand perturbs the cluster through static quenching, as demonstrated by comparing fluorescence intensities and lifetimes. From a structural perspective, size exclusion chromatography shows that exchange of quencher and target strands accompanies an enhancement in fluorescence intensity. Enumeration via fluorescence correlation spectroscopy demonstrates that this signal amplification arises from a proportional increase in the number of emitting species. These results indicate that the quencher sequence invades and disrupts the favored binding site in the host C3AC3AC3GC3A, and this perturbation is relieved by exchange with the target strand. Another advantage of this fluorophore is its near-infrared electronic transition that allows probe detection in biological media. Comparable intensity changes in pristine buffer vs. buffer with serum support a common mechanism of strand exchange in these two environments. Collectively, the utility of this DNA-encapsulated silver cluster fluorophore is established through its convenient, base-programmable synthesis, its inherent brightness, a tempered fluorescence response using a label-free quencher strand, and functionality in complex biological environments.
Figure 1.
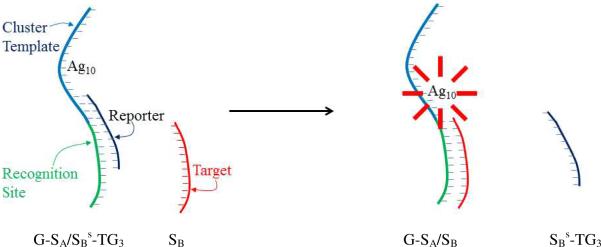
Representation of the strand exchange process using the bifunctional sensor strand with a template sequence for the silver cluster (blue) and a recognition site (green) for the target strand (red). The bridging quencher strand (black) statically quenches emission from a fraction of the clusters bound to the DNA host. After the quencher is displaced by the target, emission from the near-infrared emissive silver cluster is enhanced by a proportional increase in the number of emitting species. This population shift could be associated with a greater accessibility of the favored cluster binding site when the target binds.
Experimental Section
Oligonucleotides (Integrated DNA Technologies and Operon) were purified by desalting and dissolved in deionized water (Barnstead Nanopure). Concentrations were determined by absorbance using molar absorptivities based on the nearest-neighbor approximation. Silver clusters were synthesized by combining DNA and Ag+ solutions in a 10 mM citrate buffer at pH = 7 to give a relative concentration of 8 Ag+:oligonucleotide.18 Then, an aqueous solution of BH4− was added to give a final concentration of 4 BH4−:oligonucleotide, and the resulting solution was vigorously shaken for 1 min. The samples reacted overnight in the dark at 4 °C. Prior to forming the clusters, duplexes were formed by combining their single-stranded components in stoichiometric amounts and heating to 90 °C followed by slow (> 2 hr.) cooling at room temperature. For strand exchange, the target strand was added to yield a final concentration of 50 μM in the 400 μL solution containing a total duplex concentration of 25 μM of the cluster-conjugated quencher duplex in the citrate buffer. The resulting sample was warmed to 42 °C for 5 min followed by equilibration to room temperature. Absorption spectra were acquired on a Cary 50 (Varian), and emission spectra were acquired on a Fluoromax-3 (Jobin Yvon Horiba) using an excitation wavelength of 725 nm. Fluorescence lifetimes were measured using time-correlated single-photon counting using excitation with a pulsed diode laser (730 nm) operating at 5 MHz. Emission was collected with a microscope objective and spectrally filtered with a 735 nm long pass filter. TTL pulses from the photomultiplier tube (R928, Hamamatsu) were recorded by a TPSPC module (PicoHarp 300, PicoQuant). Fluorescence decays accounted for convolution of the fluorescence decay with the instrument response function. Fluorescence correlation spectroscopy studies were conducted using diode laser excitation at 690 nm with current and temperature control (LTC100, Thorlabs).30 A 63× 1.2 NA water immersion microscope objective (Zeiss) was used in a laser epi-illuminated geometry to excite the sample and collect the fluorescence. A long-pass dichroic (Semrock) was used to reflect the laser and transmit cluster emission, which was passed through a long pass filter and coupled into a 50-μm diameter fiber located at the image plane of the microscope. The fiber was connected to actively-quenched single-photon counting avalanche photodiode detectors (SPCMAQR14, Perkin Elmer) using a Hanbury Brown-Twiss setup.31 The resulting TTL signal outputs were cross correlated (Flex02-01D, Correlator.com) to give an autocorrelation free of afterpulsing artifacts, thereby improving time resolution. To determine the probe volume and hence cluster concentrations, dilute reference solutions of Cy7 with known concentrations were used.32,33 To fully model the correlation function, coupling of a dark electronic state was included in the analysis.30 Size exclusion chromatography used a 300 × 7.8 mm i.d. column (BioSep, Phenomenex) on an HPLC system (Prominence, Shimadzu) using a 10 mM citrate buffer at pH = 7 with 300 mM NaClO4 to minimize matrix adsorption.34 Absorbance and fluorescence measurements of the separated species were made using the SPD-M20A and RF-10XL, respectively. For the thymine oligonucleotides dT5, dT12, dT30, the averages and standard deviations were used in the linear fit to relate the retention times to the hydrodynamic radii, from which the radii of the cluster conjugates were determined using standard error propagation methods.35
Results
Bifunctional Oligonucleotide
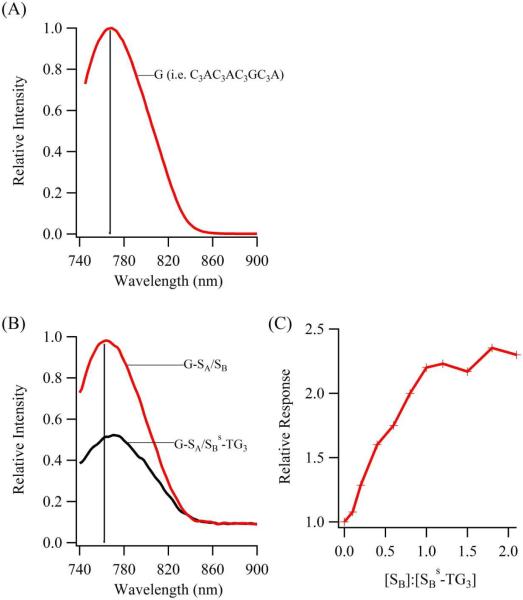
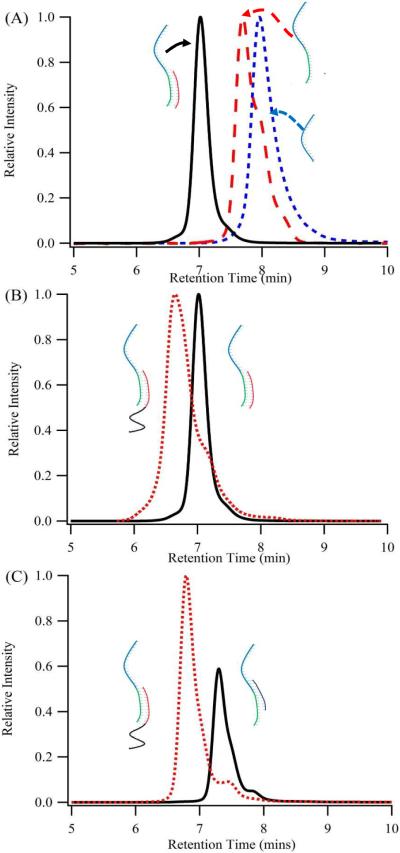
DNA detection is accomplished with a sensing oligonucleotide that integrates synthesis of a specific silver cluster and association with a particular DNA strand (Fig. 1). For the cluster encoding region, C3AC3AC3GC3A complexes 10 silver atoms, and the resulting aggregate emits at ~770 nm (λex = 720 nm) with a comparable brightness (ε × ϕf = 24,000 M−1cm−1) to spectrally similar organic and quantum dot fluorophores (Fig. 2).29,33,36 Base substitutions indicate that the cluster associates with the 3' region of C3AC3AC3GC3A, thus the recognition component for DNA binding was appended to this terminus.30 The cluster bound to C3AC3AC3GC3A-duplexes display similar spectra in relation to C3AC3AC3GC3A alone, thus supporting integrity of the cluster binding site with the proximal duplex (Figs. 2A/2B and 1S). To establish secondary structure within the DNA-cluster assemblies, chromatographic and spectroscopic efforts were directed towards the G–SA/SB construct, in which the mixed-base, 12 base-pair SA/SB duplex is derived from the bacteriophage lambda genome (Table 1).37 Size exclusion chromatography coupled with fluorescence detection demonstrates that progression from C3AC3AC3GC3A (i.e.G) to G–SA to G–SA/SB correlates with shorter retention times for the DNA-cluster complexes, indicative of their increasing size due to oligonucleotide lengthening and hybridization (Fig. 3A). Because silver clusters can perturb DNA conformation, duplex formation within the cluster complex with G–SA/SB is established using a dT10 size reporter attached to SB (Fig. 3B).38 This thymine tail is innocuous because it has a low affinity for silver clusters at neutral pH and because its 5' terminal positioning does not interfere with hybridization.39 Based on five thymine oligonucleotide standards, addition of the thymine tail to G–SA/SB increases the overall hydrodynamic radius of the cluster-host complex by 1.45 ± 0.06 nm, which is similar to the 1.6 nm hydrodynamic diameter of dT10 alone.40 This correspondence supports a structure in which C3AC3AC3GC3A hosts the near infrared emitting cluster and has a duplex appendage.
Figure 2.
(A) Emission spectra collected for cluster bound to C3AC3AC3GC3A, with the vertical line depicting the emission maximum. (B) Emission spectra for the clusters conjugated with G–SA before (black) and after (red) exchange of SBs-TG3 with SB. The vertical line has the same spectral position as the emission maximum in (A), and this spectral similarity between the G and G–SA/SB bound clusters indicates their comparable environments. A two- to three-fold enhancement in the emission was determined from multiple measurements with different sample preparations. (C) Relative enhancement of the near infrared emission when increasing amounts of SB are added to the cluster conjugate of G–SA/SBs-TG3. The plateau in the fluorescence intensity indicates saturation of the recognition site through hybridization with the complementary sequence.
Table 1.
Oligonucleotides used for Strand Exchange Studiesa
| DNA Sequence | Denoted by |
|---|---|
| C3AC3AC3GCCCA | G |
| C3AC3AC3GCCCA - AGGTCGCCGCCC + T C CAGCGGCGGG | G-SA + SB |
| C3AC3AC3GCCCA - AGGTCGCCGCCC + GGGT TCCAGCGGCGGG | G-SA + SB-TG3 |
| C3AC3AC3GCCCA - AGGTCGCCGCCC + GGGT T C CAGCG | G-SA + SBs-TG3 |
| C3AC3AC3GCCCA - GGGCGGCGACCT + C C CGCCGCTGGA | G-SB + SA |
| C3AC3AC3GCCCA - GGGCGGCGACCT + GGGT CCCGCCG | G-SB + SAs-TG3 |
| C3AC3AC3GCCCA - TCAA CATC AGTC TGAT AAGC TA + AGTT GTAG TCAG ACTA TTCGAT | G-SC + SD |
| C3AC3AC3GCCCA - TCAA CATC AGTC TGAT AAGC TA + GGGT AGTT GTAG TCAG A | G-SC + SDs-TG3 |
The first column provides the sequences used for the strand exchange reactions.
Consistent with the color scheme in Figure 1, the silver cluster template has blue lettering, the recognition component has green lettering, the quencher has black lettering, and the target sequence has red lettering. The second column provides the abbreviations used in the text to designate these constructs.
Figure 3.
(A) Size-exclusion chromatograms derived from the near-infrared emission of the clusters associated with G–SA/SB, G–SA, and G, in order of increasing retention times. This trend supports the cluster bound to structurally-distinct DNA hosts. (B) Duplex formation within the cluster complex with G–SA is supported by size-exclusion chromatograms of G–SA/SB and GSA/SB-dT10 that differ by addition of the thymine tail. This shift in retention time is arises from the size of single-stranded dT10. (C) Using a dT10 tail appended to SB, the differences in retention times between G–SA/SBs-TG3 and G–SA/SB–dT10 based clusters allowed strand exchange to be followed chromatographically. A small peak following strand exchange is not attributed to inefficient strand exchange based on the lack of change in the relative intensities of the two peaks with up to 5 equivalents of SB>s-dT10 and the same relative pattern of intensities and retention times for the two peaks associated with the G–SA/SB template.
Fluorescence Quenching
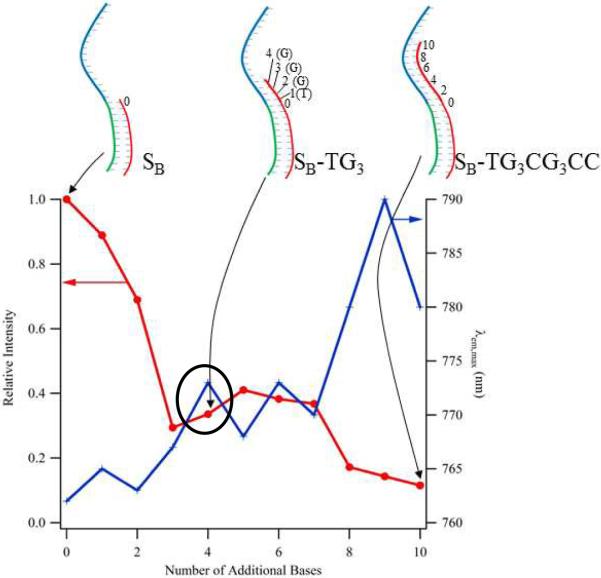
To spectroscopically detect hybridization of the target sequence SB with G–SA, a label-free quencher sequence was designed to modify the cluster emission. Earlier studies of the pH dependence of cluster emission showed that higher degrees of protonation of the N3 sites in cytosine and thymine inhibit cluster affinity, and these results provided the basis for considering how the cluster is influenced by base pairing.39,41 Through a series of 3' additions to the anchoring SB sequence, complementary oligonucleotides progressively encroach on the cluster binding region of C3AC3AC3GC3A and correspondingly diminish the fluorescence intensity (Fig. 4). The steepest diminution over the range of 1 to 4 additional bases is consistent with perturbation of the favored cluster binding site on the 3' end of C3AC3AC3GC3A. Our subsequent studies focus on the oligonucleotide with 4 additional bases, SB-TG3, which causes a three-fold reduction in the emission intensity. Residual emission accompanying further extension suggests that the cluster binds to alternate binding sites within the repeated motifs of the cluster template where it emits less efficiently. These intensity changes are linked with environmental changes, as the emission shifts to longer wavelengths with lengthening of the complementary sequences (Fig. 4). This effect of base pairing in the cluster binding region provides the foundation for recognition of specific DNA sequences.
Figure 4.
Dependence of the fluorescence intensity (left axis, crosses) and emission maximum (right axis, circles) on the length of the oligonucleotides that are anchored in the recognition site and encroach into the cluster template. The oligonucleotides having 0, 4, and 10 additional bases appended to SB are specifically depicted using the inset structures. The sequence SBs-TG3 provided the basis for the quenching strand used in our studies (circle).
Quencher Sequence
A quencher sequence was constructed that bridges the cluster template and target recognition regions and consequently both reduces cluster emission and inhibits target binding. Two design factors were balanced: the quencher should have a 3' terminus that induces measurable quenching and should be competitively displaced by the target strand (Fig. 1). Analogous to SB-TG3, the shortened variant SBs-TG3 also has the 4 base sequence TGGG that complements the 3' terminus of the cluster template and comparably quenches the emission (Fig. 2B and 4). Relative its analog, SBs-TG3 is truncated by 5 bases on its 5' terminus to give an 11-base strand whose affinity is lower relative to the 12-base target sequence SB. Conservation of the 3' terminus that is responsible for the diminished fluorescence yields similar fluorescence spectra for the unmodified SB-TG3 and truncated SBs-TG3 quenching sequences. Furthermore, these similarities demonstrate that the run of 5 unpaired bases at the terminus of SA are not effective binding sites for the clusters and therefore do not interfere with cluster binding on C3AC3AC3GC3A.
Spectroscopic and chromatographic studies establish that the target strand can be detected by displacing the quencher strand. Starting with an overall duplex concentration of 25 μM, cluster conjugates produced with G–SA/SBs-TG3 exhibit similar spectra and lifetimes in relation to clusters that form with C3AC3AC3GC3A alone (Fig. 2 and 5). These analogous properties support the formation of the same 10-atom Ag cluster with both templates.30 The spectral differences indicate that the electronic environment for the cluster depends on the DNA context, and the impact of the nucleobase environment is also evident in the two- to three-fold enhancement in the emission when SBs-TG3 is replaced by SB (Fig. 2B). Comparable relative enhancements are observed for overall oligonucleotide concentration of < 1 μM, where the buffer was supplemented with Mg2+ to promote duplex stability (Fig. 5B).42 An intensity increase up to 1 SB:1 SBs-TG3 is followed by a plateau, and this stoichiometric trend supports a single binding site for the strands on G–SA with a relatively greater affinity for SB (Fig. 2C). The origin of this emission amplification lies in strand exchange on G–SA, as substantiated by size exclusion chromatography using near infrared emission to track the cluster conjugates (Fig. 3C). A dT10 tail on SB was used to distinguish the two duplexes, and this 5' addition to SB acts only as a size reporter, as the fluorescence intensity changes are similar for reaction of both SB and SB-dT10 with the cluster conjugate to G–SA/SBs-TG3. Following this strand exchange using SB-dT10, the retention time of the cluster is reduced, thus supporting its association with the newly formed, SB-based duplex.
Figure 5.
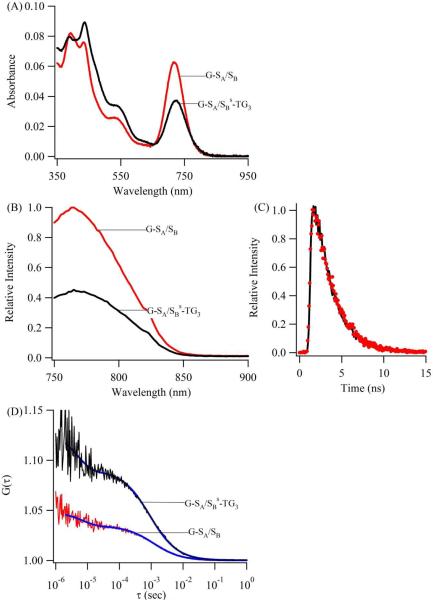
Spectroscopic-based measurements before (black) and after (red) the addition of SB to the cluster conjugate with G-SA/SBs-TG3 support relief of static quenching using strand exchange. Absorption spectra (A) show evidence of a shift in the electronic environment. Emission spectra (B) show fluorescence enhancement while the fluorescence lifetimes (C) are unchanged. The fluorescence lifetimes are 2.08 ± 0.03 ns and 2.05 ± 0.03 ns for the clusters bound to G-SA/SBs-TG3 and G-SA/SB, respectively, which are similar the lifetime of 2.11 ± 0.01 ns when using the C3AC3AC3GC3A template alone. The fluorescence enhancement (B) is accompanied by a transition from higher to lower contrast in the fluorescence correlation function (D), indicating an increase in the cluster concentration with strand exchange.
The strand exchange process was further scrutinized through the stringency of hybridization imposed by complementary base pairing. The cluster complex with G–SA/SBs-TG3 was challenged with two alternate sequences: (i) SBrr has the identical base composition but with reversed polarity in relation to SB and (ii) SA is abstracted from the bifunctional oligonucleotide G–SA (Fig. 1S). With two-fold excess amounts of SBr and SA, the cluster conjugate with G–SA/SBs-TG3 exhibits no intensity changes, and the inferred selectivity for SB supports the necessity of strand exchange for fluorescence amplification.
Target detection via the cluster-DNA sensor is modular, as the recognition component of the bifunctional oligonucleotide can be varied while retaining the C3AC3AC3GC3A template for the near-infrared emitting cluster (Fig. 1S and Table 1). Mimicking SBs-TG3, these new quencher sequences also have the 3' terminal TGGG bases that pair with latter four bases of the C3AC3AC3GC3A and are truncated on their 5' terminus to facilitate displacement by the target oligonucleotide. These new quenchers suppress cluster fluorescence, and emission is enhanced when they are expelled by the target strand. One set of studies reversed the positioning of the target and recognition sequences to detect SA using the cluster conjugate with G–SB. A second set of studies considered the reaction of SD, a 22 nucleotide DNA analog of a cancer-related microRNA, with the cluster conjugate with G–SC/SDs-TG3.43 Both exchanges produced two-fold increases in the fluorescence intensity, comparable to the changes observed for the reaction of SB with the cluster complex with G–SA/SBs-TG3.
Similar fluorescence enhancements among the above three systems support their common basis in strand exchange. Absorption and correlation spectroscopy and fluorescence lifetime measurements indicate that a target relieves perturbation of the cluster environment by the quencher strand. In the absorption spectra of the cluster conjugate with G–SA/SBs-TG3, interchange of the quencher SBs-TG3 and target SB strands shifts the absorption maximum and increases the absorbance (Fig. 5A). These spectral signatures of the electronic environment provide a context for interpreting the fluorescence intensity and lifetime measurements of the DNA-bound clusters (Fig. 5B/5C). When SBs-TG3 is replaced with SB on G–SA, enhanced emission contrasts with the similar fluorescence lifetimes of 2.08 ± 0.03 ns and 2.05 ± 0.03 ns for the clusters bound to G–SA/SBs-TG3 and G–SA/SB, respectively. Thus, the spectral shifts and the higher intensities while maintaining identical fluorescence lifetimes support static quenching that is relieved by SB.15 Further substantiation of a population shift that accompanies strand exchange is provided by direct cluster enumeration using fluorescence correlation spectroscopy. Using confocal detection with diode laser excitation, the probe volume occupancy and hence cluster concentrations were determined from the amplitude of the slow component in the correlation function.32 Over the >10 fold concentration range 2–27 nM for the cluster conjugate with G–SA/SBs-TG3, addition of SB is accompanied by a consistent 2–3 fold increase in the number of emitting species that proportionally tracks the fluorescence enhancement (Figure 5 and Table 2). This correspondence indicates that exclusive association of the target with the recognition site restores the favored binding environment of the emissive cluster, thereby facilitating the increase in the number of emissive clusters.
Table 2.
Comparison of the Intensity and Cluster Ratios Derived from the Strand Exchange Reaction of G-SA/SBs-TG3 with SBa
| [Cluster-G-SA/SBs-TG3] (nM) | Intensity Ratio | Number Ratio |
|---|---|---|
| 1.8 ± 0.1 | 1.7 ± 0.4 | 1.8 ± 0.2 |
| 5.0 ± 0.3 | 2.5 ± 0.4 | 2.0± 0.1 |
| 9.8 ± 0.4 | 2.4 ± 0.3 | 2.7 ± 0.1 |
| 11.5 ± 0.6 | 2.0 ± 0.2 | 2.5 ± 0.1 |
| 27.0 ± 0.3 | 2.7 ± 0.2 | 3.2 ± 0.1 |
Based on FCS studies, the first column provides the concentration of near-infrared emissive clusters associated with G-SA/SBs-TG3.
The last two columns provide the ratio of the integrated emission intensity (second column) and the numbers of emitting species (third column) before vs. after the reaction SB with the cluster conjugate with G-SA/SBs-TG3.
Serum Solutions
With electronic transitions in the near infrared spectral region, this silver cluster is well-suited for direct analysis in biological samples. In the 650 – 900 nm window, solutions with blood and serum exhibit reduced emission from endogenous chromophores as well as diminished scattering and absorption of the excitation light.44–46 A resulting low spectral background in conjunction with the high molecular brightness of this fluorophore favors sensitive analysis in spectrally-challenging biological environments. However, the cluster is less stable in buffer with 10% (v/v) serum, as indicated by the decay in emission intensity over a 60 min time period (Fig. 2S). The integrity of the DNA-bound clusters could be compromised by competing interactions with the electron-rich functional groups of the serum proteins, as supported by the blue-shift in the emission band with serum (Fig. 2S). Through the addition of Ag+ to the serum samples prior to adding the reduced silver clusters, this instability and spectral alternation are reversed (Fig. 2S). Enhanced stability and spectral shifts are accompanied by fluorescence quenching, but the reaction of SB with the cluster conjugate with G–SA/SBs-TG3 still produces the same relative fluorescence enhancement observed in buffer. This correspondence suggests that supplementing with Ag+ stabilizes the clusters and does not interfere with strand exchange.
Discussion
This sensor has two overarching characteristics: a DNA recognition strand integrated with a fluorogenic silver cluster and a quencher strand that signals analyte binding. Near-infrared emission is elicited by association of a 10 silver atom complex with the host sequence C3AC3AC3GC3A, and this favored cluster environment is preserved with 3'-appended duplexes. Hybridization of a specific target sequence is directed through base pairing with the recognition component, and this binding event is transduced using a competing quencher strand that bridges the cluster template and strand recognition moieties within the overall sensor. Transformation from lower to higher emission intensity that accompanies the displacement of the quencher by the target is directly attributable to an increase in the number of emitting clusters. Such a detection mechanism in which the binding event is linked with a change in the number of fluorophores may be a general feature of DNA-bound silver clusters. Fluorescence lifetime and brightness measurements show that thrombin diminishes the net fluorescence intensity by reducing the number of red emitting clusters associated with a cluster template – aptamer strand.25 Two methods of DNA detection are based on how guanine-rich strands elicit enhanced red emission from proximal cluster-laden strands.26 The fractional enhancement directly correlates with the number of emitting clusters and the high 500-fold enhancement demonstrates the potential utility of silver clusters for sensitive DNA detection.26 Shifting the cluster population may also be accomplished using smaller analytes, as Hg2+ statically quenches emission from red emitting clusters bound to dC12.23 Differences in how particular clusters respond to perturbations may be a manifestation of large variations in their chemical behavior due to their small sizes.47
Key to our studies is the use of both fluorescence correlation spectroscopy and fluorescence lifetime measurements to demonstrate a population shift that accompanies strand exchange. Static quenching is described by an equilibrium in which the florescence intensity depends on the association constant for complex formation.15 In our case, this equilibrium describes the extent to which the clusters are perturbed by base pairing, with a fraction remaining in the favored binding site and retaining their inherent fluorescence lifetime. The balance of clusters is not emissive and the increased concentration of clusters that accompanies strand exchange is directly measured using fluorescence correlation spectroscopy. Progress in understanding this mechanism of signal enhancement may lie in the repeated C3X (X = A or G) motif of the silver cluster template. Quencher invasion into the 3' region of C3AC3AC3GC3A may drive the cluster to a closely related binding site having a different electronic environment, as suggested by the residual emission and the shift in the spectra observed with longer complementary sequences (Fig. 4). Single base changes within this particular repeated motif have been shown to impact the photophysical properties of the clusters, and efforts are now directed to sequence variations to decipher the significance of multiple binding sites. An alternative suggested by changes in the absorption spectra is that other cluster species may be involved in this transformation, and we are currently exploring chromatographic separation to understand the potential impact of these cluster-DNA conjugates.18 Evaluating these possibilities will advance the use of this silver cluster-DNA conjugate for sensing.
One such application is DNA detection in biological environments. This fluorophore has a high molecular brightness when compared with its spectral counterparts, thus facilitating strand detection in solutions with serum, which exhibit a relatively low fluorescence background in the near-infrared spectral region. Similar magnitudes of the fluorescence enhancements in pristine buffer relative to buffers with 10% serum suggest a common mechanism based on strand exchange. However, with serum, chemical stability of the cluster-DNA conjugate is compromised, as indicated by the loss of emission and the blue-shift in the emission spectrum. This decomposition and electronic perturbation are reversed by supplementing the solutions with Ag+, which may poison the electron-rich sites on the serum proteins that could competitively complex with the clusters. Alternatively, Ag+ may complex open coordination sites on C3AC3AC3GC3A, thereby modifying the host DNA conformation and hence the solvent exposure of the cluster.48,49 Support for this interaction is provided by fluorescence quenching that is associated with Ag+. Current efforts are focused on chemically stabilizing the clusters while maintaining the fluorescence enhancement.
Conclusion
Our central finding is that an oligonucleotide for a silver cluster can be covalently modified to recognize a DNA analyte, and modular detection is achieved using specific recognition sites appended to the cluster template. A bridging quencher strand between the template and recognition components within the composite sensor statically quenches emission and its displacement by the target increases the number of emitting clusters. A key question is whether other cluster templates can be likewise modified. Because minor variations in the base sequence dictate the types of clusters and hence their emission spectra, multiplex analysis of a broader range of analytes could be achieved. Such parallel analysis, along with the ability to modify the regional DNA environment for fluorogenic silver clusters, may open new strategies for sensitive, convenient, and specific analyte detection.
Supplementary Material
Acknowledgement
We thank the National Science Foundation (CBET-0853692) for support of this work. We are grateful to the National Institutes of Health (R15GM071370) for primary support during the initial stages of this work. In addition, we thank the National Science Foundation (CHE-0718588), Henry Dreyfus Teacher-Scholar Awards Program, and the National Institutes of Health (P20 RR-016461 from the National Center for Research Resource) for their support throughout this project. JTP received partial sabbatical support through matching commitments to an NSF RII Cooperative Agreement, EPS-0903795.
Footnotes
Briefs: Bifunctional oligonucleotide templates silver clusters and detects DNA strands.
Supporting Information Available: Supplemental figures describing fluorescence changes accompanying strand exchange. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Liu J, Cao Z, Lu Y. Chem. Rev. 2009;109(5):1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Lubin AA, Plaxco KW. Acc. Chem. Res. 2010;43(4):496–505. doi: 10.1021/ar900165x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Kostrikis LG, Tyagi S, Mhlanga MM, Ho DD, Kramer FR. Science. 1998;279(5354):1228–1229. doi: 10.1126/science.279.5354.1228. [DOI] [PubMed] [Google Scholar]
- (4).Tyagi S, Kramer FR. Nat. Biotechnol. 1996;14(3):303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- (5).Bao G, Rhee WJ, Tsourkas A. Annual Review of Biomedical Engineering. 2009;11(1):25–47. doi: 10.1146/annurev-bioeng-061008-124920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Tyagi S. Nat. Methods. 2009;6(5):331–338. doi: 10.1038/nmeth.1321. [DOI] [PubMed] [Google Scholar]
- (7).Cho EJ, Lee JW, Ellington AD. Annu Rev Anal Chem (Palo Alto Calif) 2009;2:241–264. doi: 10.1146/annurev.anchem.1.031207.112851. [DOI] [PubMed] [Google Scholar]
- (8).Sefah K, Phillips JA, Xiong X, Meng L, Van Simaeys D, Chen H, Martin J, Tan W. Analyst. 2009;134(9):1765–1775. doi: 10.1039/b905609m. [DOI] [PubMed] [Google Scholar]
- (9).Bunka DHJ, Stockley PG. Nature Reviews Microbiology. 2006;4(8):588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- (10).Stoltenburg R, Reinemann C, Strehlitz B. Biomol. Eng. 2007;24(4):381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- (11).Hermann T, Patel DJ. Science. 2000;287(5454):820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- (12).Fang X, Tan W. Acc. Chem. Res. 2009;43(1):48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wang H, Yang R, Yang L, Tan W. ACS Nano. 2009;3(9):2451–2460. doi: 10.1021/nn9006303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Nutiu R, Li Y. Methods. 2005;37(1):16–25. doi: 10.1016/j.ymeth.2005.07.001. [DOI] [PubMed] [Google Scholar]
- (15).Lakowicz JR. Principles of Fluorescence Spectroscopy. Third ed. Springer; New York: 2006. [Google Scholar]
- (16).Petty JT, Zheng J, Hud NV, Dickson RM. J. Am. Chem. Soc. 2004;126(16):5207–5212. doi: 10.1021/ja031931o. [DOI] [PubMed] [Google Scholar]
- (17).Gwinn EG, O'Neill P, Guerrero AJ, Bouwmeester D, Fygenson DK. Adv. Mater. 2008;20(2):279–283. [Google Scholar]
- (18).Petty JT, Fan C, Story SP, Sengupta B, St. John Iyer A, Prudowsky Z, Dickson RM. J. Phys. Chem. Lett. 2010;1(17):2524–2529. doi: 10.1021/jz100817z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Richards CI, Choi S, Hsiang J-C, Antoku Y, Vosch T, Bongiorno A, Tzeng Y-L, Dickson RM. J. Am. Chem. Soc. 2008;130(15):5038–5039. doi: 10.1021/ja8005644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Sharma J, Yeh HC, Yoo H, Werner JH, Martinez JS. Chem. Commun. 2010;46(19):3280–3282. doi: 10.1039/b927268b. [DOI] [PubMed] [Google Scholar]
- (21).Patel SA, Cozzuol M, Hales JM, Richards CI, Sartin M, Hsiang J-C, Vosch T, Perry JW, Dickson RM. J. Phys. Chem. C. 2009;113(47):20264–20270. doi: 10.1021/jp9079537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Richards CI, Hsiang J-C, Senapati D, Patel S, Yu J, Vosch T, Dickson RM. J. Am. Chem. Soc. 2009;131(13):4619–4621. doi: 10.1021/ja809785s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Guo W, Yuan J, Wang E. Chem. Commun. 2009;23:3395–3397. doi: 10.1039/b821518a. [DOI] [PubMed] [Google Scholar]
- (24).Su Y-T, Lan G-Y, Chen W-Y, Chang H-T. Anal. Chem. 2010;82(20):8566–8572. doi: 10.1021/ac101659d. [DOI] [PubMed] [Google Scholar]
- (25).Sharma J, Yeh HC, Yoo H, Werner JH, Martinez JS. Chem. Commun. 2011;47:2294–2296. doi: 10.1039/c0cc03711g. [DOI] [PubMed] [Google Scholar]
- (26).Yeh HC, Sharma J, Han JJ, Martinez JS, Werner JH. Nano Lett. 2010;10(8):3106–3110. doi: 10.1021/nl101773c. [DOI] [PubMed] [Google Scholar]
- (27).Knemeyer JP, Marme N, Sauer M. Anal. Chem. 2000;72(16):3717–3724. doi: 10.1021/ac000024o. [DOI] [PubMed] [Google Scholar]
- (28).Guo W, Yuan J, Dong Q, Wang E. J. Am. Chem. Soc. 2010;132(3):932–934. doi: 10.1021/ja907075s. [DOI] [PubMed] [Google Scholar]
- (29).Lavis LD, Raines RT. ACS Chem. Biol. 2008;3(3):142–155. doi: 10.1021/cb700248m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Petty JT, Fan C, Story SP, Sengupta B, Sartin M, Hsiang J-C, Perry JW, Dickson RM. J. Phys. Chem. B. 2011;115(24):7996–8003. doi: 10.1021/jp202024x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Vosch T, Antoku Y, Hsiang J-C, Richards CI, Gonzalez JI, Dickson RM. Proc. Natl. Acad. Sci. U. S. A. 2007;104(31):12616–12621. doi: 10.1073/pnas.0610677104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Rigler R, Mets U, Widengren J, Kask P. Eur. Biophys. J. Biophys. Lett. 1993;22(3):169–175. [Google Scholar]
- (33).Texier I, Goutayer M, Da Silva A, Guyon L, Djaker N, Josserand V, Neumann E, Bibette J, Vinet F. J. Biomed. Opt. 2009;14(5):054005. doi: 10.1117/1.3213606. [DOI] [PubMed] [Google Scholar]
- (34).Phan AT, Gueron M, Leroy JL. Methods in Enzymology. 2001;338:341–371. doi: 10.1016/s0076-6879(02)38228-4. [DOI] [PubMed] [Google Scholar]
- (35).Bevington P, Robinson DK. Data Reduction and Error Analysis for the Physical Sciences. McGraw-Hill; 2002. [Google Scholar]
- (36).Kim S, Lim YT, Soltesz EG, De Grand AM, Lee J, Nakayama A, Parker JA, Mihaljevic T, Laurence RG, Dor DM, Cohn LH, Bawendi MG, Frangioni JV. Nat. Biotechnol. 2004;22(1):93–97. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Nichols BP, Donelson JE. J. Virol. 1978;26(2):429–434. doi: 10.1128/jvi.26.2.429-434.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Sengupta B, Springer K, Buckman JG, Story SP, Abe OH, Hasan ZW, Prudowsky ZD, Rudisill SE, Degtyareva NN, Petty JT. J. Phys. Chem. C. 2009;113(45):19518–19524. [Google Scholar]
- (39).Sengupta B, Ritchie CM, Buckman JG, Johnsen KR, Goodwin PM, Petty JT. J. Phys. Chem. C. 2008;112(48):18776–18782. doi: 10.1021/jp804031v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Doose S, Barsch H, Sauer M. Biophys. J. 2007;93(4):1224–1234. doi: 10.1529/biophysj.107.107342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Ritchie CM, Johnsen KR, Kiser JR, Antoku Y, Dickson RM, Petty JT. J. Phys. Chem. C. 2007;111(1):175–181. doi: 10.1021/jp0648487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Manning GS. Q. Rev. Biophys. 1978;11(2):179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- (43).Li J, Schachermeyer S, Wang Y, Yin Y, Zhong W. Anal. Chem. 2009;81(23):9723–9729. doi: 10.1021/ac901983s. [DOI] [PubMed] [Google Scholar]
- (44).Chance B. Ann. N. Y. Acad. Sci. 1998;838(Advances in Optical Biopsy and Optical Mammography):29–45. [PubMed] [Google Scholar]
- (45).Frangioni JV. Curr. Opin. Chem. Biol. 2003;7(5):626–634. doi: 10.1016/j.cbpa.2003.08.007. [DOI] [PubMed] [Google Scholar]
- (46).Campbell RE, Chang CJ. Curr. Opin. Chem. Biol. 2010;14(1):1–2. doi: 10.1016/j.cbpa.2009.12.001. [DOI] [PubMed] [Google Scholar]
- (47).Henglein A, Mulvaney P, Linnert T. Faraday Discuss. 1991;92:31–44. [Google Scholar]
- (48).Loo K, Degtyareva N, Park J, Sengupta B, Reddish M, Rogers CC, Bryant A, Petty JT. J. Phys. Chem. B. 2010;114(12):4320–4326. doi: 10.1021/jp908085s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Wang Y, Li J, Wang H, Jin J, Liu J, Wang K, Tan W, Yang R. Anal. Chem. 2010;82(15):6607–6612. doi: 10.1021/ac101114w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.