Abstract
There are over 10,000 species of venomous marine molluscs, the vast majority of these, which are generally referred to as “turrids”, are traditionally assigned to a single family, Turridae (Powell 1966). Here, we provide an initial molecular analysis of the type genus of the family, Turris Röding, 1798, thought to be among the most well characterized groups in the family. We show that the type genus is not monophyletic.
We analyzed specimens conventionally assigned to 9 different Turris species using molecular markers, combined with the shell morphology and radular anatomy whenever feasible. The results suggest that species assigned to the genus Turris, provisionally assigned to two different subgenera are not monophyletic. Five previously described species belong to the subgenus Turris (s.s.) Röding 1798: T. babylonia,(Linne, 1758), T. grandis, (J. E. Gray, 1834), T. dollyae, (Olivera, 1999), T. normandavidsoni (Olivera, 1999) and T. spectabilis (Reeve, 1843). With a change in species designation, T. assyria (formerlyT. babylonia1010) is added to a well-defined clade, which is in turn more closely related to Lophiotoma and Gemmula species than to the other five Turris species.
We show that these five species conventionally assigned to Turris do not belong in the same subgenus, and form a clade provisionally designated as Annulaturris Powell, 1966: T. annulata, (Reeve, 1843), T. undosa, (Lamarck, 1816), T. cristata, (Vera-Peláez, Vega-Luz, and Lozano-Francisco 2000) T. cryptorrhaphe (G. B. Sowerby, 1825) and T. nadaensis (Azuma, 1973). Implications of the molecular phylogenetic results and its correlation with radular morphology are discussed.
Keywords: Turris, Gemmula, Lophiotoma, radulae, molecular phylogeny, shell morphology, morphospecies
Introduction
Traditionally comprising three major families: the Conidae (“cone snails”), the Terebridae (“auger snails”), and the Turridae s.l. (Powell, 1966; McLean, 1971; Ponder, 1973; Ponder and Waren, 1988), the superfamily Conoidea is an extremely biodiverse group of venomous marine snails. Venoms of conoideans are a complex mixture, with more than a hundred individual peptide components, comprising a largely untapped pharmacological resource (Olivera et al., 1990; Olivera, 1999).
Members of the Turridae include the first conoideans that appear abundantly at the Eocene/Oligocene boundary about 34 million years ago (Tucker, 2004). The extant species traditionally assigned to Turridae encompass a huge morphological diversity. Recent fieldwork in the tropical Pacific (Bouchet et al., 2002) has led to an estimate of over 10,000 recent species of “turrids”.
The last comprehensive treatment of the genus Turris was that of Powell (1964); eight species were defined, with one, Turris crispa, divided into 4 subspecies. Subsequently, Powell (1966) recognized that two of these species, Turris annulata and Turris amicta, diverged significantly from the six, and for these he designated a subgenus, Annulaturris,. This treatise on turrids summarized the nine subfamilies and more than 500 genera proposed to belong to the family Turridae. A later in-depth study of foregut anatomy and radular morphology in a number of conoideans (Taylor, et al. 1993) led to a proposal for the complete revision of traditional taxonomy of Conoidea. Taylor and co-workers demonstrated that some of the subfamilies previously included in the Turridae were more closely allied to Conus and they assigned these taxa to the family Conidaem demanding a narrower definition of Turridae.
The first molecular phylogenetic investigation of Conoidea suggested an even more complex situation. Several subfamilies of Turridae (e.g. Crassispirinae and Cochlespirinae), were found to be polyphyletic or paraphyletic groups (Puillandre et al., 2008) suggesting that continuous reassessment of Conoidean taxonomy will be required as newer data are acquired. Recently, it was proposed (Tucker and Tenorio, 2009) that the species conventionally in Turridae be placed in two separate superfamilies, some to a newly proposed superfamily, Turroidea, (with the terebrids), with other lineages remaining in the superfamily Conoidea (with the cone snails).
Genus Turris Roding, 1798, the type genus of the family Turridae consists of approximately 20 recent and 20 extinct species. (Tucker, 2004). The genus Turris defines the subfamily Turrinae (type species, Turris babylonia), the only subfamily of Turridae for which monophyly has not been rejected. Previously published studies on the molecular phylogeny of Turrinae include the analysis of species in Unedogemmula/Lophiotoma (Heralde et al., 2007), and Xenuroturris (Kantor et al., 2008). Although a number of new species have been described in the genus Turris in the last decade (Olivera et al., 1999; Vera-Peláez, Vega-Luz, and Lozano-Francisco, 2000; Bozzetti,2006) neither an in-depth evaluation of the genus nor an examination of phylogeny using any criteria except shell morphology has been carried out. Here our goal is to provide a framework for understanding intrageneric relationships within Turris by as we correlate shell and radular morphology with molecular data.
Materials and method
Specimens
Material for present study was collected at different localities within Philippines with most specimens from either the Danajon Bank near Olango Island, or from Sogod, Cebu Island, both in Cebu province in the central Visayas. The specimens analyzed are summarized in Table 1. We include Turridrupa, Lophiotoma and Gemmula species to determine how each of the Turris species fit into the subfamily Turrinae: we do not assume monophyly of the genus Turris. Most specimens were collected by hookah at depths between 10–30 meters, and by gill nets at depths of 70–150 meters. The specimens were kept alive, and dissected within 1–2 days. Shells were photographed, and foot samples preserved in 95% alcohol for molecular analysis. Samples of the buccal mass with surrounding tissues were preserved in 70% alcohol for preparation of the radula.
Table I.
Summary of Specimens Analyzed
| Species assignment based on shell morphology | Specimen ID | Radula | Locality | Genbank | Accession | Numbers |
|---|---|---|---|---|---|---|
| 12S | 16S | COI | ||||
| A. Species generally assigned to Turris: | ||||||
| Turris annulata | 271 | Aliquay | GU300000 | GU345750 | GU299967 | |
| Turris babylonia | 1008 | + | Danajon Bank, off Olango Is. | GU300001 | GU345751 | GU299968 |
| Turris babylonia* | 1010 | Danajon Bank, off Olango Is. | GU300002 | GU345752 | GU299969 | |
| Turris babylonia | 1012 | + | Sogod | GU300003 | GU345753 | GU299970 |
| Turris grandis | 761 | Philippines | GU300004 | GU345754 | GU299971 | |
| Turris cristata | 1014 | + | Danajon Bank, off Olango Is. | GU300005 | GU345755 | GU299972 |
| Turris cristata | 1015 | + | Danajon Bank, off Olango Is. | GU300006 | GU345756 | GU299973 |
| Turris cristata | 1021 | + | Danajon Bank, off Olango Is. | GU300007 | GU345757 | GU299974 |
| Turris cryptorrhaphae | 1016 | + | Danajon Bank, off Olango Is. | GU300008 | GU345758 | GU299975 |
| Turris dollyae | 845 | Sogod | GU300009 | GU345759 | GU299976 | |
| Turris dollyae | 1013 | + | Danajon Bank, off Olango Is. | |||
| Turris nadaensis | 599 | Philippines | GU300010 | GU345760 | GU299977 | |
| Turris nadaensis | 849 | Sogod | GU300011 | GU345761 | GU299978 | |
| Turris nadaensis | 850 | Danajon Bank, off Olango Is. | GU300012 | GU345762 | GU299979 | |
| Turris nadaensis | 1020 | + | Danajon Bank, off Olango Is. | GU300013 | GU345763 | GU299980 |
| Turris nadaensis | 590 | Sogod | GU300014 | GU345764 | GU299981 | |
| Turris nadaensis | 589 | + | Danajon Bank, off Olango Is. | GU300015 | GU345765 | GU299982 |
| Turris normandavidsoni | Heralde | Philippines | GU300016 | GU345766 | GU299983 | |
| Turris normandavidsoni | 1024 | + | Danajon Bank, off Olango Is. | |||
| Turris spectabilis | 600 | Philippines | GU300017 | GU345767 | GU299984 | |
| B. Species not assigned to Turris: | ||||||
| Lophiotoma acuta | 513 | Panglao | GU300018 | GU345768 | GU299985 | |
| Lophiotoma cerithiformis | 351 | Oahu, Hawaii | GU300020 | EU682298 | GU299987 | |
| Lophiotoma cf jickelli | 230 | Philippines | GU300021 | GU345770 | GU299988 | |
| Lophiotoma olangoensis | 315 | Olango | GU300023 | J868138 | GU299990 | |
| Gemmula diomedea | 515 | Panglao | GU300027 | GU345775 | GU299994 | |
| Gemmula lisajoni | 757 | Sogod | GU300028 | GU345776 | GU299995 | |
| Gemmula monilifera | 432 | Batangas | GU300029 | GU345777 | GU299996 | |
| Gemmula speciosa | 434 | Batangas | GU300031 | GU345778 | GU299997 | |
| Turridrupa cf bijubata | 700 | Mactan | GU300032 | GU345780 | GU299999 |
This form has been designated Turris assyria, new species in an another paper (Olivera, Seronay, Fedosov, 2010) — see text.
Radular morphology
Buccal complexes were dissected to isolate the radular sac, then soft tissues were dissolved in solution of potassium hypochlorite (5%) and cleaned radulae were prepared for further SEM examination.
DNA extraction, amplification and sequencing
Genomic DNA was prepared from 10 mg footpad tissue using the Gentra PUREGENE DNA Isolation Kit (Gentra Systems, Minneapolis, MN) according to the manufacturer’s standard protocol.
Ten ng of genomic DNA was used as a template for polymerase chain reaction (PCR) with oligonucleotide primers corresponding to 12S-I (5′ TCG CAG CAG YCG CGG TTA) and 12S-III (5′ AGA GYG RCG GGC GAT GTG T) mitochondrial rRNA segments (Simon et. al., 1991) and 16SH (5′ CCG GTC TGA ACT CAG ATC ACT G) and 16LC (5′ GTT TAC CAA AAA CAT GGC TTC) mitochondrial rRNA segments (Palumbi, 1996), and oligonucleotides corresponding to COI dgLsimiCO-1490 (5′ GGT CAA CAA ATC ATA AAG AYA TGY G 3′) and COI dgHCO-2198 gene segments (5′ TAA ACT TCA GGG TGA CCA AAR AAY CA 3′). The PCR cycling profile was as follows: Initial denaturation (95°C, 60s); followed by 40 cycles of denaturation (95°C, 20s); annealing (55°C, 20s) and extension (72°C, 30s).
The resulting PCR products were purified by gel electrophoresis, recovered from agarose using High Pure PCR Product Purification Kit (Roche Diagnostics, Indianapolis, IN). A number of recent studies (e.g. Buhay, 2009) show that active mitochondrial genes, encoding cytochrome c oxidase subunit 1 often incorporate into the nuclear genome where they become inactive and start to accumulate multiple substitutions. Thus typical PCR product may contain a mixture of molecules of different sequences clearly decreasing the quality of the sequences. The preferred method of obtaining reliable sequence is to clone PCR products and sequence multiple clones in order to determine the mitochondrial consensus sequence. Consequently, the eluted DNA fragments were annealed to pNEB206A vector using the USER Friendly Cloning kit (New England BioLabs, Inc., Beverly, MA) following manufacturer’s suggested protocol and the resulting products transformed into DH5a competent cells. The nucleic acid sequences of these 12S, 16S and COI-encoding clones were determined according to the standard protocol for automated sequencing.
The voucher specimens used in this analysis will be deposited at the Philippine Biodiversity Resources Center, Marine Science Institute, University of the Philippines, Quezon City, Philippines.
Phylogenetic Analysis
Individual 12SrRNA, 16SrRNA and COI equences were aligned with Clustal and then refined by eye using the graphical interface of MacClade4.08 (Maddsion and Maddison 2005) to correct obviously homologous regions that Clustal failed to recognize. These include cases which required a simple shift in the position of a gap to avoid a stop codon not present in other sequences or to complete a codon whose members flanked the Clustal inferred gap. We refined the rRNAs alignments with Rcoffee (Notredame et al. 2000) to account for secondary structure. Because Rcoffee runs are restricted to 50 taxa and 1000 base pairs, the Clustal alignments were divided into smaller subsets, aligned with Rcoffee and then concatenated for further analysis. Alignments within Rcoffee are guided by secondary structure characteristics (Notredame et al. 2000). The color-coded CORE indices were used to identify the best among alternative alignments that include the Clustal and Rcoffee alignments and hand refined versions of each.
Sequences were concatenated with MacClade4 (Maddsion and Maddison 2005) and then optimized using MrBayes (Huelsenbeck and Ronquist, 2001) with GTR+I+G maximum likelihood model parameters estimated separately for each gene. Each analysis comprised two simultaneous runs with four chains each. Two million generations reduced the average standard deviation of the split frequencies below 0.01. Plots of the number of generations against the maximum likelihood scores indicated equilibrium. Further diagnostics included the potential scale reduction factor (PSRF) that measures the fit of branch length and all parameters. Trees and parameters from the first 25% of the generations were discarded (the burn in) after completion of the MCMC (Markov Chain Monte Carlo) search.
For the maximum likelihood analyses of the individual genes, we estimated parameters for GTR+I+G models of sequence evolution and optimized the tree and using Phylml (Guindon and Gascuel, 2003). Our analysis of 1000 bootstrap replicates provided measures of support on the clades of the maximum likelihood trees. For the concatenated sequences of several genes we used RAxML (Stamatakis, et al. 2008) to optimise the maximum likelihood tree using the GTR+I+G model with parameters estimated separately for each gene and to analyse the bootstrap replicates
Finally we tested various alternative topologies (e.g. monophyly of the Turris species) using Shimodaira-Hasegawa tests with GTR+I+G optimised parameters in PAUP4b10 (Swofford, 2002) to determine whether any differed significantly from the optimal tree. Rather than simply moving the two intact Turris clades together, we optimized with maximum likelihood the relationships among all the Turris species simultaneously within the monophyly constraint. To ensure that these were tests of topology alone, we constrained the topologies but not the branch length estimates.
Results
Radular morphology
The radulae of the specimens indicated in Table IA are illustrated in Figures 3–4. The most striking feature is the presence, absence, or state of reduction of a distinctive central tooth. As previously reported by Powell, in many species of Turris, there is either no central tooth or only a rudimentary tooth. The most reduced central teeth are characteristic of Turris babylonia, Turris normandavidsoni, and Turris spectabilis (see Fig 3). In these specimens, the base of the central tooth is indistinct and the cusp is rudimentary. In Turris dollyae, the base of the central teeth is triangular or bow-shaped, with a triangular, short and blunt cusp. Finally, a strong central tooth was found in Turris cryptorrhaphe, Turris cristata, Turris nadaensis and Turris undosa. In these species, the central tooth has a distinct, wide base, especially in Turris cryptorrhaphe, and a long, pointed awl-shaped cusp (Figure 4).
Figure 3.
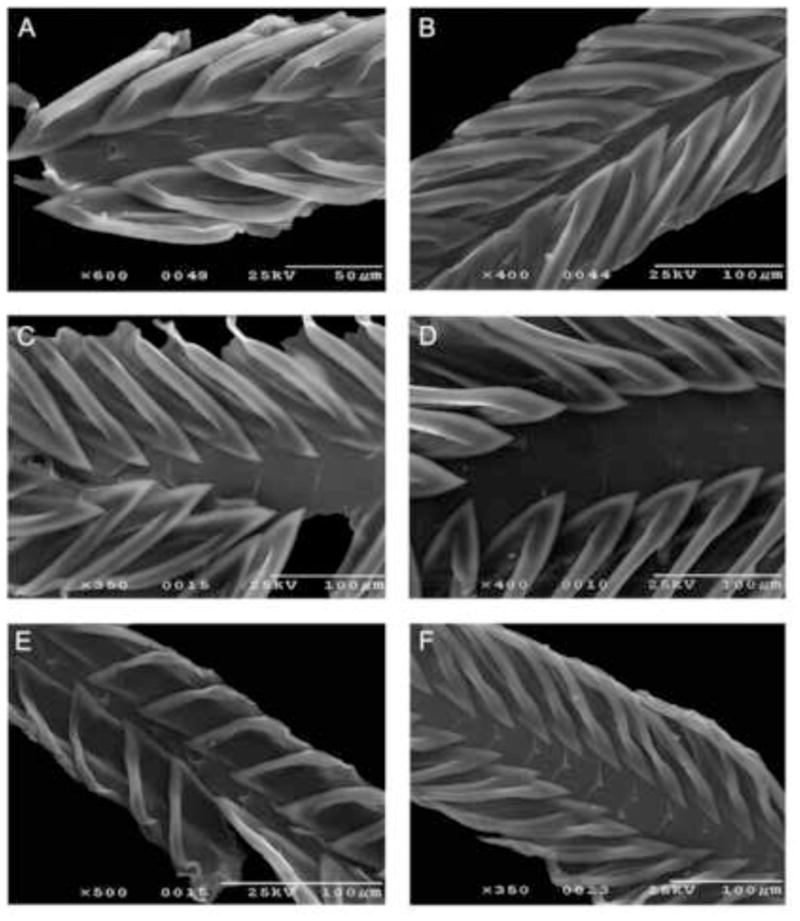
Radular morphology of some Turris species. A – T. babylonia (#1012); B – T. dollyae (#1013); C – Turris garnonsi (#1019); D – T. normandavidsoni (#1024); E – T. spectabilis (#1031); F – T. totiphylis (#1028)
Figure 4.
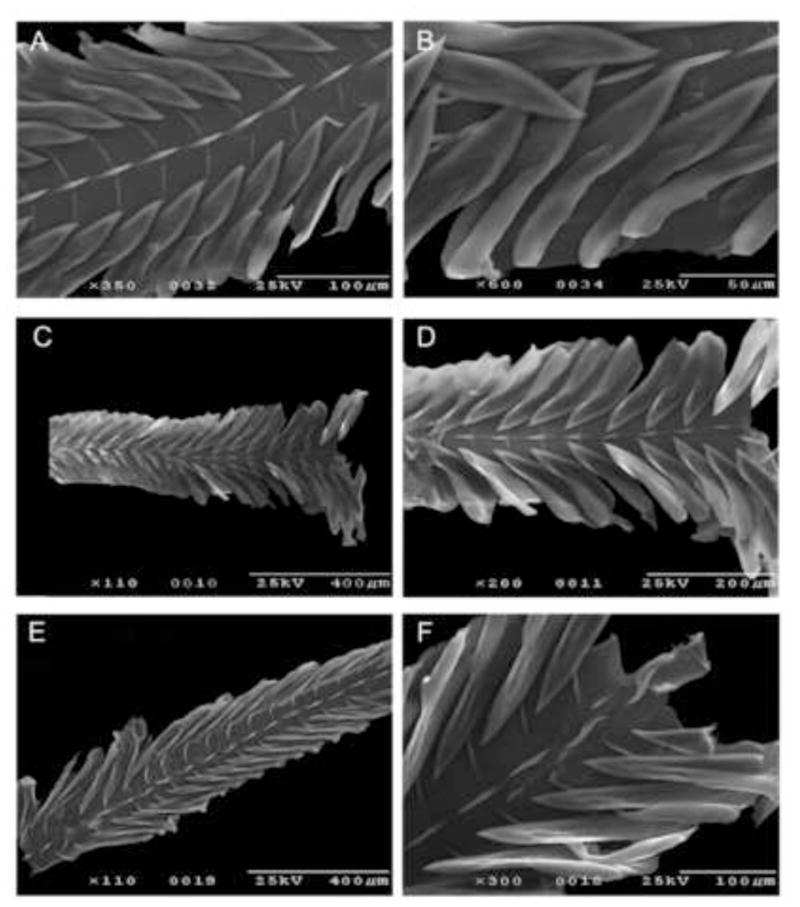
Radular morphology of some Turris species. A, B – T. cryptorrhaphe (#1016); C, D – T. cristata (#1015); E, F – T. nadaensis (#1022)
Another important radular character is the structure of the marginal teeth. In general, the wishbone teeth, characteristic of the Turrinae, have strongly thickened margins, which form a major axial element and the accessory limb of each tooth (Kantor and Taylor, 2000). In most species of the genus Turris that have been examined for this study, both axial elements are well pronounced, spread through the entire length of the tooth and become confluent at its tip. In specimens of Turris cryptorrhaphe, Turris cristata, Turris nadaensis and Turris undosa examined, the accessory limb is weakly developed and the major axial elements do not coalesce at the tip of the tooth, so the margins of the tooth at the tip are not thickened and instead form a kind of blade.
Molecular analysis
The trees inferred from the three individual genes, COI, 12S and 16S are poorly resolved (Supplementary Figure 1). On the other hand, the tree inferred from the concatenated sequence of the three genes (Figure 5) is comparatively well resolved. Contrary to traditional taxonomy, the tree suggests that the genus Turris is not monophyletic. This tree comprises a clade, hereafter referred to as Annulaturris, that includes Turris annulata, Turris cristata, Turris cryptorrhaphe, Turris nadaensis and Turris undosa to the exclusion of a clade comprising Lophiotoma and Gemmula species with the other Turris species (T. babylonia, T. dollyae, T. grandis, T. normandavidsoni, T. spectabilis T. assyria). The hypothesis that the Turris species are monophyletic is rejected (Shimodaira-Hasegawa test, ln likelihood difference=62.3, p≤0.0001).
Figure 5.
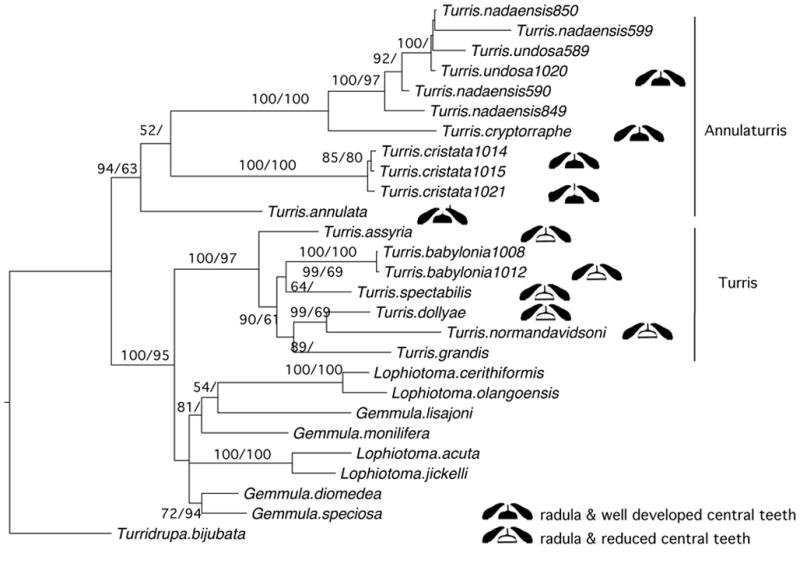
Optimal tree inferred using gene-partitioned Bayesian methods from the concatenated alignments of 12SrRNA, 16S rRNAs and COI genes. The phylogeny describes the evolutionary relationships among Turris species and selected Lophiotoma and Gemmula species. Turridrupa bijubata is the out-group species. Some Turris taxa are labeled to indicate radulae and central tooth type relative to the optimal molecular tree. Branches are labeled with posterior probabilities greater than 50% from the Bayesian analysis of the three genes. Maximum likelihood bootstrap values over 50% appear to the right of the posterior probabilities. Clade A – Annulaturris, provisionally given subgeneric status. Clade B – Turris s.s., provisionally given subgeneric status.
Discussion
Turris phylogeny: recent literature
Following the discoveries of nine new species in the Philippines by Olivera (1999) and Vera-Pelaes et al. (2000), Olivera (1999) considered several species of the genus Turris to diverge from the major clade (“Clade I”) that includes Turris babylonia. These include in Clade II Turris cryptorrhaphe, Turris nadaensis, Turris “undosa” (= cristata) and in Clade III Turris annulata. The various hypotheses discussed earlier, suggesting the division of Turris species into widely divergent clades (e.g. Powell, 1966), are refined by the evidence from radular anatomy and molecular phylogeny that we present here. Two recent books for shell collectors illustrate many species of Turris (Poppe, 2008; Robin, 2008) and facilitate comparison of molecular and radular traits with shell traits.
Radular mophology
In his 1964 analysis of the genus, Powell indicated that species in Turris were characterized by having only marginal teeth, and that these were wishbone shaped. The radular anatomy was one characteristic feature of species in this genus. In later separating Turris amicta and Turris annulata into the Annulaturris, Powell (1966) noted that in contrast to the marginals-only pattern observed for Turris babylonia and Turris crispa, Turris amicta had a large well-formed central tooth, with a long slender central cusp on a broadly rectangular base that is recurved at the edges, and that the pair of marginals were “of considerably modified wishbone type”.
We have demonstrated that the presence of a central tooth with a distinct central cusp is a character of several of the Turris species analyzed (T. cristata, T. cryptorrhaphe, and T. nadaensis and T. undosa). Thus, the radular morphology presented in this study provides support for the division of the genus Turris into infrageneric groups. Relatively few species of the Turrinae have been analyzed with respect to their radular anatomy. However, the available data suggests that the characteristic radular morphology observed for the species in Annulaturris, with a large central tooth, is a more primitive condition, and that a marked reduction or loss of the central tooth that is observed in otherTurris speces is the more derived condition. However, this interpretation is based on a fairly sparse sampling of taxa within the subfamily Turrinae.
Reconciling molecular and radular evidence
Our findings are consistent with our optimal molecular tree (Figure 5) supporting separation of the Turris species into two distinct clades within the Turrinae. The molecular tree supports a clade including Turris annulata, T. cristata, T. cryptorrhaphe, T. nadaensis and T. undosa (94% posterior probability, 63% ML bootsrap support) as a distinct clade within the Turris. A provisional taxonomic solution is to use a name for this clade previously provided by Powell, Annulaturris (Powell, 1966). Together with topology tests (Kishino-Hasegawa and Shimodaira-Hasegawa) that reject monophyly of the Turris species, it is clear that the proposed Annulaturris species should be considered to be well seaparated from the other Turris species.
The tree is a striking example of how misleading shell morphology can be; Turris cristata, with similar color pattern to the forms assigned to the Turris nadaensis complex is often mistaken for Turris nadaensis, while Turris cryptorrhaphe is appears unmistakably different. Nevertheless, based on all three genes, it is clear that Turris cryptorrhaphe is more closely related to the various forms in the nadaensis/undosa clade than to Turris cristata.
Based on shell morphology, three specimens conventionally assigned to Turris babylonia fall into two divergent branches within the larger clade comprising T. babylonia, T. dollyae, T. grandis, T. normandavidsoni, T. spectabilis. Powell (1964) regarded Turris babylonia 1008 and 1012 as the “typical form”. The specimen 1010 belongs to a separate species, which has been regarded as a distinct form of T. babylonia (“niniveh form”, Olivera,, 1999). The type material for Turris babylonia, stored in the Linnaean society in London, consists of two syntypes. One is conspecific with our specimens 1008 and 1012 while the other is rather similar to our Turris assyria (or babylonia, specimen 1010). The fact that the type appears to be a mixture of two species is taxonomically problematic with resolution remaining for future studies. And yet while the true identity of the type specimen for Turris babylonia remains controversial (R. Kilburn, personal communication), the two morphospecies both widely assigned to Turris babylonia clearly belong in the same clade (Turris s.s. and not Annulaturris).
Nevertheless, further molecular, anatomical and shell morphological analyses are indicated. For example, the type species of Annulaturris, T. amicta (E.A. Smith, 1877) was not available for this analysis. Although supported by morphological data, the inference that T. amicta species falls in the clade (Figure 5) that includes Turris cristata and Turris annulata is speculative. The relationships of subtropical species, such as Turris orthopleura and Turris ruthae, also needs to be assessed. As discussed by Kilburn (1983), these species have shell morphological and radular characters that suggest that they are not in Turris s.s.
Compared to the relatively unreliable shell morphology, the combination of differences in radular morphology and multiple molecular markers provides a firmer definition for the type species of the family Turridae, and the clade to which the type species belong, defined here as Turris, excluding the Annulaturris ssp. We predict that these new insights into the phylogeny of Turris and its close relatives will be reflected in the analysis of the gene products expressed in their venoms, an analysis that we have begun. A phylogenetic framework has been an extremely useful guide to the discovery of unique pharmacological agents from Conus venoms. Similarly, an understanding of evolutionary history, summarized by our phylogeny of the Turridae,, will extend future venom research.
Supplementary Material
Figure 1.
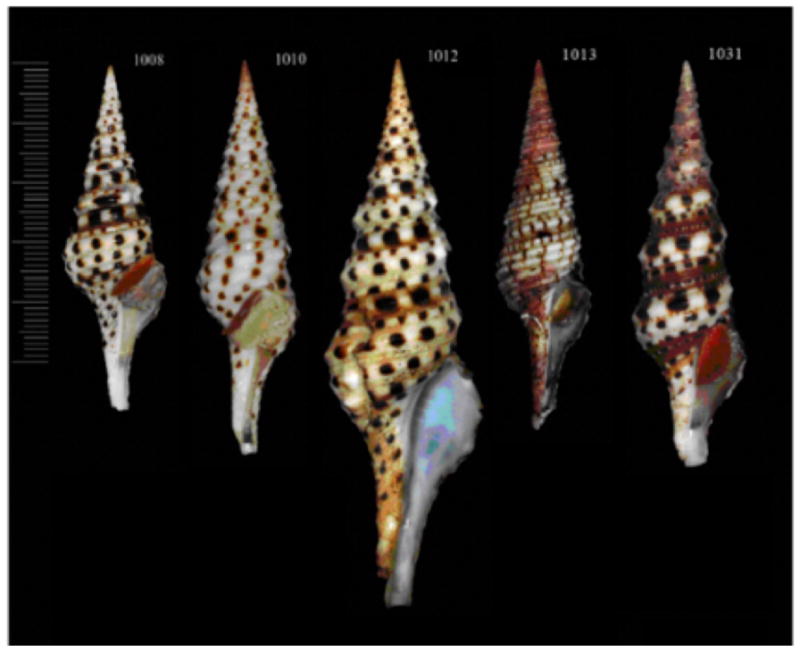
Shells of some studied specimens of the genus Turris. A–C - T. babylonia #1008, T. babylonia (#1010, assyria or the “niniveh” form, Olivera, 1999), T. babylonia #1012 correspondingly) D – T. dollyae (#1013); E – T. garnonsii (#1019); F – T. normandavidsoni (#1024); G – T. spectabilis (#1031); H – T. totiphylis (#1028)
Figure 2.
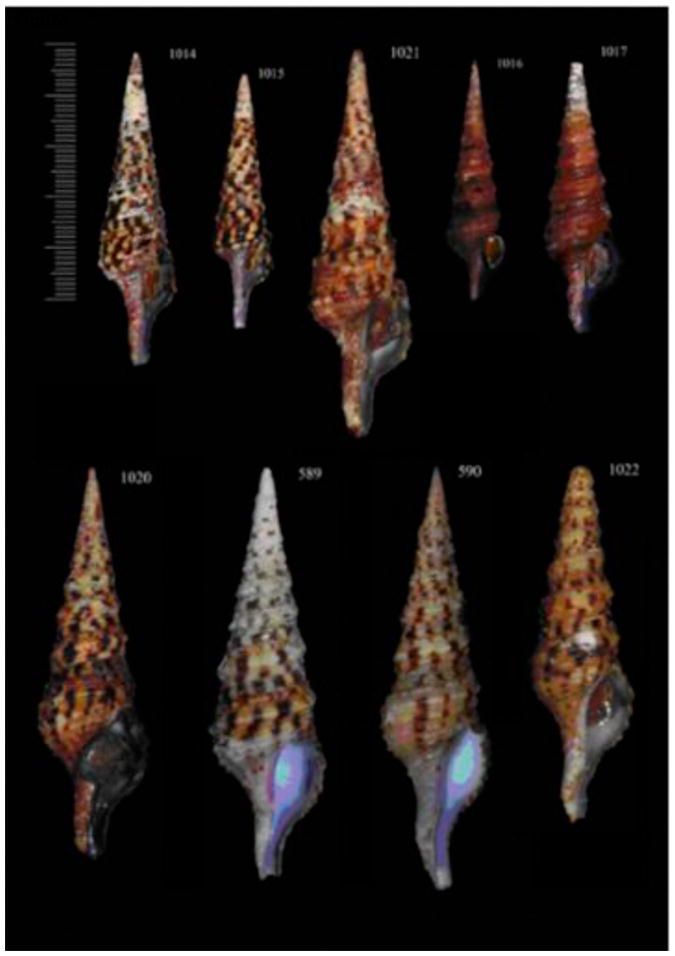
Shells of some studied specimens of the genus Turris. A – T. cristata (#1014); B – T. cristata (#1015); C – T. cristata (#1021); D – T. cryptorraphe (#1016); E - T. cryptorraphe (#1017); F – T. undosa (#1020); G – T. undosa (#589); H – T. nadaensis (#590); I – T. nadaensis (#1022); J - T. nadaensis (#1023)
Acknowledgments
We are grateful to Yuri Kantor for his advice during the course of this work. We thank anonymous reviewers and editors for suggestions that greatly improved the manuscript. This study was supported by PharmaSeas, a program funded by the Department of Science and Technology, Republic of the Philippines (to GPC), by Grant GM48677 from the National Institutes of General Medical Sciences, U.S. Public Health Service (to BMO), by an International Cooperative Biodiversity Grant from the Fogarty Center, National Institutes of Health and by a grant from the Russian Fund for basic research RFBR-09-04-00911.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bouchet P, Lozouet P, Maestrati P, Heros V. Assessing the magnitude of species richness in tropical marine environments: high numbers of molluscs at a New Caledonia site. Biological Journal of the Linnean Society. 2002;75:421–436. [Google Scholar]
- Bozzetti L. Turris ankaramanyensis (Gastropoda: Hypsogastropoda: Turridae) nuova specie dal Madagascar meridionale. Malacologia Mostra Mondiale. 2006;52:8–9. [Google Scholar]
- Buhay JE. “COI-like” Sequences are Becoming Problematic in Molecular Systematic and DNA Barcoding Studies. Journal of Crustacean Biology. 2009;29:96–110. [Google Scholar]
- Heralde FM, 3rd, Concepcion GP, Watkins M, Ownby J-P, Bandyopadhyay P, Olivera BM, Santos AD. Molecular phylogeny of some Indo-Pacific genera in the subfamily Turrinae H. Adams and A. Adams, 1853 (1838) (Gastropoda: Neogastropoda) The Nautilus. 2007;121:131–138. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Kantor YI, Puillandre N, Olivera BM, Bouchet P. Morphological proxies for taxonomic decision in turrids (mollusca, neogastropoda): a test of the value of shell and radula characters using molecular data. Zoolog Sci. 2008;25:1156–1170. doi: 10.2108/zsj.25.1156. [DOI] [PubMed] [Google Scholar]
- Kantor YI, Taylor JD. Formation of marginal radular teeth in Conoidea (Neogastropoda) and the evolution of the hypodermic envenomation mechanism. Journal of Zoology. 2000;252(2):251–262. [Google Scholar]
- Kilburn RN. Turridae (Mollusca: Gastropoda) of southern Africa and Mozambique. Part 1. Subfamily Turrinae. Ann Natal Mus. 1983;25:549–585. [Google Scholar]
- Kohn AJ. Food specialization in Conus in Hawaii and California. Ecology. 1966;47:1041–1043. [Google Scholar]
- Maddison D, Maddison W. MacClade 4.08. Sinauer Assoc; Sunderland, MA: 2005. [Google Scholar]
- McLean JH. A revised classification of the family Turridae with the proposal of new subfamilies, genera and subgenera from the eastern Pacific. The Veliger. 1971;14:114–130. [Google Scholar]
- Olivera BM. The subfamily Turrinae in the Philippines: the genus Turris (Röding, 1798) Philippine J Sci. 1999;128:295–318. [Google Scholar]
- Olivera BM, Rivier J, Clark C, Ramilo CA, Corpuz GP, Abogadie FC, Mena EE, Woodward SR, Hillyard DR, Cruz LJ. Diversity of Conus neuropeptides. Science. 1990;249:257–263. doi: 10.1126/science.2165278. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Seronay RA, Fedosov AE. Turris babylonia; re-evaluation of a species complex and description of Turris assyria, new species. Philippine Science Letters. 2010 [PMC free article] [PubMed] [Google Scholar]
- Olivera BM, Teichert RW. Diversity of the neurotoxic Conus peptides: a model for concerted pharmacological discovery. Molecular Interventions. 2007;7:251–260. doi: 10.1124/mi.7.5.7. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Walker C, Cartier GE, Hooper D, Santos AD, Schoenfeld R, Shetty R, Watkins M, Bandyopadhyay P, Hillyard DR. Speciation of cone snails and interspecific hyperdivergence of their venom peptides. Potential evolutionary significance of introns. Ann NY Acad Sci. 1999;870:223–237. doi: 10.1111/j.1749-6632.1999.tb08883.x. [DOI] [PubMed] [Google Scholar]
- Palumbi SR. Nucleic acids II: the polymerase chain reaction. In: Hillis DM, Moritz C, Mable BK, editors. Molecular Systematics. Sinauer & Associates Inc; Sunderland, Massachusetts: 1996. pp. 205–247. [Google Scholar]
- Ponder WF. The origin and evolution of Neogastropoda. Malacologia. 1973;12:295–338. [PubMed] [Google Scholar]
- Ponder WF, Waren A. Classification of Caenogastropoda and Heterostropha – A list of family-group names and higher taxa. Malacological review Suppl. 1988:288–317. [Google Scholar]
- Poppe GT. ConchBooks. 2008. Philippine Marine Mollusks. [Google Scholar]
- Powell AWB. The molluscan families Speightiidae and Turridae. Bulletin of the Auckland Institute and Museum. 1966;5:1–184. + 123 plates. [Google Scholar]
- Puillandre N, Samadi S, Boisselier MC, Sysoev AV, Kantor YI, Cruaud C, Couloux A, Bouchet P. Starting to unravel the toxoglossan knot: molecular phylogeny of the “turrids” (Neogastropoda: Conoidea) Molecular Phylogenetic Evolution. 2008;47:1122–1134. doi: 10.1016/j.ympev.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Robin A. Xenophora and Conchbooks. Hackenheim, Germany: 2008. Encyclopedia of Marine Gastropods. [Google Scholar]
- Stamatakis, Hoover P, Rougemont J. A Rapid Bootstrap Algorithm for the RAxML Web-Servers. Systematic Biology. 2008;75(5):758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Simon C, Franke A, Martin A. The Polymerase Chain Reaction: DNA Extraction and Amplification. In: Hewitt, Johnson AWB, Young JPW, editors. Molecular Techniques in Taxonomy, G. New York: Springer-Verlag; 1991. pp. 329–355. [Google Scholar]
- Swofford DL. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates; Sunderland, Massachusetts: 2002. PAUP*. [Google Scholar]
- Taylor JD, Kantor Y, Sysoev AV. Bulletin. 2001. Natural History Museum; London: 1993. Foregut anatomy, feeding mechanisms, relationships and classifiction of the Conoidea (=Toxoglossa) (Gastropoda) pp. 125–170. [Google Scholar]
- Tucker JK. Catalog of recent and fossil turrids (Mollusca: Gastropoda) Zootaxa. 2004;682:1–1295. [Google Scholar]
- Tucker JK, Tenorio MJ. Conchbooks. 2009. Systematic classification of Recent and fossil conoidean gastropods. [Google Scholar]
- Vera-Peláez JL, Vega-Luz R, Lozano-Francisco MC. Five new species of the genus Turris Röding, 1798 (Gastropoda; Turridae; Turrinae) of the Philippines and one new species of the Southern Indo-Pacific. Makakos: Monografía. 2000;2:1–29. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


