Abstract
Since their discovery in 1986, Ral (Ras-like) GTPases have emerged as critical regulators of diverse cellular functions. Ral-selective guanine nucleotide exchange factors (RalGEFs) function as downstream effectors of the Ras oncoprotein, and the RalGEF-Ral signaling network comprises the third best characterized effector of Ras-dependent human oncogenesis. Because of this, Ral GTPases as well as their effectors are being explored as possible therapeutic targets in the treatment of RAS mutant cancer. The two Ral isoforms, RalA and RalB, interact with a variety of downstream effectors and have been found to play key and distinct roles in both normal and neoplastic cell physiology including regulation of vesicular trafficking, migration and invasion, tumor formation, metastasis, and gene expression. In this review we provide an overview of Ral biochemistry and biology, and we highlight recent discoveries.
Keywords: Exocyst, GTPase activating protein, guanine nucleotide exchange factor, Ras, Rho, small GTPase
1. Introduction
Identified initially as Ras-like (Ral) proteins, the Ral small GTPases are members of the Ras branch of the Ras superfamily of small GTPases [1]. RALA was identified initially using oligonucleotide probes to identify RAS-related genes in a cDNA library established from immortalized simian B-lymphocytes [2]. Three years later, using the simian RALA cDNA as a probe, human RALA and a related RALB gene were identified from a human pheochromocytoma cDNA library [3]. Subsequently, single RAL orthologs were identified in C. elegans (RAL-1) [4] and Drosophila (RalA) [5] (Fig. 1). Interestingly, although there are well-conserved RAS orthologs in yeast, no RAL orthologs are present in S. cerevisiae or S. pombe.
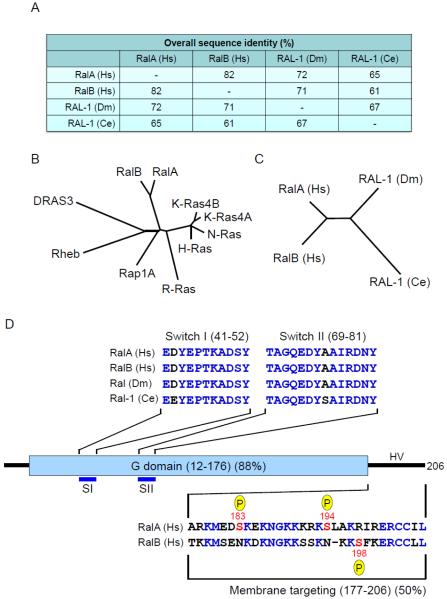
Fig. 1.
Evolutionary conservation of Ral small GTPases. A. Human and invertebrate Ral orthologs exhibit strong sequence identity. The RalA and RalB isoforms are found in all vertebrate species [17]. There is one Ral ortholog in C. elegans (Ce) and D. melanogaster (Dm). Overall sequence identity was determined by CLUSTALW multiple sequence alignment. B. Ral GTPases are members of the Ras branch of the Ras superfamily. Shown here is a comparison with the four Ras proteins and representative members of the Ras family. The dendrogram was generated by CLUSTALW multiple sequence alignment. C. Dendrogram showing sequence relationship of human and invertebrate Ral proteins. D. Ral domain structure. Human RalA and RalB G domains (12-176) shares 88% sequence identity and contain the SI and SII domains that change in conformation during GDP-GTP cycling and are involved in interaction with regulators and effectors. The switch regions are conserved between human Ral proteins and Drosophila Ral and differ by a single residue in each switch in C. elegans Ral (identical residues indicated in blue text). The hypervariable (HV) C-terminus (50% identity) consists of the membrane targeting region and contains key post-translational phosphorylation sites that regulate Ral subcellular localization and effector interaction. Multiple sequence alignment was done by ClustalW analyses and domain topology by SMART analyses. Numbers correspond to the human Ral amino acid sequences.
The three human RAS genes (HRAS, KRAS and NRAS) comprise one of the most frequently mutated gene families in human cancers [6]. Consequently, they have been the subject of intense research scrutiny and cancer drug discovery. Initially, the discovery of Ral proteins simply added to a rapidly growing roster of proteins that now comprise a large superfamily of >150 Ras-related small GTPases [1]. However, with discoveries that Ral GTPases are key regulators of vesicular trafficking and are effectors of Ras oncoprotein-driven growth transformation, Ral proteins stepped into the spotlight in 2003 to bask in their “15 minutes of fame” [7]. Since those initial findings, more discoveries on the role of Ral in normal and cancer cell physiology have ensured that their “fame” will last considerably more than 15 minutes. In this review, we summarize our current knowledge on Ral GTPases and we highlight recent findings in Ral function.
2. Ral proteins
2.1. Ral protein structure
The highly related human RalA and RalB isoforms share 82% overall amino acid sequence identity (Fig. 1A) and are members of the Ras branch of the Ras superfamily (Fig. 1B). They share 46-51% sequence identity and domain architecture with Ras proteins [8]. However, Ral proteins contain an N-terminal 11 amino acid extension not found in Ras, accounting for the 11 residue shift in numbering compared with Ras residue numbering (Fig. 1C). This is followed by the G domain, involved in GTP binding and hydrolysis, and the C-terminal membrane targeting sequence. The majority of sequence divergence occurs within the C-terminal hypervariable regions (50% shared identity) (Fig. 1C).
Like Ras, Ral proteins cycle between inactive GDP-bound and active GTP-bound states (Fig. 2A). RalA and RalB share complete sequence identity in the switch I (SI) and II (SII) sequences that change conformation during GDP-GTP cycling [8] (Fig. 2B). As described below, SI and SII are involved in recognition by both regulators and effectors. The conservation of SI and SII sequences in Drosophila and C. elegans Ral proteins support their interaction with conserved regulators and effectors.
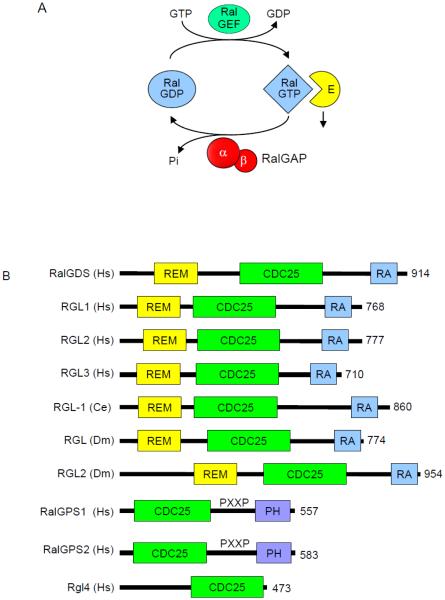
Fig. 2.
Regulators of the Ral GDP-GTP cycle. A. Regulation of Ral GDP-GTP cycling. Ral-selective GEFs and GAPs accelerate the low intrinsic exchange and GTP hydrolysis activities to promote formation of active GTP-bound and inactive GDP-bound Ral. B. The RalGEFs are highly conserved across species. All RalGEFs contain a CDC25 homology domain, which is responsible for catalytic activity. There are four human isoforms of RalGEF that contain Ras association (RA) domain. These isoforms also contain a Ras exchanger motif (REM) that likely stabilizes the CDC25 homology domain and is essential for RalGEF catalytic activity. There is one homolog in C. elegans and two in Drosophila. The RalGEF homolog in C. elegans is most similar to RalGDS. The RalGPS RalGEFs lack a REM domain and do not associate with Ras, but instead contain a pleckstrin homology (PH) domain. RGL4 contains a CDC25 homology domain, but lacks a REM, RA or PH domain. C. The RalGAPs are heterodimeric complexes formed by either a RalGAPα1 or RalGAPα2 catalytic subunit with the regulatory RalGAPβ subunit. The RalGAPβ subunit serves to regulate the catalytic activity of the RalGAPα subunits, similar to TSC1 regulation of TSC2. Percentages indicate sequence identity with the RalGAPα1 catalytic domain. Orthologs of the human RalGAPα and RalGAPβ subunits are present in C. elegans and Drosophila. Multiple sequence alignment and sequence identity was determined by ClustalW analyses and domain topology by SMART analyses.
Similar to Ras, the intrinsic GDP-GTP exchange and GTP hydrolysis activities of Ral GTPases are very weak, with each activity accelerated by Ral-selective guanine nucleotide exchange factors (RalGEFs) and GTPase activating proteins (RalGAPs), respectively (Fig. 2A). RalGEFs stimulate guanine nucleotide exchange. With intracellular levels of GTP approximately 10-fold higher than GDP, RalGEF stimulation favors formation of Ral-GTP. Ral GTPase-activating proteins (RalGAPs) catalyze the hydrolysis of the bound GTP, returning Ral to an inactive conformation. When bound to GTP, RalA and RalB can interact with the same array of downstream effector proteins and mediate numerous cellular processes.
2.2. RalGEFs
The first RalGEF identified, Ral guanine nucleotide dissociation stimulator (RalGDS) (Fig. 2B), was found by yeast two-hybrid screens performed in the early 1990s to identify Ras effectors [9-11]. RalGDS was found to catalyze nucleotide exchange on both RalA and RalB but not on other small GTPases including members of the Ras, Rho, and Rab families. Subsequent yeast two-hybrid library screening studies using H-Ras, R-Ras, TC21/R-Ras2, and Rit as baits identified three additional RalGEF proteins that were named Rgl (RalGDS-like), Rgl2/Rlf, and Rgl3 [12-14] [15]. These RalGEFs contain a common domain architecture including an N-terminal Ras exchanger motif (REM) domain followed by a CDC25 homology domain (RasGEF) and a C-terminal Ras-association (RA) domain (Fig. 2A) [16]. The CDC25 homology domain shares sequence identity with the catalytic domains of RasGEFs [17]. In addition to the three Ras isoforms, other Ras family small GTPases can also bind and activate the RA domain-containing RalGEFs [18].
RalGPS1 and RalGPS2 (Ral GEF with PH domain and SH3-binding motif) comprise a second distinct family of RalGEFs [19-21] (Fig. 2A). These two related proteins (63% identity) contain an N-terminal CDC25 homology RasGEF but lack a REM and RA domain. Instead, they contain a C-terminal pleckstrin homology (PH) domain. Additionally, they possess is a central proline-rich sequence with PxxP motifs recognized by Src homology 3 (SH3) domain-containing proteins.
The absence of an RA domain uncouples these RalGEFs from direct association with Ras family small GTPases. Instead, the PH domain has been shown to be sufficient for membrane targeting and necessary for Ral activation [19]. The regulation of these RalGEFs is poorly understood, but some evidence suggests that RalGPS2 plays a role in regulating the actin cytoskeleton [21]. Interestingly, members of both RalGEF subclasses have been implicated in cytokinesis [22].
Another RalGEF, now designated RGL4, was identified originally as a RalGDS-related (Rgr) oncogene in a DMBA (7,12-dimethylbenz[α]anthracene)-induced rabbit squamous cell carcinoma [23]. However, while RGL4 does contain a CDC25 homology domain, it lacks a well-defined RA or PH domain (Fig. 2B). Furthermore, while the other RalGEFs described above are highly selective activators of Ral, RGR has also been described to activate other Ras family small GTPases [24].
2.3. RalGAPs
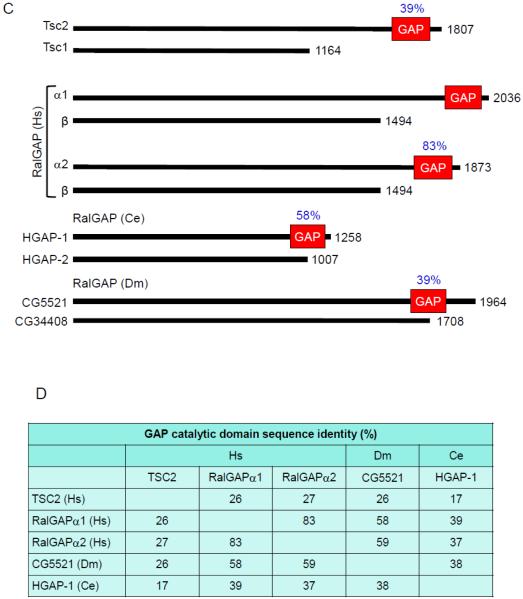
Although the existence of RalGAPs was first reported in 1991 [25], only recently has the molecular identification of RalGAPs been achieved (Fig. 2C). Work done by Feig and colleagues in the early 1990s detected and characterized RalGAP activity in brain and testes cytosolic extracts, and the putative RalGAP activity was distinct in size from Ras or Rho GAPs [25]. Subsequently, using a GTPase-deficient, persistently GTP-bound mutant of RalA for affinity chromatography, two distinct RalGAP complexes were identified in brain cytosol [26]. Each heterodimeric complex consists of a shared regulatory RalGAPβ subunit and one of two related catalytic RalGAPα1 and α2 subunits (53% overall sequence identity) (Figs. 2A,C). Independently, Saltiel and colleagues used a similar RalA affinity purification approach and identified RalGAPα2 (RGC2) and RalGAPβ (RGC1) as components of a Ral-selective GAP [27].
RalGAPα1 (GARNL1/TULIP1) and RalGAPα2 (AS250; Akt substrate of 250 kDa) were identified previously as proteins with sequence identity with the GAP catalytic domain of TSC2 (also known as tuberin) [28, 29] (Fig. 2B) and distinct from the RasGAP catalytic domain [17]. RalGAPα2 was also identified to form a complex with RalGAPβ (KIAA1219). TSC2 is the catalytic subunit of a GAP selective for the Rheb small GTPases, another member of the Ras branch of the Ras superfamily [1]. However, TSC2 alone is not sufficient for RhebGAP activity and requires heterodimer formation with TSC1 (also known as hamartin). Hence, the active RalGAPα/β complexes share both sequence and structural similarities with the heterodimeric tuberous-sclerosis (TSC) complex [30]. Although RalGAPβ lacks sequence similarity with Tsc1, it serves an analogous role in stabilizing RalGAPα and is required for RalGAP activity. RalGAPβ is expressed ubiquitously, whereas more variable expression profiles are seen for the two RalGAPα subunits. RalGAPs are conserved in evolution, with orthologs of both subunits found in C. elegans and Drosophila (Fig. 2B) [26, 31]. The RalGAPα GAP catalytic domains of C. elegans (HGAP-1) and Drosophila (CG5521) share 37-39% and 58-59% sequence identity, respectively, with the GAP catalytic domains of their human counterparts.
RalGAP accelerates the GTPase activity of both RalA and RalB but not for other small GTPases tested (H-Ras, Rap1, Rheb, RhoA, Ran and Rab27) [26, 27, 31]. The RalGAPα subunits share 53% overall sequence identity and 83% sequence identity in their GAP domains (Figs. 2C,D). Additionally, RalGAPα2 is subject to insulin-stimulated phosphorylation by the AKT serine/threonine kinase, analogous to a similar mechanism of AKT regulation of TSC2 [32]. As with TSC2, AKT phosphorylation of RalGAPα2 impaired the ability of the RalGAP complex to catalyze RalA GTP hydrolysis. This is not due to altered intrinsic GAP activity but to a reduced RalA interaction with RalGAPα2.
3. Ral effectors
Like Ras and other small GTPases, Ral interacts with a number of effector proteins when bound to GTP (Fig. 3). However, unlike with Ras, the Ral binding domains (RBD) lack primary sequence identity. The best characterized Ral effectors are RalBP1/RLIP76 and the Sec5 and Exo84 subunits of the octameric exocyst complex. The evolutionarily conserved exocyst complex mediates the tethering of post-Golgi secretory vesicles to the plasma membrane prior to exocytic fusion [33]. The exocyst subunits may also exist as monomers or subcomplexes, and can possess non-exocyst functions. Ral interaction with each subunit occurs in distinct subcellular locations, interacting with Sec5 at the plasma membrane and with Exo84 with intracellular vesicles [34, 35]. Although RalA and RalB can interact with the same set of effectors in vitro, as described below, the distinct biological functions of RalA and RalB are mediated by differences in subcellular localization, leading to their interaction with distinct subsets of effectors.
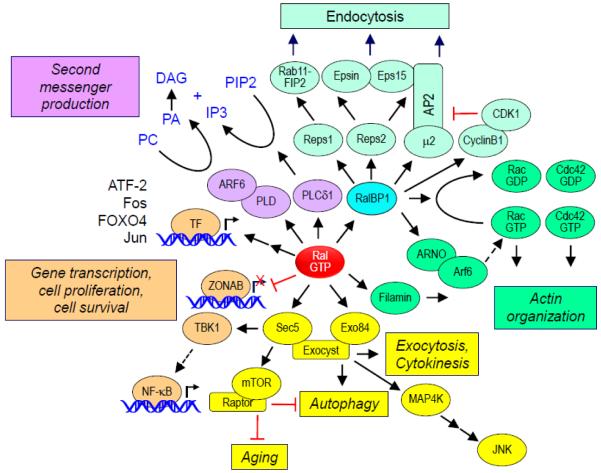
Fig. 3.
Ral effectors and effector functions. Active Ral can bind to a variety of downstream effectors and modulate numerous cellular activities. RalBP1 acts as a RhoGAP as well as a scaffold for other proteins that regulate endocytosis and other cellular processes. Ral association with Sec5 or Exo84 can regulate exocyst-dependent and –independent processes. Other effector processes include regulation of cell cycle progression through PLD1-dependent cytokinesis and cytoskeletal changes through filamin A, IP3 signaling through PLCδ1, and gene transcription through ZONAB. Ral activation also stimulates signaling pathways that lead to the activation of various transcription factors (TF), stimulating gene expression.
The structures of Ral in complex with the RBDs of these three effectors have been determined. Whereas the Sec5 RBD interacts with SI alone [36], Exo84 [37] and RalBP1 [38] RBD interaction involves both SI and SII [39]. Consistent with 100% conservation of SI and SII residues, and residues involved with effector binding, where studied, RalA and RalB interact with the same set of effectors in vitro.
3.1. RalBP1/RLIP76
The first Ral effector described, RalBP1 (Ral binding protein 1; also called RLIP76 or RIP1), was identified in screens for proteins that bound preferentially to activated RalA [40-42]. RalBP1 orthologs are found in Drosophila and C. elegans. RalBP1 contains a RhoGAP catalytic domain that has activity for the Cdc42 and Rac small GTPases, members of the Rho branch of the Ras superfamily [1]. Cdc42 stimulates filopodia formation whereas Rac stimulates lamellipodia formation. Thus, RalBP1 provides a link between Ral and modulation of the actin cytoskeleton changes that drive these cellular activities [40].
In additional to its RhoGAP domain, RalBP1 has additional functions (Fig. 3). Two ATP binding motifs have been identified in RalBP1 and shown to be important for transport function involving glutathione conjugates of electrophilic compounds [43, 44]. This transport function may facilitate the cellular export of chemotherapeutic drugs and radiation-induced oxidative damage byproducts [45]. RalBP1 overexpression has been found in a spectrum of human cancers, and suppression of RalBP1 expression can impair tumorigenic growth in vivo [46]. However, phenotypes attributed to RalBP1 do not necessarily implicate their role in Ral signaling [47].
RalBP1 also functions as a scaffold and interacts with a spectrum of functionally distinct proteins that regulate endocytosis and signal transduction (Fig. 3). The AP2 adaptor complex, a regulator of clathrin-mediated endocytosis from the plasma membrane, associates with the N-terminal region of RalBP1 [48]. The Eps homology (EH) domain-containing proteins Reps1 and Reps2 (POB1) were identified as proteins that interacted with the C-terminus of RalBP1 [49, 50]. These proteins are known to be important for receptor tyrosine kinase-regulated endocytosis, with Reps1 interacting with Rab11-FIP2 and Reps2 binding Epsin and Eps15 [51, 52].
Another protein that associates with the RalBP1 C-terminus is cyclin B1 [53]. In turn, the RalBP1-bound cyclin B1 complexes with Cdk1, with Cdk1 phosphorylation of Epsin preventing endocytosis during mitosis. This activity was shown to be mediated by RalA activation.
RalBP1 has been implicated as a key effector for several Ral-driven processes. In these studies, the typical approach has been the utilization of mutants of Ral that are selectively impaired in effector interaction. The D49N substitution impairs RalBP1 but not Sec5 or Exo84 effector binding, whereas the D49E mutation has the opposite consequence [40, 54, 55]. For example, shRNA silencing analyses determined that RalB but not RalA was required for invadopodia formation in pancreatic cancer cell lines [56]. RalB D49E but not D49N could rescue loss of endogenous RalB and restore invadopodia formation, indicating that RalBP1 was a critical effector for this RalB activity. This RalBP1 function was GAP-independent but abolished by mutation of the ATP binding motifs [56].
RalA was shown to utilize RalBP1 to regulate mitochondrial fission at mitosis [57]. Mitochondria exist as dynamic interconnected networks that are maintained through a balance of fusion and fission. Fission facilitates equal distribution of mitochondria to daughter cells during mitosis. Fission is controlled by the GTPase DRP1 on the outer mitochondrial membrane. RalA was found to recruit RalBP1 to mitochondria, where RalBP1 acts as a scaffold to facilitate cyclin B/Cdk1 phosphorylation of Drp1 to promote mitochondrial fission. Suppression of either RalA or RalBP1 expression caused a loss of mitochondrial fission at mitosis.
Recently, RalBP1 was shown to be necessary and sufficient for RalA-driven mislocalization of the cyclin-dependent kinase inhibitor p27 KIP1, leading to inhibition of TGF-β–mediated growth arrest in epithelial cells [58]. This function appeared to require an intact RhoGAP domain.
3.2. The Sec5 and Exo84 subunits of the exocyst
The best-characterized Ral effectors are two components of the exocyst complex, Sec5 and Exo84 [54, 55, 59]. The association of Ral with both Sec5 and Exo84 has been found to be important in exocytosis. Ral regulates the subcellular localization of the exocyst through mediating Sec5-paxillin association and the assembly of the octameric exocyst complex by interacting with Sec5 and Exo84 [55, 60]. Ral interaction with Sec5 may also regulate exocyst-independent functions.
Recent evidence suggests that Ral engages exocyst subunits to perform a variety of cellular processes independent of their roles in exocytosis. White and colleagues found that the association of RalB with Sec5 is critical in the innate immune response [61]. RalB binding to Sec5 leads to an interaction of Sec5 with TBK1, a protein kinase known to regulate NF-κB signaling. Intriguingly, TBK1 has recently been identified in siRNA screens as a synthetic lethal partner of activated K-Ras [62], although a subsequent study failed to support this relationship [63].
Recently, a mechanism where the integrin αvβ3 recruited a K-Ras-RalB complex to the plasma membrane to activate TBK1 and NF-κB signaling was identified (Fig. 3) [64]. This signaling mechanism regulated tumor initiation and growth.
The association of RalB with the exocyst has also been shown to regulate macroautophagy [34]. When cells are grown in nutrient-rich conditions, RalB engages Sec5. Upon nutrient starvation, RalB then engages Exo84 and the exocyst, leading to an upregulation of autophagosome formation. This process is mediated through the assembly of the ULK1 serine/threonine kinase and Beclin1-VPS34 complexes on the exocyst. Autophagy has emerged as a key component of Ras-driven transformation in a variety of cell types, perhaps highlighting an underlying importance of Ras-RalGEF signaling in tumor cell autophagy.
3.3. Other effectors
One lesser-characterized Ral effector is phospholipase D1 (PLD1) [65, 66]. However, unlike other effectors, the association with Ral is not GTP-dependent and instead the association is with the N-terminal 11 amino acid extension (Fig. 1D). PLD1 is best known for its role in converting phosphotidylcholine to phosphatidic acid and choline in response to G-protein coupled receptor (GPCR) stimulation. Recent evidence shows that RalA is necessary for the PLD1-mediated stimulation of mTORC1 signaling [67]. Furthermore, the RalA-PLD1 interaction has been shown to promote proper p27 localization, thus allowing for proper TGF-β signaling [58]. The interaction of both RalA and RalB with PLD1 has been shown to be critical for HeLa cell cytokinesis [22].
Filamin is an important component of the actin cytoskeleton and is involved in actin crosslinking and lamellipodia formation. The association of RalA with filamin was found to be important for filopodia formation in Swiss-3T3 cells [68]. Additionally, RalA did not induce filopodia in a human melanoma cell line that lacks expression of filamin.
Lastly, active RalA has been shown to engage the transcription factor ZONAB (zonula occludens 1-associated nucleic acid binding protein) in a cell density dependent manner in MDCK cells [69]. At high cell densities, RalA engages ZONAB, unlocking the transcription of ZONAB targets, but it is unclear which genes are turned on [69]. While a direct role for Ral association with these lesser-studied effectors has not been found in Ral-driven cancers, their important roles in mitosis, motility, and gene regulation make them intriguing targets as Ral studies progress.
4. Post-translational modification and regulation of Ral function
RalA and RalB exhibit the most significant sequence divergence in their C-terminal membrane targeting sequences (50% identity) (Fig. 1C). This sequence divergence results in their distinct subcellular localization that contributes to the functional differences described for RalA and RalB by regulating effector utilization in vivo [56, 70-73]. Both isoforms can be found at the plasma membrane as well as in endomembranes, with cell type differences seen. In this section we summarize the role of posttranslational modifications that regulate Ral subcellular localization.
4.1. CAAX modifications
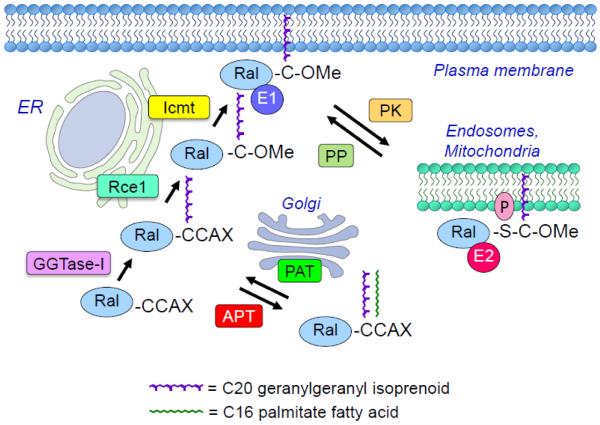
Like the majority of Ras family small GTPases, RalA and RalB terminate in a CAAX (C = cysteine, A = aliphatic amino acid; X = terminal amino acid) tetrapeptide motif (Fig. 4). The CAAX motif signals for a series of posttranslational modifications that increase hydrophobicity and promote membrane anchoring, where the terminal X residue determines protein prenyltransferase specificity [74]. When X = S, A, Q, and M, the protein is preferentially recognized by farnesyltransferase (FTase)-catalyzed addition of a C15 farnesyl isoprenoid lipid; when X = L or I, it signals for geranylgeranyltransferase type I (GGTase-I)-catalyzed addition of a C20 geranylgeranyl isoprenoid.
Fig. 4.
Regulation of Ral subcellular localization and membrane association. CAAX motif-signaled posttranslational modifications. Ral is geranylgeranylated by GGTase-1 on the first cysteine residue of the CAAX (RalA: CCIL; RalB: CCLL). By the canonical CAAX processing pathway, Ral is then modified at the endoplasmic reticulum (ER) by Ras converting enzyme (Rce1) which cleaves between the two cysteine residues. Isoprenyl cysteine carboxyl methyltransferase (ICMT) then catalyzes methylation of the isoprenylated free cysteine residue, facilitating recruitment to the plasma membrane. A second non-canonical pathway has been described by which Ral is palmitoylated at the cysteine residue at the A1 position by Golgi-associated protein acetyltransferase (PAT) after geranylgeranylation. Palmitate addition is reversible and depalmitoylation is catalyzed by acylprotein thioesterase (APT). Whether this double lipid modified form is associated with a different membrane compartment has not been determined. Ral subcellular localization is also regulated by a dynamic and reversible protein kinase (PK)-mediated phosphorylation and protein phosphatase (PP)-mediated dephosphorylation cycle. RalA and RalB possess distinct C-terminal phosphorylation sites for different protein kinases (PK). Phosphorylation causes dissociation from the plasma membrane and translocation to specific endomembrane compartments, resulting in a switch in effector (E) interaction.
For Ral, the initial step is catalyzed by covalent addition of geranylgeranyl to the cysteine residue of the CAAX motif by cytosolic GGTase-I [75]. This is followed by endoproteolytic removal of the AAX residues, catalyzed by endoplasmic reticulum-associated Ras converting enzyme 1 (RCE1), and subsequent carboxymethylation of the now terminal prenylated cysteine residue, catalyzed by isoprenylcysteine carboxyl methyltransferase (ICMT).
The CAAX-signaled modifications are critical for both RalA and RalB function. Mutation of the cysteine residue to prevent all CAAX-signaled modifications disrupts Ral membrane association and function [75]. Similarly, treatment with a pharmacologic inhibitor of GGTase-I also disrupted Ral membrane association and signaling. Since inhibition of the GGTase-I modification prevents all subsequent modifications, the role of the Rce1 and ICMT catalyzed modifications in Ral function remain to be addressed.
Recently, the Ral CAAX motifs were identified as members of a distinct subset of CA1A2X motifs where the A1 residue is a second cysteine residue (CCAX). CCAX motifs can undergo an alternative modification pathway (Fig. 4). As shown for a Rho family small GTPase (Cdc42), this motif can signal for dual lipid modification: prenylation followed by covalent addition of a palmitate fatty acid [76]. For Cdc42, after the initial GGTase-I catalyzed prenylation step, a Golgi-associated protein acetyltransferase (PAT) catalyzes covalent addition of palmitate to the adjacent cysteine residue rather than the conventional modification by Rce1 and ICMT. For Cdc42, this alternative modification prevented its recognition by RhoGDI, a protein that masks the prenyl lipid and disrupts membrane association, resulting in a cytosolic pool of Cdc42. Since there is no known RalGDI, the consequences of this palmitate modification on Ral subcellular localization and membrane association, and function, have not been determined.
4.2. Phosphorylation regulation of subcellular localization and effector interaction
An emerging theme in the regulation of small GTPases is reversible post-translational modifications that dynamically regulate subcellular localization, thereby influencing effector interaction and biological activity [77]. In particular, recent studies have highlighted protein kinase-mediated phosphorylation of small GTPases in their C-terminal membrane-targeting regions. For example, K-Ras4B phosphorylation by protein kinase C (PKC) on S181 in its C-terminal membrane targeting sequence altered K-Ras4B subcellular localization [78]. Nonphosphorylated K-Ras4B was plasma membrane associated, whereas S181 phosphorylation K-Ras4B caused translocation to mitochondrial and endoplasmic reticulum (ER) membranes. S181 is positioned within a polybasic amino acid stretch in K-Ras4B that serves as a second signal that together with the CAAX modifications promote full plasma membrane association. The negative charge caused by phosphorylation reduces the positive charge of the polybasic stretch. The ER-associated K-Ras4B then associated with inositol trisphosphate receptors (InsP3) on the ER in a Bcl-xL-dependent fashion, blocking the ability of Bcl-xL to potentiate the InsP3 regulated flux of calcium from ER to mitochondria that is required for respiration, inhibition of autophagy, and cell survival [79].
The Ral proteins are also regulated by similar mechanisms, with distinct protein kinases phosphorylating serine residues distinct for the C-termini of RalA and RalB (Fig. 4). Aurora-A kinase and protein kinase A (PKA) have been found to phosphorylate RalA on S194 [80, 81] and protein phosphatase 2A dephosphorylates RalA at S194 as well as S183 [82]. Counter and colleagues showed that phosphorylation of RalA on S194 was critical for RalA to promote the anchorage-independent growth in vitro and tumorigenic growth in nude mice of pancreatic ductal adenocarcinoma (PDAC) cell lines [71]. This phosphorylation event dramatically altered RalA subcellular localization from the plasma membrane to internal membranes, where it had an enhanced interaction with RalBP1. More recently, Aurora-A phosphorylation of RalA has been found to promote RalA translocation to the outer face of mitochondria, where it then recruits RalBP1 to stimulate mitochondrial fission [57].
Studies by our lab and others have found that RalB is similarly regulated by PKCα phosphorylation of S198 in the C-terminal membrane targeting sequence [72, 81]. In one study, it was found that S198 phosphorylation caused RalB translocation from the plasma membrane to endocytic vesicles [72]. Associated with this change in subcellular localization was a switch in effector utilization. Whereas unphosphorylated RalA preferentially associated with Sec5, S198 phosphorylation caused preferential association with RalBP1. Phosphorylation of RalB S198 was necessary for proper exocytic vesicle trafficking and fusion at the plasma membrane, with delivery of surface alpha-5 integrin being regulated by dynamic RalB phosphorylation. Independently, Theodorescu and colleagues found that phosphorylation of RalB S198 was critical in regulating the ability of RalB to promote the metastatic growth of bladder cancer cells in a nude mouse model [81].
4.3. Ubiquitination
In the past few years, regulation of small GTPases by ubiquitination has gained recognition [77]. For example, monoubiquitination of K-Ras on K147 reduces GAP sensitivity, thus allowing K-Ras to remain active and signaling in the absence of upstream input [83]. Ubiquitination of the Ral proteins has also been shown to influence their activity and function. Regulation of the ubiquitination of RalA modulated RalA activity as well as lipid raft exposure [84]. Furthermore, ubiquitination of RalB promoted binding to Sec5 to regulate innate immunity, whereas deubiquitination allowed for binding to Exo84 and subsequent induction of autophagy [85].
5. Ral function in invertebrates – lessons learned from worms and flies
The conservation of Ral GTPases, their regulators, and effectors in Drosophila melanogaster and C. elegans has allowed genetic dissection of Ral function. Consistent with the probable double genome duplication and subsequent winnowing of vertebrate relative to invertebrate genomes [86], in Drosophila and C. elegans there exist single genes for most Ras and Ral signaling components. For example, Drosophila and C. elegans each harbor single Ral GTPase (two in mammals) and RalGEF/RalGDS (four in mammals; Drosophila but not C. elegans encodes a RalGPS ortholog), RalGAP α and β (two and one in mammals, respectively) genes. This trend is consistent with reduced gene complexity of other invertebrate Ras system signaling genes, with three mammalian Ras, three Raf, and four type I PI3K catalytic subunit encoding genes [87]. Generally, functional similarity between an invertebrate protein and a specific vertebrate isoform is difficult to extrapolate, largely because biological assays usually differ between invertebrates and mammals. But this difference also highlights the benefit of invertebrate studies, since they provide unique in vivo perspectives that complement mammalian studies. All key Ral effectors are conserved in C. elegans and Drosophila but TBK1, which is not encoded in the C. elegans genome. Below, we summarize findings from studies on C. elegans and Drosophila Ral signaling.
5.1. Toggling Ras between Raf and RalGEF regulation
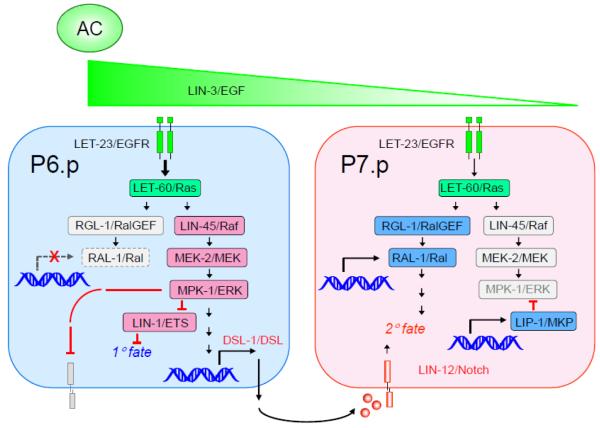
Since RalGEFs and other Ras effectors are widely expressed, how Ras effector utilization is regulated has been an unresolved issue. The C. elegans vulva is patterned by epidermal growth factor (EGF) activation of Ras (LET-60) and the Raf-MEK-ERK MAP kinase cascade to control 1° fate, and presumptive 1° cells secrete DSL Notch ligands to induce 2° fate in neighboring cells. Furthermore, a spatial EGF gradient, in addition to inducing 1° fate, contributes to 2° fate via an unknown pathway. 1° and 2° cell fates are antagonistic and mutually exclusive. Reiner and colleagues found that in addition to its canonical effector, Raf, vulval Ras utilized RalGEF-Ral 2°-promoting activity to antagonize Ras-Raf 1°-promoting activity, and that Ral promoter activity was excluded from presumptive 1° cells, thus blocking inappropriate Ral activation in 1° cells [88] (Fig. 5). Consistent with its restricted expression pattern, they found that Ras-RalGEF-Ral mediated the 2°-promoting activity of the EGF gradient. These findings delineated a Ras effector-switching mechanism whereby cell position within the morphogen gradient dictates that LET-60/Ras effector usage switched from Raf to RalGEF to promote 2° instead of 1° fate. This dynamic developmental switching in effector use may reflect diversity in tumorigenic processes that results in heterogeneity of effector predominance in tumors [89]. Other mechanisms that control Ras effector utilization will likely exist.
Fig. 5.
EGFR signaling toggles C. elegans developmental output by effector switching. The C. elegans Ras ortholog (LET-60) can interact with orthologs of human Raf (LIN-45) and RalGEF (RGL-1). The nearby anchor cell (AC) secretes EGF/LIN-3, creating a concentration gradient, inducing vulval precursor cell (VPC) development. This concentration gradient, in combination with sequential induction, patterns vulval cell fates. Active pro-1° signaling is shown in blue, with active pro-2° signaling shown in red, and quenched signaling is in gray. In presumptive 1° cells, typically P6.p, EGF activates Ras to utilize Raf to promote 1° cell fate. Pro-2° signaling through Notch is quenched. RGL-1→RAL-1 pro-2° quenching is based on RAL-1 exclusion from presumptive 1° cells. Additionally, presumptive 1° cells express and secrete notch ligand (DSL; Delta/Serrate/LAG-2) that then induces neighboring vulval precursor cells via the Notch/LIN-12 receptor to assume a 2° fate. In presumptive 2° cells, Notch induces expression of an ERK MAPK phosphatase, and other 2°-specific proteins to quench the Raf pro-1° signal. EGF activates Ras to utilize the RGL-1 RalGEF effector to promote 2° fate.
5.2. Ral and innate immunity
A recent study in mammalian cells showed that RalA mediates nuclear translocation and activation of FOXO in response to reactive oxygen species (ROS) by activating a JNK cascade scaffolded by JIP1, and that in C. elegans RAL-1 and JIP-1 mediate DAF-16/FOXO ROS-dependent nuclear translocation [90]. Additionally, transportin-1 mediates ROS-dependent nuclear translocation of DAF-16/FOXO in both mammals and C. elegans [91]. C. elegans harbors a single FOXO gene, daf-16, compared to four mammalian genes. These observations suggest that Ral-mediated stress response to ROS is conserved across evolution.
As noted above, mammalian RalB harnesses the TBK1 IκB kinase family member for tumor cell survival. These results are echoed in Drosophila, where Sec5 or Ral haploinsufficiency reduces anti-fungal response [61]. The absence of TBK1 and other IκB–related proteins in the worm genome may reflect the absence of NF-κB in the worm genome and attendant differences in innate immunity [92].
5.3. Fly insights into Ral signaling partners
Yeast two-hybrid interaction studies in Drosophila identified the Ral effectors Sec5 and RalBP1/RLIP, and RalBP1/RLIP binds with known partner REPS1 [93]. Upstream of Ral, both fly Ras and fly Rap bind RalGEF in yeast two-hybrid assays, consistent with their identical core effector binding regions. However, in mammalian systems Ras and Rap1 are localized to mostly non-overlapping subcellular compartments, and it is expected that they encounter distinct sets of effectors [94, 95]. Overexpression studies with the two splice variants of the fly RalGEF suggest that one alternative isoform, differing in N-terminal sequence, harbors Ral-independent functions. This putative GEF-independent function of RalGEF may correspond to putative N-terminally encoded GEF-independent functions of mammalian RalGDS, where RalGDS is thought to scaffold PDK and Akt, thus potentiating Akt output in certain circumstances [96, 97]. Genetic analyses, mostly using ectopically expressed constructs, implicates the Rap1 small GTPase, rather than Ras, as a putative activator of RalGEF. However, these results are also consistent with Rap1 functioning in parallel, and illustrate the potential complexities of dominant-negative mutations as experimental tools for small GTPases, as well as the difficulties of analyzing the functions of essential genes in vivo [98]. The Ras, Rap1 and Ral small GTPases act in multiple developmental events during Drosophila development, so there is ample opportunity to study their functions further.
Drosophila genetic analysis of a sensory organ apoptotic event, coupled with yeast two-hybrid gene discovery and mammalian biochemical experiments, identified an interesting and otherwise unidentified Ral effector cascade [99]. Ral signals through Sec5 of the exocyst complex, which in turn interacts with the Msn MAP4 kinase in flies and humans. Msn is thought to signal through a basket/JNK MAP kinase cascade in flies, and its ortholog HGK signals through a JNK MAP kinase cascade in human cells. The fly Msn and human HGK proteins are members of the CNH domain-containing MAP4 kinases, which contain ancient Ste20-like kinase domains thought to activate JNK and p38 MAP kinases cascades. Drosophila encodes two family members (Msn and Hpy), C. elegans encodes two family members (MIG-15 and GCK-2), and mammals contain eight distinct genes encoding CNH domain-containing MAP4 kinases, with four corresponding to each invertebrate subgroup [100]. This relatively under-investigated group of kinases may represent a new and druggable Ral signaling output.
5.4. RalGAPs connect Ral with the mTORC1 signaling network
An unexpected crosstalk between the Ral and mTOR signaling networks was identified through the sequence relationship shared with the GAPs that control each network. mTOR signaling regulates a spectrum of major cellular processes and is implicated in cancer and other pathologic conditions [30]. Despite the sequence identity with TSC2 (26-27% identity in RalGAP domains) (Fig. 2D), RalGAPs do not exhibit GAP activity for Rheb in vitro or in vivo [31]. Nevertheless, the unexpected observation that C. elegans possesses orthologs for the Rheb and Ral GTPases and for RalGAPα/β, but not Tsc1/2, led to the discovery of an unexpected signaling interplay between Ral and Rheb signaling [31]. It was determined that C. elegans RalGAP loss caused decreased lifespan, consistent with a Tsc-like function. Additionally, RalGAP suppression in mammalian cells caused RalB-selective activation and Sec5- and exocyst-dependent engagement of the mTORC1 complex and suppression of autophagy (Fig. 3). Surprisingly, it was also found that Tsc1-Tsc2 loss activated RalA/B independently of Rheb-mTOR signaling. Finally, RalGAP suppression caused mTORC1-dependent pancreatic tumor cell invasion. These findings identify an unexpected crosstalk and integration of the Ral and mTORC1 signaling networks.
Interestingly, as noted above mammalian RalB also activates autophagy, an activity supported by Drosophila experiments [34]. Since TORC1 inhibits autophagy [101, 102], the observations that RalB both promotes and inhibits autophagy are potentially contradictory. However, RalB-TORC1 signaling occurs at the plasma membrane while RalB-autophagosome signaling occurs at the lysosome/autophagolysosome. The aforementioned observations of phosphorylation-dependent subcellular trafficking and effector switching of K-Ras [78] and RalA [82] , raise the possibility that RalB can orchestrate seemingly antagonistic signaling outcomes under different conditions and in different cellular compartments.
6. Ral and cancer
Since RalGEFs participate in downstream signaling from activated Ras proteins, it was initially speculated that Ral protein activation may contribute to Ras-driven cellular transformation. However, when explored initially in NIH 3T3 mouse fibroblasts, a critical and significant role for Ral GTPases in Ras-driven cancer seemed unlikely [103, 104]. However, when Counter and colleagues explored the role of Ral in Ras-mediated growth transformation of immortalized human astrocytes, fibroblast or epithelial cells, a more significant role for Ral GTPases as effectors of Ras in human cancer was observed, suggesting species differences in the effectors that are important in Ras oncogene function [105].
That Ral GTPases serve critical roles in human cancer cell growth gained greater traction when White and colleagues found that RalB was critical for tumor but not normal cells for survival, while RalA was necessary for the anchorage-independent growth of cancer cells [106]. Importantly, this also marked the first time RalA and RalB were found to have non-overlapping functions. Since these key studies, a major theme of Ral proteins is their significant and often divergent roles in numerous cancer types. In the following section we review some of the key findings made with regards to the role of the two Ral isoforms as drivers in different human cancers. Since the RA domain-containing RalGEFs can be activated by other Ras family small GTPases, as well as by non-Ras mechanisms, and since some RalGEFs are regulated by non-Ras mechanisms, an involvement of Ral in cancers where RAS mutations are not common is not surprising.
6.1. Bladder carcinoma
Evaluation of a panel of human bladder cancer cell lines found preferentially increased levels of activated RalA and RalB in RAS-mutant [107] or invasive cell lines [108]. Using RNAi or ectopic expression of activated Ral mutants, Theodorescu and colleagues found that RalA and RalB played antagonistic roles in the migratory activity of the KRAS-mutant UM-UC-3 bladder cancer cell line, with RalA suppressing and RalB enhancing motility [109].
Activating RAS mutations occur in a low percentage (~10%) of bladder cancers. Therefore, a Ras-RalGEF mechanism may be less relevant for Ral activation in this cancer type. Consistent with this possibility, a recent study found RalGAPα2 expression in normal bladder urothelium, but reduced expression associated with advanced clinical stage and poor patient survival [108]. Furthermore, genetic depletion of Ralgapα2 in mice did not cause any apparent abnormalities but did enhance the invasive phenotype of chemically-induced bladder tumors. Thus, loss of RalGAP function may be an important mechanism for Ral activation in bladder cancer.
6.2. Colorectal carcinoma
Oncogenic KRAS and NRAS mutations occur in 45% and 8%, respectively, of colorectal cancer (CRC) tumors. Ral signaling has been shown to be a critical regulator of the anchorage-independent growth properties of CRC tumor cells [110]. Martin et al found that RNAi-mediated suppression of RalA resulted in a decrease in soft agar colony growth while loss of RalB had the opposite effect, leading to an enhancement of anchorage-independent growth. They found that RalA and RalB modulated this phenotype by utilizing both common and distinct effector proteins. Using Ral effector binding mutants that are selectively uncoupled from Exo84, Sec5, or RalBP1, they showed that RalA required Exo84 and RalBP1 binding to promote the anchorage-independent growth of CRC cells. Conversely, RalB required Sec5 and RalBP1 to suppress soft agar colony formation. Intriguingly, loss of one Ral isoform was found to increase the activation of the other isoform suggesting compensatory crosstalk between RalA and RalB. What specifically mediates this crosstalk between RalA and RalB is unknown, but it could be through either enhanced RalGEF accessibility for the remaining Ral protein or a downregulation of RalGAP activity upon single Ral isoform depletion. Depletion of RalB has also been shown to cause apoptosis in colorectal cancer cells [61].
6.3. Hepatocellular carcinoma
RAS mutations are rare (>2%) in hepatocellular carcinoma (HCC). RalA was found to be significantly overactivated in hepatocellular carcinoma (HCC) cells and tissues compared to nonmalignant samples. Suppression of RalA expression caused a significant decrease in the viability and invasiveness of HCC cells. A role for RalB was not addressed. Finally, in a transgenic mouse model for HCC (farnesoid X receptor–deficiency induced) elevated RalA-GTP was detected in the liver tumors [111].
6.4. Lung adenocarcinoma
KRAS mutations are found in 30% of lung adenocarcinomas and several studies have addressed the role of Ral in lung cancer. In one study, variable levels of RalA-GTP, independent of KRAS mutation status, were detected in a panel of lung adenocarcinoma or squamous carcinoma cell lines [112]. shRNA suppression of RalA expression in the KRAS mutant A549 lung adenocarcinoma cell line reduced the proliferation and invasion in vitro. In a second more comprehensive study, immunohistochemistry analyses of non-small cell lung cancers (NSCLC), it was found that high RalA and RalB protein expression was associated with poor survival. The levels of activated RalA but not RalB were higher in KRAS-mutant NSCLC cell lines [113]. Depletion of RALA or RALB or both reduced anchorage-dependent and –independent growth for either KRAS mutant and wild type cell lines. Depletion of RALA, RALB, or both also impaired the tumorigenic growth of KRAS-mutant NSCLC cells. Interesting, very limited analyses in this and another study suggested mutation-selective involvement of Ral in NSCLC, where KRAS G12C mutant NSCLC cell lines showed greater activation and/or dependence on Ral for growth [114].
In contrast to human lung tumor cell line studies, RalA and RalB were found to exhibit redundant functions when assessed in mouse development and in a Kras G12D-driven mouse model of lung adenocarcinoma [73]. Ralb deficient mice were viable with no overt phenotype whereas a Rala deficiency caused embryonic lethality that was further exacerbated by a combined Ralb deficiency. Neither a Rala nor a Ralb deficiency impaired Kras-driven lung tumor development. However, a combined loss of both Rala and Ralb significantly reduced lung tumor development. These results suggest redundancy in RalA and RalB function for tumor development. One possible explanation for this different conclusion may be that the human lung tumor cell line studies addressed the role of Ral in tumor maintenance whereas the mouse study addressed the role of Ral in tumor initiation and progression.
6.5. Malignant peripheral nerve sheath tumors
Loss of the NF1 RasGAP, rather than RAS mutational activation, is seen in neurofibromatosis type 1 and malignant peripheral nerve sheath tumors (MPNST) [115]. When compared with a nontransformed mouse Schwann cell line, RalA-GTP levels were elevated in a panel of MPNST cell lines established from tumors that arose from a NF1- and Tp53-deficient genetically-engineered mouse model. When evaluated in one cell line, RalA suppression impaired proliferation and invasion in vitro and tumorigenic growth in vivo [116]. RalA activation was also seen in human MPNST cell lines and tissue, and restoration of NF1 GAP activity reduced RalA activity, indicating that this was associated with Ras activation.
6.6. Melanoma
RAS mutations, predominantly NRAS, occur in 28% of skin cutaneous melanomas. With BRAF mutations seen in 60% of melanomas in a non-overlapping frequency with RAS mutations, activation of the canonical Raf-MEK-ERK mitogen-activated protein kinase (MAPK) pathway alone may seem to be sufficient for Ras-driven melanoma growth. However, analysis of Ral activation in a panel of human melanoma cells showed a consistently high level of total and activated RalA, but not RalB, activation that was independent of NRAS or BRAF mutation status [117]. Additionally, RalA and to a lesser degree RalB are necessary for the tumorigenic growth of melanomas, also regardless of BRAF and NRAS mutation status.
Studies using tumor suppressor Arf-deficient immortalized mouse melanocytes to investigate the contributions of Ras downstream signaling to melanomagenesis also indicated a role for Ral signaling [118]. Ectopic expression of the RalGEF Rgl2 engineered to contain a membrane localization sequence (to mimic Ras activation of RalGEF) was sufficient to promote the anchorage-independent growth and Matrigel invasion of these melanocytes similar to that caused by oncogenic N-Ras. Surprisingly, in contrast, activated BRAF V600E did not enhance proliferation or invasion. Finally, ectopic expression of a dominant negative mutant of RalB that blocks RalGEF function partially impaired the growth of NRAS-transformed melanocytes. Thus, together with the findings of Zipfel et al. [117], Ral GTPases can act as drivers of melanoma cancer growth in both RAS wild type and mutant cancer cells.
6.7. Ovarian carcinoma
One study has now revealed that Ral signaling has a role in ovarian cancer. Specifically, higher levels of RalA activity was found in human tumor samples compared to benign samples. Furthermore, depletion of RalA decreased proliferation and invasion of ovarian cancer cell line OVCAR-5 in vitro and decreased tumor genesis [119].
6.8. Pancreatic ductal adenocarcinoma
A significant requirement for activated Ral signaling in pancreatic adenocarcinoma (PDAC) cell line tumorigenic and invasive growth has been established. Human PDAC has a high frequency or activating KRAS mutations and Ral activation is seen in both human tissue samples and tumor cell lines [120-122]. Interestingly, activation of RalA was found at a higher frequency than the activation of either ERK or AKT in PDAC cells, suggesting a critical role for the RalGEF-Ral pathway downstream of oncogenic K-Ras.
Depletion of RalA and RalB via RNAi has elucidated roles for RalA in anchorage-independent and tumorigenic growth and RalB in invasive and metastatic growth of PDAC cells [122]. PDAC cells with stable RNAi depletion of RalA results in reduced subcutaneous tumor formation upon injection into immune compromised mice. These same cells expressing RalB RNAi do not form lung metastases post-injection into the tail-vein of nude mice. In addition to playing a role in tumor initiation, RalA has also been shown to be necessary for PDAC tumor maintenance. The use of inducible RNAi to stably deplete RalA from established primary tumors resulted in regression of the tumor, indicating a necessity for persistent RalA signaling in established PDAC tumors.
There is also recent evidence that active K-Ras signaling to RalB but not RalA plays a critical role in the formation of invadopodia in PDAC cells [56]. Invadopodia are actin-rich membrane protrusions that are known to be involved in the secretion of matrix metalloproteases (MMP) during tumor cell invasion. RalB requires the ability to interact with RalBP1 to mediate this process and RalBP1 itself is necessary for the formation of invadopodia in PDAC cells. Surprisingly, the RhoGAP activity of RalBP1 is not necessary for invadopodia formation while the ATPase activity is required. Why the ATPase activity is necessary for RalBP1 to mediate invadopodia formation is unclear.
RalGEFs have also been found to play a role in PDAC. Rgl2 is overexpressed in PDAC patient tumors and has been shown to be necessary for both the anchorage-independent and invasive growth of PDAC tumor cells [123]. RNAi-mediated depletion of Rgl2 results in a significant decrease in both RalA and RalB activation. Interestingly, expression of constitutively active RalA could not rescue soft agar growth after the loss of Rgl2 indicating that Rgl2 may have non-Ral regulatory functions or that the RalA interaction with Rgl2 is critically important for the regulation of anchorage-independent growth. Rgl2 was found to be co-localized with RalB but not RalA at the leading edge of migrating CFPac-1 PDAC cells. Loss of Rgl2 results in a loss of RalB from the leading edge, perhaps giving insight into how the migratory and invasive activity of PDAC cells relies on Rgl2/RalB signaling.
6.9. Prostate carcinoma
Increased RalA-GTP levels were observed in the RAS wild type human prostate carcinoma cell line PC3. Suppression of RalA did not impair tumor formation but did abolish bone metastasis [124]. In contrast, suppression of RalB expression did not impair metastasis.
6.10. quamous cell carcinoma
Squamous cell carcinoma is the second most common type of skin cancer. Using an in vitro model of Ras-induced human squamous cell carcinoma (SCC) of the skin, it was found that RalA suppressed rather than promoted progression [125]. Suppression of RalA but not RalB stimulated the progression of HRAS-transformed human keratinocytes to a more invasive state.
In contrast to the in vitro observations, different roles for Ral were observed in a mouse model of carcinogen-induced SCC [73]. Single application of the mutagen DMBA, followed by repeated applications of phorbol ester 12-O-tetradecanoylphorbol 13-acetate (TPA) causes Hras mutation and the development of benign papillomas, with a subset progressing to SCC. Neither a Rala nor a Ralb deficiency impaired papilloma development. As described above for Kras-driven lung adenocarcinoma formation, only combined loss of both Rala and Ralb significantly reduced papilloma development. Genetic ablation of Ralgds in this same carcinogenesis model also significantly reduced tumor incidence, size, and progression [126].
7. Conclusions and Future Prospects
Ral GTPase signaling has emerged as being critically important in both normal and neoplastic cell physiology. Over the last two and a half decades we have learned a great deal about how Ral proteins regulate many biological processes. From these studies one of the most striking observations has been the very distinct functions observed for RalA and RalB despite similar structural and biochemical properties and shared effector utilization. Do these distinct functions simply reflect to spatially distinct interactions with the same set of effectors or are their Ral isoform selective effectors that remain to be discovered. Only recently have RalGAPs been discovered and much more remains to be learned regarding their roles in regulation of Ral activity and signaling. With increasing evidence for key roles for Ral GTPases as drivers in cancer growth, it will be important to identify pharmacologic approaches for targeting aberrant Ral function for cancer treatment. Like Ras, Ral proteins are not tractable therapeutic targets, although recent progress in the identification of direct Ras binders suggests that small GTPases may yet be targeted directly. If not, indirect approaches need to be explored. With kinases implicated as downstream effectors or key regulators of Ral GTPases, can these be exploited for anti-Ral drug development? In summary, with more still to be learned regarding Ral function, it is quite certain that there will be more than 15 minutes remaining in their fame.
Highlights.
Ral proteins are members of the Ras superfamily of small GTPases
Ral functions as GTP-GDP regulated binary on-off switch in signaling
Ral proteins are key downstream effectors of Ras oncoprotein-mediated oncogenesis
Ral regulates vesicular transport and actin organization
Acknowledgements
Our Ral research studies have been supported by grants from the National Institutes of Health. Leanna R. Gentry and Timothy D. Martin were supported by a T32 training grant.
Abbreviations
- CAAX
cysteine-aliphatic-aliphatic-terminal amino acid
- DMBA
7,12-dimethylbenz[α]anthracene
- EGF
epidermal growth factor
- EH
Eps homology
- ER
endoplasmic reticulum
- GAP
GTPase activating protein
- GEF
guanine nucleotide exchange factor
- ICMT
isoprenylcysteine carboxyl methyltransferase
- NSCLC
non-small cell lung cancer
- PDAC
pancreatic ductal adenocarcinoma
- PH
pleckstrin homology domain
- PKA
protein kinase A
- PKC
protein kinase C
- RA
Ras-association domain
- RBD
Ral-binding domain
- RCE1
Ras converting enzyme 1
- REM
Ras exchanger motif
- SI/II
SwitchI/II
- SCC
squamous cell carcinoma
- SH3
Src homology 3 domain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- [2].Chardin P, Tavitian A. The ral gene: a new ras related gene isolated by the use of a synthetic probe. EMBO J. 1986;5:2203–2208. doi: 10.1002/j.1460-2075.1986.tb04485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chardin P, Tavitian A. Coding sequences of human ralA and ralB cDNAs. Nucleic Acids Res. 1989;17:4380. doi: 10.1093/nar/17.11.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Frische EW, Pellis-van Berkel W, van Haaften G, Cuppen E, Plasterk RH, Tijsterman M, Bos JL, Zwartkruis FJ. RAP-1 and the RAL-1/exocyst pathway coordinate hypodermal cell organization in Caenorhabditis elegans. EMBO J. 2007;26:5083–5092. doi: 10.1038/sj.emboj.7601922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ghiglione C, Devergne O, Cerezo D, Noselli S. Drosophila RalA is essential for the maintenance of Jak/Stat signalling in ovarian follicles. EMBO Rep. 2008;9:676–682. doi: 10.1038/embor.2008.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cox AD, Der CJ. Ras history: The saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Feig LA. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–425. doi: 10.1016/s0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- [8].Nicely NI, Kosak J, de Serrano V, Mattos C. Crystal structures of Ral-GppNHp and Ral-GDP reveal two binding sites that are also present in Ras and Rap. Structure. 2004;12:2025–2036. doi: 10.1016/j.str.2004.08.011. [DOI] [PubMed] [Google Scholar]
- [9].Albright CF, Giddings BW, Liu J, Vito M, Weinberg RA. Characterization of a guanine nucleotide dissociation stimulator for a ras-related GTPase. EMBO J. 1993;12:339–347. doi: 10.1002/j.1460-2075.1993.tb05662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hofer F, Fields S, Schneider C, Martin GS. Activated Ras interacts with the Ral guanine nucleotide dissociation stimulator. Proc Natl Acad Sci U S A. 1994;91:11089–11093. doi: 10.1073/pnas.91.23.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Spaargaren M, Bischoff JR. Identification of the guanine nucleotide dissociation stimulator for Ral as a putative effector molecule of R-ras, H-ras, K-ras, and Rap. Proc Natl Acad Sci U S A. 1994;91:12609–12613. doi: 10.1073/pnas.91.26.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kikuchi A, Demo SD, Ye ZH, Chen YW, Williams LT. ralGDS family members interact with the effector loop of ras p21. Mol Cell Biol. 1994;14:7483–7491. doi: 10.1128/mcb.14.11.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Isomura M, Okui K, Fujiwara T, Shin S, Nakamura Y. Isolation and mapping of RAB2L, a human cDNA that encodes a protein homologous to RalGDS. Cytogenet Cell Genet. 1996;74:263–265. doi: 10.1159/000134431. [DOI] [PubMed] [Google Scholar]
- [14].Shao H, Andres DA. A novel RalGEF-like protein. RGL3, as a candidate effector for rit and Ras, J Biol Chem. 2000;275:26914–26924. doi: 10.1074/jbc.M002241200. [DOI] [PubMed] [Google Scholar]
- [15].Wolthuis RM, Bauer B, van 't Veer LJ, de Vries-Smits AM, Cool RH, Spaargaren M, Wittinghofer A, Burgering BM, Bos JL. RalGDS-like factor (Rlf) is a novel Ras and Rap 1A-associating protein. Oncogene. 1996;13:353–362. [PubMed] [Google Scholar]
- [16].Ferro E, Trabalzini L. RalGDS family members couple Ras to Ral signalling and that's not all. Cell Signal. 2010;22:1804–1810. doi: 10.1016/j.cellsig.2010.05.010. [DOI] [PubMed] [Google Scholar]
- [17].van Dam TJ, Bos JL, Snel B. Evolution of the Ras-like small GTPases and their regulators. Small GTPases. 2011;2:4–16. doi: 10.4161/sgtp.2.1.15113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rodriguez-Viciana P, Sabatier C, McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol Cell Biol. 2004;24:4943–4954. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rebhun JF, Chen H, Quilliam LA. Identification and characterization of a new family of guanine nucleotide exchange factors for the ras-related GTPase Ral. J Biol Chem. 2000;275:13406–13410. doi: 10.1074/jbc.c000085200. [DOI] [PubMed] [Google Scholar]
- [20].de Bruyn KM, de Rooij J, Wolthuis RM, Rehmann H, Wesenbeek J, Cool RH, Wittinghofer AH, Bos JL. RalGEF2, a pleckstrin homology domain containing guanine nucleotide exchange factor for Ral. J Biol Chem. 2000;275:29761–29766. doi: 10.1074/jbc.M001160200. [DOI] [PubMed] [Google Scholar]
- [21].Ceriani M, Scandiuzzi C, Amigoni L, Tisi R, Berruti G, Martegani E. Functional analysis of RalGPS2, a murine guanine nucleotide exchange factor for RalA GTPase. Exp Cell Res. 2007;313:2293–2307. doi: 10.1016/j.yexcr.2007.03.016. [DOI] [PubMed] [Google Scholar]
- [22].Cascone I, Selimoglu R, Ozdemir C, Del Nery E, Yeaman C, White M, Camonis J. Distinct roles of RalA and RalB in the progression of cytokinesis are supported by distinct RalGEFs. EMBO J. 2008;27:2375–2387. doi: 10.1038/emboj.2008.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].D'Adamo DR, Novick S, Kahn JM, Leonardi P, Pellicer A. rsc: a novel oncogene with structural and functional homology with the gene family of exchange factors for Ral. Oncogene. 1997;14:1295–1305. doi: 10.1038/sj.onc.1200950. [DOI] [PubMed] [Google Scholar]
- [24].Osei-Sarfo K, Martello L, Ibrahim S, Pellicer A. The human Rgr oncogene is overexpressed in T-cell malignancies and induces transformation by acting as a GEF for Ras and Ral. Oncogene. 2011;30:3661–3671. doi: 10.1038/onc.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Emkey R, Freedman S, Feig LA. Characterization of a GTPase-activating protein for the Ras-related Ral protein. J Biol Chem. 1991;266:9703–9706. [PubMed] [Google Scholar]
- [26].Shirakawa R, Fukai S, Kawato M, Higashi T, Kondo H, Ikeda T, Nakayama E, Okawa K, Nureki O, Kimura T, Kita T, Horiuchi H. Tuberous sclerosis tumor suppressor complex-like complexes act as GTPase-activating proteins for Ral GTPases. J Biol Chem. 2009;284:21580–21588. doi: 10.1074/jbc.M109.012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen XW, Leto D, Xiong T, Yu G, Cheng A, Decker S, Saltiel AR. A Ral GAP complex links PI 3-kinase/Akt signaling to RalA activation in insulin action. Mol Biol Cell. 2011;22:141–152. doi: 10.1091/mbc.E10-08-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Schwarzbraun T, Vincent JB, Schumacher A, Geschwind DH, Oliveira J, Windpassinger C, Ofner L, Ledinegg MK, Kroisel PM, Wagner K, Petek E. Cloning, genomic structure, and expression profiles of TULIP1 (GARNL1), a brain-expressed candidate gene for 14q13-linked neurological phenotypes, and its murine homologue. Genomics. 2004;84:577–586. doi: 10.1016/j.ygeno.2004.04.013. [DOI] [PubMed] [Google Scholar]
- [29].Gridley S, Chavez JA, Lane WS, Lienhard GE. Adipocytes contain a novel complex similar to the tuberous sclerosis complex. Cell Signal. 2006;18:1626–1632. doi: 10.1016/j.cellsig.2006.01.002. [DOI] [PubMed] [Google Scholar]
- [30].Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Martin TD, Chen XW, Kaplan RE, Saltiel AR, Walker CL, Reiner DJ, Der CJ. Ral and Rheb GTPase activating proteins integrate mTOR and GTPase signaling in aging, autophagy, and tumor cell invasion. Mol Cell. 2014;53:209–220. doi: 10.1016/j.molcel.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Leto D, Uhm M, Williams A, Chen XW, Saltiel AR. Negative regulation of the RalGAP complex by 14-3-3. J Biol Chem. 2013;288:9272–9283. doi: 10.1074/jbc.M112.426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].He B, Guo W. The exocyst complex in polarized exocytosis. Curr Opin Cell Biol. 2009;21:537–542. doi: 10.1016/j.ceb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, Maiti M, Hazelett CC, Wauson EM, Balakireva M, Camonis JH, Yeaman C, Levine B, White MA. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell. 2011;144:253–267. doi: 10.1016/j.cell.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hazelett CC, Sheff D, Yeaman C. RalA and RalB differentially regulate development of epithelial tight junctions. Mol Biol Cell. 2011;22:4787–4800. doi: 10.1091/mbc.E11-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Fukai S, Matern HT, Jagath JR, Scheller RH, Brunger AT. Structural basis of the interaction between RalA and Sec5, a subunit of the sec6/8 complex. EMBO J. 2003;22:3267–3278. doi: 10.1093/emboj/cdg329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jin R, Junutula JR, Matern HT, Ervin KE, Scheller RH, Brunger AT. Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase. EMBO J. 2005;24:2064–2074. doi: 10.1038/sj.emboj.7600699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fenwick RB, Campbell LJ, Rajasekar K, Prasannan S, Nietlispach D, Camonis J, Owen D, Mott HR. The RalB-RLIP76 complex reveals a novel mode of ral-effector interaction. Structure. 2010;18:985–995. doi: 10.1016/j.str.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mott HR, Owen D. RLIP76 (RalBP1): The first piece of the structural puzzle. Small GTPases. 2010;1:157–160. doi: 10.4161/sgtp.1.3.14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cantor SB, Urano T, Feig LA. Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol Cell Biol. 1995;15:4578–4584. doi: 10.1128/mcb.15.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jullien-Flores V, Dorseuil O, Romero F, Letourneur F, Saragosti S, Berger R, Tavitian A, Gacon G, Camonis JH. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J Biol Chem. 1995;270:22473–22477. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- [42].Park SH, Weinberg RA. A putative effector of Ral has homology to Rho/Rac GTPase activating proteins. Oncogene. 1995;11:2349–2355. [PubMed] [Google Scholar]
- [43].Awasthi S, Cheng J, Singhal SS, Saini MK, Pandya U, Pikula S, Bandorowicz-Pikula J, Singh SV, Zimniak P, Awasthi YC. Novel function of human RLIP76: ATP-dependent transport of glutathione conjugates and doxorubicin. Biochemistry. 2000;39:9327–9334. doi: 10.1021/bi992964c. [DOI] [PubMed] [Google Scholar]
- [44].Awasthi S, Cheng JZ, Singhal SS, Pandya U, Sharma R, Singh SV, Zimniak P, Awasthi YC. Functional reassembly of ATP-dependent xenobiotic transport by the N- and C-terminal domains of RLIP76 and identification of ATP binding sequences. Biochemistry. 2001;40:4159–4168. doi: 10.1021/bi002182f. [DOI] [PubMed] [Google Scholar]
- [45].Vatsyayan R, Lelsani PC, Awasthi S, Singhal SS. RLIP76: a versatile transporter and an emerging target for cancer therapy. Biochem Pharmacol. 2010;79:1699–1705. doi: 10.1016/j.bcp.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Singhal SS, Singhal J, Yadav S, Dwivedi S, Boor PJ, Awasthi YC, Awasthi S. Regression of lung and colon cancer xenografts by depleting or inhibiting RLIP76 (Ral-binding protein 1) Cancer Res. 2007;67:4382–4389. doi: 10.1158/0008-5472.CAN-06-4124. [DOI] [PubMed] [Google Scholar]
- [47].Wu Z, Owens C, Chandra N, Popovic K, Conaway M, Theodorescu D. RalBP1 is necessary for metastasis of human cancer cell lines. Neoplasia. 2010;12:1003–1012. doi: 10.1593/neo.101080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jullien-Flores V, Mahe Y, Mirey G, Leprince C, Meunier-Bisceuil B, Sorkin A, Camonis JH. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: involvement of the Ral pathway in receptor endocytosis. J Cell Sci. 2000;113:2837–2844. doi: 10.1242/jcs.113.16.2837. Pt 16. [DOI] [PubMed] [Google Scholar]
- [49].Yamaguchi A, Urano T, Goi T, Feig LA. An Eps homology (EH) domain protein that binds to the Ral-GTPase target. RalBP1, J Biol Chem. 1997;272:31230–31234. doi: 10.1074/jbc.272.50.31230. [DOI] [PubMed] [Google Scholar]
- [50].Ikeda M, Ishida O, Hinoi T, Kishida S, Kikuchi A. Identification and characterization of a novel protein interacting with Ral-binding protein 1, a putative effector protein of Ral. J Biol Chem. 1998;273:814–821. doi: 10.1074/jbc.273.2.814. [DOI] [PubMed] [Google Scholar]
- [51].Cullis DN, Philip B, Baleja JD, Feig LA. Rab11-FIP2, an adaptor protein connecting cellular components involved in internalization and recycling of epidermal growth factor receptors. J Biol Chem. 2002;277:49158–49166. doi: 10.1074/jbc.M206316200. [DOI] [PubMed] [Google Scholar]
- [52].Morinaka K, Koyama S, Nakashima S, Hinoi T, Okawa K, Iwamatsu A, Kikuchi A. Epsin binds to the EH domain of POB1 and regulates receptor-mediated endocytosis. Oncogene. 1999;18:5915–5922. doi: 10.1038/sj.onc.1202974. [DOI] [PubMed] [Google Scholar]
- [53].Rosse C, L'Hoste S, Offner N, Picard A, Camonis J. RLIP, an effector of the Ral GTPases, is a platform for Cdk1 to phosphorylate epsin during the switch off of endocytosis in mitosis. J Biol Chem. 2003;278:30597–30604. doi: 10.1074/jbc.M302191200. [DOI] [PubMed] [Google Scholar]
- [54].Moskalenko S, Henry DO, Rosse C, Mirey G, Camonis JH, White MA. The exocyst is a Ral effector complex. Nat Cell Biol. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- [55].Moskalenko S, Tong C, Rosse C, Mirey G, Formstecher E, Daviet L, Camonis J, White MA. Ral GTPases regulate exocyst assembly through dual subunit interactions. J Biol Chem. 2003;278:51743–51748. doi: 10.1074/jbc.M308702200. [DOI] [PubMed] [Google Scholar]
- [56].Neel NF, Rossman KL, Martin TD, Hayes TK, Yeh JJ, Der CJ. The RalB small GTPase mediates formation of invadopodia through a GTPase-activating protein-independent function of the RalBP1/RLIP76 effector. Mol Cell Biol. 2012;32:1374–1386. doi: 10.1128/MCB.06291-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kashatus DF, Lim KH, Brady DC, Pershing NL, Cox AD, Counter CM. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat Cell Biol. 2011;13:1108–1115. doi: 10.1038/ncb2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Tazat K, Harsat M, Goldshmid-Shagal A, Ehrlich M, Henis YI. Dual effects of Ral-activated pathways on p27 localization and TGF-beta signaling. Mol Biol Cell. 2013;24:1812–1824. doi: 10.1091/mbc.E13-01-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sugihara K, Asano S, Tanaka K, Iwamatsu A, Okawa K, Ohta Y. The exocyst complex binds the small GTPase RalA to mediate filopodia formation. Nat Cell Biol. 2002;4:73–78. doi: 10.1038/ncb720. [DOI] [PubMed] [Google Scholar]
- [60].Spiczka KS, Yeaman C. Ral-regulated interaction between Sec5 and paxillin targets Exocyst to focal complexes during cell migration. J Cell Sci. 2008;121:2880–2891. doi: 10.1242/jcs.031641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, Balakireva MG, Romeo Y, Kopelovich L, Gale M, Jr., Yeaman C, Camonis JH, Zhao Y, White MA. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- [62].Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, Frohling S, Chan EM, Sos ML, Michel K, Mermel C, Silver SJ, Weir BA, Reiling JH, Sheng Q, Gupta PB, Wadlow RC, Le H, Hoersch S, Wittner BS, Ramaswamy S, Livingston DM, Sabatini DM, Meyerson M, Thomas RK, Lander ES, Mesirov JP, Root DE, Gilliland DG, Jacks T, Hahn WC. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Muvaffak A, Pan Q, Yan H, Fernandez R, Lim J, Dolinski B, Nguyen TT, Strack P, Wu S, Chung R, Zhang W, Hulton C, Ripley S, Hirsch H, Nagashima K, Wong KK, Janne PA, Seidel-Dugan C, Zawel L, Kirschmeier PT, Middleton RE, Morris EJ, Wang Y. Evaluating TBK1 as a Therapeutic Target in Cancers with Activated IRF3. Mol Cancer Res. 2014 doi: 10.1158/1541-7786.MCR-13-0642. [DOI] [PubMed] [Google Scholar]
- [64].Seguin L, Kato S, Franovic A, Camargo MF, Lesperance J, Elliott KC, Yebra M, Mielgo A, Lowy AM, Husain H, Cascone T, Diao L, Wang J, Wistuba, Heymach JV, Lippman SM, Desgrosellier JS, Anand S, Weis SM, Cheresh DA. An integrin beta3-KRAS-RalB complex drives tumour stemness and resistance to EGFR inhibition. Nat Cell Biol. 2014;16:457–468. doi: 10.1038/ncb2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Luo JQ, Liu X, Frankel P, Rotunda T, Ramos M, Flom J, Jiang H, Feig LA, Morris AJ, Kahn RA, Foster DA. Functional association between Arf and RalA in active phospholipase D complex. Proc Natl Acad Sci U S A. 1998;95:3632–3637. doi: 10.1073/pnas.95.7.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kim JH, Lee SD, Han JM, Lee TG, Kim Y, Park JB, Lambeth JD, Suh PG, Ryu SH. Activation of phospholipase D1 by direct interaction with ADP-ribosylation factor 1 and RalA. FEBS Lett. 1998;430:231–235. doi: 10.1016/s0014-5793(98)00661-9. [DOI] [PubMed] [Google Scholar]
- [67].Xu L, Salloum D, Medlin PS, Saqcena M, Yellen P, Perrella B, Foster DA. Phospholipase D mediates nutrient input to mammalian target of rapamycin complex 1 (mTORC1) J Biol Chem. 2011;286:25477–25486. doi: 10.1074/jbc.M111.249631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP. The small GTPase RalA targets filamin to induce filopodia. Proc Natl Acad Sci U S A. 1999;96:2122–2128. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Frankel P, Aronheim A, Kavanagh E, Balda MS, Matter K, Bunney TD, Marshall CJ. RalA interacts with ZONAB in a cell density-dependent manner and regulates its transcriptional activity. EMBO J. 2005;24:54–62. doi: 10.1038/sj.emboj.7600497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Shipitsin M, Feig LA. RalA but not RalB enhances polarized delivery of membrane proteins to the basolateral surface of epithelial cells. Mol Cell Biol. 2004;24:5746–5756. doi: 10.1128/MCB.24.13.5746-5756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lim KH, Brady DC, Kashatus DF, Ancrile BB, Der CJ, Cox AD, Counter CM. Aurora-A phosphorylates, activates, and relocalizes the small GTPase RalA. Mol Cell Biol. 2010;30:508–523. doi: 10.1128/MCB.00916-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Martin TD, Mitin N, Cox AD, Yeh JJ, Der CJ. Phosphorylation by protein kinase Calpha regulates RalB small GTPase protein activation, subcellular localization, and effector utilization. J Biol Chem. 2012;287:14827–14836. doi: 10.1074/jbc.M112.344986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Peschard P, McCarthy A, Leblanc-Dominguez V, Yeo M, Guichard S, Stamp G, Marshall CJ. Genetic deletion of RALA and RALB small GTPases reveals redundant functions in development and tumorigenesis. Curr Biol. 2012;22:2063–2068. doi: 10.1016/j.cub.2012.09.013. [DOI] [PubMed] [Google Scholar]
- [74].Reid TS, Terry KL, Casey PJ, Beese LS. Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity. J Mol Biol. 2004;343:417–433. doi: 10.1016/j.jmb.2004.08.056. [DOI] [PubMed] [Google Scholar]
- [75].Falsetti SC, Wang DA, Peng H, Carrico D, Cox AD, Der CJ, Hamilton AD, Sebti SM. Geranylgeranyltransferase I inhibitors target RalB to inhibit anchorage-dependent growth and induce apoptosis and RalA to inhibit anchorage-independent growth. Mol Cell Biol. 2007;27:8003–8014. doi: 10.1128/MCB.00057-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Nishimura A, Linder ME. Identification of a novel prenyl and palmitoyl modification at the CaaX motif of Cdc42 that regulates RhoGDI binding. Mol Cell Biol. 2013;33:1417–1429. doi: 10.1128/MCB.01398-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol. 2012;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, Miura J, Wiener HH, Wright L, Saba SG, Yim D, Fein A, Perez de Castro I, Li C, Thompson CB, Cox AD, Philips MR. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell. 2006;21:481–493. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- [79].Sung PJ, Tsai FD, Vais H, Court H, Yang J, Fehrenbacher N, Foskett JK, Philips MR. Phosphorylated K-Ras limits cell survival by blocking Bcl-xL sensitization of inositol trisphosphate receptors. Proc Natl Acad Sci U S A. 2013;110:20593–20598. doi: 10.1073/pnas.1306431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Wu JC, Chen TY, Yu CT, Tsai SJ, Hsu JM, Tang MJ, Chou CK, Lin WJ, Yuan CJ, Huang CY. Identification of V23RalA-Ser194 as a critical mediator for Aurora-A-induced cellular motility and transformation by small pool expression screening. J Biol Chem. 2005;280:9013–9022. doi: 10.1074/jbc.M411068200. [DOI] [PubMed] [Google Scholar]
- [81].Wang H, Owens C, Chandra N, Conaway MR, Brautigan DL, Theodorescu D. Phosphorylation of RalB is important for bladder cancer cell growth and metastasis. Cancer Res. 2010;70:8760–8769. doi: 10.1158/0008-5472.CAN-10-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Sablina AA, Chen W, Arroyo JD, Corral L, Hector M, Bulmer SE, DeCaprio JA, Hahn WC. The tumor suppressor PP2A Abeta regulates the RalA GTPase. Cell. 2007;129:969–982. doi: 10.1016/j.cell.2007.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Baker R, Lewis SM, Sasaki AT, Wilkerson EM, Locasale JW, Cantley LC, Kuhlman B, Dohlman HG, Campbell SL. Site-specific monoubiquitination activates Ras by impeding GTPase-activating protein function. Nat Struct Mol Biol. 2013;20:46–52. doi: 10.1038/nsmb.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Neyraud V, Aushev VN, Hatzoglou A, Meunier B, Cascone I, Camonis J. RalA and RalB proteins are ubiquitinated GTPases, and ubiquitinated RalA increases lipid raft exposure at the plasma membrane. J Biol Chem. 2012;287:29397–29405. doi: 10.1074/jbc.M112.357764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Simicek M, Lievens S, Laga M, Guzenko D, Aushev VN, Kalev P, Baietti MF, Strelkov SV, Gevaert K, Tavernier J, Sablina AA. The deubiquitylase USP33 discriminates between RALB functions in autophagy and innate immune response. Nat Cell Biol. 2013;15:1220–1230. doi: 10.1038/ncb2847. [DOI] [PubMed] [Google Scholar]
- [86].Gu X, Wang Y, Gu J. Age distribution of human gene families shows significant roles of both large- and small-scale duplications in vertebrate evolution. Nat Genet. 2002;31:205–209. doi: 10.1038/ng902. [DOI] [PubMed] [Google Scholar]
- [87].Shaye DD, Greenwald I. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS One. 2011;6:e20085. doi: 10.1371/journal.pone.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Zand TP, Reiner DJ, Der CJ. Ras effector switching promotes divergent cell fates in C. elegans vulval patterning. Dev Cell. 2011;20:84–96. doi: 10.1016/j.devcel.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Reiner DJ. Ras effector switching as a developmental strategy. Small GTPases. 2011;2:109–112. doi: 10.4161/sgtp.2.2.15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].van den Berg MC, van Gogh IJ, Smits AM, van Triest M, Dansen TB, Visscher M, Polderman PE, Vliem MJ, Rehmann H, Burgering BM. The small GTPase RALA controls c-Jun N-terminal kinase-mediated FOXO activation by regulation of a JIP1 scaffold complex. J Biol Chem. 2013;288:21729–21741. doi: 10.1074/jbc.M113.463885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Putker M, Madl T, Vos HR, de Ruiter H, Visscher M, van den Berg MC, Kaplan M, Korswagen HC, Boelens R, Vermeulen M, Burgering BM, Dansen TB. Redox-dependent control of FOXO/DAF-16 by transportin-1. Mol Cell. 2013;49:730–742. doi: 10.1016/j.molcel.2012.12.014. [DOI] [PubMed] [Google Scholar]
- [92].Ewbank JJ. Signaling in the immune response. WormBook. 2006:1–12. doi: 10.1895/wormbook.1.83.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Mirey G, Balakireva M, L'Hoste S, Rosse C, Voegeling S, Camonis J. A Ral guanine exchange factor-Ral pathway is conserved in Drosophila melanogaster and sheds new light on the connectivity of the Ral, Ras, and Rap pathways. Mol Cell Biol. 2003;23:1112–1124. doi: 10.1128/MCB.23.3.1112-1124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Raaijmakers JH, Bos JL. Specificity in Ras and Rap signaling. J Biol Chem. 2009;284:10995–10999. doi: 10.1074/jbc.R800061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Gloerich M, Bos JL. Regulating Rap small G-proteins in time and space. Trends Cell Biol. 2011;21:615–623. doi: 10.1016/j.tcb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- [96].Tian X, Rusanescu G, Hou W, Schaffhausen B, Feig LA. PDK1 mediates growth factor-induced Ral-GEF activation by a kinase-independent mechanism. EMBO J. 2002;21:1327–1338. doi: 10.1093/emboj/21.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Hao Y, Wong R, Feig LA. RalGDS couples growth factor signaling to Akt activation. Mol Cell Biol. 2008;28:2851–2859. doi: 10.1128/MCB.01917-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding Ras: 'it ain't over 'til it's over'. Trends Cell Biol. 2000;10:147–154. doi: 10.1016/s0962-8924(00)01740-2. [DOI] [PubMed] [Google Scholar]
- [99].Balakireva M, Rosse C, Langevin J, Chien YC, Gho M, Gonzy-Treboul G, Voegeling-Lemaire S, Aresta S, Lepesant JA, Bellaiche Y, White M, Camonis J. The Ral/exocyst effector complex counters c-Jun N-terminal kinase-dependent apoptosis in Drosophila melanogaster. Mol Cell Biol. 2006;26:8953–8963. doi: 10.1128/MCB.00506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- [101].Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- [102].Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Urano T, Emkey R, Feig LA. Ral-GTPases mediate a distinct downstream signaling pathway from Ras that facilitates cellular transformation. EMBO J. 1996;15:810–816. [PMC free article] [PubMed] [Google Scholar]
- [104].White MA, Vale T, Camonis JH, Schaefer E, Wigler MH. A role for the Ral guanine nucleotide dissociation stimulator in mediating Ras-induced transformation. J Biol Chem. 1996;271:16439–16442. doi: 10.1074/jbc.271.28.16439. [DOI] [PubMed] [Google Scholar]
- [105].Hamad NM, Elconin JH, Karnoub AE, Bai W, Rich JN, Abraham RT, Der CJ, Counter CM. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 2002;16:2045–2057. doi: 10.1101/gad.993902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Chien Y, White MA. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 2003;4:800–806. doi: 10.1038/sj.embor.embor899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Smith SC, Oxford G, Baras AS, Owens C, Havaleshko D, Brautigan DL, Safo MK, Theodorescu D. Expression of ral GTPases, their effectors, and activators in human bladder cancer. Clin Cancer Res. 2007;13:3803–3813. doi: 10.1158/1078-0432.CCR-06-2419. [DOI] [PubMed] [Google Scholar]
- [108].Saito R, Shirakawa R, Nishiyama H, Kobayashi T, Kawato M, Kanno T, Nishizawa K, Matsui Y, Ohbayashi T, Horiguchi M, Nakamura T, Ikeda T, Yamane K, Nakayama E, Nakamura E, Toda Y, Kimura T, Kita T, Ogawa O, Horiuchi H. Downregulation of Ral GTPase-activating protein promotes tumor invasion and metastasis of bladder cancer. Oncogene. 2013;32:894–902. doi: 10.1038/onc.2012.101. [DOI] [PubMed] [Google Scholar]
- [109].Oxford G, Owens CR, Titus BJ, Foreman TL, Herlevsen MC, Smith SC, Theodorescu D. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–7120. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- [110].Martin TD, Samuel JC, Routh ED, Der CJ, Yeh JJ. Activation and involvement of Ral GTPases in colorectal cancer. Cancer Res. 2011;71:206–215. doi: 10.1158/0008-5472.CAN-10-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Ezzeldin M, Borrego-Diaz E, Taha M, Esfandyari T, Wise AL, Peng W, Rouyanian A, Kermani A. Asvadi, Soleimani M, Patrad E, Lialyte K, Wang K, Williamson S, Abdulkarim B, Olyaee M, Farassati F. RalA signaling pathway as a therapeutic target in hepatocellular carcinoma (HCC) Mol Oncol. 2014 doi: 10.1016/j.molonc.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Male H, Patel V, Jacob MA, Borrego-Diaz E, Wang K, Young DA, Wise AL, Huang C, Van Veldhuizen P, O'Brien-Ladner A, Williamson SK, Taylor SA, Tawfik O, Esfandyari T, Farassati F. Inhibition of RalA signaling pathway in treatment of non-small cell lung cancer. Lung Cancer. 2012;77:252–259. doi: 10.1016/j.lungcan.2012.03.007. [DOI] [PubMed] [Google Scholar]
- [113].Guin S, Ru Y, Wynes MW, Mishra R, Lu X, Owens C, Barn AE, Vasu VT, Hirsch FR, Kern JA, Theodorescu D. Contributions of KRAS and RAL in non-small-cell lung cancer growth and progression. J Thorac Oncol. 2013;8:1492–1501. doi: 10.1097/JTO.0000000000000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, Tsao A, Liu S, Larsen JE, Wang J, Diao L, Coombes KR, Chen L, Zhang S, Abdelmelek MF, Tang X, Papadimitrakopoulou V, Minna JD, Lippman SM, Hong WK, Herbst RS, Wistuba, Heymach JV, Powis G. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104:228–239. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Basu TN, Gutmann DH, Fletcher JA, Glover TW, Collins FS, Downward J. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature. 1992;356:713–715. doi: 10.1038/356713a0. [DOI] [PubMed] [Google Scholar]
- [116].Bodempudi V, Yamoutpoor F, Pan W, Dudek AZ, Esfandyari T, Piedra M, Babovick-Vuksanovic D, Woo RA, Mautner VF, Kluwe L, Clapp DW, De Vries GH, Thomas SL, Kurtz A, Parada LF, Farassati F. Ral overactivation in malignant peripheral nerve sheath tumors. Mol Cell Biol. 2009;29:3964–3974. doi: 10.1128/MCB.01153-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Zipfel PA, Brady DC, Kashatus DF, Ancrile BD, Tyler DS, Counter CM. Ral activation promotes melanomagenesis. Oncogene. 2010;29:4859–4864. doi: 10.1038/onc.2010.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Mishra PJ, Ha L, Rieker J, Sviderskaya EV, Bennett DC, Oberst MD, Kelly K, Merlino G. Dissection of RAS downstream pathways in melanomagenesis: a role for Ral in transformation. Oncogene. 2010;29:2449–2456. doi: 10.1038/onc.2009.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Wang K, Terai K, Peng W, Rouyanian A, Liu J, Roby KF, Wise AL, Ezzeldin M, Larson J, Woo RA, Lialyte K, Farassati F. The role of RalA in biology and therapy of ovarian cancer. Oncotarget. 2013 [Google Scholar]
- [120].Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Lim KH, Baines AT, Fiordalisi JJ, Shipitsin M, Feig LA, Cox AD, Der CJ, Counter CM. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7:533–545. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- [122].Lim KH, O'Hayer K, Adam SJ, Kendall SD, Campbell PM, Der CJ, Counter CM. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol. 2006;16:2385–2394. doi: 10.1016/j.cub.2006.10.023. [DOI] [PubMed] [Google Scholar]
- [123].Vigil D, Martin TD, Williams F, Yeh JJ, Campbell SL, Der CJ. Aberrant overexpression of the Rgl2 Ral small GTPase-specific guanine nucleotide exchange factor promotes pancreatic cancer growth through Ral-dependent and Ral-independent mechanisms. J Biol Chem. 2010;285:34729–34740. doi: 10.1074/jbc.M110.116756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Yin J, Pollock C, Tracy K, Chock M, Martin P, Oberst M, Kelly K. Activation of the RalGEF/Ral pathway promotes prostate cancer metastasis to bone. Mol Cell Biol. 2007;27:7538–7550. doi: 10.1128/MCB.00955-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Sowalsky AG, Alt-Holland A, Shamis Y, Garlick JA, Feig LA. RalA suppresses early stages of Ras-induced squamous cell carcinoma progression. Oncogene. 2010;29:45–55. doi: 10.1038/onc.2009.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Gonzalez-Garcia A, Pritchard CA, Paterson HF, Mavria G, Stamp G, Marshall CJ. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer Cell. 2005;7:219–226. doi: 10.1016/j.ccr.2005.01.029. [DOI] [PubMed] [Google Scholar]