Abstract
Messenger RNAs lacking a stop codon trap ribosomes at their 3′ ends, depleting the pool of ribosomes available for protein synthesis. In bacteria, a remarkable quality control system rescues and recycles stalled ribosomes in a process known as trans-translation. Acting as a tRNA, transfer-messenger RNA (tmRNA) is aminoacylated, delivered by EF-Tu to the ribosomal A site, and accepts the nascent polypeptide. Translation then resumes on a reading frame within tmRNA, encoding a short peptide tag that targets the nascent peptide for degradation by proteases. One unsolved issue in trans-translation is how tmRNA and its protein partner SmpB preferentially recognize stalled ribosomes and not actively translating ones. Here, we examine the effect of the length of the 3′ extension of mRNA on each step of trans-translation by pre-steady-state kinetic methods and fluorescence polarization binding assays. Unexpectedly, EF-Tu activation and GTP hydrolysis occur rapidly regardless of the length of the mRNA, although the peptidyl transfer to tmRNA decreases as the mRNA 3′ extension increases and the tmRNA·SmpB binds less tightly to the ribosome with an mRNA having a long 3′ extension. From these results, we conclude that the tmRNA·SmpB complex dissociates during accommodation due to competition between the downstream mRNA and the C-terminal tail for the mRNA channel. Rejection of the tmRNA·SmpB complex during accommodation is reminiscent of the rejection of near-cognate tRNA from the ribosome in canonical translation.
Keywords: trans-translation, SmpB, tmRNA, ribosome, translation
INTRODUCTION
Messenger RNA transcripts lacking stop codons pose a threat to the viability of all living organisms. Nonstop mRNAs not only encode aberrant proteins with potentially toxic activities but also deplete the pool of available ribosomes, because ribosomes stall for extended periods of time at the 3′ end of an mRNA when they cannot recruit release factors. In response to this threat, several mechanisms have evolved to detect and destroy nonstop mRNAs and release and recycle stalled ribosomes. In the nonstop decay (NSD) pathway in yeast, for example, translation past the end of an open reading frame into the poly-A tail recruits Ski7, leading to degradation of the transcript by the exosome, release of the ribosome by Dom34/Hbs1, and targeting of the nascent polypeptide to the proteasome by the ubiquitin ligases Not4 and Ltn1 (Shoemaker and Green 2012). A similar mechanism appears to operate in many if not all eukaryotes.
In bacteria, ribosomes stall not on a poly-A tail but at the 3′ end of a nonstop mRNA. The most common and effective system for rescuing stalled ribosomes in bacteria consists of transfer-messenger RNA (tmRNA) and its protein partner, SmpB. Genetic deletion of tmRNA is lethal in some species (Huang et al. 2000; Ramadoss et al. 2013), inhibits pathogenesis in others (Thibonnier et al. 2008), and conveys susceptibility to antibiotics and stresses (Muto et al. 2000; Luidalepp et al. 2005; Thibonnier et al. 2008). In a subset of bacteria, there are also redundant mechanisms for releasing stalled ribosomes, including the alternative release factors ArfA, found in β- and γ-proteobacteria (Chadani et al. 2010), and ArfB (YaeJ), found in many Gram-negative bacteria (Handa et al. 2010; Chadani et al. 2011). The fact that tmRNA and SmpB constitute the predominant mechanism, however, is underscored by their presence in all sequenced bacterial genomes (Gueneau de Novoa and Williams 2004).
tmRNA and SmpB rescue stalled ribosomes through a remarkable template-swapping mechanism known as trans-translation (Keiler et al. 1996; Karzai et al. 1999). As its name implies, tmRNA has a dual function (tRNA function and mRNA function). First, it acts like a transfer-RNA, its terminal regions folding into a tRNA-like structure that is aminoacylated with alanine (Komine et al. 1994; Ushida et al. 1994). With SmpB bound to its tRNA-like domain (TLD), Ala-tmRNA is delivered to the A site of stalled ribosomes by EF-Tu (Barends et al. 2001; Hanawa-Suetsugu et al. 2002). The tmRNA·SmpB complex mimics the structure of a canonical tRNA, with the TLD of tmRNA forming the acceptor stem and SmpB forming the anticodon stem–loop, so that upon A-site-binding SmpB engages the decoding center in the 30S subunit (Gutmann et al. 2003; Bessho et al. 2007; Kurita et al. 2007). After the nascent polypeptide is transferred to tmRNA, the ribosome dissociates from the nonstop mRNA template and resumes translation on a short open reading frame in tmRNA. Acting now as a messenger RNA, tmRNA encodes 10 additional amino acids and the nascent polypeptide is released at a stop codon (Keiler et al. 1996; Himeno et al. 1997). The 11 amino acid tag added by tmRNA targets the truncated protein for degradation by cellular proteases (Keiler et al. 1996; Gottesman et al. 1998; Herman et al. 1998; Choy et al. 2007).
One aspect of the trans-translation mechanism that remains controversial is how the tmRNA·SmpB complex selectively reacts with stalled ribosomes and not actively translating ones in order to avoid aborting productive protein synthesis. Initially, tmRNA·SmpB was assumed to target ribosomes stalled at the 3′ end of truncated mRNA (Keiler et al. 1996). Indeed, in vitro kinetic studies confirmed that the efficiency of peptidyl transfer to Ala-tmRNA rapidly decreased as the length of the mRNA increased downstream from the P-site codon (Ivanova et al. 2004; Asano et al. 2005). In contrast, several in vivo studies seemed to suggest that trans-translation occurs even in the middle of mRNA at several contiguous rare codons, inefficient stop codons, and certain nascent peptide sequences, all of which pause or arrest translation (Roche and Sauer 1999, 2001; Collier et al. 2002; Hayes et al. 2002). One solution to this paradox is that, regardless of the mechanism, prolonged translational arrest in the middle of an mRNA induces endonucleolytic cleavage in or around the A-site codon, generating nonstop mRNA (Hayes and Sauer 2003; Sunohara et al. 2004; Li et al. 2007; Garza-Sánchez et al. 2008). On the other hand, some in vivo studies observe robust tmRNA activity on transcripts with 15–21 nt downstream from the P-site codon, following degradation of the mRNA by 3′–5′ exonucleases to the 3′ boundary protected by the ribosome (Garza-Sánchez et al. 2009; Janssen et al. 2013). The disparity between the in vitro and in vivo data and the diversity of sequences used to induce stalling have led to confusion over whether A-site cleavage is necessary for trans-translation or what the mRNA length requirements might be.
The recent crystal structure of the Thermus thermophilus tmRNA·SmpB complex bound in the A site of the 70S ribosome sheds new light on how the trans-translation machinery achieves selectivity for stalled ribosomes (Neubauer et al. 2012). Trapped by kirromycin, this complex represents the preaccommodation state following activation of EF-Tu and GTP hydrolysis but prior to the dissociation of EF-Tu. The body of SmpB is positioned in the decoding center in the 30S subunit, near the conserved nucleotides A1492, A1493, and G530 that are essential to canonical decoding. The structure confirms earlier biochemical findings that the C-terminal tail of SmpB, residues 133–160 in the Escherichia coli protein, lies within the A-site and the mRNA channel downstream from the A site (Kurita et al. 2007, 2010). The residues after 142, although unstructured in solution, form a helix within the mRNA channel, making interactions with 16S rRNA and the S5 protein. Taken together, the biochemical and structural evidence supports a model in which selectivity for stalled ribosomes is the result of competition for the A-site and mRNA channel, where mRNA blocks the binding of the SmpB C-terminal tail and inhibits tmRNA activity on actively translating ribosomes.
Given that the tail is bound in the channel prior to release of EF-Tu in the crystal structure, it seems reasonable to hypothesize that long mRNAs might block binding of tmRNA·SmpB in the A site and subsequent activation of EF-Tu. In this work, we test this hypothesis directly by examining the effect of the length of the 3′ extension of mRNA on tmRNA·SmpB binding, GTPase activation, and peptidyl transfer. We show that tmRNA·SmpB can enter ribosomes and trigger GTP hydrolysis even in the middle of mRNA and that rejection of tmRNA·SmpB from elongating ribosomes occurs during the accommodation step, following GTP hydrolysis by EF-Tu but prior to peptidyl transfer.
RESULTS AND DISCUSSION
Long 3′ extensions in mRNA block peptidyl transfer to tmRNA
Transfer of the nascent peptide from peptidyl-tRNA to Ala-tmRNA occurs more efficiently in vitro when the 3′ end of the mRNA template does not extend far beyond the P-site codon (Ivanova et al. 2004; Asano et al. 2005). It remains unclear, however, whether mRNAs with longer 3′ extensions specifically block peptidyl transfer or inhibit another step upstream of peptidyl transfer, including tmRNA·SmpB complex binding, activation of EF-Tu, and accommodation into the A site. We have developed in vitro trans-translation assays to measure the activities of initial steps of trans-translation up to and including peptidyl transfer (Konno et al. 2007; Takada et al. 2007; Kurita et al. 2010; Miller et al. 2011; Miller and Buskirk 2014). In this study, we used pre-steady-state kinetic methods to determine the rate constants for GTP hydrolysis by EF-Tu and for peptidyl transfer to tmRNA.
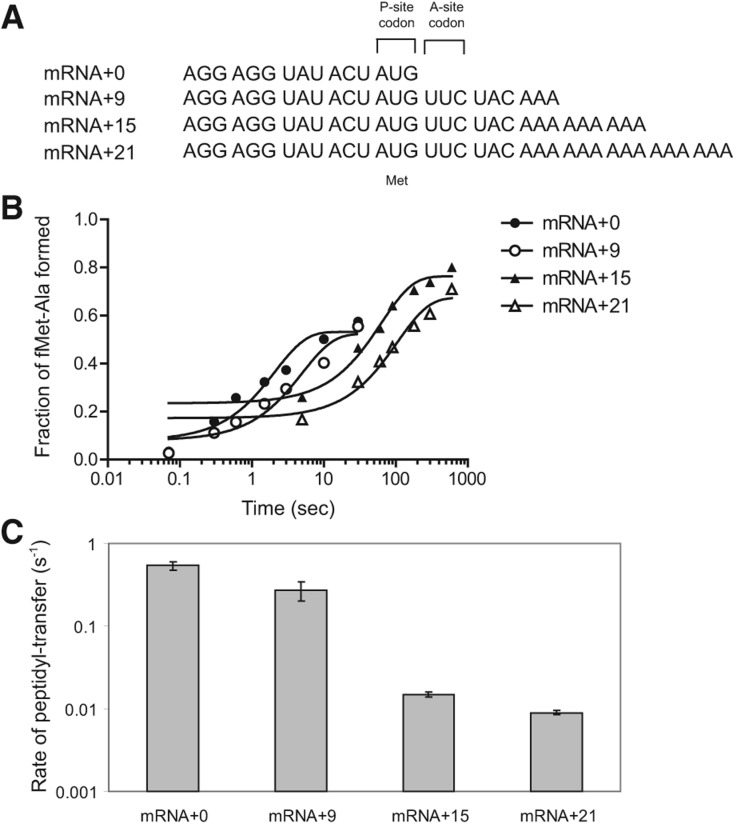
We replicated the findings of Ehrenberg and coworkers that rates of peptidyl transfer to Ala-tmRNA are substantially reduced by mRNA extending 12 or more nucleotides after the P-site codon (Ivanova et al. 2004). We formed translation initiation complexes by incubating formyl-[35S]methionyl-tRNAfMet (fMet-tRNAfMet) with E. coli ribosomes and an mRNA having a Shine–Dalgarno (SD) sequence, an AUG start codon, and varying lengths of the 3′ extension (Fig. 1A). The 3′ ends of the mRNA templates extended 0, 9, 15, or 21 nt from the P-site codon into the A-site and the mRNA-binding channel. Initiation complexes were reacted with a quaternary complex containing Ala-tmRNA, SmpB, EF-Tu, and GTP. The rate constant of trans-transfer was determined by measuring the amount of f[35S]Met-Ala dipeptide at various time points (Fig. 1B).
FIGURE 1.
Effect of the length of the 3′ extension of mRNA on trans-transfer. (A) mRNAs used in the in vitro trans-translation system and reaction scheme for trans-transfer. Each mRNA has an SD sequence, AUG codon and +0, +9, +15, or +21 nt after the AUG codon. (B) Time course of formation of the dipeptide fMet-Ala with an mRNA+0 (closed circle), an mRNA+9 (open circle), an mRNA+15 (closed triangle), or an mRNA+21 (open triangle). Stalled ribosome that contains 70S ribosome, mRNA, and f[35S]Met-tRNAfMet was mixed with the quaternary complex of Ala-tmRNA, SmpB, EF-Tu, and GTP. (C) The rate constants of peptidyl transfer determined by monitoring the rate of formation of the dipeptide fMet-Ala. All reactions were performed at least twice and the SE is given.
As expected from previous studies (Ivanova et al. 2004; Asano et al. 2005), the peptidyl-transfer rate decreased significantly when the mRNA length past the P-site codon reached a certain threshold. Although the rate decreased only twofold with a 9-nt extension, it decreased >30-fold with the mRNA having a 15-nt extension (Fig. 1C; Table 1). This is consistent with a model in which short mRNA extensions are free to move out of the way of the tmRNA·SmpB complex, but longer mRNA extensions are held in place by the downstream mRNA channel in the ribosome, blocking peptidyl transfer to the tmRNA·SmpB complex.
TABLE 1.
Rate constants of GTP hydrolysis and trans-transfer

mRNA in the A site does not block EF-Tu activation
Given the structural data showing that SmpB binds in the A-site and mRNA channel where mRNA is normally positioned, even while EF-Tu is still bound to the complex in the preaccommodation state, we expected that EF-Tu activation and GTP hydrolysis would also be inhibited on complexes with mRNAs with long extensions after the P-site codon. To measure GTP hydrolysis by EF-Tu, nonlabeled initiation complexes were combined with the quaternary complex of Ala-tmRNA, SmpB, EF-Tu, and [γ-32P]GTP. The levels of free radioactive inorganic phosphate were monitored at various time points (Fig. 2A) and the GTP hydrolysis rate constant was determined by fitting the data to a first-order exponential equation. We confirmed that GTP hydrolysis depends on the presence of SmpB as described in our previous report (Miller et al. 2011) and that no significant spontaneous GTP hydrolysis was observed in the absence of the initiation complex. Surprisingly, the rate constants for GTP hydrolysis were roughly the same regardless of the mRNA length (Fig. 2B; Table 1), indicating that GTP hydrolysis is not inhibited by the downstream mRNA. We conclude that delivery of the tmRNA·SmpB complex and activation of EF-Tu are not compromised by the presence of mRNA in the A-site or downstream mRNA channel. Although binding of tmRNA·SmpB to actively translating ribosomes induces GTP hydrolysis in an apparently wasteful manner, it is likely that this does not significantly impact cellular energy resources, because the concentration of tmRNA is low relative to canonical tRNAs (Lee et al. 1978; Dong et al. 1996) and tmRNA also has a lower affinity for EF-Tu (Barends et al. 2000).
FIGURE 2.
Effect of the length of the 3′ extension of mRNA on GTP hydrolysis. Stalled ribosome that contains 70S ribosome, mRNA, and fMet-tRNAfMet was mixed with the quaternary complex of Ala-tmRNA, SmpB, EF-Tu, and 32P-labeled GTP. (A) Time course of GTP hydrolysis with mRNA+0 (closed circle), mRNA+9 (open circle), mRNA+15 (closed triangle), or mRNA+21 (open triangle). (B) The rate constants of GTP hydrolysis determined by monitoring the rate of production of 32P-labeled inorganic phosphates. All reactions were performed at least twice and the SE is given.
With mRNA bound in the A-site and the downstream channel, how does the tmRNA·SmpB complex activate EF-Tu? In the crystal structure of the preaccommodation complex, SmpB residue His136 stacks with G530 of 16S rRNA, a key nucleotide in the canonical decoding mechanism (Neubauer et al. 2012). Mutation of His136 dramatically reduces the rate constant of GTP hydrolysis (Miller and Buskirk 2014), suggesting that the stacking of the histidine side chain on the guanine base plays a key role in EF-Tu activation. The fact that EF-Tu activation occurs rapidly regardless of mRNA length raises the possibility that tmRNA·SmpB places His136 in contact with G530 early on in the binding process.
The fact that long 3′ extensions of mRNA do not reduce EF-Tu activation rates is compatible with our previous kinetic work showing that the latter portion of the C-terminal tail of SmpB plays little or no role in EF-Tu activation (Kurita et al. 2010; Miller et al. 2011). Truncation of the tail after residue 153 or preventing helix formation by mutating Lys151 to Pro reduced GTP hydrolysis rates by only twofold to fourfold, whereas peptidyl-transfer rates were reduced by 30- to 60-fold (Miller et al. 2011). These defects are indicative of an inability of the tmRNA·SmpB complex, once released from EF-Tu, to be accommodated into the A site with the proper geometry for peptidyl transfer to take place. Presumably the interaction of the latter portion of the C-terminal tail with the mRNA channel stabilizes the tmRNA·SmpB complex within the A site in order to promote accommodation without dissociation of the complex. By interfering with binding of the tail in the channel, long mRNAs block accommodation and peptidyl transfer but not EF-Tu activation, giving the same kinetic defects observed when the tail is mutated or truncated.
tmRNA·SmpB binds more weakly to ribosomes translating longer mRNAs
If the tmRNA·SmpB complex is delivered to the ribosome and activates EF-Tu equally well regardless of mRNA length, then perhaps the reason that peptidyl transfer to tmRNA occurs slowly on longer mRNAs is that the complex fails to accommodate properly in the A site and dissociates from the ribosome. To directly test if the affinity of ribosomes for the tmRNA·SmpB complex is reduced by the presence of mRNA in the A-site and mRNA channel, we performed fluorescence polarization (FP) studies using labeled SmpB. FP is a method to characterize molecules by rotational diffusion and thus molecular weight. The polarization of the light used for excitation is retained by slowly rotating, larger molecules, while quickly rotating, smaller molecules lose polarization. By labeling the SmpB protein, we monitored the binding of the tmRNA·SmpB complex to the ribosome through observing the increase in the polarization signal as the ribosome slows rotation of SmpB.
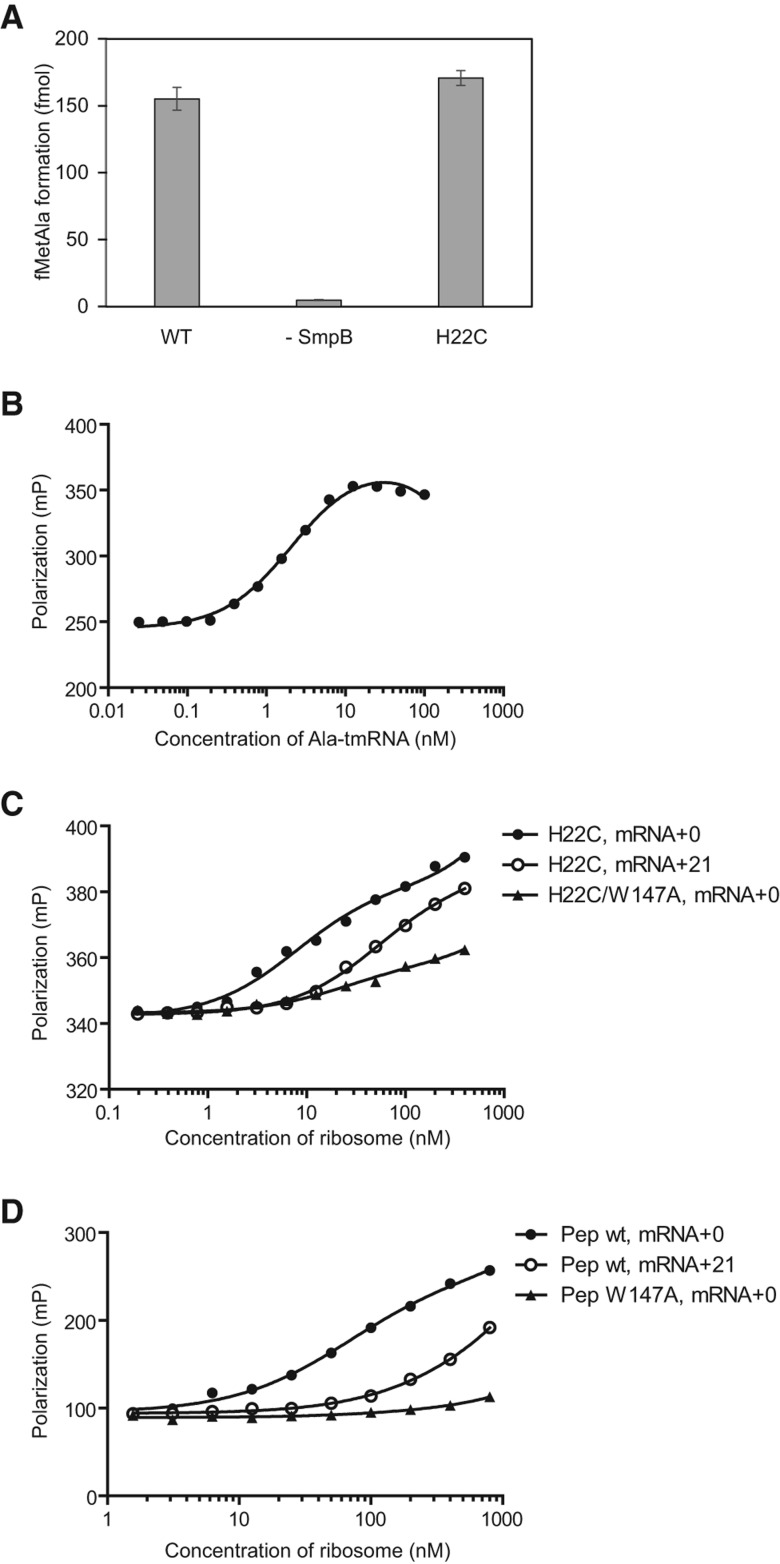
SmpB was labeled with the fluorophore Atto633 at an engineered, unique Cys residue. We previously prepared a series of SmpB mutants in which two naturally occurring cysteine residues at 82 and 123 were replaced by alanine and single cysteine mutations were introduced throughout the protein (Kurita et al. 2007). Here we used the SmpB mutant in which a cysteine residue had been introduced at position 22 (H22C) to site-specifically label the protein with Atto633. We confirmed that H22C mutant has trans-transfer activity comparable with that of wild-type SmpB (Fig. 3A). To form the quaternary complex, labeled SmpB was incubated with excess Ala-tmRNA, EF-Tu, and GTP. Using fluorescence polarization, we confirmed nearly full participation of 2 nM SmpB in a tmRNA·SmpB complex when 22 nM Ala-tmRNA was added (Fig. 3B).
FIGURE 3.
Binding of the Ala-tmRNA·SmpB complex to the stalled ribosome after GTP hydrolysis. (A) Activity of trans-transfer for H22C mutant of SmpB. N-formyl[14C]Met[3H]Ala formation was examined by bench-top assay. (B) Fluorescence polarization binding analysis of Atto633-labeled SmpB to Ala-tmRNA, EF-Tu, and GTP. Polarization was plotted against concentration of Ala-tmRNA. Measurement was repeated five times. (C) Fluorescence polarization binding analysis of the quaternary complex containing Atto633-labeled SmpB mutant (H22C) to the stalled ribosome with a short mRNA (mRNA+0, closed circle) or a long mRNA (mRNA+21, open circle). Binding of the quaternary complex containing Atto633-labeled SmpB mutant (H22C/W147A) to the stalled ribosome with a short mRNA (mRNA+0, closed triangle) was monitored. (D) Binding of the synthetic peptide corresponding to residues 133–160 of the C-terminal tail of SmpB to the stalled ribosome with a short mRNA (+0, closed circle) or a long mRNA (+21, open circle) by fluorescence polarization analysis. The peptide was labeled with TAMRA at the N-terminus. Closed triangle shows the binding of a synthetic peptide that contains W147A mutation to the ribosome stalled by mRNA+0.
The interaction between the ribosome and the tmRNA· SmpB complex was characterized by incubating the fluorescently labeled quaternary complex with various concentrations of initiation complexes. To prevent peptidyl transfer to tmRNA, which would irreversibly trap the tmRNA·SmpB complex within the ribosome and prevent an accurate determination of its apparent affinity, the initiation complexes were formed using deacyl-tRNAfMet bound to the P site.
We tested the effect of mRNA length on the binding of tmRNA·SmpB complexes to ribosomes programmed with an mRNA with no 3′ extension past the P-site codon (mRNA+0) or having a long extension (mRNA+21). As shown in Figure 3C, polarization of SmpB increased when higher concentrations of either initiation complex were added. When mRNA+21 was used, however, complex formation required an approximately sevenfold higher concentration of ribosomes than it did when mRNA+0 was used. These data confirm that the tmRNA·SmpB complex preferentially binds ribosomes programmed with mRNA having no 3′ extension. This is consistent with a previous study showing that deacyl-tmRNA binds preferentially to the complex of SmpB-ribosome programmed with mRNA having no 3′ extension (Gillet et al. 2007).
We also tried to measure the ribosome binding capacity of Ala-tmRNA·SmpB before GTP hydrolysis using the nonhydrolysable GTP analogs GDPNP, GDPCP, and GTPγS, but were unable to do so because no quaternary complex could be obtained. We also could not measure ribosome binding in the step immediately after GTP hydrolysis, but prior to release of tmRNA from EF-Tu, because kirromycin does not effectively block trans-translation mediated by tmRNA (Shimizu and Ueda 2006; Miller and Buskirk 2014).
The importance of binding of the SmpB C-terminal tail within the mRNA channel
Having observed that binding of the tmRNA·SmpB complex is weakened by mRNA with long 3′ extensions, we next asked if this loss of binding affinity resulted from displacement of the C-terminal tail of SmpB from the mRNA channel. We previously showed that a tryptophan residue within the C-terminal tail of SmpB, Trp147, is critical for trans-transfer (Kurita et al. 2010). In the preaccommodation structure of T. thermophilus, the corresponding residue contacts the S5 protein within the mRNA channel. We compared the ability of the labeled tmRNA·SmpB complex to bind to initiation complexes containing mRNA+0 using SmpB with or without the Trp147Ala mutation. We observed that the Trp147Ala mutant required >10-fold higher concentration of ribosomes for complex formation, roughly the same loss of binding affinity seen when mRNA+21 was used above. These data are consistent with the idea that the interaction of the tail and the mRNA channel contributes extra binding affinity that is essential for stable binding of the tmRNA·SmpB complex to the A site during accommodation. The hydrophobic interaction of Trp147 with the S5 protein may be an important contributor to this binding energy.
We obtained additional direct evidence that the C-terminal tail of SmpB competes with the 3′ extension of mRNA for the channel. Using the FP assay, we monitored the interaction between initiation complexes and a synthetic peptide corresponding to the C-terminal tail of SmpB (residues 133–160). The tail peptide is expected to bind to the mRNA channel in the same manner as the C-terminal tail of full-length SmpB because it blocks the binding of SmpB to the A site and inhibits EF-Tu activation and peptidyl transfer to tmRNA (Kurita et al. 2010; Miller and Buskirk 2014). When mRNA+21 was used, complex formation required an ∼10-fold higher concentration of ribosomes than when mRNA+0 was used (Fig. 3D). When we used the synthetic peptide that contains the Trp147Ala mutation, the complex formation almost completely disappeared even on mRNA+0, highlighting the importance of Trp147 to the interaction of the C-terminal tail of SmpB with the mRNA path of the ribosome. Taken together, these results demonstrate that binding of the tmRNA·SmpB complex to ribosomes is reduced when the mRNA extends into the mRNA channel. Selectivity for empty A sites arises from reduced affinity due to competition between the 3′ extension and the C-terminal tail for the mRNA channel.
How does the SmpB tail move from its disordered state in solution to form an α-helix when bound within the mRNA channel? Given the length of the tail (>20 residues), it probably moves through an opening in the mRNA entry channel rather than sliding into the channel from the decoding center. An open conformation of the mRNA channel has been observed in structures of initiation and translocation complexes (Passmore et al. 2007; Ramrath et al. 2012). Like SmpB, the ArfB protein (YaeJ) has a C-terminal tail that forms an α-helix upon binding to the mRNA channel (Gagnon et al. 2012). ArfB also contains the GGQ domain, common to release factors, that hydrolyzes the peptidyl-tRNA linkage (Handa et al. 2010; Chadani et al. 2011). Remarkably, addition of 44 residues to the C-terminus of ArfB had no effect on its activity, suggesting that even with this long extension, the tail is capable of productively binding in the channel (Kogure et al. 2014). These considerations further support a model in which the channel opens to allow the SmpB tail to enter.
A new model of the early steps in trans-translation
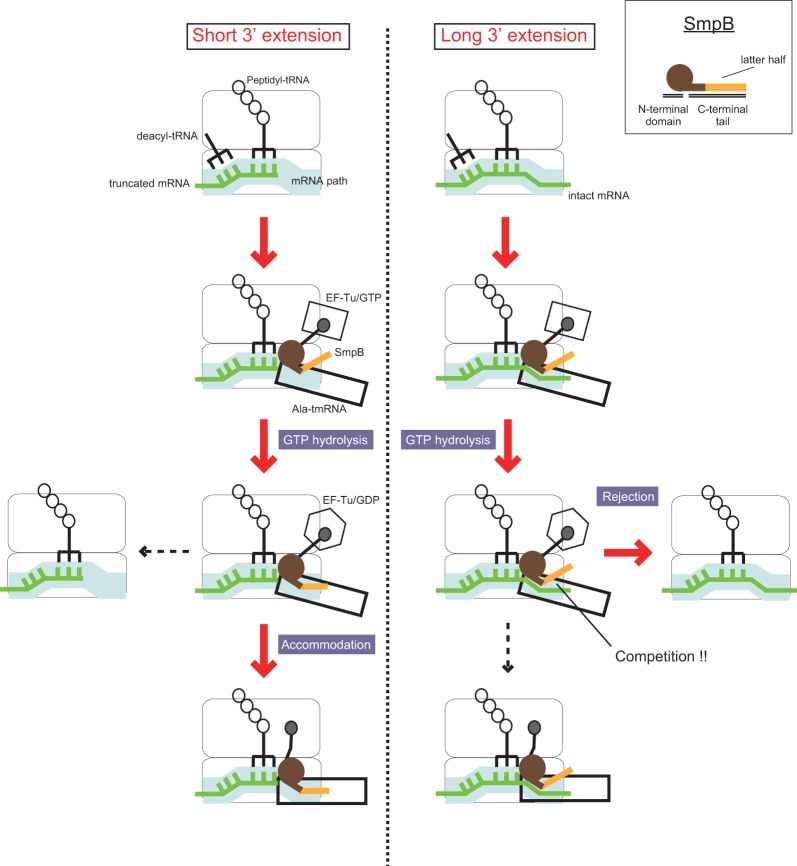
Based on our results, we propose a new model of trans-translation to explain how tmRNA·SmpB selectively recognizes stalled ribosomes (Fig. 4). Initially, Ala-tmRNA·SmpB in complex with EF-Tu and GTP binds the vacant A site of a stalled ribosome without interaction of the latter portion of the C-terminal tail of SmpB with the mRNA channel; GTP hydrolysis is triggered regardless of the length of the 3′ extension of mRNA. After GTP hydrolysis, the Ala-tmRNA·SmpB complex is released from EF-Tu. If the mRNA channel is free, and the SmpB C-terminal tail binds there, Ala-tmRNA·SmpB is accommodated in the A site of ribosome and peptidyl transfer occurs. On the other hand, if the mRNA channel is occupied by mRNA, the C-terminal tail of SmpB cannot interact with its binding site in the mRNA channel, and Ala-tmRNA·SmpB dissociates from the ribosome without peptidyl transfer. This model explains how the fate of the Ala-tmRNA·SmpB complex is determined after GTP hydrolysis through binding competition between mRNA and the latter half of the C-terminal tail of SmpB for the mRNA path. Following rejection, the stalled ribosome may accept an additional Ala-tmRNA·SmpB complex to perform peptidyl transfer, and in fact more GTP molecules are hydrolyzed per peptidyl-transfer reaction when a longer mRNA is used.
FIGURE 4.
Schematic representation of the process after the stalled ribosome is recognized by the quaternary complex. The “latter half” of the C-terminal tail of SmpB is colored orange. The quaternary complex enters the stalled ribosome and triggers GTP hydrolysis regardless of the length of 3′ extension of mRNA. After GTP hydrolysis, tmRNA·SmpB is rejected from the ribosome stalled in the middle of mRNA, while it is accommodated in the ribosome stalled at the 3′ end of mRNA for trans-translation.
It is illuminating to compare the mechanism of rejection of tmRNA·SmpB with the proofreading mechanism in canonical translation. During the decoding of canonical tRNAs, there are two consecutive selection steps, initial selection and proofreading (for review, see Zaher and Green 2009). While noncognate tRNAs are efficiently rejected during the initial selection step prior to GTP hydrolysis, some proportion of near-cognate tRNAs escape this screening process and are rejected in the second selection (proofreading step) following GTP hydrolysis. This step entails accommodation of the whole tRNA body into the A site with the CCA-end dissociating from EF-Tu and moving into the peptidyl-transferase center. This strategy of multistep discrimination is required to achieve high fidelity translation because a single selection based on base-pairing between the codon and anticodon alone is not sufficient to discriminate between the correct and incorrect substrates. In contrast, two consecutive selection steps are unnecessary for tmRNA·SmpB due to the absence of confusingly similar molecules; selectivity can be achieved in a single step, accommodation of tmRNA· SmpB into the A site.
Binding of ribosome-rescue factors to the mRNA channel is a widespread solution to the problem of discriminating between stalled and actively translating ribosomes. In bacteria, another ribosome-rescue factor, ArfB, uses its α-helical C-terminal tail to bind the mRNA channel in a manner very similar to SmpB. This implies that ArfB activity should be inhibited by long 3′ extensions in mRNA transcripts, which has been supported by a recent biochemical study (Shimizu 2012). In eukaryotes, the Dom34/Hbs1 complex rescues stalled ribosomes in an mRNA length-dependent manner. The mammalian complex discriminates against transcripts with RNA >13 nt downstream from the P-site codon (Pisareva et al. 2011). In yeast, the cutoff is not as sharply defined: Subunit splitting occurs rapidly with transcripts shorter than 23 nt after the P-site codon, at intermediate rates between 23 and 30 nt, and significantly slower on transcripts longer than 30 nt after the P-site codon (Shoemaker and Green 2011). This length dependence is thought to be mediated by the N-terminal domain of Hbs1, which binds the ribosome near the mRNA entry channel in a cryo-EM reconstruction (Becker et al. 2011). Our studies of tmRNA·SmpB suggest that although ArfB and Dom34/Hbs1 may bind transiently to actively translating ribosomes in the middle of a transcript, they probably also require significant interactions in the mRNA channel for stable and productive binding and ribosome rescue.
MATERIALS AND METHODS
Preparation of translation components
IF1, IF2, IF3, EF-Tu, MetRS, and AlaRS were purified as described (Miller et al. 2011). mRNAs were synthesized by transcription by T7 RNA polymerase using a template assembled by annealing sense and antisense oligonucleotides. tRNAfMet was purchased from Sigma-Aldrich. Formyl-[35S]Met-tRNAfMet was prepared as described (Moazed and Noller 1991). 70S ribosome, tmRNA, and His6-tagged SmpB were prepared as described previously (Kurita et al. 2010; Miller et al. 2011).
Ribosome complex formation
The 70S initiation complex was formed by incubating 4 μM ribosomes, 10 μM mRNA, 6 μM fMet-tRNAfMet, 5 μM each of IF1, IF2 and IF3, and 2 mM GTP in buffer A for 45 min at 37°C. Buffer A contains 50 mM Tris–HCl (pH 7.5), 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, and 1 mM dithiothreitol as previously described (Miller et al. 2011). To purify the complex, this solution was layered on a 1.3-mL sucrose cushion (1.1 M sucrose, 20 mM Tris–HCl [pH 7.5], 500 mM NH4Cl, 10 mM MgCl2, and 0.5 mM EDTA) followed by centrifugation at 258,000g in a TLA100.3 rotor for 2 h. The resulting pellet was resuspended in buffer A and stored at −80°C.
Peptidyl-transfer reaction
The 70S initiation complex was diluted to 100 nM. The tmRNA· SmpB quaternary complex was prepared by incubating 5 μM charged tmRNA, 30 μM SmpB, and 1 mM GTP in buffer A for 5 min at 37°C; 20 μM EF-Tu was added, and the reaction mixture was incubated for another 5 min at 37°C. Peptide-bond formation reaction was carried out at 37°C by mixing equal volumes of 70S initiation complex with the quaternary complex described above. The reaction was stopped with hydrolysis of the peptidyl-tRNA at desired time points by the addition of KOH to a final concentration of 0.3 M on a KinTek RQF-3 quench-flow instrument. Reaction products were resolved using cellulose TLC plates in pyridine acetate (pH 2.8) and analyzed by autoradiography (Youngman et al. 2004). Data were fit to a first-order exponential equation with GraphPad Prism5 software.
Peptidyl-transfer reactions were also performed by bench-top assay. One micromolar 70S ribosome was incubated with 4 µM mRNA+0 and 2 µM N-formyl[14C]Met-tRNA at 37°C for 10 min in 80 mM Tris–HCl (pH 7.8), 7 mM MgCl2, 150 mM NH4Cl, 2.5 mM dithiothreitol, 0.2 mM GTP, and 2 mM spermidine. They were subsequently incubated with 0.1 µM [3H]Ala-tmRNA, 1 µM SmpB, and 2 µM EF-Tu at 37°C for 90 sec. After incubation, aliquots (8 µL) of reaction mixture was stopped by addition of 8 µL 1.2 N NaOH and subsequently incubated at 37°C for 10 min. Then 32 µL of H2SO4 was added to reaction mixture. The peptide containing [14C]Met was extracted with ethyl acetate and radioactivity was measured by a liquid scintillation counter.
GTP hydrolysis reaction
The 70S initiation complex was formed as described above except that nonradioactive fMet-tRNAfMet was used and the complex was diluted to 500 nM. The quaternary complex was prepared by incubating 5 μM tmRNA, 20 μM SmpB, 20 μM EF-Tu, 17.5 μCi of [γ-32P]GTP (6000 Ci/mmol), 3 mM phosphoenol pyruvate, 0.1 mg/mL pyruvate kinase, 20 mM L-alanine, 2 mM ATP, and 10 μM AlaRS in buffer A for 1 h at 37°C. The mixture of quaternary complex was passed through P30 columns twice to remove excess [γ-32P]GTP. The rate constant of GTP hydrolysis was measured using a KinTek RQF-3 quench-flow instrument at 20°C, where equal volumes of the 70S initiation complex and the quaternary complex described above were mixed and quenched with 40% formic acid at the desired times. Reaction products were resolved on PEI cellulose TLC plates in 0.5 M KH2PO4 (pH 3.5) and [32P]Pi was quantified by autoradiography. Data were fit to a first-order exponential equation with GraphPad Prism5 software.
Fluorescence polarization analysis
SmpB (1 nmol) was labeled with Atto633-maleimide (10 nmol) in 50 μL of solution containing 50 mM MOPS (pH 8.2), 100 mM NaCl, 1 mM EDTA, and 5% glycerol at 37°C for 1 h. Unreacted fluorophore was removed by ultrafiltration (Amicon ultra-0.5 mL 10 kDa) and gel-filtration. The buffer was exchanged for a stock buffer containing 50 mM MOPS (pH 8.0), 120 mM NaCl, 0.1 mM EDTA, 10 mM MgCl2, and 10% glycerol. To form the quaternary complex, labeled SmpB (100 fmol) was incubated with alanyl-tmRNA (1.1 pmol), EF-Tu (25 pmol), and GTP (0.2 mM) in 25 μL of solution containing 80 mM Tris–HCl (pH 7.5), 7 mM MgCl2, 150 mM NH4Cl, 2.5 mM dithiothreitol, 2 mM spermidine, and 0.05% Tween 20 at 37°C for 10 min. Simultaneously, indicated amounts of 70S ribosomes, mRNA (mRNA+0 or mRNA+21, 100 pmol), tRNAfMet (100 pmol), and GTP (0.2 mM) were incubated in 25 μL of solution at 37°C for 10 min. Quaternary complex (or 100 fmol-labeled synthetic peptide containing TAMRA at the N-terminus) was mixed with the ribosome mixture and incubated at 37°C for 10 min. The reaction mixtures were applied to a glass-bottomed microplate and set in a MF20 single molecule fluorescence spectroscopy system (Olympus). The measurement of polarization was performed with excitation wavelength at 633 nm and laser power of 100 μW. Data acquisition time was 5 sec per measurement.
ACKNOWLEDGMENTS
We thank the staff of the Gene Research Center of Hirosaki University for the use of the facility. This work was supported by a Grant-in-Aid for Young Scientists from the Japan Society for the Promotion of Science (D.K., no. 23780099), and Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (A.M., no. 23380045; H.H., no. 23380054).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.045773.114.
REFERENCES
- Asano K, Kurita D, Takada K, Konno T, Muto A, Himeno H. 2005. Competition between trans-translation and termination or elongation of translation. Nucleic Acids Res 33: 5544–5552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barends S, Wower J, Kraal B. 2000. Kinetic parameters for tmRNA binding to alanyl-tRNA synthetase and elongation factor Tu from Escherichia coli. Biochemistry 39: 2652–2658 [DOI] [PubMed] [Google Scholar]
- Barends S, Karzai AW, Sauer RT, Wower J, Kraal B. 2001. Simultaneous and functional binding of SmpB and EF-Tu-GTP to the alanyl acceptor arm of tmRNA. J Mol Biol 314: 9–21 [DOI] [PubMed] [Google Scholar]
- Becker T, Armache JP, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R. 2011. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol 18: 715–720 [DOI] [PubMed] [Google Scholar]
- Bessho Y, Shibata R, Sekine S, Murayama K, Higashijima K, Hori-Takemoto C, Shirouzu M, Kuramitsu S, Yokoyama S. 2007. Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proc Natl Acad Sci 104: 8293–8298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadani Y, Ono K, Ozawa S, Takahashi Y, Takai K, Nanamiya H, Tozawa Y, Kutsukake K, Abo T. 2010. Ribosome rescue by Escherichia coli ArfA (YhdL) in the absence of trans-translation system. Mol Microbiol 78: 796–808 [DOI] [PubMed] [Google Scholar]
- Chadani Y, Ono K, Kutsukake K, Abo T. 2011. Escherichia coli YaeJ protein mediates a novel ribosome-rescue pathway distinct from SsrA- and ArfA-mediated pathways. Mol Microbiol 80: 772–785 [DOI] [PubMed] [Google Scholar]
- Choy JS, Aung LL, Karzai AW. 2007. Lon protease degrades transfer-messenger RNA-tagged proteins. J Bacteriol 189: 6564–6571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier J, Binet E, Bouloc P. 2002. Competition between SsrA tagging and translational termination at weak stop codons in Escherichia coli. Mol Microbiol 45: 745–754 [DOI] [PubMed] [Google Scholar]
- Dong H, Nilsson L, Kurland CG. 1996. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol 260: 649–663 [DOI] [PubMed] [Google Scholar]
- Gagnon MG, Seetharaman SV, Bulkley D, Steitz TA. 2012. Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome. Science 335: 1370–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sánchez F, Gin JG, Hayes CS. 2008. Amino acid starvation and colicin D treatment induce A-site mRNA cleavage in Escherichia coli. J Mol Biol 378: 505–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sánchez F, Shoji S, Fredrick K, Hayes CS. 2009. RNase II is important for A-site mRNA cleavage during ribosome pausing. Mol Microbiol 73: 882–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet R, Kaur S, Li W, Hallier M, Felden B, Frank J. 2007. Scaffolding as an organizing principle in trans-translation. The roles of small protein B and ribosomal protein S1. J Biol Chem 282: 6356–6363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou Y, Sauer RT. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev 12: 1338–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueneau de Novoa P, Williams KP. 2004. The tmRNA website: reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Res 1: D104–D108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann S, Haebel PW, Metzinger L, Sutter M, Felden B, Ban N. 2003. Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature 424: 5503–5509 [DOI] [PubMed] [Google Scholar]
- Hanawa-Suetsugu K, Takagi M, Inokuchi H, Himeno H, Muto A. 2002. SmpB functions in various steps of trans-translation. Nucleic Acids Res 30: 1620–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa Y, Inaho N, Nameki N. 2010. YaeJ is a novel ribosome-associated protein in Escherichia coli that can hydrolyze peptidyl-tRNA on stalled ribosomes. Nucleic Acids Res 39: 1739–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CS, Sauer RT. 2003. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol Cell 12: 903–911 [DOI] [PubMed] [Google Scholar]
- Hayes CS, Bose B, Sauer RT. 2002. Stop codons preceded by rare arginine codons are efficient determinants of SsrA tagging in Escherichia coli. Proc Natl Acad Sci 99: 3440–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman C, Thévenet D, Bouloc P, Walker GC, D'Ari R. 1998. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH). Genes Dev 12: 1348–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H, Sato M, Tadaki T, Fukushima M, Ushida C, Muto A. 1997. In vitro trans translation mediated by alanine-charged 10Sa RNA. J Mol Biol 268: 803–808 [DOI] [PubMed] [Google Scholar]
- Huang C, Wolfgang MC, Withey J, Koomey M, Friedman DI. 2000. Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J 19: 1098–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Pavlov MY, Felden B, Ehrenberg M. 2004. Ribosome rescue by tmRNA requires truncated mRNAs. J Mol Biol 338: 33–41 [DOI] [PubMed] [Google Scholar]
- Janssen BD, Garza-Sánchez F, Hayes CS. 2013. A-site mRNA cleavage is not required for tmRNA-mediated ssrA-peptide tagging. PLoS One 8: e81319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai AW, Susskind MM, Sauer RT. 1999. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J 18: 3793–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC, Waller PR, Sauer RT. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271: 990–993 [DOI] [PubMed] [Google Scholar]
- Kogure H, Handa Y, Nagata M, Kanai N, Guntert P, Kubota K, Nameki N. 2014. Identification of residues required for stalled-ribosome rescue in the codon-independent release factor YaeJ. Nucleic Acids Res 42: 3152–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. 1994. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc Natl Acad Sci 91: 9223–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno T, Kurita D, Takada K, Muto A, Himeno H. 2007. A functional interaction of SmpB with tmRNA for determination of the resuming point of trans-translation. RNA 13: 1723–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita D, Sasaki R, Muto A, Himeno H. 2007. Interaction of SmpB with ribosome from directed hydroxyl radical probing. Nucleic Acids Res 35: 7248–7255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita D, Muto A, Himeno H. 2010. Role of the C-terminal tail of SmpB in the early stage of trans-translation. RNA 16: 980–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Bailey SC, Apirion D. 1978. Small stable RNAs from Escherichia coli: evidence for the existence of new molecules and for a new ribonucleoprotein particle containing 6S RNA. J Bacteriol 133: 1015–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yokota T, Ito K, Nakamura Y, Aiba H. 2007. Reduced action of polypeptide release factors induces mRNA cleavage and tmRNA tagging at stop codons in Escherichia coli. Mol Microbiol 63: 116–126 [DOI] [PubMed] [Google Scholar]
- Luidalepp H, Hallier M, Felden B, Tenson T. 2005. tmRNA decreases the bactericidal activity of aminoglycosides and the susceptibility to inhibitors of cell wall synthesis. RNA Biol 2: 70–74 [DOI] [PubMed] [Google Scholar]
- Miller MR, Buskirk AR. 2014. An unusual mechanism for EF-Tu activation during tmRNA-mediated ribosome rescue. RNA 20: 228–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MR, Liu Z, Cazier DJ, Gebhard GM, Herron SR, Zaher HS, Green R, Buskirk AR. 2011. The role of SmpB and the ribosomal decoding center in licensing tmRNA entry into stalled ribosomes. RNA 17: 1727–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Noller HF. 1991. Sites of interaction of the CCA end of peptidyl-tRNA with 23S rRNA. Proc Natl Acad Sci 88: 3725–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Fujihara A, Ito K, Matsuno J, Ushida C, Himeno H. 2000. Requirement of transfer-messenger RNA (tmRNA) for the growth of Bacillus subtilis under stresses. Genes Cells 5: 627–636 [DOI] [PubMed] [Google Scholar]
- Neubauer C, Gillet R, Kelley AC, Ramakrishnan V. 2012. Decoding in the absence of a codon by tmRNA and SmpB in the ribosome. Science 335: 1366–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. 2007. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26: 41–50 [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. 2011. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J 30: 1804–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadoss NS, Zhou X, Keiler KC. 2013. tmRNA is essential in Shigella flexneri. PLoS One 8: e57537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramrath DJ, Yamamoto H, Rother K, Wittek D, Pech M, Mielke T, Loerke J, Scheerer P, Ivanov P, Teraoka Y, et al. 2012. The complex of tmRNA-SmpB and EF-G on translocating ribosomes. Nature 485: 526–529 [DOI] [PubMed] [Google Scholar]
- Roche ED, Sauer RT. 1999. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J 18: 4579–4589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche ED, Sauer RT. 2001. Identification of endogenous SsrA-tagged proteins reveals tagging at positions corresponding to stop codons. J Biol Chem 276: 28509–28515 [DOI] [PubMed] [Google Scholar]
- Shimizu Y. 2012. ArfA recruits RF2 into stalled ribosomes. J Mol Biol 423: 624–631 [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Ueda T. 2006. SmpB triggers GTP hydrolysis of elongation factor Tu on ribosomes by compensating for the lack of codon-anticodon interaction during trans-translation initiation. J Biol Chem 281: 15987–15996 [DOI] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. 2011. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci 108: 1392–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. 2012. Translation drives mRNA quality control. Nat Struct Mol Biol 19: 594–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunohara T, Jojima K, Tagami H, Inada T, Aiba H. 2004. Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. J Biol Chem 279: 15368–15375 [DOI] [PubMed] [Google Scholar]
- Takada K, Takemoto C, Kawazoe M, Konno T, Hanawa-Suetsugu K, Lee S, Shirouzu M, Yokoyama S, Muto A, Himeno H. 2007. In vitro trans-translation of Thermus thermophilus: Ribosomal protein S1 is not required for the early stage of trans-translation. RNA 13: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibonnier M, Thiberge JM, De Reuse H. 2008. Trans-translation in Helicobacter pylori: essentiality of ribosome rescue and requirement of protein tagging for stress resistance and competence. PLoS One 3: e3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushida C, Himeno H, Watanabe T, Muto A. 1994. tRNA-like structures in 10Sa RNAs of Mycoplasma capricolum and Bacillus subtilis. Nucleic Acids Res 22: 3392–3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman EM, Brunelle JL, Kochaniak AB, Green R. 2004. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117: 589–599 [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R. 2009. Fidelity at the molecular level: lessons from protein synthesis. Cell 136: 46–62 [DOI] [PMC free article] [PubMed] [Google Scholar]