Abstract
Many bacteria encode an ortholog of the Ro60 autoantigen, a ring-shaped protein that is bound in animal cells to noncoding RNAs (ncRNAs) called Y RNAs. Studies in Deinococcus radiodurans revealed that Y RNA tethers Ro60 to polynucleotide phosphorylase, specializing this exoribonuclease for structured RNA degradation. Although Ro60 orthologs are present in a wide range of bacteria, Y RNAs have been detected in only two species, making it unclear whether these ncRNAs are common Ro60 partners in bacteria. In this study, we report that likely Y RNAs are encoded near Ro60 in >250 bacterial and phage species. By comparing conserved features, we discovered that at least one Y RNA in each species contains a domain resembling tRNA. We show that these RNAs contain nucleotide modifications characteristic of tRNA and are substrates for several enzymes that recognize tRNAs. Our studies confirm the importance of Y RNAs in bacterial physiology and identify a new class of ncRNAs that mimic tRNA.
Keywords: noncoding RNAs, tRNA-like domain, Ro60 autoantigen
INTRODUCTION
A class of noncoding RNAs (ncRNAs) whose function is only now being elucidated is the Y class of ncRNAs. These ∼100 nt ncRNAs were first identified because they are bound by the Ro 60 kDa protein (Ro60), a common autoantigen in patients with the rheumatic diseases systemic lupus erythematosus and Sjogren's syndrome (Lerner et al. 1981). Y RNAs were identified subsequently in a variety of vertebrate species, the nematode Caenorhabditis elegans and the radiation-resistant bacterium Deinococcus radiodurans (Sim and Wolin 2011). All characterized Y RNAs are between 80 and 130 nt and fold into structures containing a large internal loop and a long stem that contains the Ro60 binding site (Kohn et al. 2013; Wolin et al. 2013). Most vertebrate species contain between two and four distinct Y RNAs (Wolin et al. 2013).
A least one role of Y RNAs is to influence the subcellular location and function of Ro60, a ring-shaped protein that is proposed to function in ncRNA quality control. Studies of Xenopus laevis Ro60 revealed that it binds the single-stranded ends of misfolded ncRNA precursors in its central cavity (Stein et al. 2005; Fuchs et al. 2006). Y RNAs bind on the outer surface of the Ro60 ring and mask a nuclear accumulation signal, thus retaining Ro60 in the cytoplasm (Stein et al. 2005; Sim et al. 2009). Also, as the misfolded ncRNA and Y RNA binding surfaces overlap, Y RNAs could regulate binding of Ro60 to misfolded ncRNAs (Stein et al. 2005; Fuchs et al. 2006).
Because Ro60 and Y RNA orthologs have not been detected in yeast, D. radiodurans has been used as a model single-celled organism for elucidating function. Consistent with a role in ncRNA surveillance, studies on this bacterium revealed that both Ro60 and Y RNA assist degradation of structured RNAs by exoribonucleases. Specifically, Y RNA tethers the Ro60 ortholog Rsr (Ro-sixty related) to the exoribonuclease polynucleotide phosphorylase (PNPase) to form RYPER (Ro60/Y RNA/PNPase Exoribonuclease RNP), an RNA degradation machine (Chen et al. 2013; Wolin et al. 2013). As in vertebrates, Rsr is a ring (Ramesh et al. 2007) and RYPER is configured such that single-stranded RNA could pass through the Rsr ring into the PNPase cavity for degradation (Chen et al. 2013). The presence of Rsr and Y RNA increases the effectiveness of PNPase in degrading structured RNAs (Chen et al. 2013). Rsr also functions with the exoribonucleases RNase II and RNase PH to mature 23S rRNA during heat stress (Chen et al. 2007). In this case, the Y-RNA-free Rsr carries out rRNA maturation, and bound Y RNA inhibits this process (Chen et al. 2007). These data have resulted in a model in which Y RNA functions to both tether Ro60 orthologs to other proteins and to regulate access of substrates to the Ro60 cavity (Chen et al. 2013).
To determine the extent to which functions identified in D. radiodurans are conserved, it is necessary to study Ro60 and Y RNA in other bacteria. However, while ∼5% of sequenced bacterial genomes and some mycobacteriophages contain recognizable Ro60 proteins (Pedulla et al. 2003; Sim and Wolin 2011), Y RNAs have not been identified by homology searching using either metazoan or D. radiodurans Y RNAs. Notably, during a recent characterization of the Ro60 ortholog in the enteric bacterium Salmonella enterica serovar Typhimurium (Chen et al. 2013), we discovered that S. Typhimurium Rsr was associated with two ncRNAs that we named YrlA (Y RNA-like) and YrlB. As in D. radiodurans, the two S. Typhimurium ncRNAs were encoded within 2 kb of Rsr (Chen et al. 2000, 2013). Consistent with a function similar to the D. radiodurans RNA, S. Typhimurium YrlA was part of a complex that also contained Rsr and PNPase (Chen et al. 2013).
Given the intimate relationship between Ro60 and Y RNAs in metazoans and the broad distribution of bacterial Ro60 proteins, it seemed likely that Y RNAs were equally widespread. To identify additional Y RNAs, we performed homology searches using the newly identified Salmonella RNAs. We report that RNAs resembling S. Typhimurium YrlA are encoded adjacent to Ro60 orthologs in >250 bacterial and phage species. Surprisingly, comparison of conserved features revealed that at least one RNA in each species contains a domain that strikingly resembles tRNA. These domains resemble tRNAs in vivo as the RNAs contain modified nucleotides characteristic of tRNAs and are substrates for enzymes that recognize tRNAs. Although the D. radiodurans Y RNA is evolutionarily distant from the newly identified RNAs, this RNA also contains some tRNA-like features. Our studies vastly expand the number of bacterial Y RNAs and reveal that members of this family constitute a new class of tRNA mimics.
RESULTS AND DISCUSSION
Potential YrlA RNAs are widely present in Ro60-containing bacteria
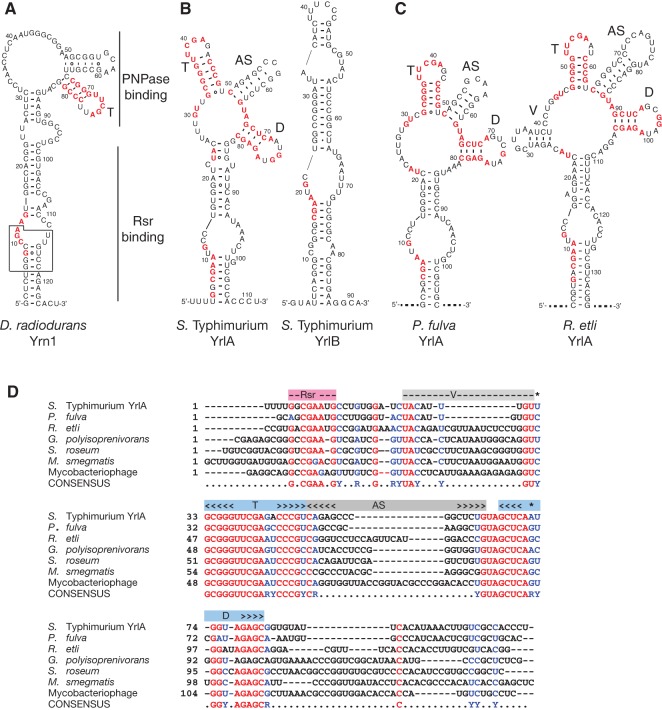
To identify additional bacterial Y RNAs, we first searched GenBank manually using the D. radiodurans and S. Typhimurium ncRNAs. RNAs used in these initial BLASTN searches were the major ncRNA bound by D. radiodurans Rsr, called Yrn1 here (Y RNA-1) (Chen et al. 2000), and two ncRNAs bound by S. Typhimurium Rsr, YrlA and YrlB (Fig. 1A,B; Chen et al. 2013). Although searches with Yrn1 did not reveal possible orthologs, and searches with YrlB only identified orthologs in other S. enterica serovars, a search with YrlA revealed possible ncRNAs in the α-proteobacterium Rhizobium etli and the γ-proteobacterium Pseudomonas fulva (Fig. 1C). By performing iterative searches with each newly identified putative RNA, we identified YrlA-related sequences in other bacterial phyla and in several mycobacteriophages that were known to encode a Ro60 ortholog (Fig. 1D).
FIGURE 1.
Newly identified YrlA-like RNAs. (A) D. radiodurans Y RNA. Regions important for Rsr and PNPase binding are indicated (Chen et al. 2013). (Box) conserved sequences required for binding of Ro60 to X. laevis and D. radiodurans Y RNAs. Sequences that may correspond to the GNCGAAN0-1G motif in YrlA RNAs are in red, as are sequences within the potential “T” stem–loop. (B) S. Typhimurium YrlA and YrlB. Nucleotides that are identical in six of the seven aligned sequences (D) are red. For clarity, only the GNCGAAN0-1G motif and nucleotides within and adjacent to the four stem–loops described in the text are colored. (C) Putative YrlA RNAs from P. fulva and R. etli. Nucleotides were colored as in B. Because the 5′ and 3′ ends of these RNAs have not been determined, they are indicated as dashed lines. Structures were predicted using Mfold (Zuker 2003) and RNAfold (Gruber et al. 2008), and adjusted manually to maximize structural homology with other YrlA RNAs. (D) Sequences of representative YrlA-like RNAs identified by iterative blast searches from P. fulva 12-X, R. etli, Gordonia polyisoprenivorans, Streptosporangium roseum, Mycobacterium smegmatis, and Mycobacteriophage ET08 were aligned with YrlA using Clustal Omega (Goujon et al. 2010; Sievers et al. 2011) (Supplemental text 1) and aligned further manually. Nucleotides that are identical in six of the seven sequences are red, while positions in which six of the seven sequences are either pyrimidines (Y) or purines (R) are blue. The “D” and “T” arms are shaded light blue, while the “V” and “AS” arms are in gray. The potential Rsr binding site is shaded pink.
To increase detection of YrlA orthologs, we used Infernal (Nawrocki and Eddy 2013) to build consensus RNA secondary structure models and search bacterial genomes systematically (see Materials and Methods and Supplemental files). Genome sequences were collected for all bacteria and bacteriophages that were annotated in GenBank as possessing a protein containing a TROVE (Telomerase, Ro, Vault) domain, the name given to the HEAT repeat domain that forms the Ro60 ring (Bateman and Kickhoefer 2003). Importantly, even when entire genomes were searched, nearly all sequences that were the best matches to the Infernal model were within 4 kb of the likely Ro60 ortholog and encoded on the same DNA strand (Supplemental Tables S1 and S2). Using these criteria, we identified YrlA-related sequences in 254 bacterial species and 22 bacteriophages.
Bacterial Y RNAs can be modeled to resemble tRNAs
By comparing the putative YrlA RNAs, we identified conserved features. One short motif near the 5′ end of each RNA, a GNCGAAN0-1G (N is any nucleotide), is also present in S. Typhimurium YrlB and overlaps sequences in D. radiodurans Yrn1 that are important for Rsr binding (Fig. 1A, box). As this motif resembles the GGUCCGA that is contacted by X. laevis Ro60 (Stein et al. 2005), we consider it likely that it is involved in Rsr recognition. Although the vertebrate motif can base-pair with conserved 3′ residues to form a bulged helix, Ro60 primarily interacts with the 5′ strand of the RNA (Stein et al. 2005), suggesting helix formation may not be needed for Ro60 recognition.
Surprisingly, the longest blocks of conserved sequences were located in the middle of the putative YrlA RNAs (Fig. 1C,D). In the MFOLD-predicted secondary structures, these sequences fold into two stem–loops that are nearly invariant between the putative YrlA RNAs (Fig. 1B–D, called T and D). Although not conserved in primary sequence, the nucleotides between these two conserved stem–loops can fold to form a third stem–loop (Fig. 1, AS). Many of the RNAs were also predicted to contain a fourth, less-conserved stem–loop upstream of stem–loop T (Fig. 1C,D, called V). Notably, the conserved “T” stem–loop was also present in D. radiodurans Yrn1 (Fig. 1A).
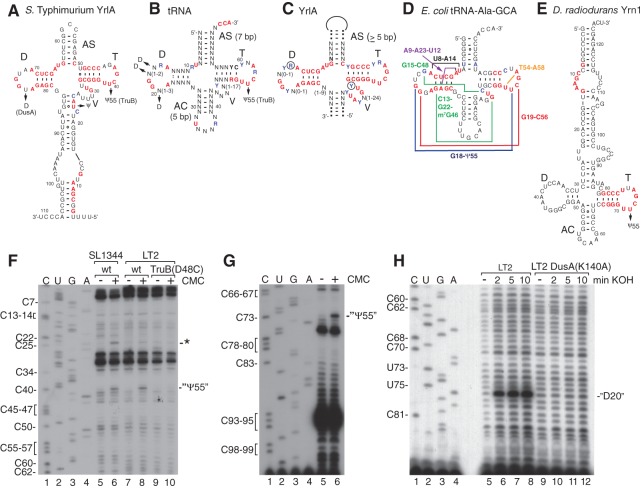
Inspection of the two most conserved stem–loops revealed similarities to the D- and T-arms of tRNAs (Fig. 2). The resemblance was most evident when S. Typhimurium YrlA or its putative orthologs were oriented such that the AS stem–loop between the “T” and “D” stem–loops of YrlA corresponds to the tRNA acceptor stem and the V region corresponds to the tRNA variable arm (Fig. 2A). In this orientation, many conserved nucleotides in YrlA RNAs correspond to invariant nucleotides in tRNA (Fig. 2B,C). Notably, at least seven tertiary interactions found in tRNA could potentially form in most YrlA orthologs (Fig. 2C,D). Importantly, for one tertiary interaction, the Levitt base pair between nucleotides 15 and 48 that stabilizes the tRNA L-shape (Levitt 1969), the various YrlA RNAs contain compensatory changes to allow formation of either a G15:C48 or A15:U8 base pair (Figs. 1D, 2C). Similar tRNA-like domains were present in the YrlA RNAs identified by Infernal (Supplemental Fig. S1). Moreover, although a full “D” arm is not evident in D. radiodurans Yrn1, this RNA can be modeled to contain some D-loop features (Fig. 2E). When modeled in this orientation, D. radiodurans Yrn1 contains a potential “anticodon arm” that resembles tRNAs in containing a 5-bp stem (Fig. 2E, AC); however the “anticodon loop” contains 6 nt, rather than the usual 7, and a V arm is not present.
FIGURE 2.
YrlA RNAs contain a tRNA-like domain. (A) S. Typhimurium YrlA drawn to resemble the tRNA cloverleaf. Nucleotides shown to be modified in vivo are indicated, as are the enzymes that carry out modification. (B) A generic tRNA. Nucleotides that are invariant, or nearly invariant, in tRNA are red, while nucleotides that are usually pyrimidine (Y) or purine (R) are blue (Dirheimer et al. 1995). The positions of the ψ55 and dihydrouridine modifications are indicated. (C) The YrlA tRNA-like domain. Nucleotides that are conserved in YrlA RNAs (Fig. 1D) are represented in red, while conserved purines and pyrimidines are blue. In most YrlA RNAs, the conserved YUAY (nucleotides 22–25 in S. Typhimurium, panel A) is predicted to begin the V loop. The proposed secondary structure is supported by enzymatic probing (Supplemental Fig. S2). (D) Escherichia coli tRNA-Ala-GCA. Tertiary interactions are depicted by lines connecting the involved bases. Nucleotides that are conserved in YrlA orthologs are colored as in (C). All seven tertiary interactions shown can potentially form in YrlA RNA. (E) D. radiodurans Yrn1 drawn to illustrate similarities with the tRNA cloverleaf. The modified uridine that may correspond to ψ55 is indicated. (F) Total RNA from S. Typhimurium strains SL1344 and LT2 was subjected to CMC modification and primer extension to detect pseudouridines in YrlA RNA (lanes 5–8). RNA extracted from S. Typhimurium LT2 carrying a mutation in the TruB catalytic domain was also assayed (lanes 9,10). To generate markers, primer extension was carried out on S. Typhimurium LT2 RNA in the presence of dideoxy nucleotides (lanes 1–4). (G) D. radiodurans RNA was subjected to primer extension as in (E) to detect pseudouridines in Yrn1 (lanes 5,6). Lanes 1–4, dideoxy sequencing. (H) RNA from S. Typhimurium LT2 was incubated with alkali for the indicated times and subjected to primer extension to detect dihydrouridines. A strain carrying a mutation that inactivates DusA catalytic activity was also assayed.
Bacterial Y RNAs contain nucleotide modifications characteristic of tRNAs
As a test of our model that some bacterial Y RNAs resemble tRNAs, we determined whether these RNAs contain a characteristic nucleotide modification, the pseudouridine at position 55 (Ψ55) in the T-loop of nearly all tRNAs (Fig. 2B; Bjork et al. 1987). Total RNA from S. Typhimurium and D. radiodurans was incubated with N-cyclohexyl-N-β-(4-methylmorpholinum)-ethylcarbodiimide p-tosylate (CMC), which reacts irreversibly with pseudouridines. Afterward, primer extension was performed because reverse transcriptase halts at the nucleotide 3′ to the modification (Bakin and Ofengand 1993). In both YrlA and Yrn1, a strong block to primer extension was detected corresponding to modification at the predicted site (Fig. 2F, lanes 6 and 8; Fig. 2G, lane 6). Pseudouridylation at this site was not detected when S. Typhimurium carried a mutation (D48C) that disrupts catalysis by TruB, the enzyme responsible for Ψ55 in tRNA (Fig. 2F, lane 10; Gutgsell et al. 2000). We also detected pseudouridine at nucleotide 23 in YrlA from the virulent strain S. Typhimurium SL1344 (Fig. 2F, lane 6), but not in YrlA from the attenuated LT2 strain (lane 8).
We also tested whether YrlA and Yrn1 contain dihydrouridine (D), which is present in one or more conserved positions in all tRNA D-loops (Fig. 2B; Phizicky and Hopper 2010). Using a primer extension assay based on the finding that mild alkaline hydrolysis opens the dihydrouracil ring and blocks primer extension (Xing et al. 2004), we detected dihydrouridine in the “D loop” of S. Typhimurium YrlA, but not that of D. radiodurans Yrn1 (Fig. 2H, lanes 6–8 and data not shown). Importantly, a mutation that inactivates S. Typhimurium DusA, the enzyme that modifies D20a in the E. coli tRNAfMet D-loop (Bishop et al. 2002; Savage et al. 2006), eliminates dihydrouridine formation in YrlA (K140A, analogous to E. coli K153A) (Fig. 2H, lanes 10–12).
To determine whether the predicted tRNA-like structures form in vitro, we carried out enzymatic probing on Y RNAs synthesized with T7 RNA polymerase. Using two RNases that preferentially cleave single-stranded RNA, T1 (which cleaves after G) and T2 (which cleaves after all four nucleotides), as well as V1, which cleaves double-stranded or stacked nucleotides, we obtained cleavage patterns consistent with the proposed secondary structures (Supplemental Fig. S2). In keeping with findings that X. laevis Ro60 contacts conserved bases in the 5′ strand of a helix formed by base-pairing the 5′ and 3′ ends (Stein et al. 2005), incubation of D. radiodurans Rsr with Yrn1 resulted in protection of the analogous nucleotides from all three nucleases (Supplemental Fig. S2; nucleotides G7, G8, C9, G11, and A12). In addition, many nucleotides within this stem showed enhanced V1 cleavage, suggesting Rsr binding stabilizes its formation (Supplemental Fig. S2D).
M. smegmatis YrlA RNA can be cleaved by RNase P and undergo CCA addition
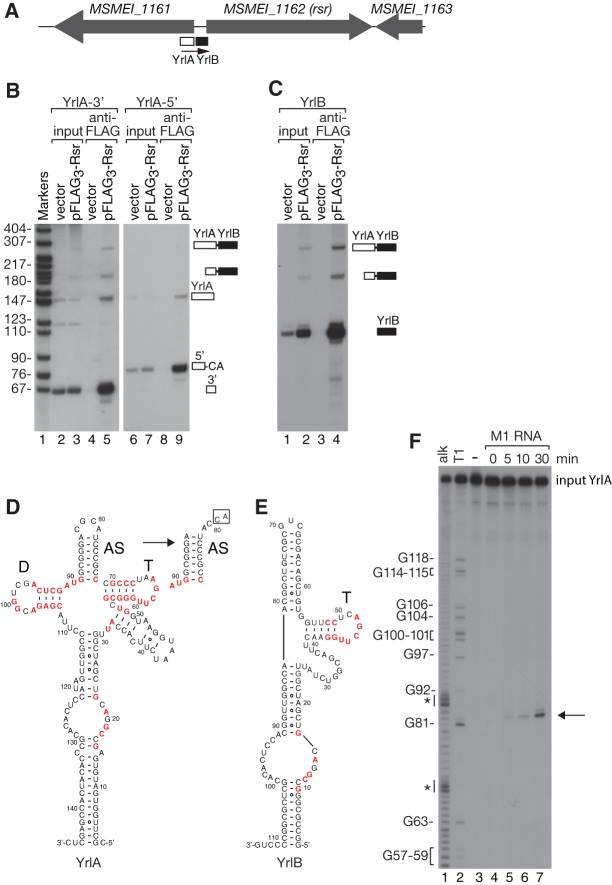
To confirm that potential YrlA RNAs identified through homology searching existed in vivo, we examined M. smegmatis. We chose this organism because the putative RNA is not under control of the RtcR transcriptional activator, a feature that prevents its transcription during normal growth in S. Typhimurium and possibly other bacteria (Chen et al. 2013; Das and Shuman 2013). In M. smegmatis, the predicted YrlA is 5′ of the Ro60 ortholog, within the annotated N terminus of the MSMEI_1161 locus (Fig. 3A).
FIGURE 3.
Mycobacterium smegmatis YrlA contains a tRNA-like module that can undergo RNase P cleavage and CCA addition. (A) Map of the M. smegmatis rsr locus. (B,C) Lysates from M. smegmatis expressing Flag3-Rsr on a plasmid, or the empty vector, were subjected to immunoprecipitation using anti-Flag. YrlA and YrlB were detected by Northern blotting. In B, the oligonucleotide probes were complementary to sequences in the 3′ (lanes 1–5) or 5′ half of the RNA (lanes 6–9). (D,E) Predicted secondary structures of YrlA and YrlB. The structure of the YrlA “acceptor stem” after cleavage and CCA addition is indicated by the arrow in D. Conserved nucleotides within the GNCGAAN0-1G motif and the tRNA-like domain are colored red as in Figure 1. (F) 5′-labeled M. smegmatis YrlA was incubated with RNase P RNA under conditions that support cleavage. At the indicated times, aliquots were removed and fractionated in denaturing gels (lanes 3–7). (Arrow) Cleavage product. Lanes 1 and 2, alkaline hydrolysis and T1 ribonuclease ladders.
To determine whether the YrlA RNA was bound by M. smegmatis Rsr, we fused three copies of Flag to Rsr and expressed it under control of the tetracycline-inducible TetRO promoter (Williams et al. 2010). Immunoprecipitation with anti-Flag, followed by Northern blotting with a probe complementary to the YrlA RNA 3′ end, revealed a band of the expected ∼147 nt (Fig. 3B, lane 5). However, the probe also detected a major band at ∼ 65 nt, far shorter than expected (Fig. 3B, lane 5). Reprobing to detect the 5′ portion of YrlA revealed another prominent ∼82-nt band in the immunoprecipitate (Fig. 3B, lane 9). By synthesizing cDNAs, we discovered that the 65 and 82 nt RNAs consisted of the 3′ and 5′ halves of YrlA, respectively, and that the 65 nt RNA corresponds to the precise species expected if YrlA undergoes cleavage by RNase P, the endoribonuclease that matures tRNA 5′ ends (Fig. 4D; Kazantsev and Pace 2006). Moreover, the 82 nt RNA contains an additional CA, such that this RNA resembles tRNA in containing a CCA tail (Fig. 3D). Reprobing the blot with oligonucleotides complementary to sequences between YrlA and Rsr revealed a second Rsr-bound RNA that we call YrlB (Fig. 3C). Both RNAs were encoded upstream of Rsr and processed from a single precursor. Notably, YrlB also contains a possible T-arm (Fig. 3E).
FIGURE 4.
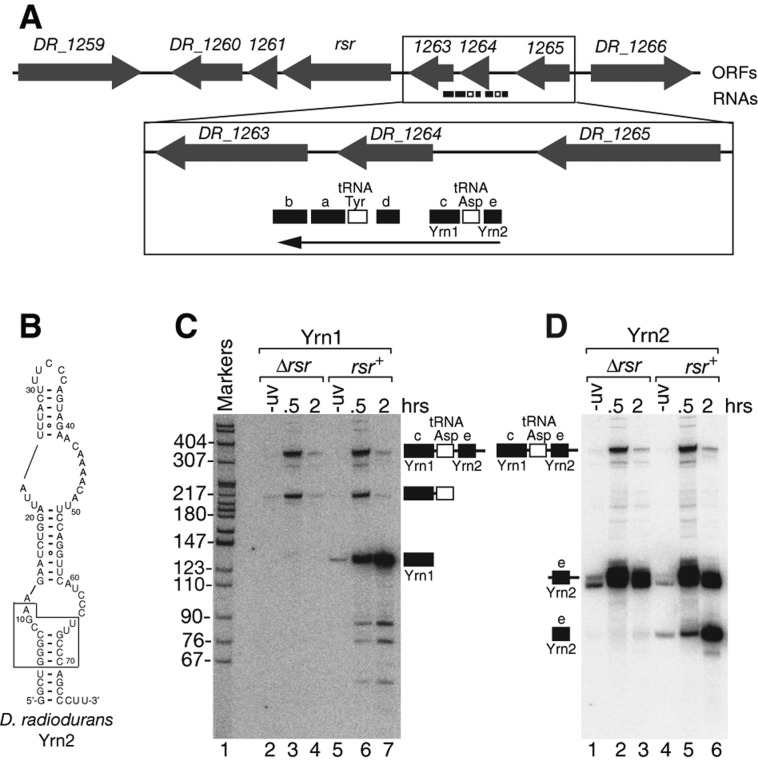
D. radiodurans contains at least one additional Y RNA. (A) The rsr locus. (B) Structure of Yrn2 predicted by Mfold. The boxed sequence, which is conserved in animal cell Y RNAs, is important for binding of Rsr to Yrn1 (Chen et al. 2013). (C,D) Wild-type and Δrsr strains were either unirradiated or irradiated with UV light as described (Chen et al. 2000). At the indicated times, aliquots were removed and the extracted RNAs subjected to Northern analysis to detect either Yrn1 (C) or Yrn2 (D).
To test if M. smegmatis YrlA could be an RNase P substrate, we incubated in vitro synthesized RNA with the catalytic subunit of E. coli RNase P under conditions that promote cleavage (Guerrier-Takada and Altman 1984). As expected if YrlA forms a tRNA-like structure, M1 RNA cleaved the RNA at the predicted site (Fig. 3F, lanes 5–7, arrow). We have not detected similar cleavage of D. radiodurans Yrn1 or S. Typhimurium YrlA either in vivo or in vitro (data not shown), most likely because only M. smegmatis YrlA contains a seven base pair “acceptor stem,” a major determinant of RNase P cleavage (Altman et al. 1993). Nonetheless, the results that M. smegmatis YrlA can be cleaved by RNase P in vitro and undergoes cleavage and CCA addition in vivo, and that YrlB also contains a T-arm, support our hypothesis that some bacterial Y RNAs contain a domain resembling tRNA.
D. radiodurans encodes at least one additional Y RNA
The finding that S. Typhimurium and M. smegmatis each contain at least two Y RNAs prompted us to reexamine D. radiodurans. Previously, we identified four small RNAs, designated a, b, c and d, that were encoded upstream of Rsr and up-regulated after ultraviolet (UV) irradiation (Fig. 4A; Chen et al. 2000). Because RNA c (here called Yrn1) was the major Rsr-bound RNA and could be folded to resemble metazoan Y RNAs, we designated this RNA as a Y RNA (Chen et al. 2000). In contrast, RNAs a, b, and d bound Rsr less strongly and their predicted structures were less similar to other Y RNAs (Supplemental Fig. S3; Chen et al. 2000), making it unclear whether they interact with Rsr in the same way as canonical Y RNAs. However, both RNA a and RNA b contain “T arms” resembling that of Yrn1 and YrlA, although the T arm of RNA a lacks the U corresponding to Ψ55 in tRNA (Supplemental Fig. S3).
Notably, during in vivo crosslinking of Rsr to RNA targets (Wurtmann and Wolin 2010), we identified another ncRNA encoded upstream of Yrn1 (RNA e) (Fig. 4A,B). Because this new RNA (unlike RNAs a, b, and d) contains a long stem, formed by base-pairing the 5′ and 3′ termini, that includes sequences important for binding of X. laevis Ro60 and D. radiodurans Rsr to Y RNAs (Fig. 4B, box; Green et al. 1998; Stein et al. 2005; Chen et al. 2013), we named this RNA Yrn2. Northern blotting revealed that Yrn2 is synthesized as a polycistronic precursor together with tRNAAsp and Yrn1 (Fig. 4C, lanes 3 and 6; Fig. 4D, lanes 2 and 5). Similar to Yrn1 (Chen et al. 2000), Yrn2 is up-regulated after UV irradiation and requires Rsr for stable accumulation (Fig. 4D, lanes 4–6). Thus, all three characterized bacterial species contain at least two Y RNAs, one of which contains a module resembling tRNA. Additionally, at least one RNA in each species folds to form a long hairpin with a small pyrimidine-rich loop (Supplemental Fig. S4, D. radiodurans Yrn2, M. smegmatis YrlB, S. Typhimurium YrlB). As in mammalian cells (Wolin et al. 2013), these other Y RNAs may allow Ro60 to form ribonucleoprotein complexes with a wider range of components.
Functional implications
Our discovery that bacterial Y RNAs contain a domain resembling tRNA both expands the inventory of tRNA mimics and raises the question of how this module contributes to YrlA and Yrn1 function. Since in both D. radiodurans and S. Typhimurium, these ncRNAs are complexed with Rsr and the PNPase exoribonuclease (Chen et al. 2013), one possibility is that the extremely stable tRNA fold protects the ncRNAs from endonucleolytic nicks that could serve as entry points for PNPase and other exoribonucleases. If M. smegmatis YrlA RNA functions similarly, CCA addition (and possibly also aminoacylation) may protect the RNase P-cleaved RNA, as mature tRNAs are not normally substrates for bacterial exoribonucleases (Li et al. 2002). In this scenario, differences between bacterial and animal cell nucleases and/or the greater subcellular compartmentalization of animal cells may obviate the need for a tRNA-like module, as this domain is not apparent in identified metazoan Y RNAs (Supplemental Fig. S4 and our observations). A general role in protecting bacterial Y RNA from nucleases would also be consistent with findings that features of the tRNA-like module differ between YrlA RNAs and Yrn1, but would not account for the observation that sequences within both the D and T stems that are not involved in maintaining tRNA tertiary structure (Dirheimer et al. 1995) are nearly invariant in YrlA RNAs (Figs. 1, 2).
A second possibility is that the tRNA module is important for interaction of these RNAs with other components, such as PNPase or ribosomes. Although biochemical and structural experiments suggest that the S1/KH domains of PNPase bind the end of Yrn1 containing the T arm (Chen et al. 2013), experiments in which we deleted the T arm from D. radiodurans Yrn1 did not reveal significant differences in complex assembly (X Chen and SL Wolin, unpubl.). Additionally, although D. radiodurans Rsr sediments with partially degraded ribosomal subunits during prolonged growth in stationary phase, this association does not require Yrn1 RNA (Wurtmann and Wolin 2010). Thus, while we favor the idea that the tRNA-like module is involved in protein–RNA or RNA–RNA interactions, we do not yet know the identity of the potential interactors.
To date, the best-characterized ncRNA that resembles tRNA is tmRNA, which rescues stalled ribosomes by functioning as both a tRNA and an mRNA (Moore and Sauer 2007; Keiler and Feaga 2014). Although tmRNA and YrlA RNAs each contain a pseudouridine-containing T arm, tmRNA also contains an acceptor stem that undergoes aminoacylation, allowing it to bind the translation elongation factor EF-Tu, enter ribosomes with the SmB protein and act as an acceptor for the stalled polypeptide (Moore and Sauer 2007; Keiler and Feaga 2014). Although M. smegmatis YrlA, which undergoes RNase P cleavage and CCA addition, could potentially be aminoacylated, we have not detected cleavage of S. Typhimurium YrlA or D. radiodurans Yrn1 at the analogous site, making it unlikely that the ability to undergo aminoacylation is a general feature of these RNAs. However, it is possible that all three Y RNAs interact with ribosomes, translation factors, or other tRNA-interacting proteins to influence cell metabolism.
Consistent with a separate evolutionary lineage, both the Ro60-binding and tRNA-like modules of D. radiodurans Yrn1 differ from those of YrlA RNAs. The Ro60-binding modules of the D. radiodurans RNAs more closely resemble metazoan Y RNAs in that both strands of the helix are retained, while only the 5′ strand is recognizable in the S. Typhimurium and M. smegmatis RNAs (Supplemental Fig. S4). Also, while both Yrn1 and YrlA RNAs contain domains resembling tRNAs, the YrlA domain mimics the tRNA D and T arms and acceptor stem, as DusA and TruB modify the expected nucleotides in S. Typhimurium YrlA, and RNase P and the CCA-adding enzyme recognize M. smegmatis YrlA. In contrast, we did not detect dihydrouridine in the Yrn1 “D loop,” making it unclear if this stem–loop forms and/or resembles a D arm in vivo. The orientation of the predicted T and D arms with respect to the RNA 5′ ends also differs between YrlA RNAs and Yrn1, as does whether the stem–loop between these arms occupies the position of the tRNA acceptor stem or anticodon arm (Fig. 2, Supplemental Fig. S4). Thus, while both YrlA and Yrn1 RNAs may have originated from tRNAs, they appear to have evolved independently.
Although our studies identified potential Y RNAs in >250 bacteria and phages, we are certain that more RNAs remain to be found. Because our characterization of Ro60-associated RNAs in all three bacteria identified RNAs that eluded bioinformatic discovery, similar immunoprecipitation experiments will likely identify additional Y RNAs in other species. As there were also Ro60-containing bacteria for which we failed to identify a strong YrlA candidate (Supplemental Table S1), we expect that Y RNAs with sequence and structural variations will be uncovered in additional searches. Conversely, some predicted RNAs, despite their homology with characterized YrlA RNAs, may not be expressed or may only be expressed under specific stress conditions. We also consider it likely that our definition of Y RNAs (which currently does not include the three other ncRNAs encoded near D. radiodurans Rsr) will expand as more RNAs are characterized.
Finally, we note that although Y RNAs were first identified in human cells, the functions of these RNAs have been best characterized in a single bacterium. Our identification of these RNAs in a wide range of bacteria, including many that are far more amenable to genetic manipulation than D. radiodurans, lays the groundwork for a host of studies that should vastly expand our understanding of these long enigmatic ncRNAs.
MATERIALS AND METHODS
Bacterial strains and growth conditions
D. radiodurans R1 was grown at 30°C in TGY (0.8% tryptone, 0.1% glucose, 0.4% yeast extract). S. Typhimurium LT2 was grown at 37°C in LB medium. M. smegmatis MC2 155 was grown at 37°C in Middlebrook 7H9 liquid media supplemented with 10% albumin-dextrose complex (ADC), 0.05% Tween 80, 0.1 M CaCl2, 100 μg/mL carbenicillin, and 100 μg/mL cycloheximide. To construct the D48C mutation in S. Typhimurium LT2 TruB, the 5′ portion of truB, with ∼160 bp of 5′ flanking sequence, was amplified from genomic DNA using primers 5′-GCTTGCTGGGTAAAGCGATG-3′ and 5′-CAACATGCCGGTCGCCAGCGGGCACAGCGCGCCGGTATGCCCGGCGCGGTTG-3′. This mutated fragment was joined with the 3′ portion of truB and 215 bp of 3′ flanking sequence amplified from genomic DNA using primers 5′-CCCGCTGGCGACCGGCATGTTG-3′ and 5′-GTGTCGTTTGCGTCACGACCG-3′. The resulting DNA was inserted into the Sma I site of pSB890 (Palmer et al. 1998). Subsequent genetic manipulations were as described (Chen et al. 2013). A similar strategy was used to generate the K140A mutation in DusA.
To construct the M. smegmatis strain carrying Flag3-Rsr, rsr (MSMEI_1162) was amplified from genomic DNA using 5′-GTGGACATCCTCAAGACCATTC-3′ and 5′-CGGGAAGCTTCTAGATGTCGCCGCGTGAG-3′, fused to three copies of Flag and cloned into the BamHI/HindIII sites of pKW08-Lx (Williams et al. 2010). The resulting plasmid pKW08-Flag-MSRsr1 (called pFlag3-Rsr in Fig. 4) was transformed into M. smegmatis, generating a strain that expressed Flag-tagged Rsr under control of the TetRO promoter. Genetic manipulations were as described (van Kessel and Hatfull 2008).
Detection of modified nucleotides
Pseudouridine mapping was performed essentially as described (Ofengand et al. 2001). Briefly, total RNA was incubated in 167 mM CMC in BEU buffer (50 mM Bicine, pH 8.5, 4 mM EDTA, 7 M urea) at 37°C for 20 min. After precipitating with ethanol and washing with 70% ethanol, the pellet was dissolved in 50 mM sodium carbonate buffer (pH 10.4), 2 mM EDTA and incubated at 37°C for 4 h. The modified RNA was subjected to primer extension using SuperScript III Reverse Transcriptase (Invitrogen) and 5′ labeled oligonucleotide primers 5′-AGGGTGGCGACAAGTTTATGTG-3′ (S. Typhimurium YrlA) and 5′-AGTGCTCTGGACAAGGGTTCGGG-3′ (D. radiodurans Yrn1). The resulting cDNAs were extracted with phenol:chloroform:isoamyl alcohol (25:24:1), precipitated with ethanol and fractionated in 8% polyacrylamide/8.3 M urea gels. Dihydrouridines were detected as described (Xing et al. 2004). Total RNA was incubated with 0.1 N KOH at 37°C for 2–30 min, and neutralized by adding 5× First-Strand Buffer (Invitrogen). The RNA was then mapped by primer extension as described above.
Northern blotting
Total RNA was isolated from bacteria using hot acid phenol as described (Chen et al. 2000). Northern blotting was as described (Chen et al. 2007). Oligonucleotide probes were as follows: 5′-GCTCGGTGATGTGGGCGTGTGAGGTAT-3′ (M. smegmatis YrlA-3′), 5′-GACCATTCCATTAGAAGTGGTAA-3′ (M. smegmatis YrlA-5′), 5′-AGTCGAACCTTGAAGTCGCCAGATA-3′ (M. smegmatis YrlB), 5′-CACCGCTTCCGCCCATTGAGGTTGG-3′ (D. radiodurans Yrn1), 5′-ATGTTTTGTTCTACTGGGAAAGTAAATAA-3′ (D. radiodurans Yrn2).
RNA structure probing
After using PCR to place S. Typhimurium YrlA and D. radiodurans Yrn1 RNA sequences behind the T7 promoter, RNAs were transcribed with T7 polymerase. For enzymatic probing, 5′-labeled RNA was first refolded by heating to 65°C for 3 min in 20 mM HEPES (pH 7.5), 50 mM NaCl and 5 mM MgCl2, followed by slow cooling to room temperature. For YrlA, 23 fmol RNA was incubated in the same buffer with 1.25 units of RNase T1 (Sigma), 0.32 units RNase T2 (Invitrogen), or 0.00037 units of RNase V1 for 10 min. For Yrn1, 2.2 pmol of D. radiodurans Rsr (purified from baculovirus [Chen et al. 2013]) was mixed with 23 fmol refolded RNA in 20 mM HEPES pH 7.5, 50 mM NaCl, and 5 mM MgCl2. After incubating at room temperature for 30 min and on ice for 30 min, RNases were added as described above and the reactions incubated on ice for 10 min. Afterward, RNAs were extracted with phenol:chloroform:isoamyl alcohol (25:24:1), precipitated with ethanol and fractionated in 8% polyacrylamide/8.3 M urea gels.
Identification of M. smegmatis Y RNAs
M. smegmatis carrying pKW08-Flag-MSRsr1 or the parent pKW08-Lx vector were grown to mid-log phase (∼1 OD600) and Rsr expression induced by adding 50 ng/mL of tetracycline to the media for 2.5 h. After harvesting, cells were lysed using a French press in 20 mM Tris–HCl pH 7.5, 100 mM NaCl, 5% glycerol, 1 mM MgCl2, 2 mM β-mercaptoethanol, 0.5 mM phenylmethanesulfonyl fluoride (PMSF), 1.25 mM vanadyl ribonucleoside complexes, and 1× protease inhibitor cocktail (Roche Applied Science). Anti-Flag immunoprecipitations were as described (Chen et al. 2013). For cDNA synthesis, RNAs were ligated to RNA linkers L5 (5′-OH AGGGAGGACGAUGCGG 3′-OH) and L3 (5′-P GTGTCAGTCACTTCCAGCGG-3′-puromycin), reverse transcribed with DNA oligomer P3 (5′-CCGCTGGAAGTGACTGACAC-3′) and amplified with P3 and P5 (5′-AGGGAGGACGATGCGG-3′) (Ule et al. 2005). RNA ends after cleavage and CCA addition were determined by amplifying with 5′-GAGCTCGGTGATGTGGGCGTGTG-3′ and P5 (to obtain the 5′ end of the YrlA 65 nt fragment) and with 5′-CACTTCTAATGGAATGGTCGC-3′ and P3 (to obtain the 3′ end of the 82 nt fragment).
RNase P cleavage of M. smegmatis YrlA
DNA containing YrlA behind the T7 promoter was amplified from genomic DNA using 5′-CATGACTAGTAATACGACTCACTATAGGGCGCTTGGTGATGTGAGCCG-3′ and 5′-GAGCTCGGTGATGTGGGCG-3′. YrlA RNA was synthesized from this template using T7 RNA polymerase (Promega) according to the manufacturer's instructions. RNase P M1 RNA was synthesized with T7 RNA polymerase using plasmid pJA2′ (a gift of Dr. S. Altman, Yale University) that was linearized with Fok I. The RNase P assay was as described (Guerrier-Takada and Altman 1984). Briefly, 187 nM of M1 RNA and 1 nM of 5′ 32P-labeled YrlA RNA were each suspended separately in RNase P cleavage buffer (50 mM Tris–HCl, pH 7.5, 100 mM NH4Cl, 100 mM MgCl2), incubated at 55°C for 5 min, and slow-cooled to 37°C. The two RNAs, together with 0.7 units/μL RNaseOUT RNase inhibitor, were then combined and incubated at 37°C.
Bioinformatic analyses
Following initial primary sequence homology searches using BLASTN, the sequences in Figure 1D were aligned using Clustal Omega (Goujon et al. 2010; Sievers et al. 2011) and the alignment used to build a consensus secondary structure model using Infernal (version 1.1) (Nawrocki and Eddy 2013) (Supplemental text 1). After collecting genome sequences for bacteria that were annotated in Genbank as possessing a TROVE-domain protein, we discarded redundant sequences and genome fragments that contained <400 bp flanking the presumptive Ro60. This yielded sequences from 303 bacterial species, 119 of which were complete genomes. Because some genomes encode two TROVE-domain proteins, a total of 307 possible Ro60 orthologs were present. Although our initial search identified YrlA RNAs in most searched phage genomes (Supplemental Table S2) relatively few YrlA RNAs were identified in cyanobacteria and δ-proteobacteria, and some of these RNAs contained a longer “acceptor stem” (e.g., Stanieria cyanosphaera) (Supplemental Fig. S1). To identify additional RNAs, a second covariance model was built that included these sequences (Supplemental text 2). Potential YrlA RNAs found using this second search with E-value <0.00005 are summarized in Supplemental Table S1. As dimeric tRNAs could potentially score as false-positives (with the T stem of the first tRNA and the D stem of the second tRNA appearing as a YrlA RNA-related sequence), all sequences that overlapped annotated tandem tRNA genes were discarded. To have the highest confidence data set, only YrlA-related sequences that were both the best matches to the Infernal model in the genome or partial genome search and were located within 4 kb of the Ro60 ortholog were reported. In those cases where there were multiple YrlA-like sequences with E-values <0.00005 within 4 kb of Rsr, all sequences were reported. In this way, 355 putative YrlAs were identified in 254 bacteria and 22 bacteriophages (Supplemental Tables S1 and S2).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Casey Fowler and Jorge Galan for S. Typhimurium SL1344 RNA and the LT2 strain, Sidney Altman for the pJA2′ plasmid, Eric Phizicky for advice, and Walter Moss and Andrei Alexandrov for comments on the manuscript. E.J.W. was supported by a National Science Foundation Predoctoral Fellowship and A.F. was supported by a Gruber Foundation Science Fellowship. This work was supported by National Institutes of Health (NIH) grant R01 GM073863 to S.L.W.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.047241.114.
REFERENCES
- Altman S, Kirsebom L, Talbot S. 1993. Recent studies of ribonuclease P. FASEB J 7: 7–14 [DOI] [PubMed] [Google Scholar]
- Bakin A, Ofengand J. 1993. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32: 9754–9762 [DOI] [PubMed] [Google Scholar]
- Bateman A, Kickhoefer VA. 2003. The TROVE module: a common element in telomerase, Ro and vault ribonucleoproteins. BMC Bioinformatics 4: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AC, Xu J, Johnson RC, Schimmel P, de Crecy-Lagard V. 2002. Identification of the tRNA-dihydrouridine synthase family. J Biol Chem 277: 25090–25095 [DOI] [PubMed] [Google Scholar]
- Bjork GR, Ericson JU, Gustafsson CE, Hagervall TG, Jonsson YH, Wikstrom PM. 1987. Transfer RNA modification. Annu Rev Biochem 56: 263–287 [DOI] [PubMed] [Google Scholar]
- Chen X, Quinn AM, Wolin SL. 2000. Ro ribonucleoproteins contribute to the resistance of Deinococcus radiodurans to ultraviolet irradiation. Genes Dev 14: 777–782 [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wurtmann EJ, Van Batavia J, Zybailov B, Washburn MP, Wolin SL. 2007. An orthologue of the Ro autoantigen functions in 23S rRNA maturation in D. radiodurans. Genes Dev 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Taylor DW, Fowler CC, Galan JE, Wang HW, Wolin SL. 2013. An RNA degradation machine sculpted by Ro autoantigen and noncoding RNA. Cell 153: 166–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das U, Shuman S. 2013. 2′-Phosphate cyclase activity of RtcA: a potential rationale for the operon organization of RtcA with an RNA repair ligase RtcB in Escherichia coli and other bacterial taxa. RNA 19: 1355–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirheimer G, Keith G, Dumas P, Westhof E. 1995. Primary, secondary, and tertiary structures of tRNAs. In tRNA: structure, biosynthesis and function (ed. Soll D, RajBhandary UL), pp. 93–126. ASM Press, Washington, DC [Google Scholar]
- Fuchs G, Stein AJ, Fu C, Reinisch KM, Wolin SL. 2006. Structural and biochemical basis for misfolded RNA recognition by the Ro protein. Nat Struct Mol Biol 13: 1002–1009 [DOI] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Res 38: W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CD, Long KS, Shi H, Wolin SL. 1998. Binding of the 60-kDa Ro autoantigen to Y RNAs: evidence for recognition in the major groove of a conserved helix. RNA 4: 750–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. 2008. The Vienna RNA Websuite. Nucleic Acids Res 36: W70–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C, Altman S. 1984. Catalytic activity of an RNA molecule prepared by transcription in vitro. Science 223: 285–286 [DOI] [PubMed] [Google Scholar]
- Gutgsell N, Englund N, Niu L, Kaya Y, Lane BG, Ofengand J. 2000. Deletion of the Escherichia coli pseudouridine synthase gene truB blocks formation of pseudouridine 55 in tRNA in vivo, does not affect exponential growth, but confers a strong selective disadvantage in competition with wild-type cells. RNA 6: 1870–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AV, Pace NR. 2006. Bacterial RNase P: a new view of an ancient enzyme. Nat Rev Microbiol 4: 729–740 [DOI] [PubMed] [Google Scholar]
- Keiler KC, Feaga HA. 2014. Resolving nonstop translation complexes is a matter of life or death. J Bacteriol 196: 2123–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn M, Pazaitis N, Huttelmaier S. 2013. Why Y RNAs? About versatile RNAs and their functions. Biomolecules 3: 143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Boyle JA, Hardin JA, Steitz JA. 1981. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science 211: 400–402 [DOI] [PubMed] [Google Scholar]
- Levitt M. 1969. Detailed molecular model for transfer ribonucleic acid. Nature 224: 759–763 [DOI] [PubMed] [Google Scholar]
- Li Z, Reimers S, Pandit S, Deutscher MP. 2002. RNA quality control: degradation of defective transfer RNA. EMBO J 21: 1132–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SD, Sauer RT. 2007. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem 76: 101–124 [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29: 2933–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofengand J, Del Campo M, Kaya Y. 2001. Mapping pseudouridines in RNA molecules. Methods 25: 365–373 [DOI] [PubMed] [Google Scholar]
- Palmer LE, Hobbie S, Galan JE, Bliska JB. 1998. YopJ of Yersinia pseudotuberculosis is required for the inhibition of macrophage TNF-α production and downregulation of the MAP kinases p38 and JNK. Mol Microbiol 27: 953–965 [DOI] [PubMed] [Google Scholar]
- Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, et al. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113: 171–182 [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK. 2010. tRNA biology charges to the front. Genes Dev 24: 1832–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh A, Savva CG, Holzenburg A, Sacchettini JC. 2007. Crystal structure of Rsr, an ortholog of the antigenic Ro protein, links conformational flexibility to RNA binding activity. J Biol Chem 282: 14960–14967 [DOI] [PubMed] [Google Scholar]
- Savage DF, de Crecy-Lagard V, Bishop AC. 2006. Molecular determinants of dihydrouridine synthase activity. FEBS Lett 580: 5198–5202 [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, Wolin SL. 2011. Emerging roles for the Ro 60 kDa autoantigen in noncoding RNA metabolism. WIREs RNA 2: 686–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, Weinberg DE, Fuchs G, Choi K, Chung J, Wolin SL. 2009. The subcellular distribution of an RNA quality control protein, the Ro autoantigen, is regulated by noncoding Y RNA binding. Mol Biol Cell 20: 1555–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM. 2005. Structural insights into RNA quality control: The Ro autoantigen binds misfolded RNAs via its central cavity. Cell 121: 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB. 2005. CLIP: a method for identifying protein–RNA interaction sites in living cells. Methods 37: 376–386 [DOI] [PubMed] [Google Scholar]
- van Kessel JC, Hatfull GF. 2008. Mycobacterial recombineering. Methods Mol Biol 435: 203–215 [DOI] [PubMed] [Google Scholar]
- Williams KJ, Joyce G, Robertson BD. 2010. Improved mycobacterial tetracycline inducible vectors. Plasmid 64: 69–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Belair C, Boccitto M, Chen X, Sim S, Taylor DW, Wang HW. 2013. Non-coding Y RNAs as tethers and gates: insights from bacteria. RNA Biol 10: 1602–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtmann EJ, Wolin SL. 2010. A role for a bacterial ortholog of the Ro autoantigen in starvation-induced rRNA degradation. Proc Natl Acad Sci 107: 4022–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing F, Hiley SL, Hughes TR, Phizicky EM. 2004. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J Biol Chem 279: 17850–17860 [DOI] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.