Abstract
Micronucleation of chromosomal DNA is an effective indicator of DNA damage and micronucleus (MN) analysis is a valuable tool for radiation biodosimetry studies. To gain a comprehensive knowledge of micronucleation process after ionising radiation (IR) exposure, whole genome-wide chromosome analysis is desirable. With this objective, multicolour fluorescence in situ hybridization (M-FISH) technique was utilised in the present study to characterise the chromosome content of spontaneous and IR-induced micronuclei in three human donors. M-FISH analysis revealed a radiation dose-dependant increase in the number of micronuclei with multi-chromosome material above 2 Gy and as many as 3–6 multicolour signals were detected in micronuclei after high γ-rays radiation doses (5–10 Gy). Involvement of each human chromosome material was more frequently detected in multicoloured micronuclei than in single-coloured micronuclei at high radiation doses (>2 Gy). Observation of dose-dependant increase in the MN frequency with multi-chromosome material may be due to misrepair of DNA double-strand breaks involving multiple chromosomes leading to asymmetric dicentric or ring chromosomes and acentric fragments. Chromosomes belonging to groups A (1, 2 and 3) and B (4 and 5) were frequently detected in 35–45% of the total micronuclei either as single entities or in combination with other chromosomes. Among the A and B groups, chromosome 1 material was consistently detected at high MN frequencies after radiation exposure in all the donors. Additionally, chromosomes 13 and 19 were more frequently observed in micronuclei than the expected frequency based on DNA content. Our whole genome approach utilising the M-FISH technique revealed that MN formation at high radiation doses might be complex involving multiple chromosome fragments. Understanding the fate and biological consequences of these multi-chromosome-containing micronuclei may provide key molecular insights for some aspects of IR-induced genomic instability and cancer development processes.
Introduction
The micronucleus (MN) assay has been extensively used both in vitro and in vivo for estimating the toxicity of aneugens and clastogens in diverse biological systems including humans (1,2). Micronuclei originate either from whole chromosomes or chromosome fragments (interstitial and terminal) due to exclusion from mitotic spindle. In case of radiation exposure, MN formation is thought to arise from asymmetric dicentric or ring chromosomes and acentric fragments which are generated by misrepair of DNA double-strand breaks (DSB). Micronuclei contain small chromatin bodies whose size ranges from 1/16 to 1/3 of the main nucleus. Current MN methodology known as cytokinesis block micronuclei, involves the use of cytochalasin B, which blocks cytokinesis after nuclear division such that micronuclei are analysed in binucleated cells after one cell cycle division (3,4). Since micronuclei are composed of either whole chromosomes or chromosome fragments, fluorescence in situ hybridization (FISH) technique using centromeric, telomeric and chromosome-specific DNA probes was successfully utilised in the past to characterise the chromosomal constitution of micronuclei.
Induction of a non-random distribution of chromosome breaks has been observed after in vitro exposure of cells to chemicals. In corroboration, preferential involvement of specific chromosomes in MN formation has been demonstrated by FISH technique. Inclusion of chromosomes 1, 9, 15, 16 and Y in MN formation has been demonstrated after 5-azacytidine treatment (5). Specific inclusion of chromosomes 9 and 16 has been reported in mitomycin C (MMC)-induced MN formation (6). Hovhannisyan et al. (7) using whole chromosome painting probes for 3, 4, 6, 7, 9, 16, 17, 18 and X, demonstrated that the chromosomes 9 and 16 were more frequently involved in MMC-induced micronuclei. MN formation with chromosome 8 was found to occur at a higher frequency than chromosome 7 after cellular exposure to 1,2,4-benzenetriol (8). Using chromosome-specific probes for 1, 7, 11, 14, 17 and 21, Fimognari et al. (9) reported that the inclusion of chromosomes into micronuclei after radiation exposure depended on the DNA content of the respective chromosomes. In the above-mentioned studies, only two colour FISH was utilised and two colour FISH has a technical limitation in detecting micronuclei containing more than two chromosomes. Multicolour FISH (M-FISH) and spectral karyotyping (SKY) techniques may be useful in detecting micronuclei involving multiple chromosomes but only a few studies have utilised these techniques. The SKY technique detected high frequency of X chromosome material in the baseline micronuclei and the frequency was found to be 12.2, 50.6 and 7.1% in 28, 42 and 72 years old female donors, respectively (10). Using the SKY technique, Lloyd et al. (11) detected the involvement of specific chromosomes in baseline micronuclei (9, 19, 22 and X) and nitrosamine 4-(methylnitrosoamine)-1-(3-pyridyl)-1 butanone (NNK)-induced micronuclei (3, 4 and 16). Interestingly, chromosomes 1, 13 and 17 were found to be involved in both baseline and NNK-induced micronuclei.
Ionising radiation (IR) induces a wide spectrum of lesions including DNA single-strand breaks, DSB, base damage and DNA–protein crosslinks. Among them, DSB are considered to be lethal as unrepaired or misrepaired DSB can either result in cell killing or in stable chromosomal aberrations depending on radiation quality, dose and dose rate. IR-induced DSB can occur either in a random or non-random fashion. A non-random distribution of DSB was demonstrated after high LET particle irradiation (12). Additionally, DSB induction also shown to be influenced by higher order chromatin organisation (13). Further, dependence of MN formation on radiation dose has made it an effective biomarker for estimating the absorbed radiation dose in large-scale population monitoring scenarios after radiological accidents. Earlier studies using single and two colour FISH have identified a few chromosomes in MN formation (5–9) but this technique is highly restrictive because it enables the analysis of only two chromosomes at a time. Identification of chromosomes that are specifically excluded into micronuclei under spontaneous and induced DNA damage conditions may provide valuable mechanistic insights for genomic instability and cancer development processes. With this objective, M-FISH technique was employed in the present study for the first time to characterize the chromosome content of micronuclei induced by different doses of γ-rays radiation in human peripheral blood lymphocytes. Such an approach may also determine whether or not there is any preferential involvement of specific chromosomes in IR-induced MN formation.
Materials and methods
Cell culture and irradiation
Blood samples were obtained from three healthy volunteers [1 female (35 years) and 2 males (25 and 27 years)] and the blood collection was performed in strict compliance with the Institutional Review Board (IRB) protocol (IRB-AAAE9551). Blood aliquots (550 μl) were irradiated with different doses of γ-rays (1–10 Gy, dose rate 0.82 Gy/min) using the Cs137 irradiator (Gamma Cell 40; Atomic Energy of Canada, Canada). Lymphocytes from unirradiated and irradiated samples were isolated using Histopaque-1077 (Sigma, St Louis, MO, USA) and the lymphocyte cultures were set up using the standard procedure (14). Cultures were treated with Cytochalasin B (6 μg/ml; Sigma) 44h after the initiation of cultures with phytohemagglutinin (PHA) stimulation and the cells were subsequently grown in the presence of cytochalasin B for an additional 26h. Cells harvested after 72h (from the start of stimulation with PHA) were treated with hypotonic solution (0.56% KCl) for 10min and fixed in three changes of ice cold acetic acid:methanol (1:3) solution. An aliquot of fixed cells (25–30 μl) was gently dropped onto glass sides and air-dried for M-FISH analysis.
Multicolour FISH
M-FISH was performed essentially according to the manufacturer’s protocol (Meta Systems, Boston, MA, USA). Briefly, slides were treated for 1min with 0.001% acidic pepsin solution (0.01 N HCl) at 37°C for 1–2min followed by two washes of 5min each in phosphate-buffered saline (PBS). The slides were post-fixed for 10min in a solution of formaldehyde/MgCl2 (1% formaldehyde/50mM MgCl2 in PBS). The slides after denaturation (2X SSC at 70°C for 20min and after cooling to ambient temperature 1min in 0.07 N NaOH) were dehydrated in graded series of ethanol (30, 70, 90 and 100%) and air dried. The M-FISH probe mixture was denatured separately by incubation at 75°C for 5min followed by incubation at 37°C for 30min to allow the annealing of repetitive DNA sequences. An aliquot of 10 μl probe was placed on the slide and covered with a coverslip. The slides were kept in a humidified hybridisation chamber at 37°C for at least 72h. The unbound probe was removed by washing the slides in pre-warmed (75°C) 1X SSC (pH 7.0–7.5) for 5min followed by incubation in 4X SSCT (4X SSC with 0.1% Tween 20) for 5min. Indirectly labeled probe (Cy5), if needed, was amplified by incubation with antibodies (biotinylated anti-streptavidin and Cy5 conjugated streptavidin; Invitrogen, Carlsbad, CA, USA) sequentially for 30min followed by two washes of 3min each in 4X SSCT and in PBS. The nuclei were counterstained with DAPI (Vectashield Laboratories, Burlingame, CA, USA). Images were captured using the Nikon epifluorescence microscope. Image analysis was performed using the ISIS software (Meta Systems, Boston, MA, USA) essentially according to the published procedure (15). Individual human chromosomes involved in MN formation were identified by unique chromosome-specific processed colour generated by ISIS software based on the combinatorial labeling of five fluorochromes (FITC, Spectrum Orange, Texas Red, DEAC and Cy5) and their pixel intensities.
FISH with whole chromosome-specific probes
Whole chromosome painting probes (WCP) directly labeled with Texas red (chromosome 1) and FITC (chromosomes 4 and 8) were purchased from Rainbow Scientific, Windsor, CT, USA. Procedures for FISH, post-hybridization washing and signal detection were performed essentially following the manufacturer’s protocol. Images were captured and analysed using a Nikon epifluorescence microscope. The WCP were used on the metaphase spreads to verify their specificity.
Results
Spontaneous and γ-rays induced MN frequency in the peripheral blood lymphocytes of healthy donors
MN frequencies observed in the peripheral blood lymphocytes of three donors after different radiation doses are summarized in Table I. The baseline frequency of micronuclei per binucleated cell was 0.049±0.01 for donor 1, 0.012±0.003 for donor 2 and 0.008±0.003 for donor 3. In corroboration with earlier studies (16,17), baseline MN frequency was more in the female donor relative to the two male donors. In all the donors, a dose-dependant increase in MN frequency was observed up to 5 Gy of γ-rays. In both male donors, MN frequency declined at 7.5 and 10 Gy of γ-rays probably owing to both impaired proliferation of lymphocytes and elimination of heavily damaged cells through apoptosis.
Table I.
Frequencies of micronuclei in cytochalasin B-blocked binucleate cells
| Dose | No. of Bi | MN (0) | MN (1) | MN (2) | MN (3) | MN (4) | MN (>4) | Total MN | MN/Bi | SEM | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor 1 | 0 Gy | 502 | 482 | 15 | 5 | 0 | 0 | 0 | 25 | 0.049 | 0.010 |
| 1 Gy | 502 | 423 | 61 | 10 | 7 | 1 | 0 | 106 | 0.211 | 0.023 | |
| 2 Gy | 502 | 365 | 106 | 25 | 6 | 0 | 0 | 174 | 0.346 | 0.030 | |
| 3 Gy | 502 | 285 | 153 | 47 | 16 | 1 | 0 | 299 | 0.595 | 0.044 | |
| 4 Gy | 502 | 221 | 172 | 71 | 28 | 9 | 1 | 439 | 0.874 | 0.057 | |
| 5 Gy | 502 | 150 | 177 | 92 | 63 | 12 | 8 | 645 | 1.284 | 0.076 | |
| Donor 2 | 0 Gy | 1253 | 1239 | 13 | 1 | 0 | 0 | 0 | 15 | 0.012 | 0.003 |
| 1 Gy | 1126 | 990 | 120 | 12 | 4 | 0 | 0 | 156 | 0.139 | 0.012 | |
| 2 Gy | 959 | 664 | 204 | 76 | 10 | 5 | 0 | 406 | 0.423 | 0.025 | |
| 3 Gy | 789 | 496 | 204 | 58 | 26 | 5 | 0 | 418 | 0.530 | 0.032 | |
| 4 Gy | 920 | 455 | 263 | 133 | 54 | 13 | 2 | 753 | 0.818 | 0.040 | |
| 5 Gy | 764 | 368 | 191 | 136 | 50 | 17 | 2 | 691 | 0.904 | 0.047 | |
| 7.5 Gy | 446 | 304 | 66 | 36 | 18 | 18 | 4 | 284 | 0.637 | 0.048 | |
| 10 Gy | 404 | 346 | 17 | 15 | 9 | 10 | 7 | 150 | 0.371 | 0.036 | |
| Donor 3 | 0 Gy | 1178 | 1169 | 9 | 0 | 0 | 0 | 0 | 9 | 0.008 | 0.003 |
| 1 Gy | 1290 | 1144 | 129 | 16 | 1 | 0 | 0 | 164 | 0.127 | 0.011 | |
| 2 Gy | 1411 | 1068 | 249 | 72 | 18 | 4 | 0 | 463 | 0.328 | 0.018 | |
| 3 Gy | 1064 | 690 | 250 | 95 | 20 | 7 | 2 | 538 | 0.506 | 0.027 | |
| 4 Gy | 834 | 455 | 183 | 110 | 63 | 15 | 8 | 693 | 0.831 | 0.043 | |
| 5 Gy | 796 | 363 | 221 | 114 | 76 | 14 | 8 | 773 | 0.971 | 0.049 | |
| 7.5 Gy | 557 | 359 | 75 | 47 | 42 | 20 | 14 | 449 | 0.806 | 0.051 | |
| 10 Gy | 520 | 463 | 27 | 6 | 8 | 8 | 8 | 141 | 0.271 | 0.026 |
M-FISH analysis of the chromosomal content of spontaneous MN in healthy donors
MN analysis performed in cytokinesis-blocked binucleated cells by M-FISH (Table II) revealed a substantial inter-individual variability in the chromosomal content of spontaneous micronuclei among the donors. Representative pictures of M-FISH are shown in Figure 1. In the female donor (donor 1), a single uniform colour was observed in 77.31% of the micronuclei involving each of the human chromosomes at varying frequencies, while the rest contained material from two or more chromosomes. In donor 1, chromosomes 4 (6.26%), 5 (8.17%), 9 (7.80%), 10 (9.80%), 13 (5.80%) and X (8.54%) were found at high frequencies in micronuclei. In one of the male donors (donor 2), 67.85% of the total micronuclei showed a uniform colour indicative of the presence of each chromosome material with the exception of chromosome 11. A total of six chromosomes [1 (12.17%), 2 (10.86%), 5 (5.48%), 6 (5.37%), 9 (5.48%) and 13 (10.86%) was detected at high frequencies in donor 2. In donor 3, only 57.14% of the total micronuclei contained single chromosome material. Further, chromosomes 3, 13, 18 and 22 were not detected in micronuclei. Chromosomes 1 (9.12%), 4 (10.25%), 6 (5.12%), 8 (5.12%), 10 (5.12%), 11 (10.25%), 12 (12.50%) and 19 (5.12%) were found at high frequencies in micronuclei. Among the chromosomes that were detected at high frequencies in micronuclei, chromosomes 1, 4 and 6 were common in both the male donors.
Table II.
Number of micronuclei analysed by M-FISH in the lymphocytes of three human donors
| Donor 1 | Donor 2 | Donor 3 | Total | |
|---|---|---|---|---|
| 0 Gy | 118 (87) | 5 (46) | 28 (26) | 203 |
| 1 Gy | 112 (88) | 107 (81) | 56 (47) | 275 |
| 2 Gy | 62 (33) | 118 (96) | 116 (71) | 296 |
| 3 Gy | 195 (110) | 119 (91) | 135 (89) | 449 |
| 4 Gy | 188 (79) | 195 (118) | 238 (182) | 621 |
| 5 Gy | 205 (76) | 208 (140) | 151 (67) | 564 |
| 7.5 Gy | 114 (65) | 186 (102) | 300 | |
| 10 Gy | 123 (71) | 124 (55) | 247 |
Numbers in parentheses indicate the micronucleated binucleate cells analysed by M-FISH.
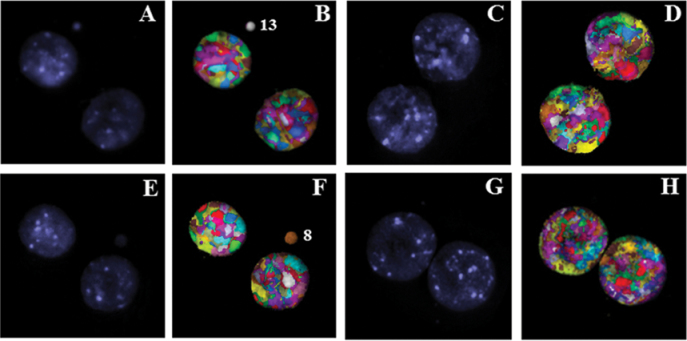
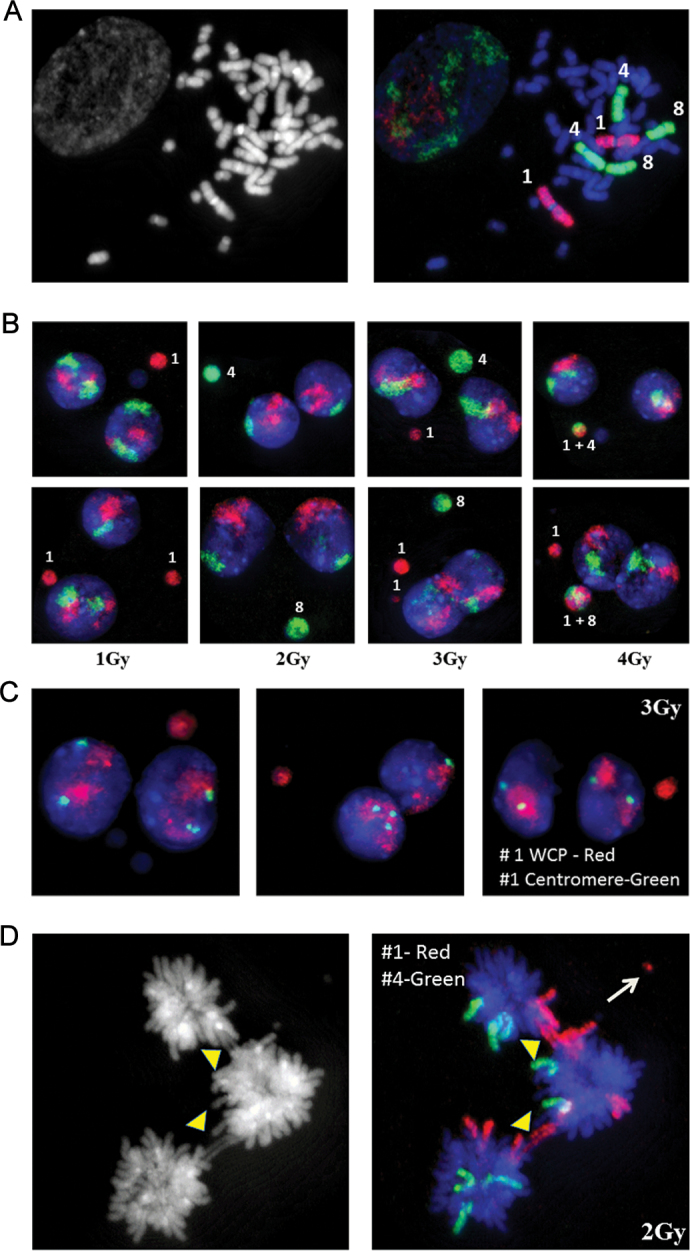
Fig. 1.
M-FISH analysis of cytokinesis blocked micronuclei in the human peripheral blood lymphocytes of donor 1 (A and B), donor 2 (C–F) and donor 3 (G and H). Note the detection of spontaneous micronucleus comprised of chromosomes 13 and 8 in donors 1 and 3. DAPI stained (A, C, E and G) and pseudo colour processed (B, D, F and H) images are shown.
M-FISH analysis of the chromosomal content of IR-induced micronuclei
M-FISH analysis detected material either from a single, double or multiple chromosomes in IR-induced micronuclei (Figure 2; Table III). Pooled data of the three donors revealed a dose-dependant increase in MN with two or more colours at radiation doses above 2 Gy (Figure 3) with a corresponding decline in micronuclei with single chromosome material. Pooled data on the actual frequencies for each of the human chromosomes observed either in single or multicoloured MN after different doses of γ-rays is shown in Table IV. Involvement of each human chromosome in multicoloured micronuclei increased at radiation doses >2 Gy (Figure 4). Interestingly, X and Y chromosomes were most exclusively found in multicoloured micronuclei. Data pooled from the three donors on the combined (single and multiple colours) percentages of micronuclei comprising all the human chromosomes are shown in Figure 5A and B. Chromosome composition of micronuclei detected by M-FISH in individual donors is shown in the supplemental section (supplementary Figure 1A–C, available at Mutagenesis Online).
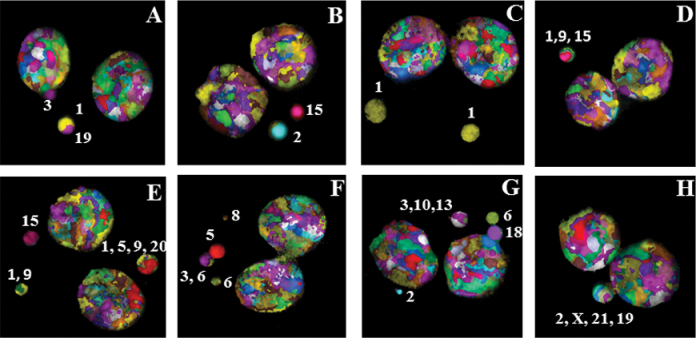
Fig. 2.
(A) M-FISH detection of chromosome content of micronuclei induced by different doses of γ-rays radiation in the peripheral blood lymphocytes of three human donors: (A) donor 2 and (B) donor 3—2 Gy; (C) donor 1 and (D) donor 2—3 Gy; (E) donor 1 and (F) donor 3—4 Gy; (G) donor 2 and (H) donor 3—5 Gy. Numbers indicate the chromosomes involved in MN formation. The percentages of micronuclei observed with each of the human chromosomes at different radiation doses in donor 1 (B), donor 2 (C) and donor 3 (D) are shown in the form of histograms. The percentages of MN detected as single and multiple colours in the mock and irradiated samples are shown in E.
Table III.
Distribution of micronuclei with single, double or multiple colours detected by M-FISH
| MN (1) | MN (2) | MN (3) | MN (4) | MN (>4) | Total MN | ||
|---|---|---|---|---|---|---|---|
| Donor 1 | 0 Gy | 93 | 23 | 2 | 0 | 0 | 118 |
| 1 Gy | 95 | 13 | 3 | 1 | 0 | 112 | |
| 2 Gy | 53 | 8 | 1 | 0 | 0 | 62 | |
| 3 Gy | 112 | 60 | 18 | 4 | 1 | 195 | |
| 4 Gy | 92 | 58 | 25 | 12 | 1 | 188 | |
| 5 Gy | 116 | 60 | 20 | 7 | 2 | 205 | |
| Donor 2 | 0 Gy | 39 | 17 | 1 | 0 | 0 | 57 |
| 1 Gy | 62 | 38 | 6 | 1 | 0 | 107 | |
| 2 Gy | 74 | 28 | 15 | 1 | 0 | 118 | |
| 3 Gy | 62 | 41 | 13 | 3 | 0 | 119 | |
| 4 Gy | 96 | 59 | 34 | 5 | 1 | 195 | |
| 5 Gy | 104 | 49 | 40 | 11 | 4 | 208 | |
| 7.5 Gy | 49 | 25 | 27 | 11 | 2 | 114 | |
| 10 Gy | 43 | 20 | 25 | 14 | 21 | 123 | |
| Donor 3 | 0 Gy | 16 | 10 | 2 | 0 | 0 | 28 |
| 1 Gy | 37 | 12 | 7 | 0 | 0 | 56 | |
| 2 Gy | 66 | 25 | 22 | 2 | 1 | 116 | |
| 3 Gy | 59 | 43 | 27 | 1 | 1 | 135 | |
| 4 Gy | 116 | 68 | 41 | 8 | 5 | 238 | |
| 5 Gy | 48 | 67 | 26 | 8 | 2 | 151 | |
| 7.5 Gy | 66 | 58 | 37 | 20 | 5 | 186 | |
| 10 Gy | 46 | 33 | 24 | 8 | 13 | 124 |
Numbers in parentheses indicate the color signals.
Fig. 3.
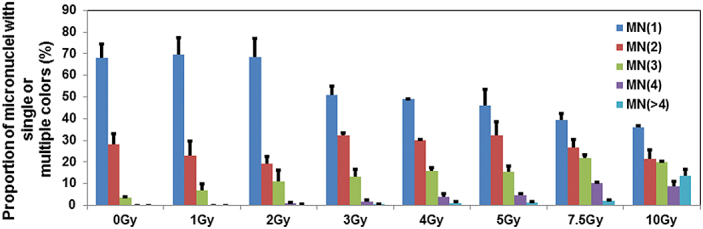
Percentages of micronuclei with single, double and multiple colours detected by M-FISH in cytochalasin B-blocked binucleated cells in mock and irradiated samples. The pooled data from the three donors are presented in the form of histograms. Bars indicate SEM.
Table IV.
Number of micronuclei positive for each of the human chromosomes as a single entity or in combination (pooled data)
| Chr. | 0 Gy | 1 Gy | 2 Gy | 3 Gy | 4 Gy | 5 Gy | 7.5 Gy | 10 Gy | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Single | Multiple | Single | Multiple | Single | Multiple | Single | Multiple | Single | Multiple | Single | Mulitple | Single | Multiple | Single | Multiple | |
| 1 | 10 | 7 | 32 | 15 | 31 | 28 | 38 | 54 | 76 | 102 | 55 | 81 | 22 | 54 | 18 | 42 |
| 2 | 7 | 5 | 8 | 10 | 20 | 23 | 15 | 39 | 20 | 38 | 23 | 48 | 15 | 57 | 18 | 36 |
| 3 | 4 | 4 | 16 | 8 | 21 | 11 | 29 | 26 | 30 | 46 | 26 | 49 | 1 | 20 | 1 | 25 |
| 4 | 9 | 6 | 5 | 7 | 9 | 17 | 22 | 29 | 32 | 51 | 16 | 48 | 11 | 29 | 6 | 22 |
| 5 | 7 | 8 | 7 | 11 | 15 | 7 | 13 | 23 | 25 | 44 | 18 | 31 | 10 | 22 | 6 | 30 |
| 6 | 7 | 3 | 3 | 9 | 8 | 7 | 13 | 26 | 8 | 25 | 21 | 28 | 4 | 29 | 2 | 19 |
| 7 | 8 | 2 | 7 | 8 | 8 | 19 | 12 | 30 | 16 | 47 | 15 | 35 | 5 | 36 | 7 | 28 |
| 8 | 9 | 4 | 11 | 9 | 8 | 19 | 10 | 35 | 11 | 44 | 15 | 51 | 6 | 19 | 3 | 30 |
| 9 | 9 | 7 | 9 | 10 | 3 | 4 | 5 | 24 | 1 | 53 | 13 | 28 | 3 | 22 | 3 | 18 |
| 10 | 8 | 9 | 7 | 5 | 7 | 9 | 7 | 20 | 11 | 36 | 6 | 39 | 3 | 22 | 4 | 18 |
| 11 | 5 | 3 | 3 | 6 | 8 | 5 | 11 | 20 | 12 | 13 | 11 | 14 | 5 | 12 | 3 | 11 |
| 12 | 11 | 2 | 9 | 10 | 7 | 3 | 7 | 17 | 14 | 31 | 15 | 19 | 1 | 10 | 0 | 14 |
| 13 | 12 | 5 | 12 | 10 | 15 | 7 | 7 | 20 | 10 | 33 | 9 | 48 | 5 | 27 | 6 | 28 |
| 14 | 3 | 4 | 2 | 5 | 1 | 3 | 0 | 11 | 2 | 9 | 1 | 24 | 0 | 8 | 1 | 11 |
| 15 | 4 | 4 | 6 | 3 | 5 | 4 | 7 | 16 | 5 | 15 | 8 | 17 | 2 | 7 | 1 | 10 |
| 16 | 4 | 3 | 11 | 7 | 4 | 12 | 5 | 16 | 4 | 40 | 6 | 30 | 2 | 34 | 3 | 22 |
| 17 | 4 | 6 | 7 | 4 | 4 | 4 | 3 | 12 | 8 | 20 | 7 | 12 | 3 | 10 | 1 | 15 |
| 18 | 6 | 1 | 6 | 2 | 2 | 8 | 3 | 20 | 6 | 23 | 5 | 21 | 2 | 15 | 0 | 20 |
| 19 | 6 | 4 | 6 | 5 | 10 | 11 | 5 | 22 | 8 | 35 | 9 | 43 | 5 | 18 | 1 | 28 |
| 20 | 4 | 4 | 4 | 4 | 1 | 6 | 2 | 11 | 2 | 14 | 0 | 16 | 5 | 15 | 1 | 13 |
| 21 | 5 | 0 | 0 | 2 | 2 | 3 | 1 | 20 | 1 | 30 | 1 | 23 | 0 | 9 | 2 | 14 |
| 22 | 3 | 3 | 4 | 7 | 3 | 3 | 3 | 13 | 0 | 18 | 1 | 19 | 0 | 14 | 0 | 11 |
| X | 1 | 12 | 14 | 10 | 5 | 8 | 7 | 18 | 9 | 41 | 5 | 28 | 3 | 11 | 0 | 19 |
| Y | 0 | 4 | 0 | 7 | 0 | 4 | 0 | 4 | 0 | 6 | 0 | 12 | 1 | 8 | 0 | 8 |
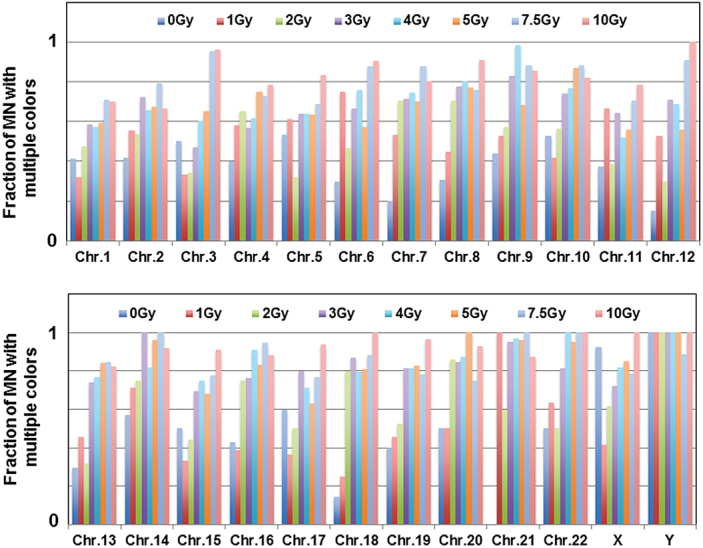
Fig. 4.
Pooled fractions of multicoloured micronuclei (double, triple or multiple colours) involving each of the human chromosomes in mock- and γ-rays-irradiated samples are shown. Note that X and Y chromosomes were exclusively found in combination with other chromosomes in MN formation.
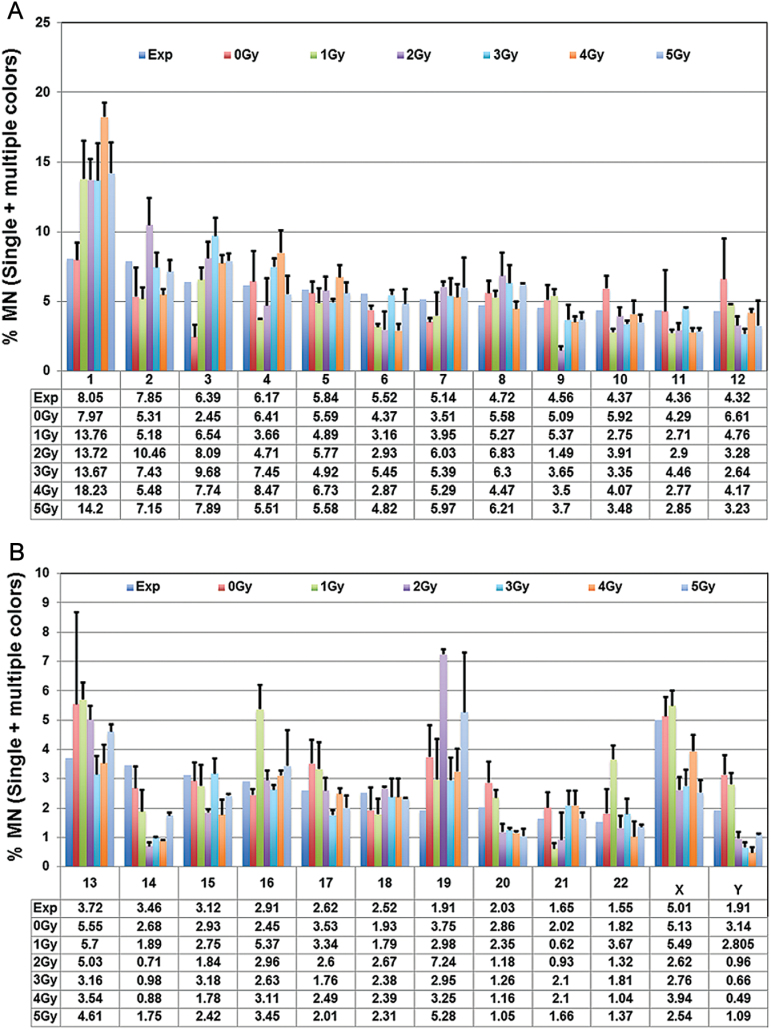
Fig. 5.
(A and B) Involvement of each of the human chromosomes in micronuclei both as a single entity or in combination with other chromosomes is shown in the form of histograms. The mean values representing the combined (single and multiple colours) percentages of micronuclei with each of the chromosomes were pooled from the three donors. Bars indicate SEM. The expected frequency of micronuclei calculated on the basis of DNA content for each of the human chromosomes is shown.
Although inter-individual variation among the three donors was detected in terms of the chromosome content of IR-induced micronuclei, the first five pairs of chromosomes were consistently detected at high frequencies (35–45% of the total micronuclei) at all radiation doses in all the donors. Chromosome 1 content in micronuclei was detected at more than the expected frequency (8.05% of the total micronuclei) on the basis of DNA content and micronuclei with chromosome 1 material occurred at varying frequencies of 9.56–21.26% in all the donors following radiation exposure (supplementary Figure 1A–C, available at Mutagenesis Online). Involvement of chromosome 1 in multicoloured micronuclei greatly increased at radiation doses >2 Gy. Pooled data from the three donors indicate that the percentage of micronuclei with three, four or more colours showed a dose-dependant increase suggestive of increased generation and fusion of acentric fragments. This is expected because chromosome fragments resulting from exchange type aberrations such as dicentrics and translocations increase almost linearly as a function of radiation dose.
After 1 Gy of radiation exposure, chromosomes 1 (12.77%), 8 (8.26%), 9 (8.56%), 22 (6.45%) and X (7.66%) in donor 1, chromosomes 1 (11.15%), 2 (7.82%), 3 (8.15%), 7 (6.96%), 13 (6.30%) and 19 (5.90%) in donor 2 and chromosomes 1 (17.37%), 2 (7.19%), 5 (7.19%) and 13 (7.65%) in donor 3 were found at high frequencies in micronuclei either as a single entity or in combination with other chromosomes. Interestingly, chromosomes 1, 2 and 13 were frequently found in micronuclei in both male donors. In donor 1, chromosomes 1 (9.88%), 2 (8.97%), 3 (9.48%), 8 (9.88%), 12 (6.76%) and 19 (15.73%) were detected at high frequencies in micronuclei after 2 Gy exposure while the chromosomes 4, 6, 14, 15, 20 and 21 were not detected in MN. In donor 2, six chromosomes (1–17.96%, 2–7.85%, 4–10.08%, 6–6.51%, 7–6.26% and 8–8.05%) were detected at high frequencies in micronuclei after 2 Gy of γ-rays. Similar to donor 2, chromosomes 1 (13.31%) and 2 (14.56%) in donor 3 were detected at high frequencies in micronuclei. Curiously, involvement of sex chromosomes in micronuclei was found at less than the expected frequency in all the three donors. Increased proportion of single and multicoloured micronuclei comprised of chromosomes 1 (12.61%), 3 (7.33%), 4 (7.93%) and 7 (5.80%) was observed in donor 1 after 3 Gy of γ-rays. Chromosomes 1 (9.56%), 2 (10.26%), 3 (12.97%), 4 (8.26%) and 8 (9.91%) were detected at high frequencies in micronuclei in donor 2 after 3 Gy. In donor 3, 59.84% of the total micronuclei contained chromosomes 1, 2, 3, 4, 6, 7 and 8 while the rest of the chromosomes were observed at varying proportions (0.27–4.78%). Among the chromosomes that were detected at high frequencies, chromosomes 1, 2, 3, 4 and 8 were common in both male donors. Chromosomes 1, 3 and 4 were found at increased frequencies in all the three donors after 4 Gy. Donor 1 showed increased proportions of chromosomes 1 (15.42%), 3 (8.27%), 4 (8.02%) and 5 (9.65%) in micronuclei. In donor 2, increased fraction of micronuclei with chromosomes 1 (18.01%), 3 (8.45%), 4 (11.46%) and 8 (5.85%) were observed. A total of 6 chromosomes [1 (21.26%), 2 (6.25%), 3 (6.51%), 4 (5.94%), 5 (6.57%) and 7 (7.35%)] were found to be more frequently involved in micronuclei relative to other chromosomes in donor 3.
Micronuclei comprised of material from chromosomes 1 (17.06%), 3 (9.08%), 4 (8.28%), 6 (7.08%), 8 (5.58%) and 9 (5.98%) were found to be higher than the rest of the chromosomes in donor 1 after 5 Gy of γ-rays exposure. In both male donors, micronuclei induced by 5 Gy of γ-rays contained chromosomes 1, 2 and 3 at high frequencies. In addition, chromosomes 4 (6.25%) and 12 (6.88%) in donor 2 and chromosomes 5 (7.39%), 7 (11.31%), 8 (6.55%) and 19 (9.83) in donor 3 were detected in micronuclei more frequently than the rest of the chromosomes. In all the three donors, the first five chromosomes (1–5) contributed to 43.97, 40.95 and 36.15% of the total micronuclei induced by 5 Gy of γ-rays.
M-FISH analysis performed on micronuclei induced by high doses of γ-rays (7.5 and 10 Gy) in both male donors 2 and 3 showed the over-representation of chromosomes 1, 2, 4, 5 and 13 in MN. Interestingly, micronuclei with Y chromosome showed the lowest frequency in both donors. Further, chromosomes 21 and 22 were mostly detected in MN as multiple chromosomes but rarely as a single entity after radiation exposure in all the donors. Pooled data of the three donors indicate that the observed frequencies for chromosomes 13 and 19 in micronuclei were much higher than the expected frequencies. Although chromosome 18 has more DNA content (~78 Mbp) than 19 (~59 Mbp), the latter was more frequently involved in MN formation.
M-FISH detection of lagging chromosomes or chromosome fragments
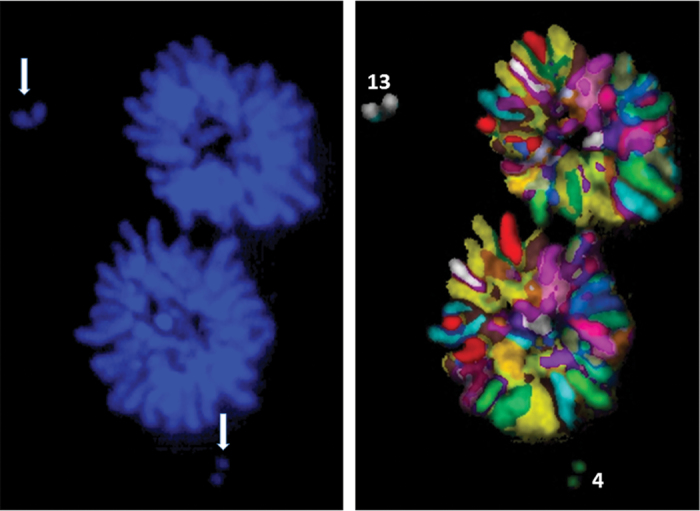
In addition to identifying the chromosome content of MN, M-FISH technique greatly enabled the precise identification of chromosomes or chromosome fragments that are excluded from the spindle during mitosis. A representative picture showing the exclusion of fragments from chromosomes 4 and 13 during anaphase/telophase in donor 2 after 2 Gy of γ-rays radiation is shown in Figure 6.
Fig. 6.
M-FISH detection of fragments from chromosomes 4 and 13 (arrows) during mitosis after 2 Gy of γ-rays radiation in the peripheral blood lymphocytes of donor 2.
MN analysis by FISH using whole chromosome-specific probes (WCP)
To verify the results of M-FISH analysis, FISH utilising WCP for chromosome 1 (Texas Red labeled), 4 and 8 (FITC labeled) were performed. Specificity of the DNA probes was verified on metaphase chromosomes prepared from primary human dermal fibroblasts (NHDF, Figure 7A). FISH experiments were performed using a pair wise comparison of chromosome 1 together with either chromosome 4 or 8. MN preparations from donor 2 and 3 were used for this purpose. Frequencies of micronuclei with chromosomes 1, 4 and 8 were analysed either as a single entity or in combination. In non-irradiated controls, M-FISH analysis detected chromosome 1 in 7.80% and 9.13% of the total micronuclei with single colour in donors 2 and 3, respectively. FISH with WCP detected chromosome 1 in 8.23% and 8.73% of the total micronuclei. In donors 1 and 2, chromosome 4 was detected in 5.98% and 10.25% of the total micronuclei. FISH with chromosome 4-specific probe detected similar frequencies in both donors (6.10 and 9.83%). Chromosome 8 was present in 4.58 and 5.12% of the total micronuclei in donors 2 and 3, respectively. FISH with chromosome 8-specific probe yielded similar number of MN detected by M-FISH technique (4.22% in donor 2 and 5.78% in donor 3). Frequencies for chromosomes 1, 4 and 8 detected by M-FISH in micronuclei after radiation exposure (1–5 Gy of γ-rays) either as a single entity or in combination were similar to that obtained by FISH with WCP probes. Representative pictures of micronuclei detected by WCP for 1, 4 and 8 are shown in Figure 7B. Since chromosome 1 was most frequently involved in MN formation, we utilised a combination of WCP together with centromeric probe specific for chromosome 1 to determine the frequency of centromere positive and negative micronuclei in both male donors. Our results indicate that 60–70% of the total micronuclei induced by different doses (1–5 Gy) of γ-rays were devoid of centromeric probe signal. Representative pictures of FISH performed with WCP and chromosome 1-specific centromeric probes after 3 Gy exposure in donor 2 are shown (Figure 7C). Combination of FISH probes specific for whole chromosome and centromere was also found useful for detecting aneuploidy between daughter nuclei. Consistent with the frequent involvement of chromosome 1 in MN formation, 15–20% of the total nucleoplasmic bridges induced by IR were also found positive by FISH using chromosome 1-specific probe (Figure 7D).
Fig. 7.
(A) FISH using whole chromosome paints for 1, 4 and 8 on human metaphase spreads. Chromosome painting probes were directly labeled with Texas Red (Chromosome 1) and FITC (Chromosome 4 and 8). (B) Detection of chromosomes 1, 4 and 8 in cytokinesis blocked MN in ex vivo γ-rays-irradiated peripheral blood lymphocytes of donor 2 (top panel) and donor 3 (bottom panel). (C) Detection of chromosome 1 in IR-induced micronuclei in donor 2 using whole chromosome painting probe (red colour) and centromere-specific probe (green colour). (D) Detection of chromosome 1 (red colour) and 4 (green colour) in IR-induced nucleoplasmic bridges (arrowheads). Chromosome 1 fragment is indicated by the arrow.
Discussion
Analysis of micronuclei has been extensively used for assessing the genotoxic effects of environmental mutagens and carcinogens. Although the precise mechanistic basis for micronuclei is still unclear, their origin is thought to be due to (i) displaced chromosomes, (ii) lagging chromosome fragments or whole chromosomes, (iii) breakage of nucleoplasmic bridges and (iv) mitotic spindle disruptions due to mitotic checkpoint deficiencies (18). In this study, M-FISH was utilised to investigate the chromosome content of micronuclei induced by different doses of γ-rays in three human donors. Our main objective was to determine whether or not exclusion of chromosomes into MN formation occurs either in a random or a non-random manner. Several earlier studies employing CREST antibody or FISH reported that the proportion of whole chromosomes involved in micronuclei varied from 30 to 80% in cultured human lymphocytes (10,16,17,19–28). In the current study, the proportion of spontaneous micronuclei with a single chromosome material varied from 57.14 to 77.13% in the three donors with the highest proportion observed in the female donor (donor 1). Inter-individual variability with regard to chromosome content of micronuclei was observed among the three donors. In the female donor, exclusion of each of the chromosomes into micronuclei was observed while chromosome 11 in donor 2 and chromosomes 3, 13, 18 and 22 in donor 3 were not detected in MN. Interestingly, involvement of chromosome 4 material in micronuclei was consistently observed at high frequency in all the three donors. In both male donors, chromosomes 1, 4 and 6 were found to be frequently involved in MN formation. Further, FISH employing a combination of chromosome 1-specific centromere and whole painting probes revealed that 60–70% of the total chromosome 1-positive micronuclei were negative for centromere in both male donors. An earlier study (10) reported a high frequency of X chromosome material in micronuclei and the frequency was found to be 12.2, 50.6 and 7.1% in 28, 42 and 72 years old female donors, respectively. These findings indicate an age-dependant inter-individual variability in the frequency of X chromosome containing micronuclei among the female donors. In the present study, an increased frequency of X chromosome involvement (8.54% of the total micronuclei) in the baseline micronuclei was noted in the 35 years old female donor which was slightly lower than that observed by Leach and Jackson-Cook (10) for 28 years old female. Analysis of blood samples from 8 newborn females and 38 adult females in the range of 19–77 years showed the X chromosome material, detected by X chromosome-specific centromeric probe, in 72.2% of the total baseline micronuclei (29). Since cumulative data on X chromosome-containing micronuclei was presented in this study, it was not clear any inter-individual variability was observed among the female donors as a function of age. Since only one female donor of 35 years old was used in the study, it is difficult to predict the reason for the variations observed between our study and that of Hando et al. (29).
Different DNA-damaging agents induce qualitatively different micronuclei with regard to their chromosome composition. Agents such as IR induce MN that predominantly contain acentric chromosome fragments while mitotic spindle disrupting agents like colchicine induce micronuclei with whole chromosome(s). Several studies were performed in the past to determine the origin of MN by relative size, DNA synthesis, kinetochores and FISH using DNA probes for telomeres, centromeres and whole chromosomes (1). Silva et al. (30) demonstrated a good correlation between radiation dose and MN size but the chromosome content was not analysed in this study. It is generally believed that small- and large-sized micronuclei are characterised by chromosome fragments and whole chromosome(s) respectively, and the size of micronuclei increases as a function of radiation dose at or above 1 Gy of γ-rays. However, M-FISH analysis performed in this study revealed the complex nature of micronuclei comprising two or more chromosomes at radiation does above 2 Gy. Further, micronuclei, irrespective of size, were found to contain material either from a single chromosome or multiple chromosomes. In all the donors, percentages of micronuclei comprised of the material from two or more chromosomes showed an increase after radiation exposure relative to unirradiated control samples. Among the human chromosomes, chromosome 1 material in micronuclei was consistently found at elevated frequencies in all the donors after radiation exposure. Among the donors, similarities in chromosome content of IR-induced micronuclei were observed in the male donors. Notably, chromosomes 1, 2 and 13 after 1 Gy, 1 and 2 after 2 Gy, 1, 2, 3, 4 and 8 after 3 Gy, 1, 2 and 4 after 4 Gy and 1, 2 and 3 after 5 Gy were detected at high frequencies in micronuclei in both male donors. Chromosomes belonging to groups A (1, 2 and 3) and B (4 and 5) in all the donors constituted around 35–45% of the total micronuclei induced by radiation exposure. These findings indicate a non-random involvement of chromosomes in the formation of IR-induced micronuclei.
An earlier study has shown the preferential involvement of chromosomes 1 and 13 in translocations in human lymphocytes after exposure to 0.1–1 Gy of γ-rays (31). In the present study, an elevated frequency of chromosome 1 was detected in micronuclei at all radiation doses in the three donors irrespective of age and gender. Inclusion of chromosome 1 in micronuclei has also been demonstrated in cells treated with 5-azacytidine and IR (5,9). Using probes specific for chromosomes 1, 7, 11, 14, 17 and 21, Fimognari et al. (9), demonstrated that the frequency of their inclusion in MN formation depends on the DNA content. In corroboration, the first five pairs of chromosomes were found to be more frequently involved in MN formation in comparison to the rest of the chromosomes. Based on the DNA content of chromosomes, the expected frequency for a random involvement of the first five chromosomes in MN formation is 34.3%. However, the observed frequency for different radiation doses ranged from 38.66 to 46.76% in the three donors. Frequencies for IR-induced stable and unstable chromosome aberrations seem to correlate well with chromosome size because of a higher probability of DSB induction owing to spatial and temporal organisation of chromatin in the interphase nuclei. Assuming a uniform distribution of DNA damage in the interphase nuclei, domains comprised of large chromosomes are expected to have a higher DSB induction relative to other domains. This is presumably the reason for the frequent involvement of larger chromosomes in MN formation.
In the present study, the presence of chromosome 1 was detected in 9–21% of the total micronuclei either as a single entity or in combination with other chromosomes after exposure to different γ-rays doses. Interestingly, involvement of chromosome 1 in both baseline and NNK-induced micronuclei was observed in an earlier study in normal and lung cancer patients (11). In addition to chromosome 1, chromosomes 13 and 17 were also detected in both baseline and NNK-induced micronuclei. In corroboration with this study, we also detected increased frequencies of chromosomes 1 and 13 in IR-induced micronuclei. Thus, it appears that chromosomes 1 and 13 are preferentially involved in MN formation induced by both IR and NNK.
Although chromosomes 1 and 2 differ only by 6Mb in DNA content, chromosome 2 was less frequently involved in micronuclei (0.52–14.56% in the three donors after different γ-rays doses) relative to chromosome 1. It is currently unclear whether the spatial or temporal organisation of chromosomes 1 and 2 in the interphase nuclei is responsible for their differential sensitivities to radiation-induced DNA damage. Using probes specific for chromosomes 1, 4, 18 and 19, an earlier study has demonstrated that the gene density-related interphase positioning of chromosomes influences the yield of radiation-induced aberrations (32). Differences in the yield of radiation-induced aberrations in chromosomes 1 and 2 have also been reported in two human donors (33). It is worth noting that chromosome 1 is highly enriched with ~2012 genes relative to ~1203 genes on chromosome 2. Therefore, elevated frequency of chromosome 1 involvement in IR-induced MN formation may be partly due to spatial organisation driven by gene density-related effects. Plan et al. (34) detected the involvement of chromosome 1 in >20% of the total dicentrics involving homologous chromosomes after radiation exposure. Mapping of γ-rays radiation-induced breaks in chromosome 1 by a combination of G-banding and FISH techniques revealed the preferential occurrence of breaks in the gene-enriched euchromatic regions (35). In the present study, frequent involvement of chromosomes 13 and 19 in micronuclei were observed at more than expected frequencies after varying doses of γ-rays suggesting that the formation of micronuclei after radiation exposure may not be always proportional to chromosome length.
Although chromosome 18 has more DNA content (~78 Mbp) than chromosome 19 (~59 Mbp), chromosome 19 is enriched with more transcriptionally active genes (1399) than chromosome 18 (268 genes). Observation of elevated frequency of chromosome 19 involvement in MN formation lends support to the idea that the gene-enriched euchromatic regions of certain chromosomes may be more prone to IR-induced DNA strand breaks. Currently, it is not clear whether the increased transcriptional activity owing to higher number of genes on chromosome 19 enhances the sensitivity to radiation exposure. These findings reinforce the idea that the positioning of chromosome territory or domain regulates the DSB induction and repair processes. In addition to group A and B chromosomes, frequent involvement of chromosomes 13 and 19 in micronuclei were observed in the three donors after varying doses of γ-rays suggesting that the formation of MN after radiation exposure may not be always proportional to chromosome length.
One distinct advantage of M-FISH and SKY techniques over the conventional dual colour FISH is that it enables the analysis of all the chromosomes simultaneously in micronuclei. Using the SKY technique, a significant correlation was found between the specific chromosomes involved in MN and lung cancer risk (11). As stated before, proportions of micronuclei with multiple fluorescent signals increased substantially at or above 2 Gy of γ-rays and micronuclei comprised of material from 2 to 5 different chromosomes were frequently observed in all the donors. It is generally assumed that the large-sized micronuclei contain whole chromosomes while the small-sized micronuclei contain acentric chromosome fragments. In this study, large-sized micronuclei were found to contain material either from single chromosome or from multiple chromosomes. Observation of increased proportions of micronuclei with multiple chromosomes at radiation doses above 2 Gy is probably due to misrepair of DNA DSB involving multiple chromosomes leading to the formation and fusion of acentric fragments from these chromosomes. Dose-dependant increase in the frequency of multicoloured micronuclei observed in the present study may be due to increased misrepair events at radiation doses higher than 2 Gy. During micro nucleation, elimination and subsequent fusion of acentric fragments from multiple chromosomes harbouring critical tumour suppressor genes may result in chromothripsis (36), a phenomenon frequently observed in a subset of cancer cells. These changes often result in complex chromosome rearrangements with alterations in copy number of vital genes involved in cell cycle regulation, DNA repair and genomic stability. In this study, M-FISH technique revealed the complexity of micronuclei involving multiple chromosome fragments after radiation exposure. Future studies are required to understand the molecular complexity of IR-induced micronuclei in the context of genomic instability and cancer development processes.
Supplementary data
Supplementary Figure 1 is available at Mutagenesis Online.
Funding
This work at the Center for High-Throughput Minimally Invasive Radiation Biodosimetry, Columbia University Medical Center, NY was supported by the National Institute of Allergy and Infectious Diseases, NIH (grant number U19 AI067773).
Supplementary Material
Acknowledgements
We thank Prof. A. T. Natarajan, Department of Ecological and Biological Sciences, University of Tuscia, Viterbo, Italy for the critical reading of the manuscript and suggestions. The content is solely the responsibility of the authors and does not necessarily represent the official views of either National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Conflict of interest statement: None declared.
References
- 1. Mateuca R. A., Decordier I., Kirsch-Volders M. (2012). Cytogenetic methods in human biomonitoring: principles and uses. Methods Mol. Biol., 817, 305–334 [DOI] [PubMed] [Google Scholar]
- 2. Samanta S., Dey P. (2012). Micronucleus and its applications. Diagn. Cytopathol., 40, 84–90 [DOI] [PubMed] [Google Scholar]
- 3. Fenech M. (2007). Cytokinesis-block micronucleus cytome assay. Nat. Protoc., 2, 1084–1104 [DOI] [PubMed] [Google Scholar]
- 4. Fenech M. (2010). The lymphocyte cytokinesis-block micronucleus cytome assay and its application in radiation biodosimetry. Health Phys., 98, 234–243 [DOI] [PubMed] [Google Scholar]
- 5. Guttenbach M., Schmid M. (1994). Exclusion of specific human chromosomes into micronuclei by 5-azacytidine treatment of lymphocyte cultures. Exp. Cell Res., 211, 127–132 [DOI] [PubMed] [Google Scholar]
- 6. Fauth E., Scherthan H., Zankl H. (2000). Chromosome painting reveals specific patterns of chromosome occurrence in mitomycin C- and diethylstilboestrol-induced micronuclei. Mutagenesis, 15, 459–467 [DOI] [PubMed] [Google Scholar]
- 7. Hovhannisyan G., Aroutiounian R., Liehr T. (2012). Chromosomal composition of micronuclei in human leukocytes exposed to mitomycin C. J. Histochem. Cytochem., 60, 316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chung H. W., Kang S. J., Kim S. Y. (2002). A combination of the micronucleus assay and a FISH technique for evaluation of the genotoxicity of 1,2,4-benzenetriol. Mutat. Res., 516, 49–56 [DOI] [PubMed] [Google Scholar]
- 9. Fimognari C., Sauer-Nehls S., Braselmann H., Nüsse M. (1997). Analysis of radiation-induced micronuclei by FISH using a combination of painting and centromeric DNA probes. Mutagenesis, 12, 91–95 [DOI] [PubMed] [Google Scholar]
- 10. Leach N. T., Jackson-Cook C. (2001). The application of spectral karyotyping (SKY) and fluorescent in situ hybridization (FISH) technology to determine the chromosomal content(s) of micronuclei. Mutat. Res., 495, 11–19 [DOI] [PubMed] [Google Scholar]
- 11. Lloyd S. M., Lopez M., El-Zein R. (2013). Cytokinesis-blocked micronucleus cytome assay and spectral karyotyping as methods for identifying chromosome damage in a lung cancer case-control population. Genes. Chromosomes Cancer, 52, 694–707 [DOI] [PubMed] [Google Scholar]
- 12. Löbrich M., Cooper P. K., Rydberg B. (1996). Non-random distribution of DNA double-strand breaks induced by particle irradiation. Int. J. Radiat. Biol., 70, 493–503 [DOI] [PubMed] [Google Scholar]
- 13. Cowell I. G., Sunter N. J., Singh P. B., Austin C. A., Durkacz B. W., Tilby M. J. (2007). GammaH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS One, 2, e1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Natarajan A. T., Boei J. J., Vermeulen S., Balajee A. S. (1996). Frequencies of X-ray induced pericentric inversions and centric rings in human blood lymphocytes detected by FISH using chromosome arm specific DNA libraries. Mutat. Res., 372, 1–7 [DOI] [PubMed] [Google Scholar]
- 15. Mackinnon R. N., Chudoba I. (2011). The use of M-FISH and M-BAND to define chromosome abnormalities. Methods Mol. Biol., 730, 203–218 [DOI] [PubMed] [Google Scholar]
- 16. Surrallés J., Falck G., Norppa H. (1996). In vivo cytogenetic damage revealed by FISH analysis of micronuclei in uncultured human T lymphocytes. Cytogenet. Cell Genet., 75, 151–154 [DOI] [PubMed] [Google Scholar]
- 17. Surrallés J., Jeppesen P., Morrison H., Natarajan A. T. (1996). Analysis of loss of inactive X chromosomes in interphase cells. Am. J. Hum. Genet., 59, 1091–1096 [PMC free article] [PubMed] [Google Scholar]
- 18. Fenech M., Bonassi S. (2011). The effect of age, gender, diet and lifestyle on DNA damage measured using micronucleus frequency in human peripheral blood lymphocytes. Mutagenesis, 26, 43–49 [DOI] [PubMed] [Google Scholar]
- 19. Bakou K., Stephanou G., Andrianopoulos C., Demopoulos N. A. (2002). Spontaneous and spindle poison-induced micronuclei and chromosome non-disjunction in cytokinesis-blocked lymphocytes from two age groups of women. Mutagenesis, 17, 233–239 [DOI] [PubMed] [Google Scholar]
- 20. Calvert G. M., Talaska G., Mueller C. A., Ammenheuser M. M., Au W. W., Fajen J. M., Fleming L. E., Briggle T., Ward E. (1998). Genotoxicity in workers exposed to methyl bromide. Mutat. Res., 417, 115–128 [DOI] [PubMed] [Google Scholar]
- 21. Carere A., Antoccia A., Cimini D., et al. (1999). Analysis of chromosome loss and non-disjunction in cytokinesis-blocked lymphocytes of 24 male subjects. Mutagenesis, 14, 491–496 [DOI] [PubMed] [Google Scholar]
- 22. Davies H. W., Kennedy S. M., Teschke K., Jenny P., Quintana E. (1998). Cytogenetic analysis of South Asian berry pickers in British Columbia using the micronucleus assay in peripheral lymphocytes. Mutat. Res., 416, 101–113 [DOI] [PubMed] [Google Scholar]
- 23. Fenech M., Morley A. A. (1989). Kinetochore detection in micronuclei: an alternative method for measuring chromosome loss. Mutagenesis, 4, 98–104 [DOI] [PubMed] [Google Scholar]
- 24. Maffei F., Fimognari C., Castelli E., Stefanini G. F., Forti G. C., Hrelia P. (2000). Increased cytogenetic damage detected by FISH analysis on micronuclei in peripheral lymphocytes from alcoholics. Mutagenesis, 15, 517–523 [DOI] [PubMed] [Google Scholar]
- 25. Norppa H., Luomahaara S., Heikanen H., Roth S., Sorsa M., Renzi L., Lindholm C. (1993). Micronucleus assay in lymphocytes as a tool to biomonitor human exposure to aneuploidogens and clastogens. Environ. Health Perspect., 101(Suppl 3), 139–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scarpato R., Landini E., Migliore L. (1996). Acrocentric chromosome frequency in spontaneous human lymphocyte micronuclei, evaluated by dual-colour hybridization, is neither sex- nor age-related. Mutat. Res., 372, 195–204 [DOI] [PubMed] [Google Scholar]
- 27. Thierens H., Vral A., Barbé M., Aousalah B., De Ridder L. (1999). A cytogenetic study of nuclear power plant workers using the micronucleus-centromere assay. Mutat. Res., 445, 105–111 [DOI] [PubMed] [Google Scholar]
- 28. Yager J. W., Eastmond D. A., Robertson M. L., Paradisin W. M., Smith M. T. (1990). Characterization of micronuclei induced in human lymphocytes by benzene metabolites. Cancer Res., 50, 393–399 [PubMed] [Google Scholar]
- 29. Hando J. C., Nath J., Tucker J. D. (1994). Sex chromosomes, micronuclei and aging in women. Chromosoma, 103, 186–192 [DOI] [PubMed] [Google Scholar]
- 30. Silva M. J., Carothers A., Dias A., Luis J. H., Piper J., Boavida M. G. (1994). Dose dependence of radiation-induced micronuclei in cytokinesis-blocked human lymphocytes. Mutat. Res., 322, 117–128 [DOI] [PubMed] [Google Scholar]
- 31. Sevan’kaev A. V., Ankina M. A., Zavitaeva T. A., et al. (1995). [A comparative study of the frequency of stable and unstable chromosome aberrations in the gamma irradiation of human lymphocytes in vitro]. Radiats. Biol. Radioecol., 35, 611–618 [PubMed] [Google Scholar]
- 32. Boei J. J., Fomina J., Darroudi F., Nagelkerke N. J., Mullenders L. H. (2006). Interphase chromosome positioning affects the spectrum of radiation-induced chromosomal aberrations. Radiat. Res., 166, 319–326 [DOI] [PubMed] [Google Scholar]
- 33. Wojcik A., Streffer C. (1998). Comparison of radiation-induced aberration frequencies in chromosomes 1 and 2 of two human donors. Int. J. Radiat. Biol., 74, 573–581 [DOI] [PubMed] [Google Scholar]
- 34. Plan Y., Hlatky L., Hahnfeldt P., Sachs R., Loucas B., Cornforth M. (2005). Full-color painting reveals an excess of radiation-induced dicentrics involving homologous chromosomes. Int. J. Radiat. Biol., 81, 613–620 [DOI] [PubMed] [Google Scholar]
- 35. Kiuru A., Lindholm C., Auvinen A., Salomaa S. (2000). Localization of radiation-induced chromosomal breakpoints along human chromosome 1 using a combination of G-banding and FISH. Int. J. Radiat. Biol., 76, 667–672 [DOI] [PubMed] [Google Scholar]
- 36. Holland A. J., Cleveland D. W. (2012). Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat. Med., 18, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.