Abstract
In pathogenic bacteria post-translationally modified proteins have been found to promote bacterial survival, replication and evasion from the host immune system. In the human pathogen Neisseria meningitidis, the protein PilE (15–18 kDa) is the major building block of type IV pili, extracellular filamentous organelles that play a major role in mediating pathogenesis. Previous reports have shown that PilE can be expressed as a number of different proteoforms, each harbouring its own set of post-translational modifications (PTMs) and that specific proteoforms are key in promoting bacterial virulence. Efficient tools that allow complete PTM mapping of proteins involved in bacterial infection are therefore strongly needed. As we show in this study, a simple combination of mass profiling and bottom-up proteomics is fundamentally unable to achieve this goal when more than two proteoforms are present simultaneously. In a N. meningitidis strain isolated from a patient with meningitis, mass profiling revealed the presence of four major proteoforms of PilE, in a 1:1:1:1 ratio. Due to the complexity of the sample, a top-down approach was required to achieve complete PTM mapping for all four proteoforms, highlighting an unprecedented extent of glycosylation. Top-down mass spectrometry therefore appears to be a promising tool for the analysis of highly post-translationally modified proteins involved in bacterial virulence.
Keywords: post-translational modification, proteoforms, Neisseria meningitidis, pili, top-down mass spectrometry
Introduction
Post-translational modification (PTM) increases the functional diversity of proteins by covalent addition of functional groups, modification of amino acid side chains and proteolysis. PTMs are implicated in almost all aspects of normal cell biology and pathogenesis. In viral or bacterial infection, pathogens often use PTMs to manipulate pathways in the host cell in order to promote their own survival, replication and evasion from the host immune system [1, 2]. For a long time PTM was considered an exclusively eukaryotic process but it is now widely accepted to also occur in bacteria and archaea. Recent evidence supports the hypothesis that acetylation broadly impacts bacterial physiology [3]. Highly phosphorylated bacterial proteins have been described as being potential intermediates of degradative pathways [4, 5] and sulfated proteins have been shown to trigger the host immune system and bacterial cell-cell communication [6]. Bacterial surface structures such as flagella (Pseudomonas aeruginosa and Campylobacter jejuni) and pili (Neisseria spp. and P. aeruginosa) have been found to be particularly rich in post-translationally modified proteins [7]. Indeed studies on these organelles have led to the description of several complete microbial glycosylation models [8]. As many of the proposed bacterial glycoproteins are surface-exposed, these modified proteins have been postulated to play important roles in pathogenicity and antigenicity.
Type IV pili (T4P) of pathogenic Neisseria are hair-like structures that protrude from the bacterial surface and are implicated in a wide variety of processes including bacterial motility and DNA uptake [9]. Since T4P are also required for host-cell adhesion, and thus play a crucial role in colonisation of the host, they are considered as a major bacterial virulence factor. T4P are protein macropolymers predominantly composed of a single protein subunit, the major pilin. This pilin protein is arranged in a helical fashion to create the long and flexible pilus fibre. In Neisseria the major pilin is the protein PilE, which is highly post-translationally modified. It is always N-terminally processed and methylated and carries a pair of oxidised cysteines close to the C-terminus. It is glycosylated by the unusual glycan 2,4-diacetamido 2,4,6-trideoxy α-D-hexose (DATDH) [10] or 2-acetamido 4-glyceramido 2,4,6-trideoxy-α-D-hexose (GATDH) [11], which can be further elaborated by up to two galactose or glucose subunits and may be O-acetylated. In addition, this protein may also harbour a number of phosphoforms such as phosphate (P) [12], phosphoethanolamine (PE) [13, 14], phosphocholine (PC) [13–15] and phosphoglycerol (PG) [16].
PilE has been reported to be concurrently expressed as number of different proteoforms each carrying an array of different PTMs. (The term proteoform has recently been proposed to describe the different molecular forms in which the protein product of a single gene can be found, including changes due to genetic variations, alternatively spliced RNA transcripts and post-translational modification) [17]. This was first suggested from X-ray crystallography data that showed a weak electron density for phosphate around Ser94 in N. gonorrhoeae strain MS11, in addition to the phosphate on Ser68 [12]. This indicated that a small proportion of the PilE population harboured an additional phosphate group. More recently proteoforms of PilE from both Neisseria meningitidis and Neisseria gonorrhoeae have been evidenced by mass spectrometry in intact mass profiling experiments. In N. gonorrhoeae, pilins modified with both PE and PC have been reported and the ratio of the different phosphoforms expressed has been shown to be dependent on the presence of the glycan and of the minor pilin PilV [13]. In N. meningitidis, proteoform variation also centres around the phosphoform [18]. Mass profiling of the 8013 strain has shown PilE to be expressed as two proteoforms in a 4:1 major/minor ratio [11]. The minor proteoform carries an extra PG on Ser93. In this strain PG has been found to be a regulated PTM that mediates bacterial pathogenesis. Increased modification of PilE with PG at Ser93, several hours after host cell contact, changes the electrostatic surface of the pilus fibre and disrupts pilus/pilus interactions. This is a prerequisite for crossing the epithelial layer and a key step in pathogenesis [19]. The extent and variation of pilin PTMs in highly pathogenic N. meningitidis strains is therefore of particular interest, as is understanding their role in bacterial virulence [20].
The tools available at present to achieve complete PTM mapping of all expressed proteoforms of a protein of interest are extremely limited. Mass spectrometry-based proteomics has proven to be powerful approach for the identification of individual PTMs, leading in some case to the global identification of hundreds to thousands of PTMs within a sample [21]. However peptide-based, bottom-up proteomics fails to provide a complete picture of PTM since the connectivity between peptides and their parent proteoforms is lost. This is particularly important when two or more modifications work together on a single protein. Moreover, bottom-up strategies do not provide proteoform level information i.e. the explicit identities of the proteoforms present in the sample and their relative abundance, which is crucial information to understand proteoform function and in vivo regulation. Top-down mass spectrometry, which is based on the analysis of intact proteins, preserves the proteoforms and thus facilitates their full characterisation including PTMs [22, 23]. Top-down approaches have been successfully applied to the characterisation of various protein PTMs [23–25] and recent improvements in intact protein chromatographic separations and high performance Fourier transform mass spectrometry (FTMS) instrumentation have greatly expanded the observable range of proteoforms in complex samples. However the analytical requirements of top-down MS remain particularly challenging due to the size of the systems under study (proteins and not peptides). Top-down MS is currently limited to high-resolution instrumental platforms (FT-ICR, Orbitrap or high resolution ToF instruments) and separation of intact proteins can be a difficult task, as can efficient protein fragmentation on an LC time scale. Finally the software options available for top-down MS data analysis are rather limited, although recent developments in the field are helping to overcome this [26, 27].
To date the PTM complement of PilE has only been investigated in two different strains of N. meningitidis (8013 and C311) [10, 11, 16, 28] and the diversity of PTMs present in the population of N. meningitidis strains found in human patients remains unknown. Because of the link between PTMs and virulence, the analysis of novel strains is of great interest, but it is limited by the lack of approaches that are sufficiently rapid and amenable to high throughput analysis. To determine the feasibility of typing clinical strains in terms of pilin PTMs, we isolated a strain from a patient hospitalised with meningitis and characterised its pilin PTMs by different approaches. Mass profiling revealed the presence of four major proteoforms of PilE, in a 1:1:1:1 ratio. Initially, a bottom-up strategy, which had proven to be efficient for the analysis of the other strains was used, but it led to an incomplete PTM mapping. We therefore employed top-down mass spectrometry to select and fragment the intact proteoforms individually. Strengths and weaknesses of both approaches for the analysis of these challenging proteins will be discussed. We will show that when more than two proteoforms of a single protein are present, bottom-up alone is fundamentally unable to map all PTMs and top-down is required to achieve this goal. Top-down mass spectrometry therefore appears a promising tool for the analysis of highly post-translationally modified proteins involved in bacterial virulence.
Materials and Methods
Bacterial strain
The strain of Neisseria meningitidis used in this study was isolated from a patient with meningitis at the Limoges hospital in the Haute Vienne County of France in 2006 (strain number 278534). It is a serogroup A capsular serotype and multilocus sequence typing revealed that it is part of the ST-5 complex/subgroup III clonal complex. N. meningitidis was grown on solid GCB Agar (Laboratorios Conda, Spain) containing Kellogg’s supplements [29]. The major pilin gene (pilE) was PCR amplified and sequenced using oligos NG1705 (GTCAAACCCGGTCATTGTCC) and NG1706 (CAGGAGTCATCCAAATGAAAGC) [30].
PilE Preparation
Pili were prepared as described previously [31]. Briefly, the content of 10–12 Petri dishes was harvested in 5 mL of 150 mM ethanolamine at pH 10.5. Pili were sheared by vortexing for 1 min. Bacteria were centrifuged at 4,000 x g, 30 min, 4 °C and the resulting supernatant further centrifuged at 15,000 x g, 30 min, ambient temperature. The supernatant was removed, pili precipitated by the addition of 10 % vol. ammonium sulfate saturated in 150 mM ethanolamine pH 10.5 and allowed to stand for 1 h. The precipitate was pelleted by centrifugation at 4,000 x g, 1 h, 20 °C. Pellets were washed twice with PBS and suspended in 100 μL distilled water.
Bottom-up Mass Spectrometry
Ten μL of crude PilE preparation was suspended in Laemmli buffer and separated by SDS-PAGE. After removal of the Bio-Safe Coomassie stain (Biorad) used for visualisation, the band corresponding to PilE was excised and digested in-gel as described elsewhere [32]. Briefly gel pieces were reduced, alkylated and digested overnight with trypsin (Promega) at 37 °C. After desalting by C18 Ziptip® samples were eluted into 10 μL spray solution of ACN:H2O:HCOOH (50:50:0.1) for mass spectrometry. The resulting digest fragments were examined in positive ion mode by direct infusion nano-ESI, using a TriVersa Nanomate (Advion Biosciences, Harlow, UK) on an Orbitrap Velos mass spectrometer, equipped with an ETD module (Thermo Fisher Scientific, Bremen, Germany). A full set of automated positive ion calibrations was performed immediately prior to mass measurement, as were the calibrations for reagent ion transfer. All spectra were acquired in full profile mode. For MS experiments ions were accumulated in the ion trap and then transferred to the Orbitrap for high resolution mass measurement. For MS/MS experiments, ions were selected with an appropriate mass window and HCD was performed at normalised collision energies of 15–25 %, with other activation parameters left as default. For ETD the reagent gas was fluoranthene and the interaction time tuned to maximise sequence coverage. Supplemental activation was also applied. The FT automatic gain control (AGC) was set at 1×106 for MS and 2×105 for MSn experiments. Spectra were acquired in the FTMS over several minutes with between one and three microscans and a resolution of 60,000 for MS and 30,000 @ m/z 400 for MS/MS before being processed with Thermo Xcalibur 2.2. Peak picking was performed manually for all spectra and fragmentation maps were generated using a home built package.
Top-Down Mass Spectrometry
Top-Down experiments were performed on a solariX 12T hybrid Qh-FT-ICR (Bruker Daltonics, Billerica, MA) equipped with a hollow dispenser cathode. Crude protein extracts were desalted by C4 Ziptip® (Millipore) and eluted into 10 μL electrospray solution MeOH:H2O:HCOOH (75:25:3). Protein was introduced into the mass spectrometer through pulled borosilicate capillaries or by using a TriVersa NanoMate® (Advion Ithaca, NY). The NanoMate was the injection method of choice over pulled capillaries with wider tips, even for samples prone to aggregation and needle (nozzle) clogging. For mass profiling experiments ions were accumulated in the hexapole for 0.1–1 s before being transferred to the Infinity™ cell for detection. Spectra were accumulated for 50–200 scans. For MS/MS spectra, since the concentration of sample, and therefore single scan intensity, was highly preparation-dependent, ions were accumulated for ≤ 4s in the hexapole in order to reach a threshold precursor ion intensity. Ions were then transferred to the Infinity™ cell where ECD was performed with a pulse length of 5–10 ms and electron energy of 1.0–1.7 eV. For each experiment, 300–450 scans were accumulated. The number of data points (1 Mega points) was chosen to have near baseline resolution without detrimentally decreasing scan speed. Calibration was performed monthly with clusters of NaI.
Top-Down Data Analysis
Data processing was performed using DataAnalysis 4.0 SP5 (Bruker Daltonics, Billerica, MA). For MS experiments spectra were deconvoluted using the maximum entropy option. This gives much cleaner deconvoluted spectra than the other options available. For MS/MS experiments, acquired spectra were internally calibrated and peak picked using the SNAP 2.0 algorithm; quality factor 0.1, S/N 2, relative intensity 1×10−5 (%), absolute intensity 0 and a maximum charge state altered to just above the maximum observed charge state. Peak picking results were saved as an XML file and peak assignment was performed by importing this data into a home built package for ion assignment and automated fragmentation map creation. PTM assignment was performed manually with this software on combined peak lists from the 14+ and 15+ charge states.
Results and Discussion
Mass profiling
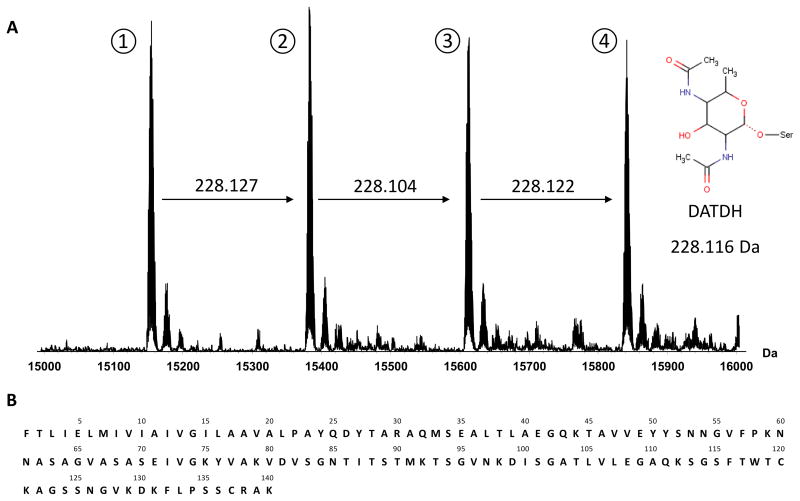
Purification of PilE from the 278534 strain produced a large amount of protein in high purity (see SDS-PAGE in Suppl. Fig. 1). When measured by FT-ICR MS the crude sample gave a complex MS spectrum exhibiting several different charge state envelopes. Deconvolution of the raw data gave four major peaks with monoisotopic neutral molecular masses (Mr) of 15,146.7058, 15,374.8325, 15,602.9369 and 15,831.0584 in an approximate 1:1:1:1 ratio (Figure 1A).
Figure 1.
To provide a reference point for further investigation the pilE gene from the N. meningitidis 278534 strain was sequenced. Surface expressed PilE is known to be post-translationally processed by the endoprotease pilD which cleaves a short N-terminal leader sequence or prepilin before a conserved phenylalanine residue [33]. Making this modification to the initial 147 amino acid sequence furnished a 140 amino acid protein with the theoretical Mr of 14,524.47 (sequence depicted in Figure 1B). Even when compared to the lowest mass major peak observed in the MS profile this represents a difference of over 620 Da and indicated that PilE from this particular clinical strain could be highly post-translationally modified.
Bottom-up analysis
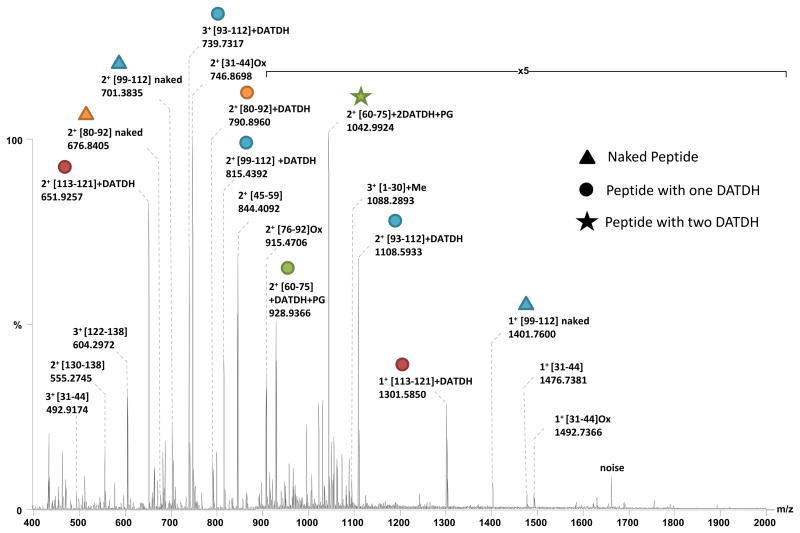
A MS strategy based on the combination of accurate high resolution intact mass measurement of proteoforms and tandem mass spectrometry experiments on peptides (bottom-up approach) had previously proven useful in identifying PTMs on PilE from N. meningitidis strain 8013 [19, 34]. Therefore a similar approach was initially employed here. A sample of PilE was subjected to SDS-PAGE followed by in-gel tryptic digestion. The digest was then analysed by nanoESI-FTMS on an Orbitrap mass spectrometer (Figure 2).
Figure 2.
Comparison of the experimental masses measured for this tryptic digest and theoretical ones calculated in silico from the PilE sequence revealed the presence of non-modified (naked) peptides spanning the ranges [31–59], [76–112] and [122–138]. A [1–30]+14.016 Da peptide was also observed, confirming the N-terminal methylation of PilE and leading to a sequence coverage of over 80% (Table 1).
Table 1.
Digest product ions of PilE from 278534 strain (from Figure 2).
| Digestion Product | Measured m/z | Charge | Measured Monoisotopic Mass [M+H]+ | Theoretical Monoisotopic Mass [M+H]+ | Error (ppm) |
|---|---|---|---|---|---|
| [1–30]+Me | 1088.2893 | 3 | 3262.8533 | 3262.8524 | 0.290 |
| [31–44] | 492.9174 | 3 | 1476.7376 | 1476.7363 | 0.912 |
| [31–44] | 738.8725 | 2 | 1476.7377 | 1476.7363 | 0.964 |
| [31–44] | 1476.7381 | 1 | 1476.7381 | 1476.7363 | 1.219 |
| [31–44]Ox | 746.8698 | 2 | 1492.7323 | 1492.7312 | 0.753 |
| [31–44]Ox | 1492.7366 | 1 | 1492.7366 | 1492.7312 | 3.618 |
| [45–59] | 844.4092 | 2 | 1687.8111 | 1687.8326 | 12.724* |
| [60–75]+DATDH+PG | 928.9366 | 2 | 1856.8659 | 1856.8637 | 1.197 |
| [60–75]+2DATDH+PG | 1042.9924 | 2 | 2084.9775 | 2084.9747 | 1.354 |
| [76–92]Ox | 915.4706 | 2 | 1829.9339 | 1829.9313 | 1.434 |
| [80–92] | 676.8405 | 2 | 1352.6737 | 1352.6726 | 0.831 |
| [80–92]+DATDH | 790.8960 | 2 | 1580.7847 | 1580.7836 | 0.711 |
| [93–112]+DATDH | 739.7317 | 3 | 2217.1805 | 2216.1769 | † |
| [93–112]+DATDH | 1108.5933 | 2 | 2216.1793 | 2216.1769 | 1.094 |
| [99–112] | 701.3835 | 2 | 1401.7597 | 1401.7584 | 0.944 |
| [99–112] | 1401.7600 | 1 | 1401.7600 | 1401.7584 | 1.141 |
| [99–112]+DATDH | 815.4392 | 2 | 1629.8711 | 1629.8694 | 1.057 |
| [113–121]+DATDH | 651.2957 | 2 | 1301.5841 | 1301.5831 | 0.786 |
| [113–121]+DATDH | 1301.5850 | 1 | 1301.5850 | 1301.5831 | 1.460 |
| [122–138] | 604.2972 | 3 | 1810.8770 | 1809.8912 | -* |
| [130–138] | 555.2745 | 2 | 1109.5417 | 1109.5408 | 0.832 |
Errors for these ions are greater than expected since these digest products contain asparagine vicinal to glycine residues and are therefore subject to facile deamidation. For the ion at m/z 844.4092 deamidation is incomplete but the monoisotopic peak is very close to the baseline. For the ion at m/z 604.2972 complete deamidation increases the observed mass by approximately 1 Da.
The monoisotopic peak for this ion is obscured by the doubly charged [31–44] at m/z 738.8725.
All peptide ions were isolated and subjected to Higher energy Collision Dissociation (HCD) in order to confirm their identity (data not shown). Despite the high sequence coverage obtained with these ions, numerous abundant, multiply charged peaks present in the MS spectrum could not be attributed. In addition, no peptide spanning the [60–75] region could be assigned. Since this region of PilE is almost always post-translationally modified, its absence encouraged us to investigate these multiply charged, non-assigned peaks in the hunt for additional PTM bearing peptides.
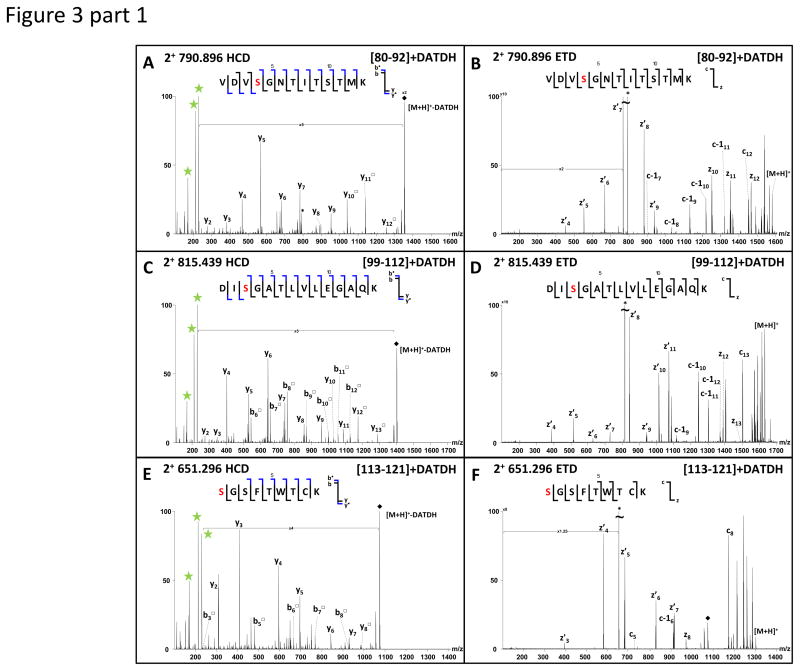
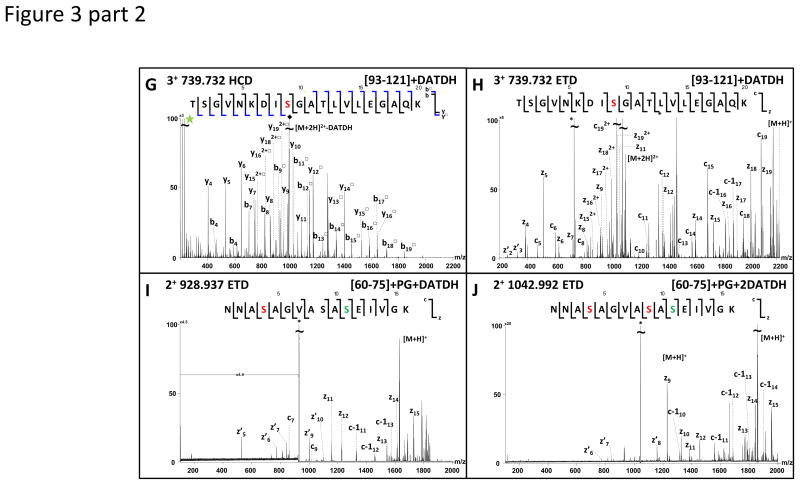
When ions observed at m/z 790.8960, 815.4392, 651.2957 and 739.7317 were subjected to HCD, three very abundant fragment ions appeared at m/z 229.118, 211.108 and 169.097 in the resulting MS/MS spectra (Figure 3 A, C, E, G).
Figure 3.
These ions correspond to the oxonium ion of the DATDH glycan, its dehydrated partner and a fragment ion characteristic of the glycan core and indicate that all fragmented peptides contain this sugar moiety. DATDH is a previously described PTM for PilE and these reporter ions have previously been used to identify glycosylated peptides [35, 36]. The analysis of the other fragment ions in the spectra (y/b ions) confirmed the sequence for the four peptides ions as [80–92], [99–112], [113–121] and [93–112] respectively, but could not be used to localise the sites of glycosylation. All four precursor ions were therefore subjected to electron transfer dissociation (ETD) in order to confidently localise the glycosylation sites (Figure 3 B, D, F, H). For the [80–92] peptide, the glycan could be easily localised on Ser83, for [99–112] and [93–112] it was localised exclusively on Ser101 and finally for [113–121], Ser113 was found to be modified with DATDH. In none of the ETD spectra was any trace of the reporter ions detected at m/z 229.118, 211.108 or 169.097 (see Supplementary Tables for all MS/MS data).
In addition, when fragmented by HCD, two other doubly charged ions, at m/z 928.934 and 1042.999, produced intense reporter ion signals for DATDH. In both cases the [60–75] tryptic peptide was identified suggesting multiple forms to be present, each modified by DATDH. ETD data identified a DATDH on Ser63 for the precursor m/z 928.934, on both Ser63 and Ser68 for m/z 1042.99 and showed that both forms were further modified by PG on Ser70 (Figure 3 I, J).
Upon inclusion of these modified peptides, the sequence coverage from the digest was extended to 98 % with only the two last amino acids of the sequence, AK, unaccounted for. These results were corroborated but not improved upon using a LC-MS approach. Interestingly, the presence of several peptides in both non-modified and modified forms confirmed that PilE was expressed as multiple proteoforms. Furthermore it appeared that these proteoforms differ in the number of DATDH glycan units. This hypothesis correlated perfectly with the pattern observed by mass profiling, where the mass difference between the four major peaks is 228.11 Da - the mass expected from addition of DATDH. PilE thus appeared to be present in multiple glycoforms each bearing a different number of DATDH subunits.
Armed with mass profiling data and the results from the bottom-up experiments, one can begin to assign peptide combinations, and thus PTMs, to specific proteoforms. Since all peptides are found in non-modified forms apart from [113–121] and [60–75], the lowest mass forms of these two peptides [113–121]+DATDH and [60–75]+PG+DATDH plus the exclusively non-modified peptides, may naturally be attributed to the lowest mass proteoform. This assignment seems satisfactory since it results in the correct 15,146.7058 Da protein mass with an experimental-theoretical mass error of only 0.03 ppm. The combination of peptides giving the protein with the heaviest mass; [60–75]+PG+2DATDH, [80–92]+DATDH, [99–112]+DATDH, [113–121]+DATDH could similarly be assigned to the highest mass proteoform. This also seems correct and is the only peptide combination possible to reach a total mass of 15,831.0584 Da. However a problem arises when considering the two intermediate proteoforms, since various combinations of peptides are possible to achieve the observed protein masses. Here, even with the mass profile as a reference, a bottom-up methodology is intrinsically unable to relate modified peptides to their parent proteoforms without making several important assumptions. One may think that examining the ion intensities of modified peptides and relating them to abundances may help solve this problem, but this is approach is unrealistic since PTM has been well documented to drastically affect peptide ionisation efficiency [37].
To achieve complete proteoform mapping, each proteoform must be investigated separately. Off-line fractionation methods require prior knowledge of the PTMs present on each proteoforms and are often difficult to implement at the protein level. A top-down MS approach allows each proteoform to be easily isolated and fragmented separately in the mass spectrometer. This top-down strategy was therefore applied to PilE. We initially sought to use the Orbitrap Velos and ETD fragmentation for this purpose but rapidly realised that in our hands this instrument was not optimised for the analysis of intact proteins and that it would not give the required sequence coverage. Therefore a 12T FT-ICR mass spectrometer was chosen for all top-down experiments, using ECD (electron capture dissociation) as the fragmentation technique.
Top-down analysis
PilE from N. meningitidis 8013 strain has previously been investigated by top-down ECD MS/MS using an FT-ICR mass spectrometer (unpublished data) and experimental parameters optimised in that study were used as a starting point here (irradiation time, energy of the electrons etc.). The PilE sample examined in this study did however exhibit some differences compared to that purified from the 8013 reference strain (PilE-8013). Clogging of nanoelectrospray needles during spectral acquisition was much more acute; despite centrifugation prior to sample injection and the use of pulled capillaries with a wider tip. A more stable spray was obtained using a Triversa NanoMate and most importantly this injection method allowed the spray signal to be monitored during spectral acquisition and the nozzle to be quickly changed if necessary. The charge state envelope was also different for the 278534 strain. It was shifted to lower charge states, with the maximum observed here 16+ compared to 19+ for the 8013 strain. This is possibly due to the difference in protein size or the different nature and number of PTMs carried by the two proteins. The envelope was also much more complex, the presence of four major proteoforms causing some proteoforms of one charge state to overlap with other proteoforms from another. Proteoforms of the highest exploitable charge states (15+ and 14+) were isolated in the hexapole of the 12T FT-ICR mass spectrometer and single charge states were subjected to top-down ECD MS/MS.
In general the 15+ ions afforded greater sequence coverage for the same precursor intensity but the 14+ charge state was more abundant and, for some proteoforms, furnished even greater sequence coverage when precursor ions were allowed to accumulate. Extensive fragmentation was observed in the top-down spectra especially in the case of the highest mass proteoform (15,831.0584 Da) (Figure 4D). Cleavage maps are also shown for the other three proteoforms (Figure 4 A–C). Interestingly the intact cysteine bridge between Cys120-Cys137 appeared to strongly inhibit fragmentation in this region for the 14+ charge state but not for the 15+ charge state.
Figure 4.
For proteoform 1, fairly extensive N- and C-terminal fragmentation enabled straightforward identification of the methylated N-terminus and a DATDH glycan on Ser113. The c62 ion at m/z 1115.4284 (6+), 1339.3085 (5+), c66 ion at m/z 1205.8037 (6+), 1446.7672 (5+) and c67 ion at m/z 1217.6449 (6+) enabled localisation of a second DATDH on Ser63. The presence of the same c ions in spectra obtained from both the 14+ and 15+ protein forms increases confidence in this assignment, as does the presence of multiple charge states for c66 in each spectrum. In addition the C-terminal fragment ions z70, z74 and y73 enabled identification of a phosphoglycerol group on Ser68 or Ser70 but the absence of ions between these residues precluded definitive localisation. This was however provided by the bottom-up data that showed Ser70 to be exclusively modified with phosphoglycerol. For proteoform 1 this gave a PTM complement of two DATDH glycans at Ser63 and Ser113 and a phosphoglycerol group at Ser70 in addition to the expected N-terminal methylation and cysteine bond. The theoretical protein mass for this assignment of 15,146.7017 Da correlated exceptionally well with the 15,146.7058 Da experimental value, giving a + 0.03 ppm error.
For proteoform 2, two DATDH subunits were also easily identified on Ser63 and Ser113; the former by the c60 and c66 ions and the latter by a large series of z ions from z26 to z52. Despite poor fragmentation in the central region of the protein, the sequence coverage was sufficient to indicate that no PTM was present between residues 71 and 112. The difference in mass between z70 and z74 (698.2519 Da) indicated the presence of both a PG and a DATDH between Ala67 and Ser70 but as for proteoform 1 the absence of fragments between these residues prevented definitive location. Taking into account the bottom-up data a PG group was assigned on Ser70 and therefore a DATDH assigned to Ser68. Again the measured mass of 15,374.8325 Da corresponded excellently to the theoretical mass for this PTM assignment (15,374.8122 Da) with an error of +1.3 ppm. In comparison to proteoform 1, proteoform 2 exhibited an additional DATDH subunit at Ser68.
For proteoform 3, a better sequence coverage in the regions of interest allowed an easy assignment of the four DATDH groups to Ser63, Ser83, Ser101 and Ser113. Again a PG could be identified either on Ser68 or Ser70 and bottom-up data were used to confirm the latter position. As the number of modifications increases so does the number of possibilities for site localisation. Deciding upon the correct PTM assignment therefore becomes increasingly difficult, especially when potential modification sites are close together in the protein sequence. In the case of proteoform 3, the large series of z type ions from z55 to z70 allowed confident assignment of DATDH on Ser83 rather than Ser68. Our results indicated that, in this proteoform, two previously unmodified serines are now glycosylated and one which was occupied in proteoform 2 is now non-modified. Similarly to proteoforms 1 and 2, the measured mass of 15,602.9369 Da corresponds excellently to the theoretical mass for this PTM assignment (15,602.9226 Da) with an error of +0.9 ppm.
Finally, for the highest mass proteoform (proteoform 4) the assignment of four DATDH was found to be similar to proteoform 3 and an additional glycan was localised on Ser68, as already observed for proteoform 2. This proteoform led to the highest sequence coverage in ECD (62%) and the presence of the c69, c70 and complementary z70 ions mean that for this proteoform no bottom-up data is required to assign PG to Ser70. The measured mass of 15,831.0584 Da again corresponds very well to the theoretical mass (15,831.0331 Da) with an error of +1.5 ppm.
These results indicated that in most cases spectra were of sufficient quality and fragmentation sufficiently extensive to allow PTMs to be unambiguously localised on the protein backbone. The only exception was for the location of the PG group which needed information from bottom-up experiments to provide the exact modification site for three of the four proteoforms. This result may be explained by a lower efficiency of electron capture at sites close to PTMs, or even capture of the electron by the modifications themselves. This would likely modify the overall reactivity of the ECD process.
Taken in the context of the mass profiling experiment, the correlation between experimental and theoretical masses for all PTM assignments is excellent, with errors at the protein level consistently below 2 ppm. The complement of post-translational modifications has been explicitly defined for each proteoform and the bottom-up and top-down data are in perfect agreement. All proteoforms are modified with a PG at Ser70 and DATDH at Ser63 and Ser113. The extra sites of glycosylation are Ser68, Ser83 and Ser101 but our results also revealed that the glycosylation process does not appear to be completely successive. This was not expected since the glycerotransferase PglO is known to transfer the glycan en-bloc to the PilE substrate and we might therefore expect sequential glycan addition based on decreasing affinity for sites on the protein backbone. Our results suggest that modification at one site may alter the affinity for the others, however the specificity of this enzyme is currently unknown. Most importantly, this level of glycosylation is a hitherto unreported phenomenon for pilin of Neisseria spp.
Strengths and weaknesses of both approaches
First of all, it is important to point out that, when aiming to characterise all PTM present on a protein or its different proteoforms, the mass profiling experiment is a key piece of information. In the case of the strain studied here, mass profiling of PilE indicated the presence of four major proteoforms, each expressed in similar abundance and each modified with phosphoglycerol plus two, three, four and five DATDH subunits respectively. This acted as primary reference for all future experiments.
The bottom-up approach was powerful for identifying post-translationally modified peptides and when coupled with an appropriate fragmentation technique such as ETD, for localising the sites of post-translational modification themselves. However, in order to achieve complete characterisation of a mixture of proteoforms and thus explicitly define the PTM content of each one, it is necessary to link these pieces of information together and to relate modified peptides to their parent proteoforms. Given the homogeneity of proteoform abundance, the strain characterised here was an ideal case to explore whether a bottom-up proteomics approach is able to achieve this goal, or not. Our results clearly show that although it is possible to completely map all PTMs on the lightest and heaviest forms of PilE, information obtained from the bottom-up approach is not sufficient for mapping of the middle forms. On the other hand, by selecting individual proteoforms and subjecting them to top-down ECD MS/MS the connectivity that was lost by the bottom-up approach was retained thus allowing the complete assignment of all glycosylation sites. This is expected to be the case in other mixtures of more than two proteoforms where each proteoform is modified by different numbers of a particular PTM. One must also point out that an important weakness of the top-down approach remains the analysis of data; from peak picking, to PTM assignment and scoring. Although currently available bioinformatics tools are sufficient for proteins with one or two known modifications, the analysis of highly modified proteins remains a challenge and manual interpretation is often needed. Improvement in this field is a requirement for more confident PTM assignment and high throughput analysis.
Conclusion
In this study the protein PilE, purified from a previously uncharacterised strain of Neisseria meningitidis isolated from a patient hospitalised with meningitis, has been analysed by different mass spectrometric approaches. Mass profiling showed that PilE exists as four major proteoforms, differing only by the number of DATDH glycans. A bottom-up strategy was initially chosen to map all PTMs of the four proteoforms. ETD on glycopeptides proved very useful for reliable assignment of glycosylation sites, however bottom-up only proved capable in characterising the PTM content of the lightest and heaviest proteoforms but not those of intermediate mass. Individual proteoforms were therefore selected and subjected to top-down ECD MS/MS on a 12T FT-ICR mass spectrometer. The sequence coverage obtained in each case allowed unambiguous identification of an increasing number of glycan subunits. Combining the top-down and bottom-up data allowed complete PTM characterisation of all proteoforms. The lightest form was found to be N-terminally processed and methylated, to carry a disulfide bridge close to the C-terminus, one phosphoglycerol on Ser70 and DATDH at Ser63 and Ser113. The additional glycosylation sites for the other proteoforms are Ser68, Ser83 and Ser101. Such an extent of glycosylation and indeed of PTM has never before been described for PilE and may be linked to increased pathogenicity or antigenicity. In general our results show that, in proteoform mixtures with more than two components, where multiple modifications of the same mass are present, a top-down mass spectrometry approach is necessary for complete proteoform mapping. In this study a 12T FT-ICR mass spectrometer was chosen for the top-down approach but it is probable that in the near future these experiments will be possible on a routine basis on other instrumental platforms such as later generation Orbitrap systems.
Supplementary Material
Acknowledgments
This work was supported by INSERM (ATIP-Avenir starting grant) and by the European Research Council (starting grant) (GD), by the CNRS and Institut Pasteur (JCR) and NIH grants P41 RR10888/GM104603 and S10 RR025082 (CEC). JCR and JG gratefully acknowledge the Monge PhD scholarship from Ecole Polytechnique which has funded the research placement of JG in Prof Costello’s group.
References
- 1.Broberg CA, Orth K. Tipping the balance by manipulating post-translational modifications. Current Opinion in Microbiology. 2010;13:34–40. doi: 10.1016/j.mib.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ribet D, Cossart P. Post-translational modifications in host cells during bacterial infection. Febs Lett. 2010;584:2748–2758. doi: 10.1016/j.febslet.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Hu LI, Lima BP, Wolfe AJ. Bacterial protein acetylation: the dawning of a new age. Molecular Microbiology. 2010;77:15–21. doi: 10.1111/j.1365-2958.2010.07204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosen R, Becher D, Buttner K, Biran D, et al. Highly phosphorylated bacterial proteins. Proteomics. 2004;4:3068–3077. doi: 10.1002/pmic.200400890. [DOI] [PubMed] [Google Scholar]
- 5.Mijakovic I. Protein phosphorylation in bacteria. Febs Journal. 2010;277:20–21. [Google Scholar]
- 6.Han S-W, Lee S-W, Bahar O, Schwessinger B, et al. Tyrosine sulfation in a Gram-negative bacterium. Nature Communications. 2012;3 doi: 10.1038/ncomms2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwashkiw JA, Vozza NF, Kinsella RL, Feldman MF. Pour some sugar on it: the expanding world of bacterial protein O-linked glycosylation. Molecular Microbiology. 2013;89:14–28. doi: 10.1111/mmi.12265. [DOI] [PubMed] [Google Scholar]
- 8.Nothaft H, Szymanski CM. Protein glycosylation in bacteria: sweeter than ever. Nature Reviews Microbiology. 2010;8:765–778. doi: 10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- 9.Giltner CL, Nguyen Y, Burrows LL. Type IV Pilin Proteins: Versatile Molecular Modules. Microbiology and Molecular Biology Reviews. 2012;76:740–772. doi: 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stimson E, Virji M, Makepeace K, Dell A, et al. Meningococcal Pilin - A Glycoprotein Substituted With Digalactosyl 2,4-Diacetamido-2,4,6-Trideoxyhexose. Molecular Microbiology. 1995;17:1201–1214. doi: 10.1111/j.1365-2958.1995.mmi_17061201.x. [DOI] [PubMed] [Google Scholar]
- 11.Chamot-Rooke J, Rousseau B, Lanternier F, Mikaty G, et al. Alternative Neisseria spp. type IV pilin glycosylation with a glyceramido acetamido trideoxyhexose residue. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14783–14788. doi: 10.1073/pnas.0705335104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forest KT, Dunham SA, Koomey M, Tainer JA. Crystallographic structure reveals phosphorylated pilin from Neisseria: phosphoserine sites modify type IV pilus surface chemistry and fibre morphology. Molecular Microbiology. 1999;31:743–752. doi: 10.1046/j.1365-2958.1999.01184.x. [DOI] [PubMed] [Google Scholar]
- 13.Hegge FT, Hitchen PG, Aas FE, Kristiansen H, et al. Unique modifications with phosphocholine and phosphoethanolamine define alternate antigenic forms of Neisseria gonorrhoeae type IV pili. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10798–10803. doi: 10.1073/pnas.0402397101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aas FE, Egge-Jacobsen W, Winther-Larsen HC, Lovold C, et al. Neisseria gonorrhoeae type IV pili undergo multisite, hierarchical modifications with phosphoethanolamine and phosphocholine requiring an enzyme structurally related to lipopolysaccharide phosphoethanolamine transferases. Journal of Biological Chemistry. 2006;281:27712–27723. doi: 10.1074/jbc.M604324200. [DOI] [PubMed] [Google Scholar]
- 15.Weiser JN, Goldberg JB, Pan N, Wilson L, Virji M. The phosphorylcholine epitope undergoes phase variation on a 43-kilodalton protein in Pseudomonas aeruginosa and on pili of Neisseria meningitidis and Neisseria gonorrhoeae. Infect Immun. 1998;66:4263–4267. doi: 10.1128/iai.66.9.4263-4267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stimson E, Virji M, Barker S, Panico M, et al. Discovery of a novel protein modification: alpha-glycerophosphate is a substituent of meningococcal pilin. Biochem J. 1996;316:29–33. doi: 10.1042/bj3160029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith LM, Kelleher NL. Consortium Top Down, P. Proteoform: a single term describing protein complexity. Nature Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jen FEC, Warren MJ, Schulz BL, Power PM, et al. Dual Pili Post-translational Modifications Synergize to Mediate Meningococcal Adherence to Platelet Activating Factor Receptor on Human Airway Cells. PLoS pathogens. 2013;9:e1003377–e1003377. doi: 10.1371/journal.ppat.1003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chamot-Rooke J, Mikaty G, Malosse C, Soyer M, et al. Posttranslational Modification of Pili upon Cell Contact Triggers N. meningitidis Dissemination. Science. 2011;331:778–782. doi: 10.1126/science.1200729. [DOI] [PubMed] [Google Scholar]
- 20.Quagliarello V. Dissemination of Neisseria meningitidis. N Engl J Med. 2011;364:1573–1575. doi: 10.1056/NEJMcibr1101564. [DOI] [PubMed] [Google Scholar]
- 21.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nature Biotechnology. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 22.Lanucara F, Eyers CE. Top-down mass spectrometry for the analysis of combinatorial post-translational modifications. Mass Spectrom Rev. 2013;32:27–42. doi: 10.1002/mas.21348. [DOI] [PubMed] [Google Scholar]
- 23.Ansong C, Wu S, Meng D, Liu X, et al. Top-down proteomics reveals a unique protein S-thiolation switch in Salmonella Typhimurium in response to infection-like conditions. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10153–10158. doi: 10.1073/pnas.1221210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Ge Y. Comprehensive Analysis of Protein Modifications by Top-Down Mass Spectrometry. Circulation-Cardiovascular Genetics. 2011;4:711. doi: 10.1161/CIRCGENETICS.110.957829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siuti N, Kelleher NL. Decoding protein modifications using top-down mass spectrometry. Nature Methods. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu XW, Sirotkin Y, Shen YF, Anderson G, et al. Protein Identification Using Top-Down. Molecular & Cellular Proteomics. 2012:11. doi: 10.1074/mcp.M111.008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zamdborg L, LeDuc RD, Glowacz KJ, Kim YB, et al. ProSight PTM 2. 0: improved protein identification and characterization for top down mass spectrometry. Nucleic Acids Research. 2007;35:W701–W706. doi: 10.1093/nar/gkm371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marceau M, Forest K, Béretti JL, Tainer J, Nassif X. Consequences of the loss of O-linked glycosylation of meningococcal type IV pilin on piliation and pilus-mediated adhesion. Molecular Microbiology. 1998;27:705–715. doi: 10.1046/j.1365-2958.1998.00706.x. [DOI] [PubMed] [Google Scholar]
- 29.Kellogg DS, Jr, Cohen IR, Norins LC, Schroeter AL, Reising G. Neisseria gonorrhoeae. II. Colonial variation and pathogenicity during 35 months in vitro. J Bacteriol. 1968;96:596–605. doi: 10.1128/jb.96.3.596-605.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kahler CM, Martin LE, Tzeng YL, Miller YK, et al. Polymorphisms in pilin glycosylation locus of Neisseria meningitidis expressing class II pili. Infect Immun. 2001;69:3597–3604. doi: 10.1128/IAI.69.6.3597-3604.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carbonnelle E, Helaine S, Prouvensier L, Nassif X, Pelicic V. Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occurring after pilus assembly, essential for fibre stability and function. Molecular Microbiology. 2005;55:54–64. doi: 10.1111/j.1365-2958.2004.04364.x. [DOI] [PubMed] [Google Scholar]
- 32.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature Protocols. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 33.Strom MS, Nunn DN, Lory S. A Single Bifunctional Enzyme, PilD, Catalyzes Cleavage and N-Methylation of Proteins Belonging to the Type-IV Pilin Family. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:2404–2408. doi: 10.1073/pnas.90.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gault JMC, Duménil G, Chamot-Rooke J. A Combined Mass Spectrometry Strategy for Complete Posttranslational Modification Mapping of N. meningitidis Major Pilin. Journal of Mass Spectrometry. 2013 doi: 10.1002/jms.3262. Accepted. [DOI] [PubMed] [Google Scholar]
- 35.Vik A, Aas FE, Anonsen JH, Bilsborough S, et al. Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4447–4452. doi: 10.1073/pnas.0809504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anonsen JH, Vik A, Egge-Jacobsen W, Koomey M. An Extended Spectrum of Target Proteins and Modification Sites in the General O-Linked Protein Glycosylation System in Neisseria gonorrhoeae. J Proteome Res. 2012;11:5781–5793. doi: 10.1021/pr300584x. [DOI] [PubMed] [Google Scholar]
- 37.Gao Y, Wang Y. A method to determine the ionization efficiency change of peptides caused by phosphorylation. Journal of the American Society for Mass Spectrometry. 2007;18:1973–1976. doi: 10.1016/j.jasms.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.