Abstract
Neutrophils are the main proinflammatory cell type in chronically infected lungs of cystic fibrosis (CF) patients; however, they fail to effectively clear the colonizing pathogens. Here, we investigated the molecular composition of non-mucoid and mucoid Pseudomonas aeruginosa-induced neutrophil extracellular traps (NETs) in vitro and compared them to the DNA-protein complexes present in the CF sputum. The protein composition of P. aeruginosa-induced NET fragments revealed that irrespective of the inducing stimuli, NET fragments were decorated with a conserved set of proteins. The DNA-protein complexes derived from CF sputum were consistent with NETosis and shared a similar protein signature, suggesting that the majority of the extracellular DNA was NET derived. The ability of polymorphonuclear leukocytes to produce NETs in response to P. aeruginosa was driven by macrophage migration-inhibitory factor (MIF) by promoting mitogen-activated protein kinase. Analysis of 132 CF patient samples revealed that elevated MIF protein levels correlated with poorer lung function. We suggest that targeting MIF by small molecular inhibitors might reduce the presence of extracellular DNA and serve as an adjunct to the use of antimicrobial drugs that could ultimately reduce bacterial fitness in the lungs during the later stages of CF disease.
Key Words: Cystic fibrosis, Infection, Inflammation, Innate immunity, Neutrophil extracellular traps, Pseudomonas aeruginosa
Introduction
Cystic fibrosis (CF) is a lung disease that features chronic inflammation of the airways associated with bacterial colonization. It is caused by mutations in the CF transmembrane regulator (CFTR), a plasma membrane channel that regulates the balance of bicarbonate and chloride secretions across the epithelial layer of the airways. Patients with CF have an elevated presence of plugs containing mucus, DNA, and proteins complexed with bacteria that obstruct airflow. These structures accumulate due to an electrolyte imbalance and a consequent inability of the epithelial cilia to beat and mediate mechanical clearance. The vast amounts of extracellular DNA (eDNA) are traditionally assumed to originate from dying cells. Notably, there is evidence that eDNA levels correlate with neutrophil counts and can be used as an index of inflammation and lung disease severity [1, 2].
The discovery of neutrophil extracellular trap (NET) release as a unique mechanism of cell death prompted us to reevaluate the nature of the eDNA found in CF patients. NETs are a neutrophil-mediated, innate immune response to close-proximity extracellular pathogens [3]. The NET-releasing event, referred to as NETosis, consists of enzyme-triggered decondensation of chromatin, rupturing of the nuclear membrane, and subsequent release of chromatin-containing structures complexed with granular or intracellular proteins into the extracellular space [4, 5, 6, 7]. Previous studies implicated NETosis as a contributor to the eDNA pool in CF [8, 9, 10]. These studies consistently identified that the eDNA is in complex with neutrophil elastase (NE) and myeloperoxidase (MPO), two proteins previously described as markers of NETosis. Interestingly, Papayannopoulos et al. [9] showed that sputum solubilization depended on the release of active NE from NETs, which facilitated DNase I activity by degrading NET-associated histones. Given the positive and negative effects that NE has during the pathogenesis of CF, Dubois et al. [8] proposed that regulating the NE activity by protease inhibitors might provide means to limit inflammation-induced damage in CF. While NE-specific inhibitors are not currently available for treatment, they are under development, and, therefore, this approach seems feasible. However, the question that remains is whether adding additional modalities to the already widely accepted DNase I treatment (e.g., Pulmozyme) is the strategy to take, or whether novel therapeutic approaches that limit the level of NETosis might be a more promising strategy to reduce eDNA burden in CF.
To this end, we were interested in defining endogenous signals or pathways that stimulate NETosis, with the expectation that NETosis should be limited or restricted to gain therapeutic benefit. Prior research showed that NETosis was triggered by activation of the mitogen-activated protein kinase (MAPK) pathway, was dependent on reactive oxygen species (ROS), and stimulated by cytokines [11]. Since these pathways phenotypically mirrored the functional activities of macrophage migration-inhibitory factor (MIF), a possible correlation between MIF and NETosis was investigated. MIF is a ‘danger’ signal released in response to pathogenic stimuli with potent proinflammatory functions. Extracellular MIF is recognized by an array of receptors, including CD74 [12], CXCR2 [13], CXCR4 [14], and CXCR7 [15], to promote cellular survival and maintain proinflammatory cytokine synthesis [16, 17]. MIF overrides the anti-inflammatory activities of corticosteroids [18], which makes it an attractive molecule to investigate in the context of neutrophil-rich corticosteroid-resistant inflammation in CF. We show evidence that NET production was promoted by MIF in an autocrine (or paracrine) manner via potentiating MAPK signaling, thus discovering a potential therapeutic target.
Experimental Procedures
Bacterial Strains
The non-mucoid Pseudomonas aeruginosa strains: 2192 nmr and the mucoid strains PA581, PA14 mucA, and 2192 were used throughout these experiments and provided by Dr. G. Pier (Brigham and Women's Hospital, Harvard Medical School, HMS) and were previously described [19, 20]. The P. aeruginosa PA14, PA14 fliC, PAO1, and PAo1 fliC strains were generously provided by Dr. S. Lory (HMS).
Mice
Ethics Statement. All studies were performed in accordance with the HMS Institutional Animal Care and Use Committee guidelines. The experimental protocols were approved by the Institutional Animal Care and Use Committee of the Harvard Medical Area Office for Research Subject Protection.
Breeding pairs of MIF knockout (KO) mice were obtained from Dr. Craig Gerard (Children's Hospital, Boston) and maintained at the Massachusetts School of Pharmacy Animal Care Facility. Control mice (C57BL6) were obtained from Charles River.
CF Serum and Sputum Samples
Serum samples from CF patients were collected at the Aarhus University Hospital during 2002-2004, as previously described [21]. The study was approved by the Ethics Committee of Aarhus County and samples were collected with signed informed consent [21]. The analysis of the CF sputum samples was performed using discarded patient material as approved by the standing Human Research Committee at Partners, Boston, Mass., USA.
Isolation of Primary Human Neutrophils
Polymorphonuclear leukocytes (PMNs) were isolated from healthy human donors. Blood (10 ml) was drawn from healthy individuals with their informed consent using a sodium-heparin blood collection kit (Becton Dickinson Vacutainer Safety-Lok blood collection set). Blood was inverted to mix with the anticoagulant agent. A density gradient was prepared using polymorph reagent (Axis-Shield) following the manufacturer's instructions. The blood was layered on the gradient and centrifuged at 500 g for 30 min at room temperature using a swing bucket centrifuge without brake. Purified PMNs were resuspended in 5 ml of HBSS buffer without Ca2+ and Mg2+ buffer (HBSS−/−; Invitrogen, Carlsbad, Calif., USA). Immediately prior to adding bacteria, PMNs were pelleted at 400 g for 10 min and resuspended in HBSS with Ca2+, Mg2+, and 0.1% gelatin (GHBSS++; Invitrogen).
Isolation of Primary Murine Neutrophils
Bone marrow was flushed out from the femurs and tibias from 6- to 8-week-old MIF KO and C57BL6 mice. Cells were resuspended in PBS and 5 mM EDTA, and spun down to pellet at 600 g for 10 min at 4°C. Cell pellets were resuspended in 45% Percoll solution and layered over a gradient composed of 3 ml 81% Percoll, 2 ml 62% Percoll, 2 ml 55% Percoll, and 2 ml 50% Percoll. The gradient was centrifuged at 1,600 g for 30 min at 10°C with no brake. Cells were collected from the interface of the 81 and 62% layers. Cells were washed and resuspended in 3 ml HBSS−/− buffer, then layered over 3 ml of Histopaque 1119 to remove the remaining red blood cells. This gradient was spun at 1,600 g for 30 min at 10°C. Supernatant was discarded and cells resuspended in HBSS−/−.
NET Trapping and Killing Assay
1 × 106 PMN cells/sample were pelleted and resuspended in 1 ml of GHBSS++ buffer in 2-ml microfuge tubes and stimulated with 20 mM of phorbol 12-myristate 13-acetate (PMA; Abcam) for 1 h at 37°C, agitating with end-over-end rotation [22]. Plated bacterium stocks were inoculated into 5 ml of HBSS−/− buffer such that OD650 = 0.45, resulting in a bacteria suspension of 1 × 109 bacteria/ml. Following the incubation, designated neutrophil cell samples were dosed with bacteria at a multiplicity of infection (MOI) of 0.1, 1, or 10. Samples were incubated for 100 or 200 min at 37°C with end-over-end rotation. 1 kU of micrococcal nuclease (MNase; Worthington Biochemical) was added for the duration of the incubation period in a separate series of samples to serve as control. Upon completion of the incubation period, aliquots were removed from each sample, treated with 100 U DNase at 37°C for 15 min, diluted in series in 96-well, V-bottom microtiter plates using dilution buffer containing DMEM/F12 media (Invitrogen) supplemented with 5% HI-FBS and 0.1% Triton X-100 (MP Biomedicals). 10 μl of the diluted samples were spotted onto MacConkey II plates (Becton Dickinson) and allowed to spread in a streak pattern. Pre-infection bacteria stocks were also diluted and plated. Plates were allowed to incubate for 10 h at 37°C before colonies were enumerated. The remaining infection supernatants were stored at −20°C for subsequent analysis.
DNA Quantification
PicoGreen dsDNA quantitation (Quant-It kit; Invitrogen) was used to determine total DNA content present in bacterial infection supernatants. The assay was performed according to the manufacturer's instructions and the reaction plate was read on a Tecan Infinite M200 multimode microplate reader. Fluorescence intensity was measured at 520 nm and a linear fit model applied to the standard curve for analysis.
Isolation of NET Fragments
Neutrophils were seeded at 2.7 × 107 cells/dish onto 10-cm tissue culture dishes in 10 ml RPMI 1640 media (Invitrogen) supplemented with 10 mM HEPES (Invitrogen). Cells were permitted to settle for 30 min at 37°C with 5% CO2. Cells were then gently washed three times with culture media and dosed drop-wise with prepared bacterial stock. Infection was conducted in a total volume of 10 ml of culture media. Infection was permitted for 100 min in a stationary tissue culture incubator at 37°C in 5% CO2. Following the infection period, NETs from each dish were liberated with appropriate concentrations of MNase enzyme (Thermo Scientific) for 30 min at 37°C. MNase was then inactivated with 5 mM EDTA (Invitrogen) per dish and 2× final concentration of protease inhibitor cocktail (Roche). Infection supernatants were collected in 15-ml conical tubes and cell debris pelleted by centrifugation at 300 g for 5 min at 4°C. The supernatants were concentrated to 2-ml volumes via spin concentration with Amicon 3,500 MWCO filter units (Millipore) in a 4°C swing bucket centrifuge at 3,500 g. Sucrose gradients were assembled at 4°C by first adding 3.2 ml of 50% sucrose (50% sucrose w/v in HBSS−/−) into a 13.2-ml thin-wall polyallomer ultracentrifuge tube (Beckman Coulter), overlaid with 2 ml of 30% sucrose (30% sucrose w/v in HBSS−/− supplemented with 2× protease inhibitor cocktail). The third sucrose layer of 2 ml of 10% sucrose was added, followed by 2 ml of infection reaction supernatants. Assembled gradient samples were ultracentrifuged at 36,000 rpm for 22 h at 4°C using a SW41 rotor in a Beckman L8-M ultracentrifuge or 1,600 g. Samples were fractionated with a peristaltic pump into ten 1-ml fractions per gradient tube. Fractions were analyzed for DNA via PicoGreen assay and for protein content via Bradford assay (Bio-Rad).
The protein in each sample was precipitated with trichloroacetic acid overnight at −20°C. Supernatants were carefully removed and washed twice with 100% acetone. The frozen pellets were processed for mass spectrometry (MS) analysis at the Taplin Biological Mass Spectrometry Core facility (HMS). The functional analysis of the liquid chromatography (LC)/MS-identified proteins was carried out using the String v9.05 software.
Immunoblot Analysis
To analyze for the presence of NET-associated proteins, protein-specific Western blots were performed on sucrose gradient fractions. Probing antibodies and blotting conditions included anti-NE (1:500; Calbiochem) and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 1 mg/ml; Abnova). All secondary antibodies were used at a final concentration of 1:10,000. Blotting controls included active, purified NE (Abcam) as well as soluble cell lysates from non-differentiated HL-60 cells, HL-60 cells differentiated with 70 mM dimethylformamide for 5 days, or PMN cell pellets. Soluble lysates were prepared by resuspending cell pellets (7.5 × 106 for PMNs) in 200 μl radioimmunoprecipitation assay buffer (RIPA buffer; Boston BioProducts) with 2× protease inhibitor cocktail. Soluble supernatants were transferred and stored at −20°C. The 4–12% Bis-Tris 15-well NuPAGE gels were loaded with non-reduced samples in 1× lithium dodecyl sulfate sample loading buffer (Invitrogen) and run for 35 min at 200 V. Gels were transferred onto nitrocellulose by running program P2 for 6 min in the iBlot transfer system (Invitrogen). The membranes were subsequently blocked at room temperature for 1 h in blocking buffer [PBS with 5% nonfat dry milk and 0.001% Tween 20 (Sigma)]. The primary antibody incubation followed using PBS-0.001% Tween 20 buffer with 0.5% milk containing the desired probing antibody at the concentration stated previously. The primary antibody was allowed to incubate with the membrane for at least 2 h with rocking at room temperature. Next, the sample was washed 5 times for 5 min with PBS-0.001% Tween 20, followed by a 1-hour incubation with the appropriate secondary antibody at room temperature with rocking. Blots were developed using the SuperSignal West Femto maximum sensitivity substrate (Thermo Scientific). Exposures were captured using an Alpha Innotech MultiImage II light cabinet and imaging system.
Analysis of CF Patient Sputum
Frozen CF patient sputum samples were thawed on ice before being centrifuged at 16,000 g at 4°C for 15 min. The soluble fraction was collected. Genomic DNA was digested with DNase I (Sigma) or MNase (Thermofisher) to yield DNA fragments. The reaction was carried out at 37°C for 5-10 min and stopped by addition of EDTA (5 mM final) and protease inhibitor cocktail (Roche). DNA-protein complexes were fractionated by sucrose density gradient ultracentrifugation as described above in the NET fragment purification section. All samples were derived from CF patients colonized with P. aeruginosa.
Agarose Gel Analysis of NETs
To visualize the isolated NET DNA, peak fractions from NET isolation gradients were loaded onto 10-well, 1% agarose/ethidium bromide gels as follows. Gel was run for 45 min at 120 V in 1× Tris acetate running buffer. Gels were then imaged on a UV light box and a CCD camera in a dark box.
Purification of Recombinant MIF
Recombinant MIF (rMIF) protein was produced in Escherichia coli BL21 Star (DE3) cells (Invitrogen) after induction with 1 mM isopropyl-β-D-thiogalactopyranoside for 3.5 h [17]. Bacterial cells were pelleted by centrifugation in a Beckman Allegra 6R centrifuge (3,000 rpm, 15 min, ambient temperature). Approximately 1 g of bacterial pellet was resuspended in 5 ml of 50 mM Tris (pH 8.5), 50 mM KCl, 5 mM magnesium acetate, and 0.1% sodium azide, and sonicated. The lysate was centrifuged at 15,000 rpm for 20 min, then filtered through a Millipore Steriflip vacuum filtration system with 0.2-µM membranes. The filtered lysate was loaded onto a 5-ml QHP HiTrap anion exchange column in a negative chromatography mode such that rMIF remained unbound. The rMIF was buffer exchanged and concentrated by loading onto a Mono S cation exchange column and eluted with a linear gradient.
Analysis of ROS
0.5 to 1 × 106 purified murine PMNs per sample were exposed to P. aeruginosa PAO1 in the presence of horseradish peroxidase (Sigma) and luminol. Released ROS were monitored for up to 1 h using a Tecan luminescence reader [23].
Analysis of MIF Levels
MIF levels in CF patient sputum or serum samples were quantified using an MIF ELISA kit (R&D Systems) according to the manufacturer's instructions.
Confocal Imaging
Purified PMNs were stimulated with PAO1 at MOI 10 and incubated at 37°C in 4-well slides (Lab Tek, ThermoFisher Scientific). Cells were fixed with 4% formalin for 15 min, washed with HBSS−/−, permeabilized with HBSS−/−, 0.2% Tween 20. DNA was visualized by SYTOX Green according to the manufacturer's instructions (Invitrogen). Samples were imaged using an LSM 510 META laser scanning confocal microscope (Zeiss). Alexa 488 was detected using the 488-nm excitation line and a 500- to 530-nm band-pass filter. Alexa 555 and Alexa 647 were excited at wavelengths of 543 and 633 nm, and detected using a 560- to 615-nm band-pass filter and a 650-nm long-pass filter, respectively.
Statistical Analysis
Data were analyzed with GraphPad Prism 5.0 software (GraphPad Software Inc.). Sample distribution was analyzed for normality. When appropriate, data were compared using Student's t test or one-way ANOVA followed by Bonferroni's post hoc test. Linear regression analysis was applied to determine potential associations between MIF serum protein levels and forced expiratory volume in 1 s (FEV1), forced vital capacity, and forced expiratory flow between 25 and 75% as dependent variables.
Results
Both Non-Mucoid and Mucoid P. aeruginosa Strains Stimulate the Release of NETs That Share a Common ‘Core’ Proteome
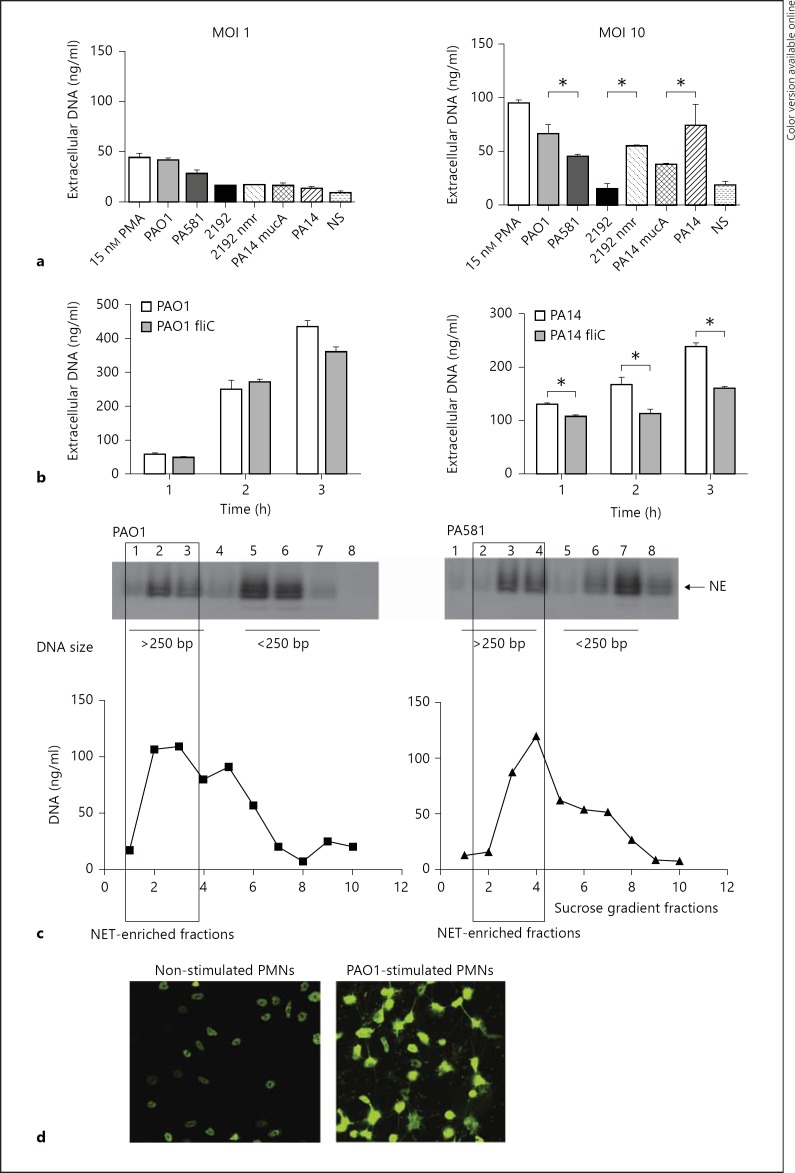
To examine whether the non-mucoid and the mucoid P. aeruginosa isolates stimulate neutrophils to NETose, human PMNs were stimulated with bacteria at MOI 1 or 10, and eDNA was released by partial MNase digest and quantified. The mucoid (2192, PA581, and PA14 mucA) and the non-mucoid strains (PAO1, 2192 nmr, and PA14) of P. aeruginosa-triggered eDNA release. The levels of the released eDNA were significantly higher when the PMNs were stimulated with the non-mucoid strains when compared to the mucoid P. aeruginosa (fig. 1a, b). Since P. aeruginosa loses flagella during colonization while acquiring mucoid characteristics, the contribution of flagellin in inducing NETosis was analyzed. Two pairs of strains were used in these experiments: PAO1 and PAO1 fliC mutant or PA14 and PA14 fliC mutant (fig. 1c). In the absence of flagellin, the PAO1 fliC mutant showed a tendency to induce less eDNA release, but this difference did not reach significance. In contrast, when the PA14 and PA14 fliC pair was compared, the flagellin-deficient mutant induced significantly lower eDNA release, suggesting that the presence of flagella triggers NETosis.
Fig. 1.
Non-mucoid or mucoid P. aeruginosa strains induce NET release. a Purified human PMNs were stimulated with non-mucoid or mucoid P. aeruginosa strains at MOI 1 or 10 and eDNA was quantified 1 h after exposure. Results are representative of 3 independent experiments. Pair-wise comparisons were carried out using Student's t test. * p < 0.05. b Representative image of P. aeruginosa PAO1-induced NETosis. Purified murine PMNs were exposed to PAO1 for 1 h at MOI 10, fixed with formalin, permeabilized with 0.3% Tween 20/PBS and stained with SYTOX green. c The levels of NETosis induced by P. aeruginosa PAO1/PAO1 fliC (MOI 6) or PA14/PA14 fliC (MOI 10) were compared. The reaction supernatants were harvested at different time points after the onset of reaction and eDNA was quantified. Results are representative of 2 independent experiments. Pair-wise comparisons were carried out using Student's t test. * p < 0.05. c NETosis was induced by P. aeruginosa PA01 and PA581 mucA strains. NET-containing fractions were defined as fractions that were rich in HMW eDNA and NE. The Western blotting analysis for NE is compared to the DNA present in the individual sucrose density gradient fractions. These data are representative of at least 3 independent experiments. d Representative images of NETs induced by PAO1 at MOI 10. PMNs were fixed and permeabilized prior to DNA staining.
To verify that the eDNA were NET fibers, the NET DNA fragments were purified by sucrose density gradient ultracentrifugation followed by Western blotting for NE, a marker for NETs [4, 9] (fig. 1c). When the individual fractions were examined for reactivity with NE-specific antibody, a positive signal correlated with the presence of DNA (fig. 1c). In addition, fluorescent microscopy analysis was carried out to observe netting PMNs in response to PAO1 stimulation (fig. 1d).
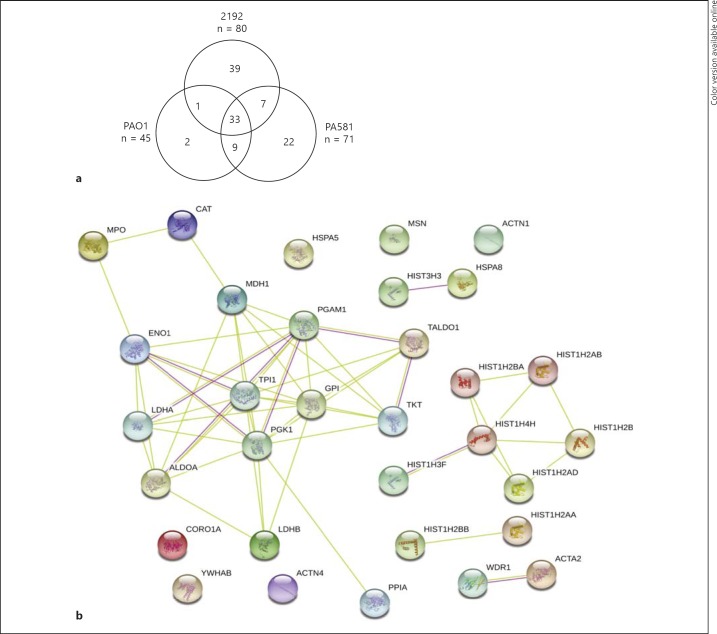
To further characterize the composition of NETs released in response to stimulation with the different P. aeruginosa strains, equal concentrations of NET DNA-containing samples were precipitated and the proteins were characterized using LC-MS/MS. Fractions enriched for high-molecular-weight (HMW) DNA and NE were pooled and analyzed. While 24 NET-associated proteins were previously reported [24, 25, 26], the number of proteins identified in this study ranged from 45 to 80, depending on the nature of the stimuli (online suppl. table 1; for all online suppl. material, see www.karger.com/doi/10.1159/000363242). There were 33 common NET-associated proteins when NETosis was triggered by the three different P. aeruginosa strains. The majority of human proteins attached to NETs were shared between the non-mucoid and the mucoid P. aeruginosa strains (χ2 analysis for trend, p = 0.0012). For example, more than 90% of the identified NET-associated host proteins that were released in response to the PAO1 stimulation were identical to the NET-associated proteins released in response to the mucoid P. aeruginosa 581 (an alginate-producing PAO1 strain) stimulation. Twenty-two proteins were unique for the PA581-stimulated NETs (fig. 2) and 39 proteins were unique for the 2192 NETs (fig. 2). Functional analysis of the proteome showed that the P. aeruginosa-induced NETs were decorated with the ‘classical’ NET members (e.g., histones, elastase, lysozyme, MPO, and superoxide dismutase), similar to the previously published PMA-induced NETs [26]. The NET proteins could be segregated into different functional categories, such as nucleic acid binding, enzymes, structural proteins, and proteins with previously reported antimicrobial activity (online suppl. table 1). String-based analysis for protein-protein interactions showed that the NET proteome is significantly enriched for protein associations (p = 0.01), illustrating a conserved core of DNA-associated proteins that represent the ‘NET signature’ (fig. 2b).
Fig. 2.
NET-associated proteins released in response to stimulation with the different P. aeruginosa strains share a high degree of commonality. a Venn diagram displaying the numbers of shared and unique NET-associated proteins. The total number of identified proteins is expressed under each stimulant. b Functional analysis of the shared NET-associated proteins or ‘NET core proteins’. Different line colors represent the types of evidence for the association. Green lines indicate interactions based on experimental data, the red lines indicate associations based on KEGG database searches (colored in the online version only). Each circle represents an individual protein identified by LC/MS-MS analysis that had >10% unique amino acid sequence coverage. The available crystal structure of the identified protein is shown inside the circle. The image is generated using the String v9.05 software.
CF Sputum Is Enriched for NETosis
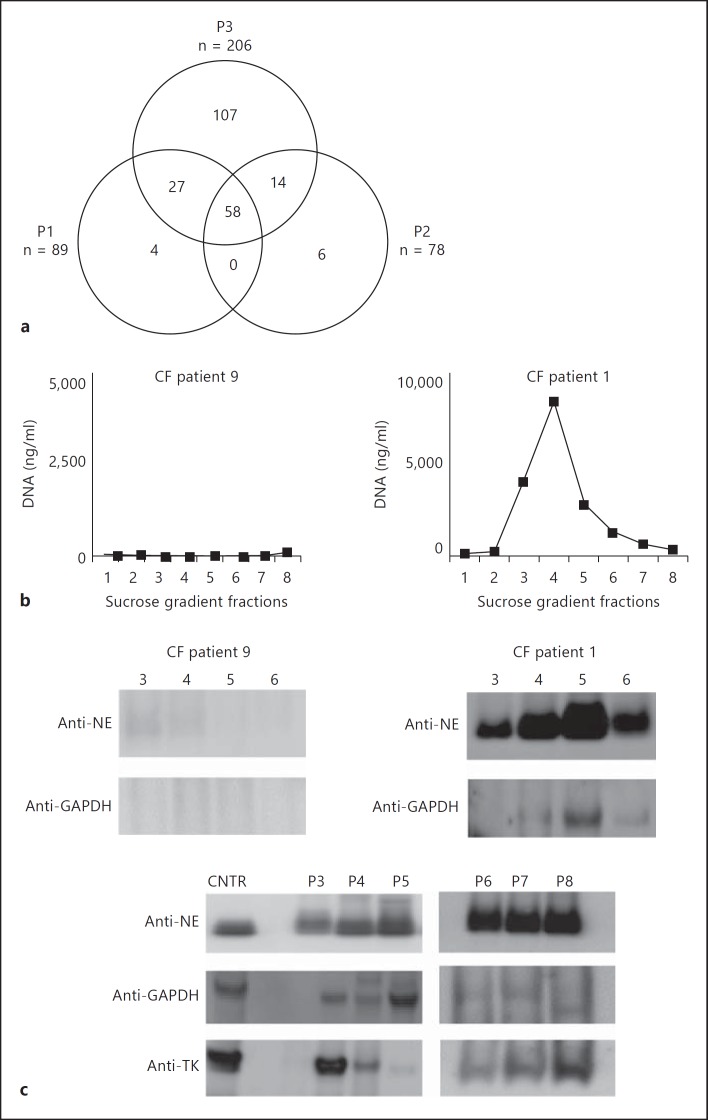
To compare the in vitro generated NETs with the DNA-protein complexes in CF sputum, LC/MS-MS analysis was executed using preparations enriched for NET structures (fig. 3). All proteins identified with more than 3 unique peptides per sequence were considered for analysis. Fifty-eight of the identified proteins that passed the inclusion criteria were shared between patients 1, 2, and 3 (χ2 analysis for trend, p = 0.001), demonstrating a significant degree of commonality (fig. 3). The NET markers elastase, MPO, and lysozyme [25] were present among the 55 proteins identified, suggesting that the eDNA in CF is organized in NETs. These findings are in agreement with the recently published observations that the CF-derived DNA carries NE and MPO [8, 9] and has a physical appearance similar to that of NETs when visualized by atomic force microscopy [10]. Our data expand the existing knowledge by providing a list of proteins that commonly present in CF sputum (table 1).
Fig. 3.
CF sputum is NET rich. a Venn diagram displaying the numbers of shared and unique NET-associated proteins identified in 3 CF patient samples. NET fragments were purified from CF sputum samples from 3 patients using sucrose gradient ultracentrifugation. DNA was fragmented by brief DNase I treatment and NET complexes enriched using sucrose gradient ultracentrifugation. HMW DNA-containing fractions were precipitated, and proteins identified by LC/MS-MS. Protein sequences which displayed 3 or more unique peptides were considered as true hits. b The presence of NET proteins depends on DNA. CF sputum samples containing high and low DNA levels were fractionated using sucrose density gradient ultracentrifugation, DNA was quantified in the individual gradients samples. Samples 3-6 were subsequently analyzed for NET markers, NE and GAPDH, using Western blotting. c Western blotting analysis for NET protein markers in samples derived from 6 additional CF patients. CF sputum samples from 6 CF patients were fractionated using sucrose density ultracentrifugation and DNA was quantified. The HMW DNA-rich fractions were precipitated and analyzed for NE, TK, and GAPDH. Purified NE or total neutrophil lysates were used as control samples for Western blotting.
Table 1.
Shared NET-associated proteins identified in CF patients
| Function | Shared proteins | ||||
|---|---|---|---|---|---|
| Glycolysis-related proteins | GAPDH (O14556) | Triosephosphate isomerase (P60174) | |||
| Structural proteins | Coronin-1A (P31146) | Myosin heavy chain (P35579) | Gelsolin (P06396) | Profilin-1 (P07737) | Α-Actinin 1 (P12814) |
| Filamin A (P21333) | Actinin 4 (O43707) | ||||
| Antimicrobial proteins | Lactotransferrin (P02788) | MPO (P05164) | Eosinophil peroxidase (P11678) | Cathepsin G (P08311) | Peptidoglycan recognition protein 1 (O75594) |
| Azurocidin (P20160) |
Cathelicidin (P49913) |
NE (P08246) | Lysozyme C (P61626) |
S100A12 (P80511) S100A4 (P26447) |
|
| BPIB2 (Q8N4F0) | |||||
| Chaperones | Annexin A1 (P04083) | Annexin A3 (P12429) | Annexin A4 (P09525) | Heat shock cognate 71-kDa (P11142) | Annexin A6 (P08133) |
| Heat shock cognate 70-kDa protein 1 (P08107) | Heat shock cognate 70-kDa protein 1 like (P34931) | Heat shock cognate 70-kDa protein 2 (P54652) | Heat shock cognate 70-kDa protein 5 (P11021) | ||
| Oxidoreductases | Catalase (CAT) (P04040) | ||||
| Other enzymes and enzyme inhibitors | Proteinase 3 (D6CHE9) | Phospholipase B-like 1 (Q6P4A8) | Chitinase-3-like protein 1 (P36222) | Matrix metalloproteinase 8 (P22894) | Matrix metalloproteinase 9 (P14780) |
| Serpin A3 (P01011) | α1-Antichymotrypsin (P01011) |
Peptidyl-prolyl-cis-transisomerase B (P23284) | Quiescin Q6 sulfhydryl oxidase 1 (O00391) | Serpin peptidase inhibitor (P48595) | |
| Serpin peptidase inhibitor clade G (P05155) | Serpin A1 (P01009) | Transketolase (P29401) | Transaldolase (P37837) | ||
| Not classified |
Vinculin (P18206) | Integrin α M (P11215) |
Zinc-α2 glycoprotein (P25311) |
Complement component C3 (P01024) | Leucine-rich α-glycoprotein (P02750) |
| Cysteine-rich secretory protein (P54108) |
Hemopexin (P02790) |
IQGAP1 (P46940) | Lymphocyte cytosolic protein (P13796) | α1-B glycoprotein (P04217) | |
| Mucin 5B (Q9HC84) | Lipocalin 2 (P80188) | Adenylyl cyclaseassociated protein 1 (Q01518) | |||
Common NET-associated proteins present in the 3 CF patient sputum samples were grouped depending on function. All listed proteins were identified using two or more unique peptide sequences. Previously reported NET-associated proteins are italicized [24, 25, 26]. P. aeruginosa-induced NET-associated proteins described in this study are in bold.
To verify the LC-MS/MS findings, sputum samples from 6 additional CF patients were fractionated to yield enriched DNA-protein complexes. Three NET-associated protein markers, namely NE, transketolase (TK), and glyceraldehyde phosphate dehydrogenase (GAPDH), were analyzed. Albeit to a different degree, all patient samples were positive for NE, TK, and GAPDH, confirming that LC-MS/MS identified NET-associated proteins were present in the fractionated CF sputum. To demonstrate that the NE and GAPDH occurrence correlated with the DNA, the CF patient sputum sample which had low levels of DNA (<8 ng/ml of sputum) was fractionated and analyzed for NET-associated proteins (fig. 3b). When DNA levels were low, there were only trace amounts of NE and GAPDH (fig. 3c), suggesting that NE and GAPDH signals correlated with DNA.
Distinct Sensitivity of the Different P. aeruginosa Strains to NET Capture and Killing
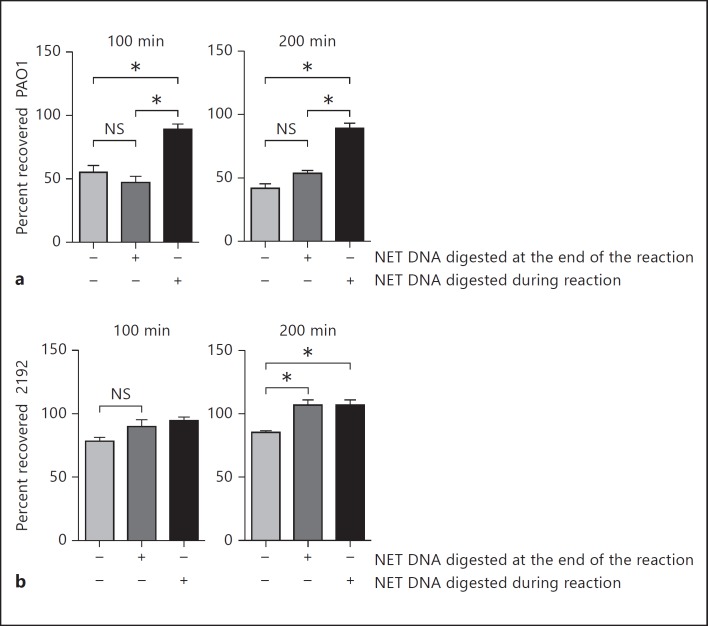
To determine the functional significance of NETs, in vitro experiments addressing NET-mediated capture of P. aeruginosa and killing were carried out. Because the previously reported PMA-induced NETs had similar core protein composition to the P. aeruginosa-induced NETs [26], we exposed neutrophils to PMA and analyzed the survivability of the non-mucoid (e.g., PAO1) versus the mucoid (e.g., 2192) P. aeruginosa strains in the presence or absence of endonuclease (e.g. MNase; fig. 4). Upon completion of the incubation period, all samples were treated briefly (15 min) with DNase I to disperse trapped bacteria (fig. 4). Aliquots of the reaction were plated before and after DNase I treatment to allow differentiation between the trapped and killed bacteria [27]. The samples that were treated with MNase for the duration of the experiment had no intact NETs and served as controls since bacterial trapping in the MNase-treated samples did not occur. These experiments suggested that NETs aggregated and killed 50% of the non-mucoid PAO1 strain (fig. 4a) but only trapped 20% of the mucoid strain 2192 and, consequently, failed to kill the mucoid strain 2192 (fig. 4b). These findings suggest that NETs were not effective at protecting against the late P. aeruginosa colonizers. While these data confirm earlier published observations [28], our experimental approach distinguishes between trapping and killing of P. aeruginosa by NETs, an issue which was not addressed in the prior research and provides insights into the resistance mechanisms, namely resistance to capture.
Fig. 4.
Non-mucoid and mucoid P. aeruginosa strains show distinct sensitivity to NET-mediated killing. Purified human PMNs were pretreated with PMA to induce NETosis and exposed to PAO1 (a) or 2192 (b) at MOI 0.1 for 100 or 200 min. The endonuclease presence in the reaction mixture is indicated (+/-). Results are representative of 2 independent experiments. Samples were compared with one-way ANOVA for variance and significant differences indicated with asterisks.
MIF Promotes NET Release
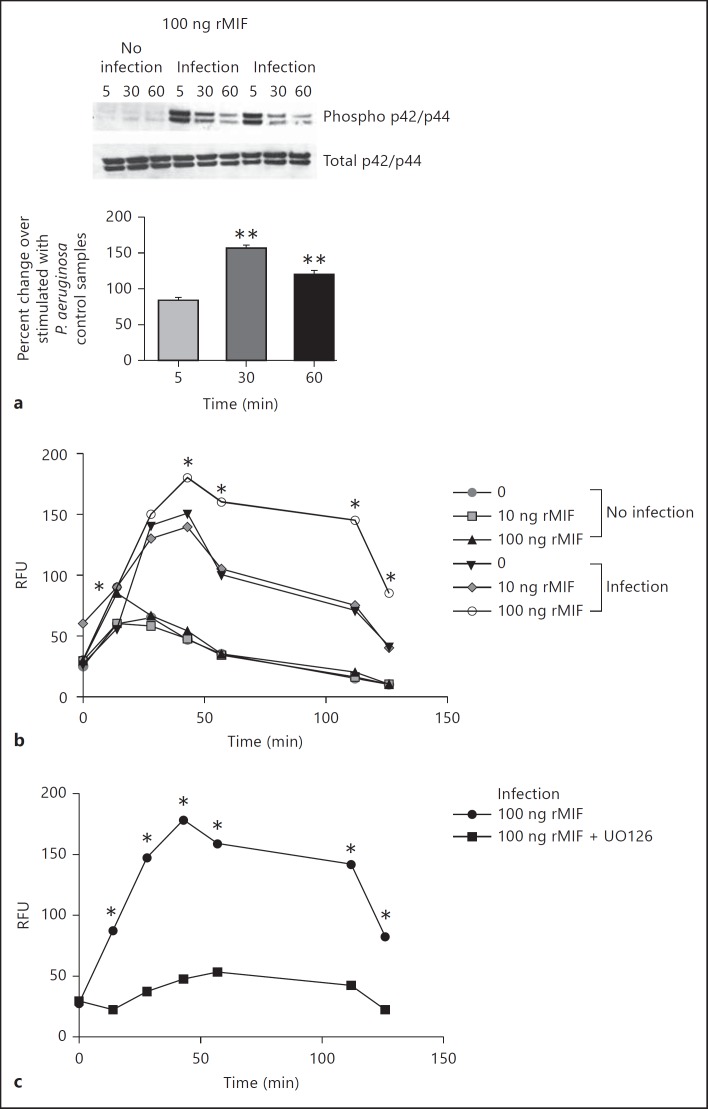
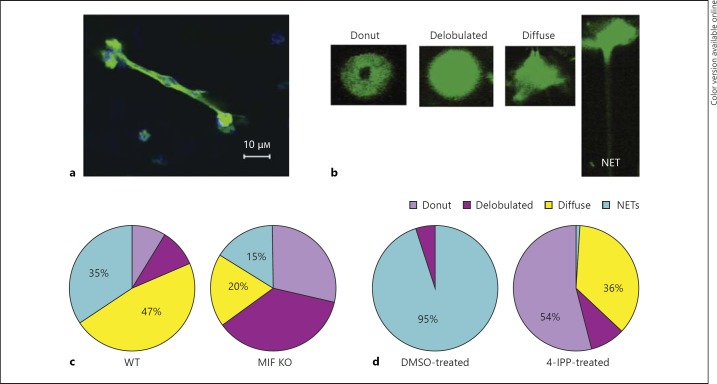
Because NETs failed to kill the mucoid P. aeruginosa, we next examined the molecular mechanism of NETosis with the objective to identify host-specific molecular targets that can be manipulated in CF to lower the levels of extracellular DNA and facilitate sputum solubilization. Recent screening for intracellular pathways that stimulated NET release showed that activation of the MAPK p44/p45 pathway promoted NETosis over apoptosis [29]. Since we and others have reported that the endogenous cytokine MIF is rapidly released in response to infectious challenge and stimulates MAPK activation [17, 30], the influence of MIF on NETosis was evaluated [31]. When MIF-deficient PMNs were purified from MIF KO mice and exposed to P. aeruginosaPAO1, the resulting MAPK activation was sustained upon reconstitution with 100 ng rMIF as evident by the elevated phosphorylation state of p42/p44 30 and 60 min after infection (fig. 5). Reconstituting rMIF of MIF-deficient PMNs promoted ROS release in response to P. aeruginosa and this was ablated by the presence of the MEK inhibitor UO126 (fig. 5). Treatment with 100 ng rMIF alone induced a mild increase in ROS over the background in the absence of infectious agent. When 100 ng rMIF was added to samples in the presence of P. aeruginosa, the released ROS was significantly elevated compared to the background. Next, MIF-deficient and MIF-sufficient [i.e. from wild-type (WT)] PMNs were stimulated with the PAO1 strain and the levels of NETing PMNs quantified. Fluorescent confocal imaging was used to evaluate the DNA morphology of P. aeruginosa-activated PMNs. WT PMNs formed DNA-rich elongated structures after stimulation with P. aeruginosa strain PAO1 at MOI 10 (fig. 5). Several PMN nuclear phenotypes could be distinguished: donut-shaped nuclei, specific for resting PMNs; delobulated nuclei, specific for the nuclei that undergo chromatin remodeling; diffuse-shaped nuclei, specific for the early stage of NETosis, and actual NETs, with elongated chromatin structures, consistent with previously described stages for DNA reorganization (fig. 6a, b) [22, 32, 33]. Upon quantification of the four distinct stages of NETosis, MIF-sufficient PMNs released significantly more NETs than MIF-deficient PMNs (fig. 6c). Infected WT PMNs that casted NETs accounted for 35% of the total PMN population whereas only 15% of the MIF-deficient PMNs released NETs (light blue sectors in the online version of fig. 6c; t test p < 0.05). To determine whether inhibition of MIF affected NETosis, human PMNs were pretreated with a small molecule, irreversible inhibitor of MIF [4-iodo-6-phenylpyrimidine (IPP)] and stimulated with P. aeruginosa PAO1 at MOI 10 [34]. Of the DMSO-treated PMNs, 95% showed signs of NET release in response to P. aeruginosa stimulation whereas 37% in the 4-IPP-treated PMNs were NET releasing (including the PMNs that presented with diffuse nuclear appearance). Of the 4-IPP-treated PMNs, 54% preserved their donut-shaped nuclei as opposed to none in the DMSO control (t test p < 0.05; fig. 6d).
Fig. 5.
MIF promotes MAPK activation in neutrophils. a Purified mouse MIF-deficient PMNs were infected with PAO1 and lyzed with RIPA buffer supplemented with protease inhibitors 5, 30, and 60 min after the infectious challenge. Separate sets of samples were collected with and without infection to monitor baseline and P. aeruginosa-induced phosphorylation of MAPK (p42/p44). 100 ng rMIF was supplemented to MIF-deficient PMNs during infection to monitor MIF-driven MAPK activation during infection. The bottom panel represents densitometry analysis of the Western blotting images. The percent change of MAPK activation was calculated for the rMIF-treated, infected samples compared to non-rMIF-treated, infected samples. The images are representative of duplicated experiments. b MIF-deficient PMNs were supplemented with different concentrations of rMIF, infected with P. aeruginosa, and ROS release monitored. Samples were compared with one-way ANOVA, and significant differences are indicated with asterisks. Data are representative of 3 independent experiments. c MIF-deficient PMNs were supplemented with 100 ng rMIF, treated with the MEK inhibitor UO126, and the levels of ROS monitored after the infectious challenge with P. aeruginosa. Samples were compared with one-way ANOVA and significant differences indicated with asterisks. Data are representative of 2 independent experiments.
Fig. 6.
MIF promotes NET release. a NETs formed by C57BL6-derived PMNs in response to P. aeruginosa PAO1 stimulation. NETs were visualized by fluorescent DNA-specific staining with SYTOX Green in stimulated neutrophils after fixing and permeabilizing. The images were acquired on a Nikon confocal microscope with ×63 and ×40 objectives. b Stages of nuclear DNA rearrangement in response to P. aeruginosa stimulation of PMNs. The ‘donut’-shaped nuclei are typical of resting PMNs. ‘Delobulated’ and ‘diffuse’ nuclei are characteristic of activated PMNs. NETs are elongated DNA-containing structures released by PMNs. c Pie graph representing the percentage of different stages of NETosis of PMNs derived from C57BL6 (WT; left pie chart) and MIF KO (right pie chart) mice responding to P. aeruginosa PAO1 stimulation. Data are representative of 2 experiments. d Pie graph representing the percentage of different stages of NETosis of human PMNs treated with 50 μM 4-IPP. The left pie chart represents the DMSO-treated PMNs, whereas the right pie chart represents the MIF inhibitor (4-IPP)-treated PMNs. Data are representative of 2 experiments.
Elevated MIF Protein Levels Correlate with Poor Lung Function in CF Patients
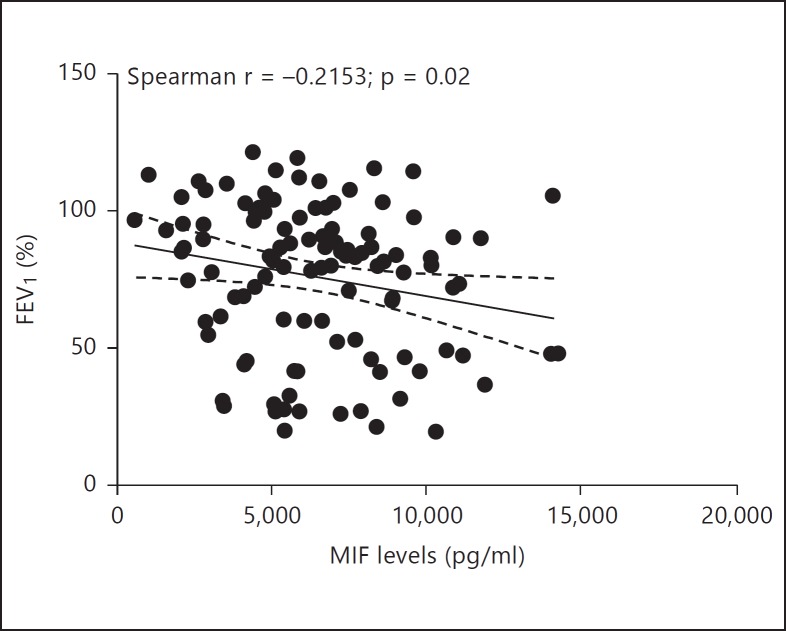
Prior reports have shown that patients who carry MIF promoter polymorphisms resulting in low MIF levels exhibit less colonization by P. aeruginosa and milder CF disease, suggesting that regulating MIF levels or MIF activity may confer therapeutic benefit for CF patients [35]. To expand on these findings, we analyzed a cohort of serum samples from 117 Caucasian CF patients [21] for MIF protein levels and correlated those with disease pathology. The mean MIF levels in CF patient sera (6,328 ± 2,891 pg/ml) were significantly increased compared to the MIF levels in pooled human serum from 10 healthy individuals (2,400 ± 900 pg/ml; p < 0.0001; fig. 7). MIF levels in CF serum samples ranged widely (from 562 to 14,299 pg/ml). MIF levels in CF patients significantly predicted FEV1 percent values, β = −0.002, t(107) = −2.18, p = 0.03. MIF levels also explained a significant proportion of variance in FEV1 values, R2 = 0.04, F(1, 107) = 4.76, p = 0.03.
Fig. 7.
Serum MIF levels correlate with decreased lung function. Serum MIF levels in 117 CF patients were measured by ELISA. FEV1 percent values are plotted against MIF levels per patient. Linear regression analysis of the data indicates a mild correlation between the increased MIF serum levels and lower FEV1 values (R2 = 0.04; p = 0.03).
Discussion
The majority of the currently available data on NET proteomes were derived from in vitro studies where neutrophils were either stimulated with PMA, cytokines, or IgG. To the best of our knowledge, there are no studies characterizing the pathogen-induced NET proteomes. Here, we provide data that NETs induced by either non-mucoid or mucoid P. aeruginosa-strains carried a conserved core of proteins that were shared, irrespectively of the inducing bacterial stimuli. The ‘core proteins’ included nucleosome-associated proteins (e.g., histones), structural proteins (e.g., coronin-1, β-actin, or α-actinin), enzymes (e.g., NE, transaldolase, TK, GAPDH, or α-enolase), and proteins with antimicrobial activity (e.g., MPO, lysozyme C, lactotransferrin, or azurocidin). While there were stimulus-specific proteins, the overall NET proteome did not differ significantly from previously reported PMA [25, 26], tumor necrosis factor, and rheumatoid factor [24] proteomes. Cumulatively, these data demonstrate that NETs harbored a conserved repertoire of innate immune molecules to mediate protection. Irrespectively of the inducing signal (cytokine vs. bacterial stimulation), NETs were decorated with a conserved set of proteins that we termed the ‘NET core signature’.
The presence of the conserved NET-associated proteins prompted us to evaluate whether NET structures were present in CF patients. Based on the proteomic findings, we found a significant overlap between the in vitro-induced NETs and the DNA-containing complexes in CF sputum (table 1). We argue that if the eDNA originated from cellular debris, the composition of the DNA-protein complexes would be highly variable and no common trends could be identified between patients. In contrast, the presence of a common protein signature would suggest a specific immune response. The presence of non-secretory proteins (table 1) within the NET ‘core’ proteome suggested that it is unlikely that the complexes were formed between pathogen-derived (naked) eDNA and secretory granules, supporting the argument that the structures in the CF sputum most likely originated from NETosing cells. These data strengthen previously published observations [8, 9] that demonstrated that 50% of the CF sputum-derived DNA is in complex with MPO and NE. Cumulatively, these data advocate that a significant percent of the eDNA in CF are NETosis derived, providing a rationale for designing alternative clinical markers to monitor CF disease and novel treatment approaches.
One of the most controversial issues since the discovery of NETosis is whether NETs kill P. aeruginosa [2, 27, 28]. The proposition that NETs have antimicrobial activity is supported by the broad array of antimicrobial proteins covering NET structures. Given the proteomic data, one may assume that the bactericidal properties of NETs are mediated by the production of ROS by MPO and QSOX1, involve the destruction of bacterial cell wall integrity by lysozymes, histones, and cathelicidin, and metabolic starvation by the creation of local environments with low divalent cation abundance (the majority of NET-associated proteins are divalent cation chelators, e.g. S100A proteins). To address the issue of whether P. aeruginosa withstands NET immunity, we carried out experiments using the non-mucoid strain PAO1 and the mucoid clinical isolate PA2192 to model the survivability of early (non-mucoid) versus late (mucoid) colonizers, utilizing an experimental approach [27] that allowed us to distinguish between the trapped and killed bacteria. Our data reproducibly showed that 50% of P. aeruginosa PAO1 was captured and killed by NETs. In contrast, the mucoid 2192 was significantly less susceptible to NET capture than the non-mucoid PAO1. These experiments produced a novel understanding of the process, as they compared bacterial trapping to bacterial killing in the presence of NETosis. The resistance of the mucoid P. aeruginosa strain to NET adhesion was not due to the overexpression of alginate, as the mutant P. aeruginosa strain that overexpressed alginate (PA581) was sensitive to NET capture and killing (data not shown).
One important inference of these studies is that, during colonization, P. aeruginosa acquires adaptive mutations that render this pathogen resistant to NET adhesion and subsequent killing. It is likely that the selective pressures elicited by MPO-coated NETs drive bacterial pathoadaptation. The long-lived NET structures in CF provide a scaffold for P. aeruginosa deposition and, while failing to effectively eradicate the pathogen, are a perfect niche to initially promote P. aeruginosa colonization. This process is driven at least partially by the pathogen as flagella promotes NETosis. Therefore, destroying NETs in CF early in the course of disease might interfere with bacterial pathoadaptation.
It is important to note that not all CF patients benefit equally well from Pulmozyme treatment. The use of the recombinant human DNase I is accompanied by the release of active elastase, since the enzyme is released from NETs upon DNA cleavage [8, 9, 36]. The primary targets for the liberated elastase are NET-associated histones and their subsequent enzymatic digest improves eDNA fragmentation [9]. However, the question remains whether the released active elastase should be controlled by addition of protease inhibitors [8, 37] or whether alternative therapies that reduce the levels of NETosis should be devised.
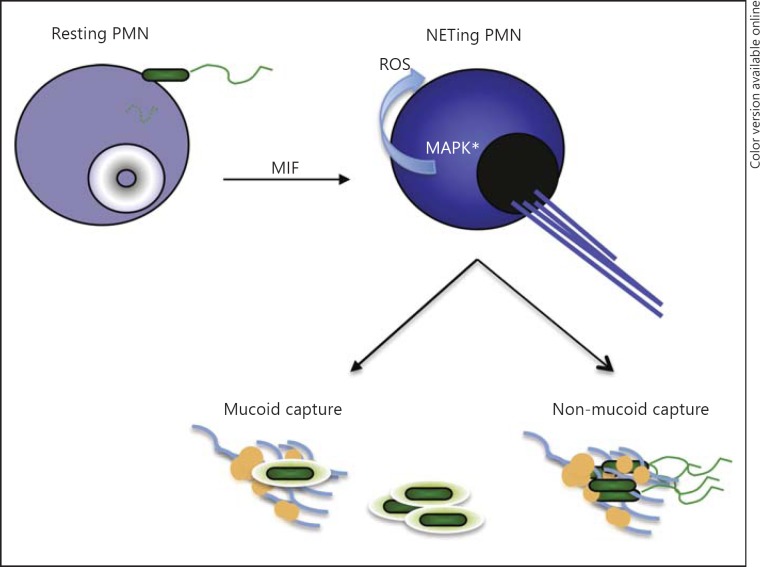
To this end, we report that NETosis was promoted by MIF, a cytokine that is released in response to P. aeruginosa stimulation (fig. 8). MIF is a proinflammatory cytokine produced by a vast majority of host cells, including neutrophils. MIF stimulates MAPK activation by binding to CD74 in macrophages or B cells. In addition to the CD74 receptor, CXCR2, CXCR4 and CXCR7 were implicated as non-cognate MIF receptors on various host tissues or cancer cells [15, 38]. Since neutrophils carry CXCR2, but not CD74 or CXCR7, it is possible that MIF may trigger CXCR2-driven responses in CF. Because the ability of neutrophils to NETose depended upon the effective blockade of apoptosis and activation of MAPK-promoted NETosis, we hypothesized that MIF prolonged this process [22]. In neutrophils, MIF-regulated pathways are largely unknown, with the exception of the interesting finding that MIF promoted neutrophil survival by inhibiting neutrophil apoptosis [31]. This finding prompted us to evaluate the role of MIF in NETosis. We demonstrated that MIF promoted MAPK activation and ROS production, potentiating NETosis (fig. 5). This observation supported our predictions that prolonged MIF signaling exacerbates CF (fig. 7). The latter concept is further corroborated by data showing that MIF KO mice are less sensitive to chronic airway infection by P. aeruginosa[39]. The elucidation of MIF as a component of CF pathogenesis provides a novel therapeutic target that may produce improved outcomes and prognoses for CF.
Fig. 8.
Summary: NETing depends on local MIF concentrations. This is achieved through MAPK-driven ROS release. NETs are extruded in response to P. aeruginosa infection in a P. aeruginosa-triggered MIF-dependent manner. The non-mucoid P. aeruginosa strains are sensitive to NET capture and killing while the mucoid strains are resistant.
Supplementary Material
Supplementary data
Acknowledgments
We would like to thank the members of the Division of Infectious Disease (Brigham and Women's Hospital) for their helpful comments and blood sampling. We would like to thank Prof. A. Zychlinsky for providing the NET purification protocol. We thank the Taplin Core Facility, HMS, for carrying out the LC-MS/MS protein identifications. R.M. is supported by NIH CA 102285 and M.G. is supported by NIH EY022054.
References
- 1.Kirchner KK, Wagener JS, Khan TZ, Copenhaver SC, Accurso FJ. Increased DNA levels in bronchoalveolar lavage fluid obtained from infants with cystic fibrosis. Am J Respir Crit Care Med. 1996;154:1426–1429. doi: 10.1164/ajrccm.154.5.8912759. [DOI] [PubMed] [Google Scholar]
- 2.Ratjen F, Paul K, van Koningsbruggen S, Breitenstein S, Rietschel E, Nikolaizik W. DNA concentrations in BAL fluid of cystic fibrosis patients with early lung disease: influence of treatment with dornase alpha. Pediatr Pulmonol. 2005;39:1–4. doi: 10.1002/ppul.20134. [DOI] [PubMed] [Google Scholar]
- 3.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 4.Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. 2010;191:677–691. doi: 10.1083/jcb.201006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med. 2010;207:1853–1862. doi: 10.1084/jem.20100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rohrbach AS, Slade DJ, Thompson PR, Mowen KA. Activation of PAD4 in NET formation. Front Immunol. 2012;3:360. doi: 10.3389/fimmu.2012.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neeli I, Radic M. Opposition between PKC isoforms regulates histone deimination and neutrophil extracellular chromatin release. Front Immunol. 2013;4:38. doi: 10.3389/fimmu.2013.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubois AV, Gauthier A, Brea D, Varaigne F, Diot P, Gauthier F, Attucci S. Influence of DNA on the activities and inhibition of neutrophil serine proteases in cystic fibrosis sputum. Am J Respir Cell Mol Biol. 2012;47:80–86. doi: 10.1165/rcmb.2011-0380OC. [DOI] [PubMed] [Google Scholar]
- 9.Papayannopoulos V, Staab D, Zychlinsky A. Neutrophil elastase enhances sputum solubilization in cystic fibrosis patients receiving DNase therapy. PLoS One. 2011;6:e28526. doi: 10.1371/journal.pone.0028526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manzenreiter R, Kienberger F, Marcos V, Schilcher K, Krautgartner WD, Obermayer A, Huml M, Stoiber W, Hector A, Griese M, Hannig M, Studnicka M, Vitkov L, Hartl D. Ultrastructural characterization of cystic fibrosis sputum using atomic force and scanning electron microscopy. J Cyst Fibros. 2012;11:84–92. doi: 10.1016/j.jcf.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Almyroudis NG, Grimm MJ, Davidson BA, Rohm M, Urban CF, Segal BH. NETosis and NADPH oxidase: at the intersection of host defense, inflammation, and injury. Front Immunol. 2013;4:45. doi: 10.3389/fimmu.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan C, Rajasekaran D, Syed MA, Leng L, Loria JP, Bhandari V, Bucala R, Lolis EJ. MIF intersubunit disulfide mutant antagonist supports activation of CD74 by endogenous MIF trimer at physiologic concentrations. Proc Natl Acad Sci USA. 2013;110:10994–10999. doi: 10.1073/pnas.1221817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber C, Kraemer S, Drechsler M, Lue H, Koenen RR, Kapurniotu A, Zernecke A, Bernhagen J. Structural determinants of MIF functions in CXCR2-mediated inflammatory and atherogenic leukocyte recruitment. Proc Natl Acad Sci USA. 2008;105:16278–16283. doi: 10.1073/pnas.0804017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz V, Lue H, Kraemer S, Korbiel J, Krohn R, Ohl K, Bucala R, Weber C, Bernhagen J. A functional heteromeric MIF receptor formed by CD74 and CXCR4. FEBS Lett. 2009;583:2749–2757. doi: 10.1016/j.febslet.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarnowski M, Grymula K, Liu R, Tarnowska J, Drukala J, Ratajczak J, Mitchell RA, Ratajczak MZ, Kucia M. Macrophage migration inhibitory factor is secreted by rhabdomyosarcoma cells, modulates tumor metastasis by binding to CXCR4 and CXCR7 receptors and inhibits recruitment of cancer-associated fibroblasts. Mol Cancer Res. 2010;8:1328–1343. doi: 10.1158/1541-7786.MCR-10-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Binsky I, Haran M, Starlets D, Gore Y, Lantner F, Harpaz N, Leng L, Goldenberg DM, Shvidel L, Berrebi A, Bucala R, Shachar I. IL-8 secreted in a macrophage migration-inhibitory factor- and CD74-dependent manner regulates B cell chronic lymphocytic leukemia survival. Proc Natl Acad Sci USA. 2007;104:13408–13413. doi: 10.1073/pnas.0701553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reidy T, Rittenberg A, Dwyer M, D'Ortona S, Pier G, Gadjeva M. Homotrimeric macrophage migration inhibitory factor (MIF) drives inflammatory responses in the corneal epithelium by promoting caveolin-rich platform assembly in response to infection. J Biol Chem. 2013;288:8269–8278. doi: 10.1074/jbc.M112.351064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calandra T, Bucala R. Macrophage migration inhibitory factor (MIF): a glucocorticoid counter-regulator within the immune system. Crit Rev Immunol. 1997;17:77–88. doi: 10.1615/critrevimmunol.v17.i1.30. [DOI] [PubMed] [Google Scholar]
- 19.Qiu D, Eisinger VM, Head NE, Pier GB, Yu HD. ClpXP proteases positively regulate alginate overexpression and mucoid conversion in Pseudomonas aeruginosa. Microbiology. 2008;154:2119–2130. doi: 10.1099/mic.0.2008/017368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldberg JB, Hatano K, Meluleni GS, Pier GB. Cloning and surface expression of Pseudomonas aeruginosa O antigen in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:10716–10720. doi: 10.1073/pnas.89.22.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olesen HV, Jensenius JC, Steffensen R, Thiel S, Schiotz PO. The mannan-binding lectin pathway and lung disease in cystic fibrosis - dysfunction of mannan-binding lectin-associated serine protease 2 (MASP-2) may be a major modifier. Clin Immunol. 2006;121:324–331. doi: 10.1016/j.clim.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 22.Hakkim A, Fuchs TA, Martinez NE, Hess S, Prinz H, Zychlinsky A, Waldmann H. Activation of the Raf-MEK-ERK pathway is required for neutrophil extracellular trap formation. Nat Chem Biol. 2011;7:75–77. doi: 10.1038/nchembio.496. [DOI] [PubMed] [Google Scholar]
- 23.Wu W, Hsu YM, Bi L, Songyang Z, Lin X. CARD9 facilitates microbe-elicited production of reactive oxygen species by regulating the LyGDI-Rac1 complex. Nat Immunol. 2009;10:1208–1214. doi: 10.1038/ni.1788. [DOI] [PubMed] [Google Scholar]
- 24.Khandpur R, Carmona-Rivera C, Vivekanandan-Giri A, Gizinski A, Yalavarthi S, Knight JS, Friday S, Li S, Patel RM, Subramanian V, Thompson P, Chen P, Fox DA, Pennathur S, Kaplan MJ. NETs are a source of citrullinated autoantigens and stimulate inflammatory responses in rheumatoid arthritis. Sci Transl Med. 2013;5:178ra140. doi: 10.1126/scitranslmed.3005580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, Nacken W, Brinkmann V, Jungblut PR, Zychlinsky A. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 2009;5:e1000639. doi: 10.1371/journal.ppat.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Donoghue AJ, Jin Y, Knudsen GM, Perera NC, Jenne DE, Murphy JE, Craik CS, Hermiston TW. Global substrate profiling of proteases in human neutrophil extracellular traps reveals consensus motif predominantly contributed by elastase. PLoS One. 2013;8:e75141. doi: 10.1371/journal.pone.0075141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker H, Albrett AM, Kettle AJ, Winterbourn CC. Myeloperoxidase associated with neutrophil extracellular traps is active and mediates bacterial killing in the presence of hydrogen peroxide. J Leukoc Biol. 2012;91:369–376. doi: 10.1189/jlb.0711387. [DOI] [PubMed] [Google Scholar]
- 28.Young RL, Malcolm KC, Kret JE, Caceres SM, Poch KR, Nichols DP, Taylor-Cousar JL, Saavedra MT, Randell SH, Vasil ML, Burns JL, Moskowitz SM, Nick JA. Neutrophil extracellular trap (NET)-mediated killing of Pseudomonas aeruginosa: evidence of acquired resistance within the CF airway, independent of CFTR. PLoS One. 2011;6:e23637. doi: 10.1371/journal.pone.0023637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keshari RS, Verma A, Barthwal MK, Dikshit M. Reactive oxygen species-induced activation of ERK and p38 MAPK mediates PMA-induced NETs release from human neutrophils. J Cell Biochem. 2013;114:532–540. doi: 10.1002/jcb.24391. [DOI] [PubMed] [Google Scholar]
- 30.Bernhagen J, Krohn R, Lue H, Gregory JL, Zernecke A, Koenen RR, Dewor M, Georgiev I, Schober A, Leng L, Kooistra T, Fingerle-Rowson G, Ghezzi P, Kleemann R, McColl SR, Bucala R, Hickey MJ, Weber C. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. 2007;13:587–596. doi: 10.1038/nm1567. [DOI] [PubMed] [Google Scholar]
- 31.Baumann R, Casaulta C, Simon D, Conus S, Yousefi S, Simon HU. Macrophage migration inhibitory factor delays apoptosis in neutrophils by inhibiting the mitochondria-dependent death pathway. FASEB J. 2003;17:2221–2230. doi: 10.1096/fj.03-0110com. [DOI] [PubMed] [Google Scholar]
- 32.Brinkmann V, Goosmann C, Kuhn LI, Zychlinsky A. Automatic quantification of in vitro net formation. Front Immunol. 2012;3:413. doi: 10.3389/fimmu.2012.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brinkmann V, Laube B, Abu Abed U, Goosmann C, Zychlinsky A. Neutrophil extracellular traps: how to generate and visualize them. J Vis Exp. 2010;36:1724. doi: 10.3791/1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winner M, Meier J, Zierow S, Rendon BE, Crichlow GV, Riggs R, Bucala R, Leng L, Smith N, Lolis E, Trent JO, Mitchell RA. A novel, macrophage migration inhibitory factor suicide substrate inhibits motility and growth of lung cancer cells. Cancer Res. 2008;68:7253–7257. doi: 10.1158/0008-5472.CAN-07-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plant BJ, Gallagher CG, Bucala R, Baugh JA, Chappell S, Morgan L, O'Connor CM, Morgan K, Donnelly SC. Cystic fibrosis, disease severity, and a macrophage migration inhibitory factor polymorphism. Am J Respir Crit Care Med. 2005;172:1412–1415. doi: 10.1164/rccm.200412-1714OC. [DOI] [PubMed] [Google Scholar]
- 36.Cantin AM. DNase I acutely increases cystic fibrosis sputum elastase activity and its potential to induce lung hemorrhage in mice. Am J Respir Crit Care Med. 1998;157:464–469. doi: 10.1164/ajrccm.157.2.9608033. [DOI] [PubMed] [Google Scholar]
- 37.Hansen G, Hoffjan S, Mosler K, Schuster A. Effect of recombinant human DNase on alpha1-proteinase inhibitor function: an experimental approach to the combined clinical use of rhDNase and alpha1-PI in CF patients. Lung. 2001;179:185–194. doi: 10.1007/s004080000060. [DOI] [PubMed] [Google Scholar]
- 38.Lue H, Dewor M, Leng L, Bucala R, Bernhagen J. Activation of the JNK signalling pathway by macrophage migration inhibitory factor (MIF) and dependence on CXCR4 and CD74. Cell Signal. 2011;23:135–144. doi: 10.1016/j.cellsig.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adamali H, Armstrong ME, McLaughlin AM, Cooke G, McKone E, Costello CM, Gallagher CG, Leng L, Baugh JA, Fingerle-Rowson G, Bucala RJ, McLoughlin P, Donnelly SC. Macrophage migration inhibitory factor enzymatic activity, lung inflammation, and cystic fibrosis. Am J Respir Crit Care Med. 2012;186:162–169. doi: 10.1164/rccm.201110-1864OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data