Abstract
Neutrophil extracellular traps (NETs) have been described as a fundamental innate immune defence mechanism. They consist of a nuclear DNA backbone associated with different antimicrobial peptides (AMPs) which are able to engulf and kill pathogens. The AMP LL-37, a member of the cathelicidin family, is highly present in NETs. However, the function of LL-37 within NETs is still unknown because it loses its antimicrobial activity when bound to DNA in the NETs. Using immunofluorescence microscopy, we demonstrate that NETs treated with LL-37 are distinctly more resistant to S. aureus nuclease degradation than nontreated NETs. Biochemical assays utilising a random LL-37-fragment library indicated that the blocking effect of LL-37 on nuclease activity is based on the cationic character of the AMP, which facilitates the binding to neutrophil DNA, thus protecting it from degradation by the nuclease. In good correlation to these data, the cationic AMPs human beta defensin-3 and human neutrophil peptide-1 showed similar protection of neutrophil-derived DNA against nuclease degradation. In conclusion, this study demonstrates a novel role of AMPs in host immune defence: beside its direct antimicrobial activity against various pathogens, cationic AMPs can stabilise neutrophil-derived DNA or NETs against bacterial nuclease degradation.
Key Words: Neutrophil extracellular traps, Antimicrobial peptides, Nucleases, Cathelicidin
Introduction
Classically, two strategies by which neutrophils serve as a first line of defence against invading pathogens are understood: the secretion of antimicrobial peptides (degranulation) and the engulfment of bacteria (phagocytosis) [1, 2]. More recently, Brinkmann et al. [3] characterised neutrophil extracellular traps (NETs) as a novel additional antimicrobial function of these specialised leukocytes. NET formation (NETosis) can be activated by pathogens, such as bacteria, pro-inflammatory cytokines or different substances like LPS, M protein-fibrinogen complexes or phorbol 12-myristate 13-acetate (PMA) [4].
The backbone of NETs is nuclear DNA, as nuclease treatment results in the disruption of their structure [5]. As a mechanism of innate immune evasion, nucleases secreted by Gram-positive bacterial pathogens such as group A streptococci (GAS), Streptococcus pneumoniae and Staphylococcus aureus degrade NETs to promote neutrophil resistance and the spread of infection in vivo [6, 7, 8, 9].
Besides DNA, NETs are comprised of histones, several granule proteins and antimicrobial peptides (AMPs), such as the cationic pro-inflammatory peptide cathelicidin LL-37 [10, 11, 12]. Human LL-37 is a member of the cathelicidin family of mammalian cationic AMPs. It derives its name from the amino acid sequence (37 amino acids) starting with two leucine residues. It gains antimicrobial activity after maturation through cleavage of the protein hCAP-18 by the serine protease proteinase 3 and adoption of the alpha-helical, amphipathic configuration of the mature peptide [13]. LL-37 is constitutively expressed in neutrophils [14], mast cells, natural killer cells and epithelial cells; during infection it can also be expressed by keratinocytes. During inflammatory responses, the expression of LL-37 is increased several-fold [15].
Previous studies have observed a correlation between bacterial resistance to LL-37 killing and NET-mediated killing [16, 17]. Consequently, it was inferred that the high local concentration of LL-37 in NETs could be a critical contributor to the antimicrobial activity of the NETs [16, 17]. However, Weiner et al. [18] showed that LL-37 can lose its antimicrobial activity when bound to DNA. Thus, even though LL-37 is found within NETs, its precise function here remains unclear. The goal of this study was to investigate its role in NET formation and stability.
Methods
Isolation of Neutrophils and Neutrophil-Derived DNA
Primary blood-derived neutrophils were isolated from fresh blood of healthy donors by density gradient centrifugation using PolymorphPrep™ (Progen Biotechnik) as previously described [19]. DNA was isolated with the NucleoSpin Blood™ kit (MACHEREY-NAGEL) according to the manufacturer's recommendation. For in vitro NET assays, the cells were seeded on poly-L-lysine-coated glass slides in 24-well plates at a concentration of 5 × 105 cells/well (250 µl/well) or on 48-well plates at a concentration of 2 × 105 cells/well (100 µl/well). RPMI without phenol red was used for cultivation of the cells at 37°C and 5% CO2.
Bacterial Strains and Nucleases
S. aureus nuclease (micrococcal nuclease, MN) was purchased from CellSystems (Troisdorf, Germany). Recombinant EndA H160G (10 nM) in combination with 20 mM Tris, 5 mM MgCl2 and 50 mM imidazole (pH 8) was used as previously described [20]. We used a panel of nuclease (nuc)-deficient mutants and nuclease-producing control strains of S. aureus USA300 LAC strain: S. aureus LAC wild-type (WT) empty vector control (WT + pCM28), nuc-mutant empty vector control (nuc + pCM28) and complemented mutant strain (nuc + pCM28nuc) [9]. Furthermore, the following three different nuclease-producing GAS strains isolated from infected patients in Portugal were used: 2004V0695P (emm type 1, GAS 1), SH1131A (emm type 1, GAS 2) and 2003V1350P (emm type 4, GAS 3) [21]. The GAS were grown in Todd-Hewitt broth and the S. aureus in brain-heart infusion broth up to the mid-log growth phase, centrifuged at 3,000 g for 10 min, and then 1 ml of the supernatant was sterile-filtered with a 0.4-µm filter and transferred to a new reaction tube. These supernatant samples were stored at −20°C until further use in DNA degradation assays.
NET Degradation by S. aureus Nuclease
For NET induction, neutrophils were stimulated with 25 nM PMA in the presence or absence of 40 µg/ml aprotinin (Sigma) and incubated for 4 h at 37°C 5% and CO2. Next, LL-37 was added to a final concentration of 5 µM to each well followed by incubation for 30 min. Finally, 50 µl MN from S. aureus was added to obtain a final concentration of 0.01 U/ml per well after which the plate was incubated for 1 h at 37°C and 5% CO2. Cells were fixed with 4% PFA and NETs were visualised as described below.
Visualisation of NETs
Fixed cells were washed 3 times with PBS, permeabilised and blocked with 2% BSA in 0.2% Triton X-100/PBS for 45 min at RT. Incubation with a mouse monoclonal anti-H2A-H2B-DNA complex (clone PL2-6) [22], 0.5 µg/ml in 2% BSA in 0.2% Triton X-100/PBS) was carried out overnight at 4°C, followed by washing (3 times with PBS) and subsequent incubation with an Alexa-Fluor-488-labelled goat-anti-mouse antibody for 45 min at RT. After washing, slides were mounted in ProlongGold® antifade with DAPI (Invitrogen) and analysed by confocal fluorescence microscopy using a Leica TCS SP5 microscope with an HCX PL APO 40× 0.75-1.25 oil immersion objective. Settings were adjusted with control preparations using an isotype control antibody. For each preparation, three randomly selected images were acquired and used for the quantification of the NETs. Data were expressed as area covered with NETs. The mean value derived from 6 images for each condition per experiment was used for statistical analysis.
Immunostaining of LL-37
Immunostaining of LL-37 was performed using a similar protocol to that described above for the visualisation of NETs. A polyclonal rabbit-anti-LL-37 antibody was used at a final concentration of 1.6 µg/ml [23]. Subsequently, samples were incubated with Alexa-Fluor-488 goat-anti-mouse and goat-anti-rabbit antibody (both 1:1,000; Invitrogen), respectively. Again, microscope settings were adjusted with control preparations using respective isotype control antibodies.
Entrapment Assay
The entrapment assay was performed as previously described [9]. Briefly, NET-producing neutrophils incubated in the presence or absence of LL-37 for 30 min (as described above) were infected with FITC-labelled S. aureus USA300 LAC strain for 30 min. Percentage of entrapment was calculated compared to total amount of surviving CFU under the respective conditions in the presence or absence of LL-37.
Negative Staining and Transmission Electron Microscopy
Human primary blood-derived neutrophils were isolated and treated as described above. The binding between LL-37 and DNA from neutrophil NETs was visualised by negative staining and electron microscopy as previously described [24]. LL-37 was conjugated with 5 nm colloidal gold particles according to routine protocols [25]. LL-37-Au conjugates were incubated with DNA for 30 min at RT and negatively stained with 0.75% uranyl formate. Specimens were examined under a Philips/FEI CM100 BioTwin transmission electron microscope (×100,000).
DNA Degradation Assay with DNA-Intercalating Dyes
The Quant-iT™ PicoGreen® dsDNA reagent and SYTOX® Green nucleic acid stain (Invitrogen) were used to measure degradation of host DNA. Calf thymus DNA (Sigma) or blood-derived neutrophil DNA at a final concentration of 5 µg/ml was added to a pureGrade™ black 96-well BRANDplate. Next, LL-37 (final concentration: 5 µM) and MN (final concentration: 100 U/ml) or the bacterial supernatants were added. The plate was incubated for 1 h at 37°C. Quant-iT PicoGreen dsDNA reagent or SYTOX Green nucleic acid stain was then added as recommended by the manufacturer. Fluorescence was measured with the Optima FluoStar (BMG Labtech) with excitation at 485 nm and emission at 520 nm with optimised gain settings that were used throughout a similar set of experiments. Human neutrophil peptide-1 (HNP-1) and human beta defensin-3 (hBD3) (both from Anaspec), full-length LL-37, a fragment library of 8 different LL-37 fragments with overlapping sequences and varying biochemical properties [26], and a scrambled form of LL-37 (Anaspec) were used.
Ethidium Bromide Gel
A total amount of 10 µg/ml calf thymus DNA was incubated with 5 µM LL-37 for 1 h at 37°C. Ethidium bromide (Sigma) was added to obtain a concentration of 0.5 µg/ml. Lastly, 6× loading dye was added and 20 μl of each sample was run on 1% agarose gel for visual examination of DNA.
Immunodotblot Determination of LL-37 in NETs
Neutrophils were stimulated with 25 nM PMA for 4 h, centrifuged for 5 min at 370 g and then incubated in the presence of 0.01 U/ml MN to degrade NETs. The supernatant was harvested before (released LL-37) and after nuclease treatment (LL-37 associated with NETs) and 200 µl were spotted on a PVDF membrane using a vacuum dotblotting system (Carl Roth). After blocking of the membrane in 5% non-fat dry milk in PBS-Tween (0.1% Tween 20) for 45 min at constant agitation, the membrane was incubated with 0.49 µg/ml polyclonal rabbit anti-LL-37 antibody overnight at 4°C with agitation. After washing with Tween-PBS, the membrane was incubated with the secondary goat-anti-rabbit HRP-conjugated IgG (BioRad) for 45 min at RT under agitation. After subsequent washing, the LL-37 signal was detected using the SuperSignal® West Femto maximum sensitivity substrate (Thermo Scientific) as recommended by the manufacturer.
Statistical Analysis
Data were analysed using Excel 2003 (Microsoft) and GraphPad Prism 5.0 (GraphPad software). All experiments were performed at least 3 times. NET experiments were performed with blood from three different blood donors. Differences between the 2 groups were analysed by using a one-tailed Student t test. The significance is indicated as * p < 0.05, ** p < 0.005 and *** p < 0.001.
Results and Discussion
LL-37 Protects Host DNA and NETs against Degradation by Bacterial Nucleases
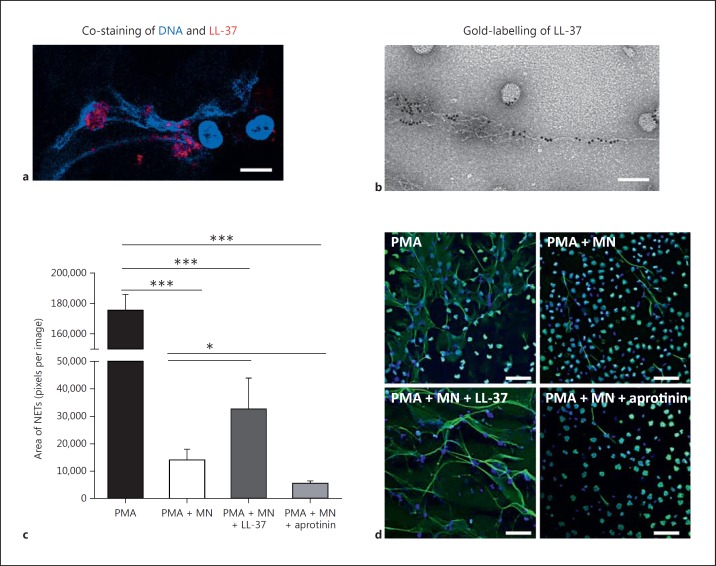
LL-37 has been shown to be associated with DNA fibres of NETs [10, 11, 12]. Immunofluorescence and electron microscopy confirmed that a high amount of LL-37 is present in NETs (fig. 1a, b). Four Immunodotblot experiments revealed that an average of 42.5 ± 12.2% of the LL-37 that is released by PMA-activated neutrophils is associated with the NET fibres. Previously, LL-37 has been shown to protect bacterial plasmid DNA and mammalian DNA against degradation by serum nucleases [12, 27]. Therefore, we analysed the effects of LL-37 on the stability of neutrophil DNA and/or NETs when treated with S. aureus nuclease. Human blood-derived neutrophils were treated with 25 nM PMA for 4 h to induce NET formation [5]. Subsequently, the induced NETs were treated with 0.01 U/ml S. aureus nuclease in the presence or absence of externally added LL-37. The addition of LL-37 significantly protected NETs against degradation (fig. 1c, d). When treating the cells with aprotinin to block the proteinase-3-mediated activation of endogenous, neutrophil-derived LL-37 [28], a significantly higher degree of NET degradation by S. aureus nuclease was detectable (fig. 1c, d).
Fig. 1.
LL-37 protects NETs against degradation by S. aureus nuclease. Confocal immunofluorescence (a) and electron micrograph (b) of structures, e.g. NETs and nanoparticles released by PMA-activated neutrophils which are associated with a high amount of LL-37. Bar: 8 µm (a), 100 nm (b). c Human blood-derived neutrophils were stimulated with 25 nM PMA for 4 h to induce 100% NET formation. Aprotinin (40 µg/ml) was used to block formation of endogenous active LL-37. The NETs were then treated with 0.01 U/ml S. aureus nuclease (MN) in the presence or absence of externally added LL-37 (5 µM). d Representative fluorescent micrographs displaying the results of the column bar graph (c). Bars: 50 µm. NETs were visualized using an Alexa 488-labelled antibody against H2A-H2B-DNA complexes (green) in combination with DAPI to stain the nuclei in blue. The area covered with NETs was quantified using Image J. The graph represents the mean ± SEM of a minimum of 18 images derived from 3 independent experiments. * p < 0.05, *** p < 0.001 by t test.
To study the effect of LL-37 on the stabilisation of host DNA in more detail, we adopted a biochemical assay using the fluorescent dye PicoGreen to provide a precise method of monitoring the nuclease activity in vitro. PicoGreen is a sensitive fluorescent dye widely used in analytical protocols in which double-stranded DNA detection is needed [29]. The dye associates with double-stranded DNA through intercalation, minor-groove binding and electrostatic interactions, resulting in a significant increase in fluorescence compared to the fluorescence of the free dye in solution [30]. Small fragments of DNA (like when double-stranded DNA is treated with nuclease) interact only weakly with PicoGreen, resulting in a decrease in fluorescence.
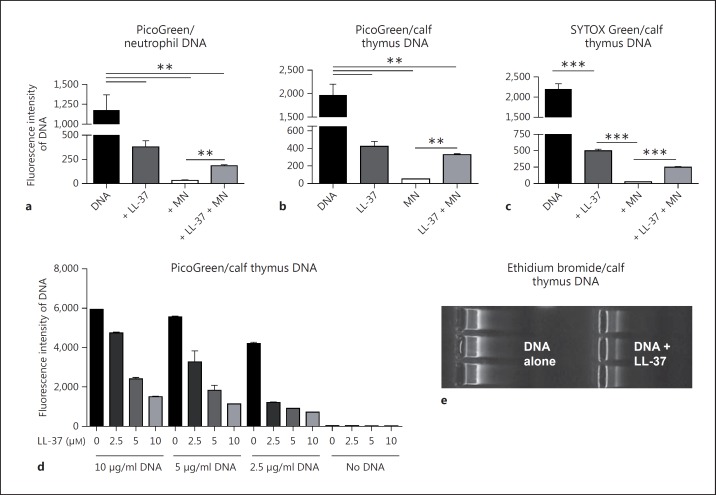
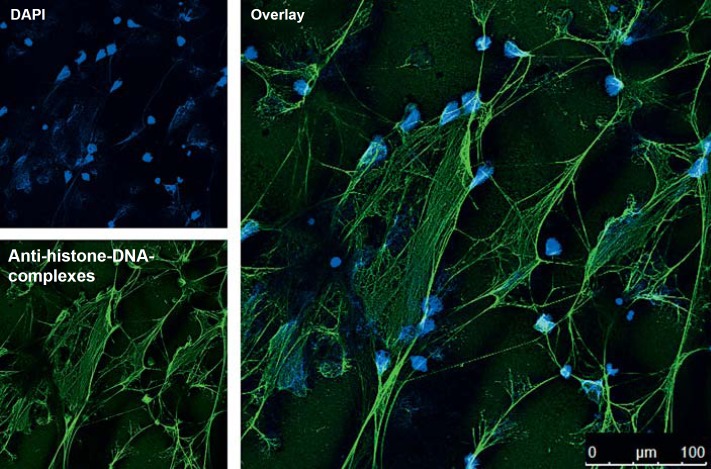
Correlating well with our microscopy data, the addition of LL-37 significantly protected neutrophil- and calf-thymus-derived DNA against degradation by S. aureus nuclease and resulted in a higher fluorescence signal than its LL-37-free control (fig. 2a, b). Interestingly, LL-37 alone decreased the PicoGreen signal, indicating that it binds to DNA and thereby blocks the DNA-PicoGreen interaction and subsequent fluorescence signal. This indicated that protection of DNA against nuclease degradation was the result of LL-37-mediated sequestration of the DNA. This effect is concentration-dependent: the more LL-37 that is added to the DNA, the lower the DNA-PicoGreen signal that is detected (fig. 2d). Similar results were observed with other DNA-intercalating dyes. The presence of LL-37 decreased the ethidium bromide signal in an agarose gel-based DNA detection system (fig. 2e) and DNA detection with the fluorescent dye SYTOX Green using a fluorimeter (fig. 2c). LL-37 thus interferes with DNA-intercalating dyes and so this might not be the method of choice to visualise NETs. A DNA-intercalating dye like DAPI is not efficient enough to visualise the complete structure of a NET (fig. 3). In contrast, an antibody-mediated method, e.g. an antibody against histone-DNA complexes as it was used here, gives a more complete image of the structure of the NETs.
Fig. 2.
LL-37 protects host DNA against degradation by S. aureus nuclease. Quantification of DNA and its degradation using different DNA-intercalating dyes: PicoGreen (a, b, d), SYTOX Green (c) and ethidium bromide (e) in the presence or absence of 5 µM LL-37 ± 100 U/ml S. aureus nuclease (MN). All graphs represent the mean ± SEM of minimum 3 independent experiments. * p < 0.05, ** p < 0.005, *** p < 0.001 by t test.
Fig. 3.
DAPI and immunostaining of NETs. Confocal immunofluorescent micrograph of NETs. NETs were visualized using an Alexa 488-labelled antibody against H2A-H2B-DNA complexes (green) and with DAPI to stain the extra- and intracellular DNA in blue.
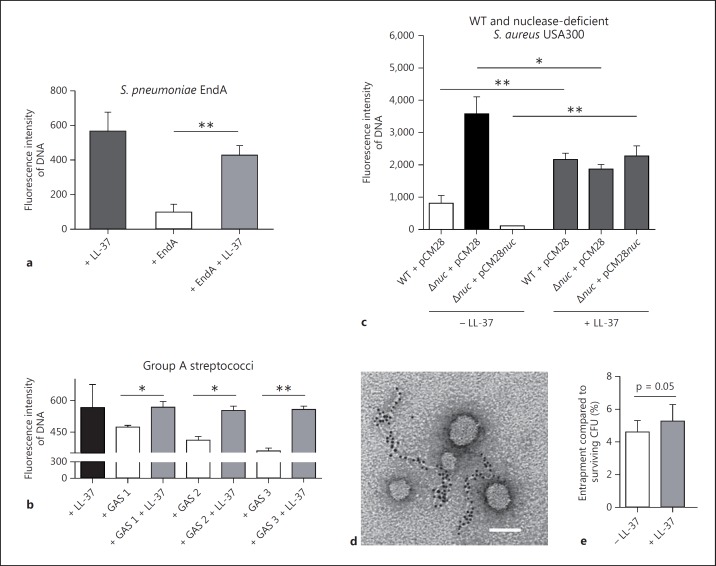
Despite the interference by LL-37 of the PicoGreen signal, the assay was still sensitive enough to determine the degradation of DNA by bacterial nucleases and its protection by LL-37 (fig. 2a-c). Next, we determined if LL-37 was also able to protect the DNA against degradation by nucleases derived from other bacterial species. It was able to protect the DNA against degradation by purified EndA from S. pneumoniae (fig. 4a), by nucleases from three different nuclease-producing GAS strains (fig. 4b) and by the prototype community-acquired MSRA (methicillin-resistant S. aureus) strain, USA300 LAC (fig. 4c). In this experiment, the supernatant of an isogenic nuclease mutant strain (nuc) was used as a negative control. In the absence of LL-37, the supernatant of the nuclease-producing WT MRSA strain showed significantly greater DNA degradation than that of the nuclease-deficient strain. In the presence of LL-37, the nuclease activity was blocked and all supernatants yielded similar DNA levels (fig. 4c).
Fig. 4.
LL-37 protects host DNA against degradation by nucleases derived from different Gram-positive bacteria: DNA degradation in the presence or absence of 5 µM LL-37 by purified EndA (a), supernatants of three different GAS strains (b) and S. aureus USA300 LAC (c) using PicoGreen as a marker. c A panel of nuclease (nuc)-deficient mutants and nuclease-producing control strains of S. aureus USA300 LAC strain was used: S. aureus LAC WT empty vector control (WT + pCM28), nuc-mutant empty vector control (nuc + pCM28) or complemented mutant strain (nuc + pCM28nuc). Electron micrograph of NETs after degradation with purified MN: only small left-over NET-structures that are decorated with gold-labelled LL-37 are visible. All NETs that are not decorated with LL-37 (partially seen in fig. 1b) have been enzymatically degraded and are not detectable anymore. Bar: 100 nm (d). Entrapment of S. aureus USA300 LAC strain by NET-releasing neutrophils in the presence or absence of 5 µM LL-37: percentage of entrapment was calculated compared to total amount of surviving CFU under the respective conditions in the presence or absence of LL-37 (e). All graphs represent the mean ± SEM of a minimum of 3 independent experiments. * p < 0.05, ** p < 0.005 by t test.
We demonstrated a novel role of the AMP LL-37, i.e. the protection of neutrophil DNA and NETs against degradation by bacterial nucleases. Accordingly, an electron micrograph of NETs after degradation with purified and highly concentrated MN revealed shortened left-over NET structures decorated with gold-labelled LL-37 (fig. 4d). All parts of the NETs that had not been decorated with LL-37 (fig. 1b) were enzymatically degraded and were no longer visible. Importantly, a functional assay analysing the entrapment of MRSA USA300 LAC in the presence or absence of LL-37 revealed a distinct (p = 0.05) increase of entrapment of surviving CFU in the presence of 5 µM of LL-37. Thus, LL-37 might not only efficiently kill bacteria [31] or increase the antimicrobial activities of neutrophils [32] as previously shown, but also enhance NET-mediated entrapment and the subsequent immobilization of bacteria.
The Positive Overall Charge of LL-37 Is Responsible for LL-37-Mediated Stabilisation of Host DNA against Bacterial Nucleases
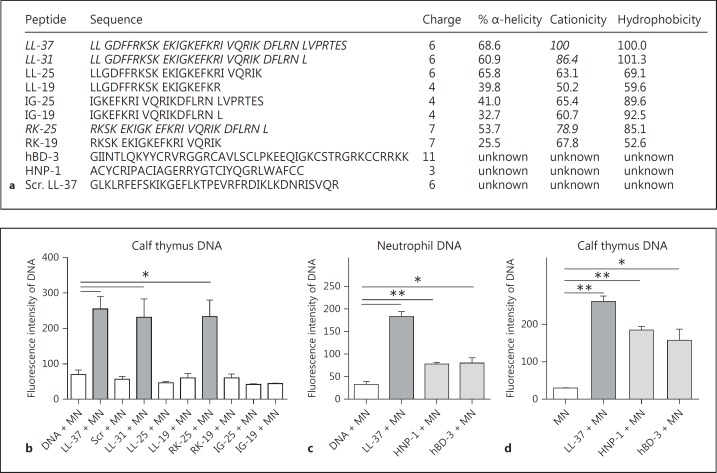
With our next experiments, we aimed to determine the biochemical features that cause LL-37-mediated protection of host DNA against bacterial nuclease degradation. The PicoGreen assay was conducted using the library of LL-37 fragments with overlapping sequences (fig. 5a) and a scrambled form of the peptide with the same amino acid composition serving as a control. In addition to the full-length LL-37 peptide, fragments LL-31 and RK-25 were able to stabilise the DNA (fig. 5b). A statistical analysis using the Pearson correlation test confirmed that the cationicity correlates with the ability to protect the DNA against nuclease degradation.
Fig. 5.
Cationicity is involved in the protection of host DNA against bacterial nuclease-mediated degradation. a Sequences and biochemical characteristics of LL-37, LL-37 fragments, scrambled LL-37 (Scr. LL-37), hBD-3 and HNP-1. Quantification of DNA-degradation by 100 U/ml S. aureus nuclease (MN) in the presence or absence of 5 µM LL-37, LL-37 fragments, scrambled LL-37 (b) or 5 µM HNP-1 and hBD-3 (c, d). All graphs represent the mean ± SEM of minimum 3 independent experiments. * p < 0.05, ** p < 0.005 by t test.
To verify that cationicity is involved in the phenomenon of DNA stabilisation, two other well-characterised human cationic AMPs, HNP-1 and hBD-3, were tested in an identical assay. Both peptides significantly diminished DNA degradation by S. aureus nuclease; however, neither peptide protected the DNA against nuclease degradation to the degree afforded by LL-37 (fig. 5c, d). These data imply that a cocktail of different host-derived cationic peptides may function in concert to stabilise NETs against bacterial nucleases.
Conclusions
We showed that LL-37 protected NETs, purified neutrophil DNA and calf thymus DNA against degradation mediated by nucleases derived from bacterial pathogens including S. aureus, S. pneumoniae and GAS. Nuclease production by these pathogens can protect against NET-mediated entrapment and immobilisation in S. pneumoniae as well as NET-mediated bacterial killing in GAS and S. aureus [6, 7, 8, 9]. However, residual NETs can still be detected in the tissue of mice infected with nuclease-producing bacteria, as shown for S. aureus[9], indicating an incomplete degradation by bacterial nucleases in vivo based on a stabilisation by host factors.
The abundance of the cathelicidin AMP within NETs may not be an accidental by-product of simultaneous NETosis and degranulation but may reflect an integral role of the peptide in NET formation and longevity.
Acknowledgements
We wish to thank Mário Ramirez, Faculty of Medicine, University of Lisbon, Portugal, for providing the GAS strains. This work was supported in part by a DFG grant (KO 3552/4-1) to M.v.K.-B., NIH grants (AI052453 and AR052728) to R.L.G. and V.N. and a Vidi grant from the Dutch Scientific Organisation NWO (to S.H.M.R., E.T.M.B. and D.A.C.S.).
Footnotes
A.N. and L.V. contributed equally to this work.
References
- 1.Lehrer RI, Ganz T. Antimicrobial peptides in mammalian and insect host defence. Curr Opin Immunol. 1999;11:23–27. doi: 10.1016/s0952-7915(99)80005-3. [DOI] [PubMed] [Google Scholar]
- 2.Tauber A. The birth of immunology. III. The fate of the phagocytosis theory. Cell Immunol. 1992;139:505–530. doi: 10.1016/0008-8749(92)90089-8. [DOI] [PubMed] [Google Scholar]
- 3.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 4.von Köckritz-Blickwede M, Nizet V. Innate immunity turned inside-out: antimicrobial defense by phagocyte extracellular traps. J Mol Med. 2009;87:775–783. doi: 10.1007/s00109-009-0481-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, Weinrauch Y, Brinkmann V, Zychlinsky A. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. 2007;176:231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sumby P, Barbian KD, Gardner DJ, Whitney AR, Welty DM, Long RD, Bailey JR, Parnell MJ, Hoe NP, Adams GG, Deleo FR, Musser JM. Extracellular deoxyribonuclease made by group A streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc Natl Acad Sci USA. 2005;102:1679–1684. doi: 10.1073/pnas.0406641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beiter K, Wartha F, Albiger B, Normark S, Zychlinsky A, Henriques-Normark B. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr Biol. 2006;16:401–407. doi: 10.1016/j.cub.2006.01.056. [DOI] [PubMed] [Google Scholar]
- 8.Buchanan JT, Simpson AJ, Aziz RK, Liu GY, Kristian SA, Kotb M, Feramisco J, Nizet V. DNase expression allows the pathogen group A streptococcus to escape killing in neutrophil extracellular traps. Curr Biol. 2006;16:396–400. doi: 10.1016/j.cub.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 9.Berends ET, Horswill AR, Haste NM, Monestier M, Nizet V, von Köckritz-Blickwede M. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J Innate Immun. 2010;2:576–586. doi: 10.1159/000319909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow OA, von Köckritz-Blickwede M, Bright AT, Hensler ME, Zinkernagel AS, Cogen AL, Gallo RL, Monestier M, Wang Y, Glass CK, Nizet V. Statins enhance formation of phagocyte extracellular traps. Cell Host Microbe. 2010;8:445–454. doi: 10.1016/j.chom.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, Nacken W, Brinkmann V, Jungblut PR, Zychlinsky A. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 2009;5:e1000639. doi: 10.1371/journal.ppat.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lande R, Ganguly D, Facchinetti V, Frasca L, Conrad C, Gregorio J, Meller S, Chamilos G, Sebasigari R, Riccieri V, Bassett R, Amuro H, Fukuhara S, Ito T, Liu YJ, Gilliet M. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci Transl Med. 2011;3:73ra19. doi: 10.1126/scitranslmed.3001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sörensen OE, Follin P, Johnsen AH, Calafat J, Tjabringa GS, Hiemstra PS, Borregaard N. Human cathelicidin, hCAP-18, is processed to the antimicrobial petide LL-37 by extracellular cleavage with proteinase 3. Blood. 2001;97:3951–3959. doi: 10.1182/blood.v97.12.3951. [DOI] [PubMed] [Google Scholar]
- 14.Zanetti M, Gennaro R, Romeo D. Cathelicidins: a novel protein family with a common proregion and a variable C-terminal antimicrobial domain. FEBS Lett. 1995;374:1–5. doi: 10.1016/0014-5793(95)01050-o. [DOI] [PubMed] [Google Scholar]
- 15.Lai Y, Gallo RL. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–140. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole JN, Pence MA, von Köckritz-Blickwede M, Hollands A, Gallo RL, Walker MJ, Nizet V. M protein and hyaluronic acid capsule are essential for in vivo selection of covRS mutations characteristic of invasive serotype M1T1 group A streptococcus. MBio. 2010;1:e00191–10. doi: 10.1128/mBio.00191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauth X, von Köckritz-Blickwede M, McNamara CW, Myskowski S, Zinkernagel AS, Beall B, Ghosh P, Gallo RL, Nizet V. M1 protein allows group A streptococcal survival in phagocyte extracellular traps through cathelicidin inhibition. J Innate Immun. 2009;1:202–214. doi: 10.1159/000203645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiner DJ, Bucki R, Janmey PA. The antimicrobial activity of the cathelicidin LL37 is inhibited by F-actin bundles and restored by gelsolin. Am J Respir Cell Mol Biol. 2003;28:738–745. doi: 10.1165/rcmb.2002-0191OC. [DOI] [PubMed] [Google Scholar]
- 19.von Köckritz-Blickwede M, Chow O, Ghochani M, Nizet V. Visualization and functional evaluation of phagocyte extracellular traps. In: Kaufmann S, Kabelitz D, Immunology of Infection, editors. Series: Methods in Microbiology. ed 3. Maryland Geights: Elsevier Science; 2010. [Google Scholar]
- 20.Midon M, Schäfer P, Pingoud A, Ghosh M, Moon AF, Cuneo MJ, London RE, Meiss G. Mutational and biochemical analysis of the DNA-entry nuclease EndA from Streptococcus pneumoniae. Nucleic Acids Res. 2011;39:623–634. doi: 10.1093/nar/gkq802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friães A, Ramirez M, Melo-Cristino J, Portuguese Group for the Study of Streptococcal Infections Nonoutbreak surveillance of group A streptococci causing invasive disease in Portugal identified internationally disseminated clones among members of a genetically heterogeneous population. J Clin Microbiol. 2007;45:2044–2047. doi: 10.1128/JCM.00496-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Losman MJ, Fasy TM, Novick KE, Monestier M. Monoclonal autoantibodies to subnucleosomes from a MRL/Mp (-)+/+ mouse. Oligoclonality of the antibody response and recognition of a determinant composed of histones H2A, H2B, and DNA. J Immunol. 1992;148:1561–1569. [PubMed] [Google Scholar]
- 23.Dorschner RA, Pestonjamasp VK, Tamakuwala S, Ohtake T, Rudisill J, Nizet V, Agerberth B, Gudmundsson GH, Gallo RL. Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group A streptococcus. J Invest Dermatol. 2001;117:91–97. doi: 10.1046/j.1523-1747.2001.01340.x. [DOI] [PubMed] [Google Scholar]
- 24.Bober M, Enochsson C, Collin M, Mörgelin M. Collagen VI is a subepithelial adhesive target for human respiratory tract pathogens. J Innate Immun. 2010;2:160–166. doi: 10.1159/000232587. [DOI] [PubMed] [Google Scholar]
- 25.Baschong W, Wrigley NG. Small colloidal gold conjugated to Fab fragments or to immunoglobulin G as high-resolution labels for electron microscopy: a technical overview. J Electron Microsc Tech. 1990;14:313–323. doi: 10.1002/jemt.1060140405. [DOI] [PubMed] [Google Scholar]
- 26.Molhoek EM, den Hertog AL, de Vries AM, Nazmi K, Veerman EC, Hartgers FC, Yazdanbakhsh M, Bikker FJ, van der Kleij D. Structure-function relationship of the human antimicrobial peptide LL-37 and LL-37 fragments in the modulation of TLR responses. Biol Chem. 2009;390:295–303. doi: 10.1515/BC.2009.037. [DOI] [PubMed] [Google Scholar]
- 27.Sandgren S, Wittrup A, Cheng F, Jönsson M, Eklund E, Busch S, Belting M. The human antimicrobial peptide LL-37 transfers extracellular DNA plasmid to the nuclear compartment of mammalian cells via lipid rafts and proteoglycan-dependent endocytosis. J Biol Chem. 2004;279:17951–17956. doi: 10.1074/jbc.M311440200. [DOI] [PubMed] [Google Scholar]
- 28.Yamasaki K, Schauber J, Coda A, Lin H, Dorschner RA, Schechter NM, Bonnart C, Descargues P, Hovnanian A, Gallo RL. Kallikrein-mediated proteolysis regulates the antimicrobial effects of cathelicidins in skin. Faseb J. 2006;20:2068–2080. doi: 10.1096/fj.06-6075com. [DOI] [PubMed] [Google Scholar]
- 29.Singer VL, Jones LJ, Yue ST, Haugland RP. Characterization of PicoGreen reagent and development of a fluorescence-based solution assay for double-stranded DNA quantitation. Anal Biochem. 1997;249:228–238. doi: 10.1006/abio.1997.2177. [DOI] [PubMed] [Google Scholar]
- 30.Dragan AI, Casas-Finet JR, Bishop ES, Strouse RJ, Schenerman MA, Geddes CD. Characterization of PicoGreen interaction with dsDNA and the origin of its fluorescence enhancement upon binding. Biophys J. 2010;99:3010–3019. doi: 10.1016/j.bpj.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner J, Cho Y, Dinh NN, Waring AJ, Lehrer RI. Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils. Antimicrob Agents Chemother. 1998;42:2206–2214. doi: 10.1128/aac.42.9.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alalwani SM, Sierigk J, Herr C, Pinkenburg O, Gallo R, Vogelmeier C, Bals R. The antimicrobial peptide LL-37 modulates the inflammatory and host defense response of human neutrophils. Eur J Immunol. 2010;40:1118–1126. doi: 10.1002/eji.200939275. [DOI] [PMC free article] [PubMed] [Google Scholar]