Abstract
This work describes the coupling of the IR-MALDESI imaging source with the Q Exactive mass spectrometer. IR-MALDESI MSI was used to elucidate the spatial distribution of several HIV drugs in cervical tissues that had been incubated in either a low or high concentration. Serial sections to those analyzed by IR-MALDESI MSI were homogenized and analyzed by LC-MS/MS to quantify the amount of each drug present in the tissue. By comparing the two techniques, an agreement between the average intensities from the imaging experiment with the absolute quantities for each drug was observed. This correlation between these two techniques serves as a prerequisite to quantitative IR-MALDESI MSI. In addition, a targeted MS2 imaging experiment was also conducted to demonstrate the capabilities of the Q Exactive and to highlight the added selectivity that can be obtained with SRM or MRM imaging experiments.
Keywords: IR-MALDESI, mass spectrometry imaging, LC-MS/MS, drug distribution
Introduction
Mass spectrometry imaging (MSI) is a method by which mass spectra are generated from discrete locations in a two dimensional array across a sample surface.[1] This combination of spatial information with the specificity and sensitivity of mass spectrometric detection make MSI a valuable tool in a variety of scientific fields.[2-9] One such example is the potential that MSI has demonstrated in the analysis of pharmaceuticals in tissue sections.[10-12] Knowledge of the distribution of a drug, and in some cases its metabolites, within certain compartments or sub-compartments of a particular tissue can have numerous implications in the areas of drug pharmacokinetics and pharmacodynamics.[13-14] Whole-body autoradiography (WBA) is the predominant technique for the determination of drug distribution in tissue.[15] While WBA is a valuable quantitative technique, it shares an inherent flaw with all radiolabeled studies in that it only follows the distribution of the radiolabel and provides no information as to which form of the drug is being detected (parent or metabolite). Given that MSI is based on mass spectrometric detection, it is capable of distinguishing between the parent drug and its metabolites. Perhaps one of the most enticing attributes of MSI is its label-free nature that precludes the use of expensive radiolabels and allows for the simultaneous analysis of xenobiotics and endogenous compounds in a single global experimental approach. In some cases the additional knowledge gained from endogenous distributions can provide valuable insight on the impact of the drug on the local environment, implications for site-specific efficacy and toxicity, and can be used to identify certain histological features.[11, 16-18] MSI is befitting to drug discovery and development where the early understanding of preclinical drug distribution (and without requiring a radiolabel) can improve efficiency by narrowing the list of potential candidates.
Matrix-assisted laser desorption/ionization (MALDI) is perhaps the most common ionization technique that is currently used for MSI. Several characteristics of MALDI that make it a suitable match for MSI include its high sensitivity and small spot size which are prerequisites for high spatial resolution imaging.[19] There are, however, several issues regarding this technique including the requirement that the sample must be amenable to high vacuum as well as the extensive sample preparation that is involved prior to analysis.[20-21] Other ionization methods such as desorption electrospray (DESI), liquid extraction surface analysis (LESA), and matrix-assisted laser desorption electrospray ionization (MALDESI) have demonstrated promise for pharmaceutical tissue imaging with little to no prior sample preparation.[22-24]
MALDESI refers to any technique which involves resonant excitation of an endogenous or exogenous matrix to facilitate analyte desorption followed by post-ionization through ESI. The matrix described here refers to any molecule present in large excess that strongly absorbs the laser energy and leads to analyte ejection.[25] The first MALDESI publication was presented in 2006 by Muddiman as proof of principle using a UV laser (UV-MALDESI) and a ubiquitin solution mixed with an organic acid matrix.[26] This resulted in the detection of multiply charged ubiquitin with a charge state distribution that was indistinguishable from that obtained by direct infusion ESI which affirmed electrospray post-ionization in the MALDESI mechanism. Muddiman, as well as several other research groups, have also explored the utility of other laser wavelengths for MALDESI in each case utilizing a matrix that strongly absorbed within the wavelength region of the laser emission.[27-31] An exciting combination arose with the use of a Mid-IR laser (2.94 μm) with endogenous or exogenous water as the laser energy absorbing matrix (IR-MALDESI) which offers certain intrinsic benefits including the lack of matrix interference in the mass spectra and the natural occurrence of water in most biological samples. While the use of water as a matrix for IR laser desorption was not an entirely new concept,[32] ion yields for IR-MALDI using ice were typically low.[33] Thus, post-ionization of the larger fraction of neutral molecules in the ablation plume using ESI could result in improved ion yields.[34] MSI using laser ablation electrospray ionization (LAESI), an analogue of IR-MALDESI, has been presented using the endogenous water in the tissue as a laser energy absorbing matrix.[35] However, we have found that depositing a uniform ice matrix over the sample surface provides improved reproducibility from pixel to pixel resulting in higher image quality.[24, 36-37]
Due to the complex nature of biological tissue samples, MSI techniques rely heavily on the mass spectrometer to resolve species by m/z. Several groups have demonstrated the utility of coupling imaging sources with high resolving power mass spectrometers in order to resolve several unique species with the same nominal mass that often times have different spatial distributions.[11, 19, 36, 38-42] The primary benefits to imaging with Fourier transform (FT) mass spectrometers include not only the ability to increase the spectral peak capacity but also the ability to obtain accurate mass measurements that improve the confidence of ion identification. However, due to the relatively slow acquisition rates, FT imaging can be a time consuming process. The hybrid quadrupole orbitrap mass spectrometer (Q Exactive) has several unique features that make it highly amenable to MSI.[43] In addition to using the enhanced FT (eFT) to reduce the required transient times, the Q Exactive also allows for multiplexing capabilities to improve the overall duty cycle. Several groups demonstrated the application of the Q Exactive for MSI.[44-46]
The use of MSI to visually evaluate the distribution of drugs in tissue is of particular interest in the field of human immunodeficiency virus (HIV), an intracellular infection primarily of CD4+ T cells. In this disease, current limitations in assessing mucosal tissue antiretroviral drug distribution have slowed the progress of small molecule research in the prevention of HIV infection.[47] Moreover, an understanding of antiretroviral distribution in HIV tissue reservoirs, where low-level viral replication may still be occurring in the face of clinically effective antiretroviral therapy[48] will form the foundation of successful eradication strategies. Previous uses of MALDI in the HIV field have focused on the quantification of antiretrovirals in plasma,[49] and more recently MSI has been used to examine the disposition of a novel antiretroviral in brain tissue.[50] A reliable method of visually evaluating antiretroviral exposure in tissues relevant to HIV infection will greatly inform the development of novel therapies for HIV treatment, prevention, and cure.
Herein, we present on the coupling of the IR-MALDESI imaging source to the Q Exactive to determine the distribution of several commonly used antiretroviral drugs in incubated tissues. Imaging is performed as either a broadband acquisition to obtain a global perspective of the three drugs of interest with endogenous species or as a more selective targeted MS2 acquisition. In addition, tissue sections that are adjacent to those used for imaging were homogenized and analyzed by LC-MS/MS to quantify the total amounts of each drug present in the tissue. These values were then compared with the relative abundances from the imaging experiments to determine the quantitative capabilities of IR-MALDESI MSI.
Experimental
Materials
HPLC Grade methanol and water were purchased from Burdick and Jackson (Muskegon, MI) and formic acid was purchased from Sigma-Aldrich (St. Louis, MO). Emtricitabine, tenofovir and raltegravir were obtained from the NIH AIDS Reagent Program, directed by the Pathogenesis and Basic Research Branch, Basic Sciences Program, Division of AIDS (DAIDS), NIAID, NIH. All materials were used as purchased without further purification.
Samples
Cervical tissues were obtained from surgical waste via the University of North Carolina Tissue Procurement Facility through UNC IRB # 09-0921. Written informed consent was obtained from all patients. After harvest, tissues were placed immediately in culture media [Iscove's Modified Dulbecco's Media (Gibco, Grand Island, NY), 10% fetal bovine serum (Gibco), 240 units/mL nystatin (Sigma, St. Louis, MO), 100 units/mL penicillin-streptomycin (Gibco), and MEM vitamin solution (Sigma)] and kept on ice until receipt into the laboratory. Once received into the lab, tissues were trimmed of fat and other connective tissue with sterile scissors and cut into approximately 1cm2 pieces. Pieces were placed into individual wells of a 24-well tissue culture plate with 1mL culture media containing low or high concentrations of the nucleoside reverse transcriptase inhibitors emtricitabine (FTC) and tenofovir (TFV) and the integrase strand transfer inhibitor raltegravir (RAL). Here, low concentrations were defined as 10 times the reported plasma Cmax value in humans for each drug: 18,000 ng/mL FTC, 3,800 ng/mL TFV, and 28,880 ng/mL RAL. High concentration was defined as 100,000 ng/mL for all three drugs. These concentrations were selected to mimic mucosal tissue concentrations seen clinically in Phase I studies.[51-52] The incubation concentration along with the exact monoisotopic mass for each drug is also presented in Table 1. All tissues were incubated for 24 hours at 37°C. After 24 hours, tissues were removed from drug and rinsed with fresh culture media before being frozen with dry ice vapor and stored at -80°C. Each tissue was sectioned at -20°C using a Leica CM1950 cryomicroto me (Buffalo Grove, IL) into 10, 25, or 50 μm thick sections. The sections were then thaw-mounted onto glass microscope slides for imaging. Sections of each tissue were mounted adjacent to each other so that each glass slide contained a section of each tissue in the low and high concentrations at the same thickness. Sections that were serial cuts to those used for imaging were also collected for LC-MS quantitation.
Table 1. Incubation concentrations for the three HIV drugs.
| Drug | Abbreviation | Monoisotopic Mass of [M+H]+ | Low Conc. (μg/mL) | High Conc. (μg/mL) |
|---|---|---|---|---|
| Tenofovir | TFV | 288.0856 | 3.8 | 100.0 |
| Emtricitabine | FTC | 248.0499 | 18.0 | 100.0 |
| Raltegravir | RAL | 445.1630 | 28.88 | 100.0 |
IR-MALDESI Imaging
A more detailed description of the IR-MALDESI imaging source can be found elsewhere.[36-37] In short, the tissue sample is placed on a liquid cooled thermoelectric stage that is cooled to -10°C while under nitrogen purge and is then exposed to the ambient environment in order to deposit a thin layer of ice over the surface of the tissue. Once the ice layer has been deposited, the enclosure around the source is purged with nitrogen to maintain a relative humidity of around 10 % which we have found to be the point where ice deposition and sublimation are close to equilibrium to preserve a consistent ice matrix layer throughout the course of the imaging experiment. A mid-IR laser tuned to 2.94 μm (IR-Opolette 2371, Opotek, Carlsbad, CA) is used to resonantly excite the ice matrix layer and facilitate the desorption of neutral molecules from the tissue section. These neutrals then partition into the charged solvent droplets of an electrospray plume where ions are generated through an ESI-like process. The geometry of the IR-MALDESI source has recently been optimized for tissue imaging.[37] For the imaging experiments, 50% (v/v) aqueous methanol with 0.2% formic acid was used for the electrospray solvent as this composition has been shown to work well for small molecules and lipids.[24] All imaging experiments were performed with a spot-to-spot distance of 100 μm.
Q Exactive
The IR-MALDESI imaging source was fully synchronized with a Thermo Fisher Scientific Q Exactive (Bremen, Germany) such that ion accumulation was triggered to overlap with the pulsing of the laser resulting in a single orbitrap acquisition at each pixel. The automatic gain control (AGC) of the instrument was turned off for all imaging experiments due to its incompatibility with our pulsed ionization source. The AGC is normally used to maintain a consistent number of ions in the orbitrap. It does so by determining the rate of ion generation during a prescan and varying the amount of time that ions are accumulated to reach a target number of ions (AGC target) for the analytical scan. With the AGC off, ions are accumulated for a set period of time according to the maximum ion injection time (IT). Even with the AGC off, mass accuracy was verified to be maintained within 1 ppm by using two diisooctyl phthalate peaks (391.2843 [M+H]+ and 413.266 [M+Na]+) as lock-masses in the instrument control software.
For broadband acquisition, two laser pulses were performed at 20 Hz (50 ms/pulse) with a 150 ms IT to accumulate ions from both laser pulses in the C-trap followed by a single Orbitrap acquisition. The mass range for the orbitrap acquisition was set to 150-600 m/z. The mass resolution was set to 140,000 at m/z 200.
For MS2 acquisition, a targeted MS2 method file was created using an inclusion list for isolating the protonated ion of RAL (m/z 445.16302) with a maximum IT of 150 ms. Two IR pulses were performed at each pixel (20 Hz) where ions from each pulse were isolated with a 4 m/z window and a 1.5 m/z offset followed by ion accumulation in the C-trap. The accumulated ion packet was then fragmented in the HCD cell at a normalized collision energy of 20. All resulting fragments were analyzed in a single orbitrap acquisition. The normalized collision energy was optimized through the direct infusion of a RAL standard. Unique transitions for RAL were also determined during the direct infusion of the drug standards. The mass resolution was set to 140,000 at m/z 200 for the MS2 acquisition in the orbitrap in order to obtain high mass accuracies for the fragments.
Data Analysis
For individual ion images, the raw data (.raw) from the Thermo Q Exactive was converted to the mzXML format using the MSConvert software from Proteowizard[53] For the stacked ion images, the raw files were converted to mzML files using the MSConvert software from Proteowizard and were then converted to individual imzML files using imzMLConverter.[54] The imzML Converter was then used to stack the individual imzmL files into one master imzML file. The mzXML or imzML files were then loaded into the standalone version of MSiReader which is freely available software developed in our lab for processing MSI data.[55] In order to demonstrate the quality of the raw data, ion images presented in this manuscript were neither interpolated nor normalized (unless otherwise specified). MSiReader was used to extract peak intensities to the regions around the low and high concentration tissues in order to determine the average peak intensity for comparison with the absolute amounts determined by LC-MS/MS. A modified ‘hot’ colorscale was used to demonstrate changes in intensity. Despite its widespread use in visualizing data, the ‘rainbow’ or ‘jet’ colorscale leads to misleading and non-intuitive distinctions between intensity values and was thus not used here.[56-59]
LC-MS/MS Quantitation
Tissue sections (10, 25, and 50 μm) from the low and high concentration tissue samples were extracted and analyzed by LC-MS/MS for TFV, FTC, and RAL concentrations. Sections were homogenized and extracted in 1 mL of 70:30 acetonitrile:1 mM ammonium phosphate (pH 7.4) using a Precellys® 24 tissue homogenizer. Calibration standards were prepared at 0.3, 0.6, 1.5, 6, 15, 30, 75, 150, 255, and 300 ng/mL in 70:30 acetonitrile:1 mM ammonium phosphate (pH 7.4). Quality control (QC) samples were prepared at 0.9, 21, and 240 ng/mL in 70:30 acetonitrile:1 mM ammonium phosphate (pH 7.4). Following centrifugation, 300 μL of each standard/QC/sample was mixed with 50 μL of an internal standard solution (13C5-TFV, 13C15N2-FTC, and RAL-d3 at 50 ng/mL in 50:50 methanol:water). The resulting solutions were evaporated to dryness under nitrogen at 50°C. Samples were reconstituted in 100 μL of 1 mM ammonium phosphate (pH 7.4) and transferred to a 96-well plate for LC-MS/MS analysis.
A Shimadzu HPLC system (SIL-20AC autosampler, LC-20AD pumps, and CTO-20A column oven; Shimadzu Scientific Instruments, Columbia, MD) was used for this analysis. A Waters Atlantis T3 column (2.1 mm × 100 mm, 3 μm, Waters, Milford, MA) was utilized at 35°C. A gradient elution using water with 0.1% formic acid (Mobile Phase A) and acetonitrile with 0.1% formic acid (mobile Phase B) was used to perform chromatographic separation. A Sciex API 5000 Triple Quad mass spectrometer (AB Sciex, Foster City, CA) equipped with a Turbo spray interface was used as the detector. TFV and 13C5-TFV were detected in negative ion mode with mass transitions of 286 → 107 and 291 →111, respectively. FTC, 13C15N2-FTC, RAL, and RAL-d3 were detected in positive ion mode with mass transitions of 248 → 130, 251 →133, 445 → 361, 448 →109, respectively.
Calibration standards and QCs for all three analytes were within 15% of nominal concentrations. With the tissue samples being extracted in 1 mL of solvent, the final result (in ng/mL) was equivalent to mass extracted (in ng) from each sample. The tissues (all < 1mg) provided negligible volume to the homogenized sample allowing for the direct correlation between ng/mL and ng extracted. Since the tissue slices were too small to weigh, the final drug concentrations were not adjusted for tissue mass and are presented as ng extracted per slice.
Results and Discussion
Full Acquisition IR-MALDESI MSI
Cervical tissues were incubated in either a high or low concentration of the three HIV drugs tenofovir (TFV), emtricitabine (FTC), and raltegravir (RAL). Two adjacent 10 μm sections of the high concentration tissue were thaw-mounted onto glass microscope slides. These two tissue sections were then imaged either with or without an ice matrix in order to validate the use of ice as a matrix for IR-MALDESI MSI.[37] The optical images of the tissues for both conditions as well as the corresponding ion maps for the protonated ions of the three incubated drugs are shown in Figure 1. The ion images for each drug provide clear evidence of the degree of improvement that can be achieved when using ice as a matrix. Also, the average pixel intensity and frequency of pixels in the tissue-related area where signal was detected (number of non-zero pixels/total number of pixels for the tissue-related area) are also provided for each drug under both conditions to provide a more objective comparison. The average pixel intensity is shown to be between about 15 fold higher for the tissue with ice as a matrix compared to the tissue without a matrix. While it is likely linked to the improvement in overall intensity, there is also marked improvement in the frequency of pixels with signal in the imaging experiment with ice which is especially clear for tenofovir which is the least abundant of the three incubated drugs. The results of these experiments help to validate the use of ice as a matrix for IR-MALDESI MSI and support our previous conclusions.[37]
Figure 1.
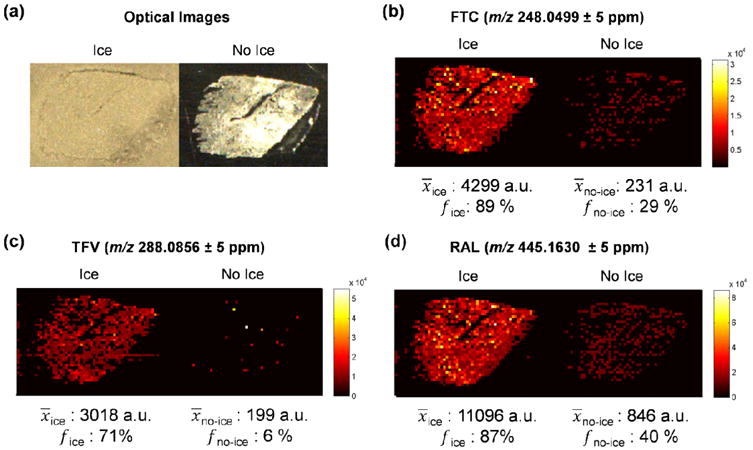
Validation of the use of ice as a matrix for IR-MALDESI MSI. Optical images (a) of two 10 μm serial sections of the high concentration tissue either with or without (respectively) a deposited layer of ice. Ion maps of (b) emtricitabine as [M+H]+, (c) tenofovir as [M+H]+, and (d) raltegravir as [M+H]+. In addition, the average intensity and frequency of the pixels with signal within the tissue area is provided for each of the ions under both conditions (with or without ice matrix) to demonstrate the degree of improvement that is achieved when using the ice matrix.
The high and low concentration incubated tissues were then cryo-sectioned into several thicknesses (10, 25, and 50 μm) to investigate the influence of tissue thickness on the imaging experiment. IR-MALDESI MSI was then performed on each set of tissues (high and low concentration at each tissue thickness) using a broadband acquisition method where at each image pixel a single full mass spectrum (m/z 150-600) was collected. This broadband acquisition allows for the observation of all three drugs as well as any endogenous species that fall within the m/z range. For each tissue thickness a single imaging experiment was conducted which encompassed both the low and high concentration tissue sections. Ion images for the protonated forms of all three drugs as well as the optical images of the tissue sections are shown in Figure 2. All of the ion maps shown in Figure 2 are on the same intensity scale to demonstrate the relative intensities of all three drugs across the different tissue thicknesses. Coupling the IR-MALDESI imaging source with the Q Exactive has demonstrated a vast improvement in the analysis time necessary to complete a high resolving power imaging experiment. On the Q Exactive, the broadband imaging experiments were acquired at a rate of 1.6 scans/second. For reference, the acquisition rate for the same experiment on our LTQ-FT is roughly 0.5 scans/second, which represents a nearly threefold improvement in acquisition speed with the Q Exactive. This improvement is likely due to the eFT which allows for shorter transients in addition to the multiplexing capabilities of the Q Exactive where ions can be accumulated for the next acquisition while the FT analysis is being performed.
Figure 2.
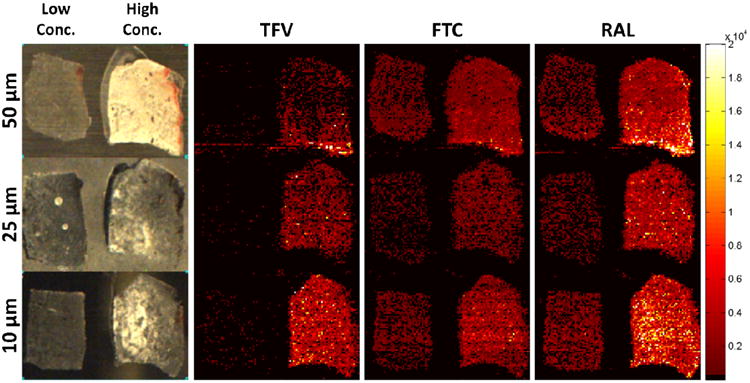
IR-MALDESI MSI analysis of cervical tissues incubated in either a low or high concentration of three HIV drugs including emtricitabine (FTC), tenofovir (TFV), and raltegravir (RAL). Three different tissue thicknesses were investigated (10, 25, and 50 μm). The ion maps for all three drugs at each tissue thickness are shown on the same intensity scale to highlight relative differences in abundance.
At each pixel, the laser ablates all the way through the tissue and therefore it may be expected that the intensity for any of the drugs from the 50 μm section should be roughly five times that of the 10 μm section since there is five times more material (this assumption is validated from the LC-MS/MS quantification presented later). This, however, was not observed and the signal from all three thicknesses appears to be relatively similar. The lack of correlation between tissue thickness and the signal abundance could be occurring for several reasons. One possible explanation is that in IR-MALDESI the ablated material interacts with the charged solvent droplets of the electrospray plume where the analyte is extracted into the solvent droplets and is later ionized through an ESI-like process. The efficiency of this neutral capture event may inversely scale with the amount of material that is ablated. In essence, the extraction of analyte may be limited because there are a finite number of droplets present that are available for interaction with the tissue material. Despite this, Figure 2 shows that for a given tissue thickness there is an observable difference in intensity between the low and high concentrations and in fact the ratio of the average intensity of the low and high concentrations is consistent for all three tissue thicknesses.
Comparison of IR-MALDESI MSI with LC-MS/MS Quantitation
Tissue sections serial to those imaged by IR-MALDESI MSI were homogenized and analyzed by LC-MS/MS to quantify the amount of each drug that was present in both the high and low concentration tissues for all three tissue thicknesses. Table 2 shows the results of this analysis and provides the absolute amount of each drug per tissue section (ng/tissue). The ratio of low to high concentration was determined for each set of tissues and these values were then compared with the corresponding average intensity ratios from the IR-MALDESI imaging experiments. This comparison is shown in Figure 3. It should be noted that differential uptake of these drugs precludes the ability to compare their incubated concentrations with ratios observed by LC-MS and IR-MALDESI (data not shown). Figure 3a shows the relative intensity ratio from the low to the high concentration tissues for each of the three drugs. It is evident that for both techniques and the different tissue thicknesses, the low/high ratio clusters together for a given drug. To compare the two methods, the low/high concentration ratios are plotted as a function of the method, thus a slope of 1 indicates agreement between the two methods (Figure 3b). The slope of the least-squares fit line through the data (forcing the intercept to 0) was around 0.74 with a correlation coefficient of close to 0.8. The deviation from an ideal slope of 1 is likely due to an underestimation of the low/high ratio by IR-MALDESI. The cause of this underestimation of the ratio is possibly due to the decreased frequency of detection of these drugs in the low concentration tissue which is a result of being near the detection limit as evidenced by Figure 2. While there obviously room for improvement, the low/high concentration ratios between LC-MS/MS and IR-MALDESI are in fairly good agreement. A correlation between MSI and LC-MS/MS has been demonstrated previously for other ionization methods including MALDI [18, 50, 60-70] and DESI,[22, 71-72] but this is the first example of agreement between IR-MALDESI MSI with LC-MS/MS. Given that LC-MS/MS is a validated quantitation method; the correlation between the average intensities from IR-MALDESI with the absolute quantities determined by LC-MS/MS provides a foundation for quantification directly from an imaging experiment. Direct quantitation from an MSI experiment would be ideal (especially for heterogeneous tissues) and is the focus of research efforts from several groups.[40, 61, 63-70, 72-77]
Table 2. Absolute drug quantities determined by LC-MS/MS.
| Incubated Conc. | Tissue Thickness | TFV (ng/Tissue) | FTC (ng/Tissue) | RAL (ng/Tissue) |
|---|---|---|---|---|
| Low | 10 | 0.877 | 3.36 | 2.29 |
| 25 | 1.91 | 6.95 | 4.90 | |
| 50 | 4.04 | 14.4 | 10.6 | |
|
| ||||
| High | 10 | 8.82 | 9.19 | 12.5 |
| 25 | 21.9 | 23.1 | 33.2 | |
| 50 | 43.3 | 44.7 | 64.6 | |
Figure 3.
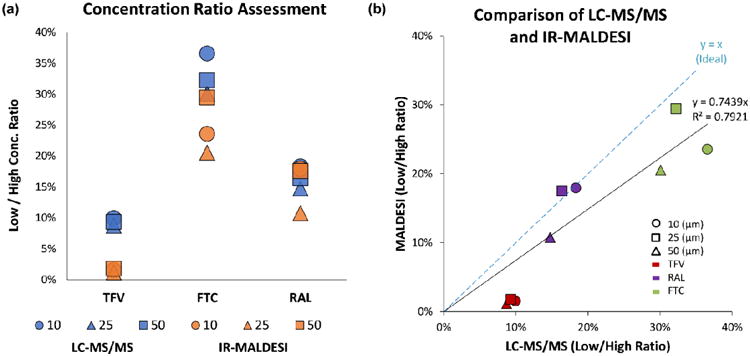
Comparison of data from LC-MS/MS and IR-MALDESI. (a) Plot of the low to high concentration ratios of all three drugs across the three tissue thicknesses that were investigated. (b) Plot of the data from both methods (LC-MS/MS vs. IR-MALDESI). A slope near 1 indicates relatively good agreement between the results from the LC-MS/MS and IR-MALDESI MSI experiments.
A potential limitation of this study is the use of mucosal tissue explants incubated in drug-containing media, rather than utilizing tissue biopsies obtained from patients receiving the drug. It is possible that antiretroviral uptake into explants ex-vivo differs from that observed in-vivo. However, the human tissue explant model has previously been validated for generating pharmacokinetic/pharmacodynamic (PK/PD) relationships that are clinically useful.[78-79] With the promising data generated from this current investigation, future drug distribution experiments evaluating tissues obtained from animals and humans dosed with these drugs are being planned.
MS2 IR-MALDESI MSI
IR-MALDESI MSI was performed as a targeted MS2 experiment for the drug RAL. Ions generated from two laser pulses were filtered for the protonated form of RAL using the selection quadrupole followed by accumulation in the C-Trap. The ions were then fragmented in the HCD cell and mass analyzed in the orbitrap at 140,000 RP (at m/z 200) to obtain accurate mass data on the fragment ions. This process was then repeated over the surface of the tissue such that a fragment ion ‘scan’ was acquired at each pixel in the image. The acquisition rate for the targeted MS2 imaging experiment was 1.5 scans/second, implying that very little overhead was incurred with the added step of fragmentation. Ion maps for the several RAL transitions are shown in Figure 4a. The colocalization of all of the transitions with the parent ion distribution at m/z 445 demonstrates the higher selectivity that can be achieved with an MRM-MSI approach. In addition, the ion abundance ratios from this pseudo MRM imaging experiment were compared with those obtained from the direct infusion ESI-MS experiment that was used to determine the optimal collision energy (Figure 4b). As shown, the ion abundance ratios are nearly identical between the two techniques. This implies also that the internal energy imparted during ionization in IR-MALDESI is comparable to the softness of ionization in ESI as has been previously mentioned.[80]
Figure 4.
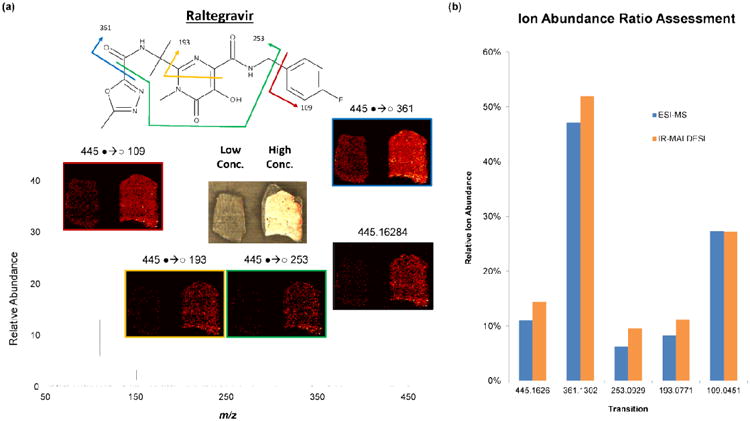
Results of MRM Imaging. (a) Ion maps for the several unique transitions for raltegravir (RAL) that were observed from the targeted MS2 IR-MALDESI MSI experiment. The colocalization of all of the transitions demonstrates the increased selectivity that can be realized with targeted MS2 imaging techniques. (b) Comparison of the ion abundance ratios for selected transitions of raltegravir for both IR-MALDESI and LC-MS/MS.
Conclusions
We have demonstrated the capabilities of IR-MALDESI MSI coupled with the Q Exactive mass spectrometer. Tissue sections that were incubated with several potent and commonly utilized antiretroviral drugs were analyzed by IR-MALDESI MSI as well as LC-MS/MS. A comparison of these two methods demonstrated that the average intensities determined from the imaging experiments agreed well with the absolute abundances determined from a validated quantitation method. These experiments serve as a foundation for direct quantitation from tissue using IR-MALDESI MSI. In addition, a targeted MS2 imaging experiment was also conducted to show the added selectivity that can be attained for this type of analysis.
Acknowledgments
The author gratefully acknowledge the assistance of Kerstin Strupat from Thermo Scientific as well as the financial support received from the National Institutes of Health (R01GM087964), U01AI095031, P30AI050410, U19AI096113, GlaxoSmithKline, the William R. Kenan, Jr. Fund for Engineering, Technology & Science, the W. M. Keck Foundation, and North Carolina State University.
References
- 1.Caprioli RM, Farmer TB, Gile J. Molecular Imaging of Biological Samples: Localization of Peptides and Proteins Using MALDI-TOF MS. Anal Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 2.Caldwell RL, Caprioli RM. Tissue Profiling by Mass Spectrometry: A Review of Methodology and Applications. Mol Cell Proteomics. 2005;4:394–401. doi: 10.1074/mcp.R500006-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Burnum KE, Frappier SL, Caprioli RM. Matrix-Assisted Laser Desorption/Ionization Imaging Mass Spectrometry for the Investigation of Proteins and Peptides. Annu Rev Anal Chem. 2008;1:689–705. doi: 10.1146/annurev.anchem.1.031207.112841. [DOI] [PubMed] [Google Scholar]
- 4.Murphy RC, Hankin JA, Barkley RM. Imaging of lipid species by MALDI mass spectrometry. J Lipid Res. 2009;50:S317–S322. doi: 10.1194/jlr.R800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svatoš A. Mass spectrometric imaging of small molecules. Trends Biotechnol. 2010;28:425–434. doi: 10.1016/j.tibtech.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Greer T, Sturm R, Li L. Mass spectrometry imaging for drugs and metabolites. J Proteomics. 2011;74:2617–2631. doi: 10.1016/j.jprot.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oppenheimer SR, Drexler DM. Tissue analysis by imaging MS. Bioanalysis. 2012;4:95–112. doi: 10.4155/bio.11.282. [DOI] [PubMed] [Google Scholar]
- 8.Schwamborn K. Imaging mass spectrometry in biomarker discovery and validation. J Proteomics. 2012;75:4990–4998. doi: 10.1016/j.jprot.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Ouyang Z. Mass spectrometry imaging for biomedical applications. Anal Bioanal Chem. 2013;405:5645–5653. doi: 10.1007/s00216-013-6916-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodwin RJ, Pitt AR. Mass spectrometry imaging of pharmacological compounds in tissue sections. Bioanalysis. 2010;2:279–293. doi: 10.4155/bio.09.180. [DOI] [PubMed] [Google Scholar]
- 11.Castellino S, Groseclose MR, Wagner D. MALDI imaging mass spectrometry: bridging biology and chemistry in drug development. Bioanalysis. 2011;3:2427–2441. doi: 10.4155/bio.11.232. [DOI] [PubMed] [Google Scholar]
- 12.Castellino S. MALDI imaging MS analysis of drug distribution in tissue: the right time!(?) Bioanalysis. 2012;4:2549–2551. doi: 10.4155/bio.12.251. [DOI] [PubMed] [Google Scholar]
- 13.Eichler HG, Müller M. Drug Distribution. Clin Pharmacokinet. 1998;34:95–99. doi: 10.2165/00003088-199834020-00001. [DOI] [PubMed] [Google Scholar]
- 14.Lanao JM, Fraile MA. Drug Tissue Distribution: Study Methods and Therapeutic Implications. Curr Pharm Des. 2005;11:3829–3845. doi: 10.2174/138161205774580679. [DOI] [PubMed] [Google Scholar]
- 15.Solon EG, Balani SK, Lee FW. Whole-Body Autoradiography In Drug Discovery. Curr Drug Metab. 2002;3:451–462. doi: 10.2174/1389200023337207. [DOI] [PubMed] [Google Scholar]
- 16.Atkinson SJ, Loadman PM, Sutton C, Patterson LH, Clench MR. Examination of the distribution of the bioreductive drug AQ4N and its active metabolite AQ4 in solid tumours by imaging matrix-assisted laser desorption/ionisation mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:1271–1276. doi: 10.1002/rcm.2952. [DOI] [PubMed] [Google Scholar]
- 17.Römpp A, Guenther S, Takats Z, Spengler B. Mass spectrometry imaging with high resolution in mass and space (HR2 MSI) for reliable investigation of drug compound distributions on the cellular level. Anal Bioanal Chem. 2011;401:65–73. doi: 10.1007/s00216-011-4990-7. [DOI] [PubMed] [Google Scholar]
- 18.Nilsson A, Forngren B, Bjurström S, Goodwin RJA, Basmaci E, Gustafsson I, Annas A, Hellgren D, Svanhagen A, Andrén PE, Lindberg J. In Situ Mass Spectrometry Imaging and Ex Vivo Characterization of Renal Crystalline Deposits Induced in Multiple Preclinical Drug Toxicology Studies. PLoS One. 2012;7:1–10. doi: 10.1371/journal.pone.0047353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Römpp A, Spengler B. Mass spectrometry imaging with high resolution in mass and space. Histochem Cell Biol. 2013;139:759–783. doi: 10.1007/s00418-013-1097-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaletaş BK, van der Wiel IM, Stauber J, Lennard JD, Güzel C, Kros JM, Luider TM, Heeren RMA. Sample preparation issues for tissue imaging by imaging MS. Proteomics. 2009;9:2622–2633. doi: 10.1002/pmic.200800364. [DOI] [PubMed] [Google Scholar]
- 21.Goodwin RJA. Sample preparation for mass spectrometry imaging: Small mistakes can lead to big consequences. J Proteomics. 2012;75:4893–4911. doi: 10.1016/j.jprot.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 22.Wiseman JM, Ifa DR, Zhu Y, Kissinger CB, Manicke NE, Kissinger PT, Cooks RG. Desorption electrospray ionization mass spectrometry: Imaging drugs and metabolites in tissues. Proc Natl Acad Sci USA. 2008;105:18120–18125. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eikel D, Vavrek M, Smith S, Bason C, Yeh S, Korfmacher WA, Henion JD. Liquid extraction surface analysis mass spectrometry (LESA-MS) as a novel profiling tool for drug distribution and metabolism analysis: the terfenadine example. Rapid Commun Mass Spectrom. 2011;25:3587–3596. doi: 10.1002/rcm.5274. [DOI] [PubMed] [Google Scholar]
- 24.Barry JA, Groseclose MR, Robichaud G, Castellino S, Muddiman DC. Assessing drug and metabolite detection in liver tissue by IR-MALDESI mass spectrometry imaging coupled to FT-ICR MS. Int J Mass Spectrom. doi: 10.1016/j.ijms.2014.05.012. Submitted 12/31/2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levis RJ. Laser Desorption and Ejection of Biomolecules From the Condensed Phase into the Gas Phase. Annu Rev Phys Chem. 1994;45:483–518. doi: 10.1146/annurev.pc.45.100194.002411. [DOI] [PubMed] [Google Scholar]
- 26.Sampson J, Hawkridge A, Muddiman D. Generation and detection of multiply-charged peptides and proteins by matrix-assisted laser desorption electrospray ionization (MALDESI) fourier transform ion cyclotron resonance mass spectrometry. J Am Soc Mass Spectrom. 2006;17:1712–1716. doi: 10.1016/j.jasms.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Sampson JS, Murray KK, Muddiman DC. Intact and Top-Down Characterization of Biomolecules and Direct Analysis Using Infrared Matrix-Assisted Laser Desorption Electrospray Ionization Coupled to FT-ICR Mass Spectrometry. J Am Soc Mass Spectrom. 2009;20:667–673. doi: 10.1016/j.jasms.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rezenom YH, Dong J, Murray KK. Infrared laser-assisted desorption electrospray ionization mass spectrometry. Analyst. 2008;133:226–232. doi: 10.1039/b715146b. [DOI] [PubMed] [Google Scholar]
- 29.Nemes P, Vertes A. Laser Ablation Electrospray Ionization for Atmospheric Pressure, in Vivo, and Imaging Mass Spectrometry. Anal Chem. 2007;79:8098–8106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 30.Sampson JS, Muddiman DC. Atmospheric pressure infrared (10.6 μm) laser desorption electrospray ionization (IR-LDESI) coupled to a LTQ Fourier transform ion cyclotron resonance mass spectrometer. Rapid Commun Mass Spectrom. 2009;23:1989–1992. doi: 10.1002/rcm.4113. [DOI] [PubMed] [Google Scholar]
- 31.Jorabchi K, Smith LM. Single Droplet Separations and Surface Partition Coefficient Measurements Using Laser Ablation Mass Spectrometry. Anal Chem. 2009;81:9682–9688. doi: 10.1021/ac901819r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson R, Rainbow M, Lohr D, Williams P. Volatilization of high molecular weight DNA by pulsed laser ablation of frozen aqueous solutions. Sci. 1989;246:1585–1587. doi: 10.1126/science.2595370. [DOI] [PubMed] [Google Scholar]
- 33.Berkenkamp S, Karas M, Hillenkamp F. Ice as a matrix for IR-matrix-assisted laser desorption/ionization: mass spectra from a protein single crystal. Proc Natl Acad Sci U S A. 1996;93:7003–7. doi: 10.1073/pnas.93.14.7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vertes A, Nemes P, Shrestha B, Barton A, Chen Z, Li Y. Molecular imaging by Mid-IR laser ablation mass spectrometry. Appl Phys A: Mater Sci Process. 2008;93:885–891. [Google Scholar]
- 35.Nemes P, Barton AA, Li Y, Vertes A. Ambient Molecular Imaging and Depth Profiling of Live Tissue by Infrared Laser Ablation Electrospray Ionization Mass Spectrometry. Anal Chem. 2008;80:4575–4582. doi: 10.1021/ac8004082. [DOI] [PubMed] [Google Scholar]
- 36.Robichaud G, Barry JA, Garrard KP, Muddiman DC. Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI) Imaging Source Coupled to a FT-ICR Mass Spectrometer. J Am Soc Mass Spectrom. 2013;24:92–100. doi: 10.1007/s13361-012-0505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robichaud G, Barry JA, Muddiman DC. IR-MALDESI Mass Spectrometry Imaging of Biological Tissue Sections using Ice as a Matrix. J Am Soc Mass Spectrom. 2014;25:319–328. doi: 10.1007/s13361-013-0787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cornett DS, Frappier SL, Caprioli RM. MALDI-FTICR Imaging Mass Spectrometry of Drugs and Metabolites in Tissue. Anal Chem. 2008;80:5648–5653. doi: 10.1021/ac800617s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manicke NE, Dill AL, Ifa DR, Cooks RG. High-resolution tissue imaging on an orbitrap mass spectrometer by desorption electrospray ionization mass spectrometry. J Mass Spectrom. 2010;45:223–226. doi: 10.1002/jms.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fehniger TE, Végvári Á, Rezeli M, Prikk K, Ross P, Dahlbäck M, Edula G, Sepper R, Marko-Varga G. Direct Demonstration of Tissue Uptake of an Inhaled Drug: Proof-of-Principle Study Using Matrix-Assisted Laser Desorption Ionization Mass Spectrometry Imaging. Anal Chem. 2011;83:8329–8336. doi: 10.1021/ac2014349. [DOI] [PubMed] [Google Scholar]
- 41.Shahidi-Latham SK, Dutta SM, Prieto Conaway MC, Rudewicz PJ. Evaluation of an Accurate Mass Approach for the Simultaneous Detection of Drug and Metabolite Distributions via Whole-Body Mass Spectrometric Imaging. Anal Chem. 2012;84:7158–7165. doi: 10.1021/ac3015142. [DOI] [PubMed] [Google Scholar]
- 42.Korte A, Lee Y. Multiplex Mass Spectrometric Imaging with Polarity Switching for Concurrent Acquisition of Positive and Negative Ion Images. J Am Soc Mass Spectrom. 2013;24:949–955. doi: 10.1007/s13361-013-0613-1. [DOI] [PubMed] [Google Scholar]
- 43.Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S. Mass Spectrometry-based Proteomics Using Q Exactive, a High-performance Benchtop Quadrupole Orbitrap Mass Spectrometer. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhandari D, Shen T, Römpp A, Zorn H, Spengler B. Analysis of cyathane-type diterpenoids from Cyathus striatusand Hericium erinaceus by high-resolution MALDI MS imaging. Anal Bioanal Chem. 2013:1–10. doi: 10.1007/s00216-013-7496-7. [DOI] [PubMed] [Google Scholar]
- 45.Janfelt C, Wellner N, Hansen HS, Hansen SH. Displaced dual-mode imaging with desorption electrospray ionization for simultaneous mass spectrometry imaging in both polarities and with several scan modes. J Mass Spectrom. 2013;48:361–366. doi: 10.1002/jms.3166. [DOI] [PubMed] [Google Scholar]
- 46.Lanekoff I, Burnum-Johnson KE, Thomas M, Short J, Carson JP, Cha J, Dey SK, Yang P, Prieto Conaway MC, Laskin J. High-Speed MS/MS in Situ Imaging by Nanospray Desorption Electrospray Ionization Mass Spectrometry. Anal Chem. 2013 doi: 10.1021/ac401760s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson CG, Cohen MS, Kashuba ADM. Antiretroviral Pharmacology in Mucosal Tissues. JAIDS J Acquired Immune Defic Syndromes. 2013;63:S240–S247. doi: 10.1097/QAI.0b013e3182986ff8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith MZ, Wightman F, Lewin SR. HIV Reservoirs and Strategies for Eradication. Current HIV/AIDS Reports. 2012;9:5–15. doi: 10.1007/s11904-011-0108-2. [DOI] [PubMed] [Google Scholar]
- 49.Meesters RJW, van Kampen JJA, Scheuer RD, van der Ende ME, Gruters RA, Luider TM. Determination of the antiretroviral drug tenofovir in plasma from HIV-infected adults by ultrafast isotope dilution MALDI-triple quadrupole tandem mass spectrometry. J Mass Spectrom. 2011;46:282–289. doi: 10.1002/jms.1897. [DOI] [PubMed] [Google Scholar]
- 50.Castellino S, Groseclose MR, Sigafoos J, Wagner D, de Serres M, Polli JW, Romach E, Myer J, Hamilton B. Central Nervous System Disposition and Metabolism of Fosdevirine (GSK2248761), a Non-Nucleoside Reverse Transcriptase Inhibitor: An LC-MS and Matrix-Assisted Laser Desorption/Ionization Imaging MS Investigation into Central Nervous System Toxicity. Chem Res Toxicol. 2012;26:241–251. doi: 10.1021/tx3004196. [DOI] [PubMed] [Google Scholar]
- 51.Patterson KB, Prince HA, Kraft E, Jenkins AJ, Shaheen NJ, Rooney JF, Cohen MS, Kashuba ADM. Penetration of Tenofovir and Emtricitabine in Mucosal Tissues: Implications for Prevention of HIV-1 Transmission. Science Translational Medicine. 2011;3:112re4. doi: 10.1126/scitranslmed.3003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patterson KB, Prince HA, Stevens T, Shaheen NJ, Dellon ES, Madanick RD, Jennings S, Cohen MS, Kashuba ADM. Differential penetration of raltegravir throughout gastrointestinal tissue: implications for eradication and cure. AIDS. 2013;27:1413–1419. doi: 10.1097/QAD.0b013e32835f2b49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Race AM, Styles IB, Bunch J. Inclusive sharing of mass spectrometry imaging data requires a converter for all. J Proteomics. 2012;75:5111–5112. doi: 10.1016/j.jprot.2012.05.035. [DOI] [PubMed] [Google Scholar]
- 55.Robichaud G, Garrard KP, Barry JA, Muddiman DC. MSiReader: An Open-Source Interface to View and Analyze High Resolving Power MS Imaging Files on Matlab Platform. J Am Soc Mass Spectrom. 2013;24:718–721. doi: 10.1007/s13361-013-0607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rogowitz BE, Treinish LA, Bryson S. How Not to Lie with Visualization. ComPh. 1996;10:268–273. [Google Scholar]
- 57.Rogowitz B, Treinish LA. Data visualization: the end of the rainbow. Spectrum, IEEE. 1998;35:52–59. [Google Scholar]
- 58.Light A, Bartlein PJ. The end of the rainbow? Color schemes for improved data graphics. Eos, Transactions American Geophysical Union. 2004;85:385–391. [Google Scholar]
- 59.Borland D, Taylor RM. Rainbow Color Map (Still) Considered Harmful. Computer Graphics and Applications, IEEE. 2007;27:14–17. doi: 10.1109/mcg.2007.323435. [DOI] [PubMed] [Google Scholar]
- 60.Reyzer ML, Hsieh Y, Ng K, Korfmacher WA, Caprioli RM. Direct analysis of drug candidates in tissue by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom. 2003;38:1081–1092. doi: 10.1002/jms.525. [DOI] [PubMed] [Google Scholar]
- 61.Hsieh Y, Casale R, Fukuda E, Chen J, Knemeyer I, Wingate J, Morrison R, Korfmacher W. Matrix-assisted laser desorption/ionization imaging mass spectrometry for direct measurement of clozapine in rat brain tissue. Rapid Commun Mass Spectrom. 2006;20:965–972. doi: 10.1002/rcm.2397. [DOI] [PubMed] [Google Scholar]
- 62.Drexler DM, Garrett TJ, Cantone JL, Diters RW, Mitroka JG, Prieto Conaway MC, Adams SP, Yost RA, Sanders M. Utility of imaging mass spectrometry (IMS) by matrix-assisted laser desorption ionization (MALDI) on an ion trap mass spectrometer in the analysis of drugs and metabolites in biological tissues. J Pharmacol Toxicol Methods. 2007;55:279–288. doi: 10.1016/j.vascn.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 63.Hsieh Y, Chen J, Korfmacher WA. Mapping pharmaceuticals in tissues using MALDI imaging mass spectrometry. J Pharmacol Toxicol Methods. 2007;55:193–200. doi: 10.1016/j.vascn.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 64.Signor L, Varesio E, Staack RF, Starke V, Richter WF, Hopfgartner G. Analysis of erlotinib and its metabolites in rat tissue sections by MALDI quadrupole time-of-flight mass spectrometry. J Mass Spectrom. 2007;42:900–909. doi: 10.1002/jms.1225. [DOI] [PubMed] [Google Scholar]
- 65.Nilsson A, Fehniger TE, Gustavsson L, Andersson M, Kenne K, Marko-Varga G, Andrén PE. Fine Mapping the Spatial Distribution and Concentration of Unlabeled Drugs within Tissue Micro-Compartments Using Imaging Mass Spectrometry. PLoS One. 2010;5:e11411. doi: 10.1371/journal.pone.0011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koeniger SL, Talaty N, Luo Y, Ready D, Voorbach M, Seifert T, Cepa S, Fagerland JA, Bouska J, Buck W, Johnson RW, Spanton S. A quantitation method for mass spectrometry imaging. Rapid Commun Mass Spectrom. 2011;25:503–510. doi: 10.1002/rcm.4891. [DOI] [PubMed] [Google Scholar]
- 67.Prideaux B, Dartois Vr, Staab D, Weiner DM, Goh A, Via LE, Barry CE, Iii, Stoeckli M. High-Sensitivity MALDI-MRM-MS Imaging of Moxifloxacin Distribution in Tuberculosis-Infected Rabbit Lungs and Granulomatous Lesions. Anal Chem. 2011;83:2112–2118. doi: 10.1021/ac1029049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hamm G, Bonnel D, Legouffe R, Pamelard F, Delbos JM, Bouzom F, Stauber J. Quantitative mass spectrometry imaging of propranolol and olanzapine using tissue extinction calculation as normalization factor. J Proteomics. 2012;75:4952–4961. doi: 10.1016/j.jprot.2012.07.035. [DOI] [PubMed] [Google Scholar]
- 69.Pirman DA, Reich RF, Kiss A, Heeren RMA, Yost RA. Quantitative MALDI Tandem Mass Spectrometric Imaging of Cocaine from Brain Tissue with a Deuterated Internal Standard. Anal Chem. 2012;85:1081–1089. doi: 10.1021/ac302960j. [DOI] [PubMed] [Google Scholar]
- 70.Takai N, Tanaka Y, Inazawa K, Saji H. Quantitative analysis of pharmaceutical drug distribution in multiple organs by imaging mass spectrometry. Rapid Commun Mass Spectrom. 2012;26:1549–1556. doi: 10.1002/rcm.6256. [DOI] [PubMed] [Google Scholar]
- 71.Kertesz V, Van Berkel GJ, Vavrek M, Koeplinger KA, Schneider BB, Covey TR. Comparison of Drug Distribution Images from Whole-Body Thin Tissue Sections Obtained Using Desorption Electrospray Ionization Tandem Mass Spectrometry and Autoradiography. Anal Chem. 2008;80:5168–5177. doi: 10.1021/ac800546a. [DOI] [PubMed] [Google Scholar]
- 72.Vismeh R, Waldon DJ, Teffera Y, Zhao Z. Localization and Quantification of Drugs in Animal Tissues by Use of Desorption Electrospray Ionization Mass Spectrometry Imaging. Anal Chem. 2012;84:5439–5445. doi: 10.1021/ac3011654. [DOI] [PubMed] [Google Scholar]
- 73.Bunch J, Clench MR, Richards DS. Determination of pharmaceutical compounds in skin by imaging matrix-assisted laser desorption/ionisation mass spectrometry. Rapid Commun Mass Spectrom. 2004;18:3051–3060. doi: 10.1002/rcm.1725. [DOI] [PubMed] [Google Scholar]
- 74.Goodwin RJA, Scullion P, MacIntyre L, Watson DG, Pitt AR. Use of a Solvent-Free Dry Matrix Coating for Quantitative Matrix-Assisted Laser Desorption Ionization Imaging of 4-Bromophenyl-1,4-diazabicyclo(3.2.2)nonane-4-carboxylate in Rat Brain and Quantitative Analysis of the Drug from Laser Microdissected Tissue Regions. Anal Chem. 2010;82:3868–3873. doi: 10.1021/ac100398y. [DOI] [PubMed] [Google Scholar]
- 75.Goodwin RJA, Mackay CL, Nilsson A, Harrison DJ, Farde L, Andren PE, Iverson SL. Qualitative and Quantitative MALDI Imaging of the Positron Emission Tomography Ligands Raclopride (a D2 Dopamine Antagonist) and SCH 23390 (a D1 Dopamine Antagonist) in Rat Brain Tissue Sections Using a Solvent-Free Dry Matrix Application Method. Anal Chem. 2011;83:9694–9701. doi: 10.1021/ac202630t. [DOI] [PubMed] [Google Scholar]
- 76.Shin Y, Dong T, Chou B, Menghrajani K. Determination of loperamide in Mdr1a/1b knock-out mouse brain tissue using matrix-assisted laser desorption/ionization mass spectrometry and comparison with quantitative electrospray-triple quadrupole mass spectrometry analysis. Arch Pharmacal Res. 2011;34:1983–1988. doi: 10.1007/s12272-011-1119-7. [DOI] [PubMed] [Google Scholar]
- 77.Källback P, Shariatgorji M, Nilsson A, Andrén PE. Novel mass spectrometry imaging software assisting labeled normalization and quantitation of drugs and neuropeptides directly in tissue sections. J Proteomics. 2012;75:4941–4951. doi: 10.1016/j.jprot.2012.07.034. [DOI] [PubMed] [Google Scholar]
- 78.Harman S, Herrera C, Armanasco N, Nuttall J, Shattock RJ. Preclinical Evaluation of the HIV-1 Fusion Inhibitor L′644 as a Potential Candidate Microbicide. Antimicrob Agents Chemother. 2012;56:2347–2356. doi: 10.1128/AAC.06108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rohan LC, Moncla BJ, Kunjara Na Ayudhya RP, Cost M, Huang Y, Gai F, Billitto N, Lynam JD, Pryke K, Graebing P, Hopkins N, Rooney JF, Friend D, Dezzutti CS. In Vitro and Ex Vivo Testing of Tenofovir Shows It Is Effective As an HIV-1 Microbicide. PLoS One. 2010;5:e9310. doi: 10.1371/journal.pone.0009310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nemes P, Huang H, Vertes A. Internal energy deposition and ion fragmentation in atmospheric-pressure mid-infrared laser ablation electrospray ionization. Phys Chem Chem Phys. 2012;14:2501–2507. doi: 10.1039/c2cp23411d. [DOI] [PubMed] [Google Scholar]


