Abstract
Melanoma causes more deaths than any other skin cancer, and its incidence in the US continues to rise. Current medical therapies are insufficient to control this deadly neoplasm, necessitating the development of new target-based approaches. The objective of this study was to determine the role and functional significance of the class III histone deacetylase SIRT1 in melanoma. We have found that SIRT1 is overexpressed in clinical human melanoma tissues and human melanoma cell lines (Sk-Mel-2, WM35, G361, A375, and Hs294T) compared to normal skin and normal melanocytes, respectively. In addition, treatment of melanoma cell lines A375, Hs294T, and G361 with Tenovin-1, a small molecule SIRT1 inhibitor, resulted in a significant decrease in cell growth and cell viability. Further, Tenovin-1 treatment also resulted in a marked decrease in the clonogenic survival of melanoma cells. Further experiments showed that the anti-proliferative response of Tenovin-1 was accompanied by an increase in the protein as well as activity of the tumor suppressor p53. This increase in p53 activity was substantiated by an increase in the protein level of its downstream target p21. Overall, these data suggest that small molecule inhibition of SIRT1 causes anti-proliferative effects in melanoma cells. SIRT1 appears to be acting through the activity of the tumor suppressor p53, which is not mutated in the majority of melanomas. However, future detailed studies are needed to further explore the role and mechanism of SIRT1 in melanoma development and progression and its usefulness in melanoma treatment.
Keywords: SIRT1, melanoma, p53, p21, Tenovin-1
Introduction
Melanoma is one of the most lethal skin cancers, and its impact on the US population continues to increase. While it accounts for as little as 5% of diagnosed skin cancers, it is responsible for an estimated 70% of skin cancer deaths. The incidence of melanoma has been rising steadily at an average rate of 3.4% per year for women, and 3.1% for men in the US [1], evidencing a growing demand for adequate treatment. In 2014, an estimated 76,100 new cases are expected to be diagnosed in the US and, under currently available treatments, 9710 people are expected to die of melanoma [2]. Therefore, there is a need to develop additional therapeutic targets to effectively treat this disease.
SIRT1 is a member of the class III histone deacetylases known as Sirtuins, which depend on NAD+ (nicotinamide adenine dinucleotide) as a cofactor for their deacetylase activity [3–5]. As perhaps the most studied member of the sirtuins to date, SIRT1 has been shown to play a role in a number of biological processes, including aging, metabolism, and the cellular stress response. SIRT1 targets mediate the DNA damage repair response, cell cycle arrest, and apoptosis (Reviewed in [6]), all of which are processes that can lead to cancer development or progression if dysregulated. Aberrant expression of SIRT1 could lead to such dysregulation, and it has been shown to be overexpressed in many different types of cancer [7]. In nonmelanoma skin cancers, including squamous and basal cell carcinomas, actinic keratosis, and especially in Bowen’s disease, SIRT1 is frequently overexpressed [8]. However, its expression status in melanoma is poorly defined and in need of further investigation. In cancers which overexpress SIRT1, inhibition through small molecule inhibitors or genetic knockdown leads to an induction of cell cycle arrest and/or apoptosis [9–14]. In breast cancer and chronic myeloid leukemia, this effect was shown to be accompanied by increased acetylation of the tumor suppressor p53 [11, 12]. Since p53 is a known SIRT1 target [15], this suggests that tumorigenesis and progression of these cancers may be due in part to SIRT1 inhibition of p53 activity through deacetylation.
P53 plays an important role in the cellular response to stresses and DNA damage primarily through transcriptional repression or activation of target genes. Under normal conditions, it is present at low levels in the cell, which are maintained by MDM2-mediated p53 degradation. In response to stress, p53 is stabilized and induces cell cycle arrest and DNA damage repair (via p21, 14-3-3), or promotes cellular senescence (via pai-1) or apoptosis (via pig, bax, puma, and noxa) [3, 16, 17]. P53 is widely studied in cancer research, as p53 function is often eliminated by genetic mutations in cancer cells, leading to increased tumor development and growth. However, in melanoma, it is estimated that its genetic coding is intact in at least 75% of tumors, yet tumor development and growth remain unchecked [18]. It is therefore reasonable to speculate that either p53 or its downstream targets might be dysfunctional in melanoma. In fact, the wild type p53 expressed in the majority of melanomas fails to induce cell cycle arrest and apoptosis [19, 20]. One possible explanation for this is the role of posttranslational modifications on the activity level of p53. Acetylation of p53 is mandatory for its function. It is required for p53 recruitment to its target transcripts, and loss of acetylation results in abolishment of p53-dependent growth arrest and apoptosis [21]. Therefore, aberrant expression or activity of SIRT1 could lead to dysregulation of p53’s function as a tumor suppressor.
In this study, we determined the status of SIRT1 expression and functional significance in melanoma. Specifically, we addressed the impact of SIRT1 on tumor cell growth and survival through the tumor suppressor p53 and its downstream effector p21.
Materials and Methods
Cell Culture
The human melanoma cell lines SK-Mel2, WM35, A375, G361, and Hs294T were obtained from American Type Culture Collection (Manassas, VA). Normal human epidermal melanocytes (NHEM) were a kind gift from Dr. V. Setaluri (University of Wisconsin-Madison, Madison, WI). Cells were maintained at standard tissue culture conditions as recommended by the vendors. Following treatment, the cells were collected using 0.25% Trypsin/2.21 mM EDTA (Cellgro, Manassas, VA), rinsed with phosphate buffered saline (pH 7.4), pelleted, and frozen at −80°C.
Immunoblot analysis
Cell lysates were prepared from frozen pellets using RIPA lysis buffer (Millipore, Billerica, MA) with freshly added PMSF (Amresco, Solon, OH) and protease inhibitor (Thermo Scientific, Waltham, MA). Protein concentration was determined by BCA Protein Assay (Thermo Scientific, Waltham, MA), and 40 µg was resolved over an 8–12% Tris-Glycine polyacrylamide gel. Protein was transferred to a 0.45 or 0.2 µm nitrocellulose membrane (Bio-Rad, Hercules, CA), then blocked with 5% non-fat dry milk or BSA (Cell Signaling Technology, Danvers, MA). Membranes were probed with the appropriate primary antibody (p53: Invitrogen, Carlsbad, CA #AHO0152; p21: Cell Signaling Technology, Danvers, MA #2947; SIRT1: Santa Cruz Biotechnology, Dallas, TX #sc-15404) and secondary horseradish peroxidase (HRP) conjugate antibody (Cell Signaling Technology, Danvers, MA). Proteins were detected by chemiluminescence using Pierce ECL Western Blotting Substrate, SuperSignal West Pico, or SuperSignal West Femto Chemiluminescent Substrates (Pierce, Rockford, IL), and the Kodak Image Station 4000 MM (Carestream Health Inc., Rochester, NY).
Real Time PCR
RNA was extracted from frozen cell pellets using the RNeasy Plus Minikit (Qiagen, Germantown, MD), then quantified and checked for RNA purity by optical density at 260 and 280 nm. cDNA was then synthesized by reverse transcription of RNA with oligo (dT), dNTPs, and M-MLV reverse transcriptase (Promega, Madison, WI). Quantitative RT-PCR (qRT-PCR) was performed with SYBR Premix Ex Taq (Takara, Tokyo, Japan) with first strand cDNA, forward and reverse primers. Primer sequences are as follows: Sirt1 Forward: CAGACCCTCAAGCCATGTTT, Sirt1 Reverse: GATCCTTTGGATTCCTGAAA, GAPDH Forward: AACTTTGGCATTGTGGAAGG, GAPDH Reverse: ACACATTGGGGGTAGGAACA. Relative target mRNA was calculated using the ΔΔCT comparative method using GAPDH as an endogenous control.
Immunohistochemistry
Human melanoma tissue microarrays were obtained from US Biomax, Inc. (Rockville, MD). The slides were deparaffinized followed by antigen retrieval. Slides were then blocked, incubated with primary SIRT1 antibody, biotin conjugated secondary antibody, then with freshly prepared ABC-Alkaline Phosphatase reagent (Vector Labs, Burlingame, CA). This was followed by an incubation with Vector Red chromagen (Vector Labs, Burlingame, CA), then hematoxylin as a nuclear counterstain. Finally, cover slips were mounted and slides evaluated via microscopic analysis by our collaborating pathologist (WZ).
Treatment with Tenovin-1
Tenovin-1 was obtained from Cayman Chemical Co. (Ann Arbor, MI) and diluted to a 10 mM concentration in DMSO. Cells were grown to 70% confluency, then incubated with 0, 2.5, 5, 10, or 25 µM of Tenovin-1. DMSO was added to achieve a final concentration of 0.5% in all sample wells, and cells were maintained for 24 or 48 hours at standard tissue culture conditions prior to collection.
Trypan blue exclusion assay
Following treatment, the cells were collected using 0.25% Trypsin/2.21 mM EDTA (Cellgro, Manassas, VA), pelleted, and resuspended in phosphate buffered saline (pH 7.4). The cells were then stained with TC10 Trypan Blue Dye (Bio-Rad, Hercules, CA), and the viable and total cells counted using the Bio-Rad TC10 cell counter (Bio-Rad, Hercules, CA).
MTT assay
Cells were grown at a density of 7.5 × 103 per well in 96-well tissue culture treated plates. After treatment, cells were rinsed with phosphate buffered saline (pH 7.4), then incubated in the dark at 37°C for 2 hours with MTT reagent (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Sigma-Aldrich, St. Louis, MO). MTT reagent was removed, and formazan crystals were solubilized through a 10 min incubation with DMSO. Optical density was read at 540 nm and normalized to the untreated control.
Colony formation assay
After treatment, 3.0 × 103 viable cells from each treatment group were plated in separate wells of a 6-well tissue culture treated plate. Cells were maintained under standard tissue culture conditions, changing media every 3 days for 10–14 days, until the cells reached optimal density. Cells were then stained at 4°C for 1 h with 0.5% crystal violet solution, rinsed with phosphate buffered saline (pH 7.4), and air dried followed by digital photography.
Annexin V/PI binding assay
Following treatments, the cells were washed with phosphate buffered saline (pH 7.4) and stained with FITC conjugated Annexin V antibody and propidium iodide (Dead Cell Apoptosis Kit with Annexin V FITC and PI, Invitrogen Corp, Carlsbad, CA) according to the manufacturer’s protocol. Twenty thousand cells for each sample were then analyzed on a FACS Calibur benchtop cytometer (BD Biosciences, San Jose, CA) at the UWCCC Flow cytometry facility and analyzed by FlowJo software (Treestar, Ashland, OR).
P53 transcriptional activity asssay
Cells were plated at a density of 1.0 × 104 (A375, Hs294T) or 1.5 × 104 (G361) cells/well in 96-well plates. Twenty-four hours later, cells were transfected with Cignal p53 Reporter (luc) DNA (SA Biosciences, CA) using Lipofectamine2000 transfection reagent (Invitrogen, CA). After 24 hours, media was replaced and cells were treated in triplicate with Tenovin-1. Twenty-four hours later, cells were collected and analyzed using the Dual Luciferase Reporter Assay (Promega, WI) according to the manufacturer’s protocol.
Results and Discussion
SIRT1 is overexpressed in human melanoma cells and tissues
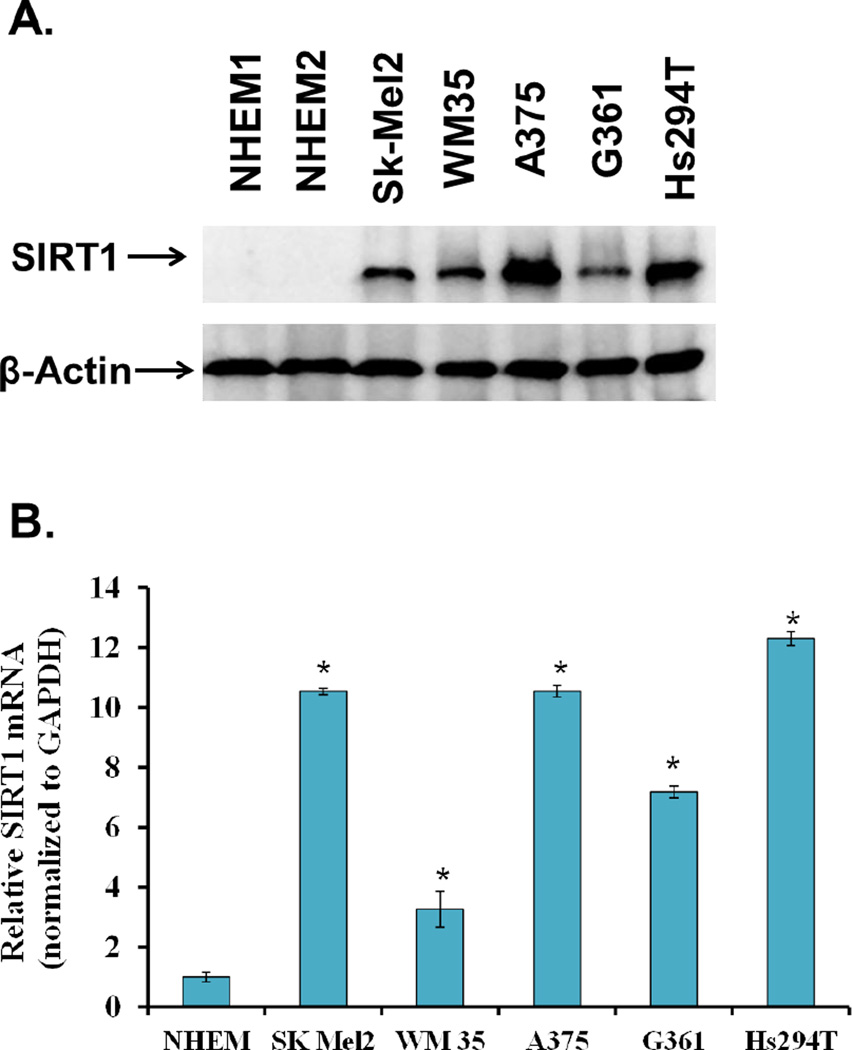
To assess the expression profile of SIRT1 in human melanoma cells, we first determined its levels in a panel of human melanoma cells relative to normal melanocytes. The melanoma cells used in this study included the early stage, radial growth phase melanoma WM35 [22], the melanotic melanoma G361 [23], and the metastatic melanomas A375 (isolated from a primary site in a malignant melanoma, reported to be highly metastatic, amelanotic) [24–28], Hs294T (amelanotic, isolated from a lymph node metastasis) [29], and Sk-Mel-2 (isolated from a metastasis on the thigh skin) [30]. The evaluation of protein levels via immunoblot analysis showed that SIRT1 protein is overexpressed across all melanoma cell lines tested (Fig 1A). Further, RT-PCR analysis revealed a similar trend of overexpression (p<0.01) of Sirt1 mRNA levels in melanoma cells (Fig 1B).
Figure 1. SIRT1 protein and mRNA levels in melanoma cells relative to normal melanocytes.
A) Cells were grown to 80% confluency prior to cell lysate preparation. SIRT1 protein levels were determined by Western Blot analysis. Equal loading was confirmed by reprobing the blot for β-actin. The data represent three separate experiments with similar results. B) SIRT1 mRNA in melanoma cells: The relative expression of the SIRT1 transcript in melanoma cell lines was determined by qRT-PCR using the ΔΔCT comparative method with GAPDH as an endogenous control. The data represent three separate experiments with similar results, and are shown as a mean +/− SEM for three samples. Statistical analysis was performed using a two-tailed student’s t-test. (*p < 0.01).
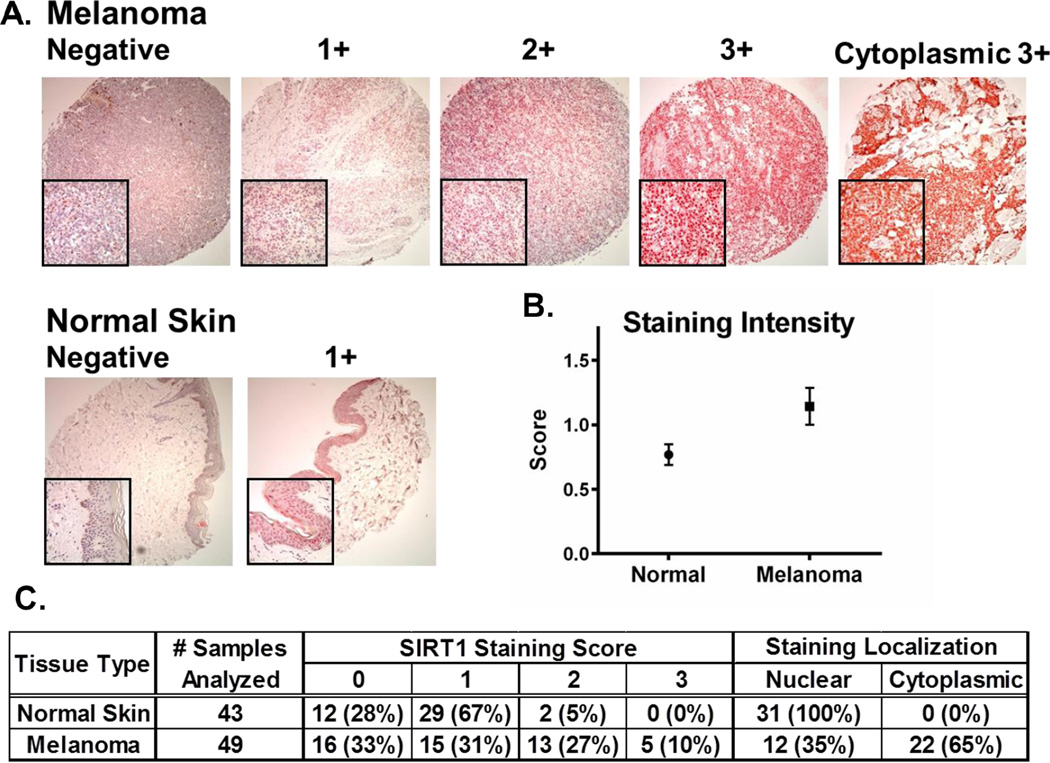
To confirm the overexpression of SIRT1 in clinical melanoma tissues, human tissue microarrays were stained and analyzed for SIRT1 levels. Results were pooled from two separate arrays consisting of 24 and 80 samples, for a total of 104 tissue cores. Of these 104 cores, 12 contained insufficient tissue for analysis leaving 49 melanoma samples and 43 normal skin samples for assessment. These samples covered an array of patient ages and genders, as well as various sites of isolation from the body. Tumor tissues were predominantly from stage II melanomas, although staging was not available for all samples. Detailed information on the tissue microarrays analyzed, as well as the score of each tissue core is provided in Supplementary Tables 1 and 2. Tissues were blindly scored by an experienced pathologist (WZ) on a scale from 0, indicating no visible staining, to 3 indicating maximal staining of SIRT1 protein within at least 50% of the melanocytes present in the sample (Representative staining is shown in Fig 2A). The results show a significant (p < 0.001) overall increase in SIRT1 staining in melanoma tissues as calculated by a Chi-squared statistical analysis (Fig 2B).
Figure 2. SIRT1 protein levels and localization in melanoma tissues relative to normal skin.
A microarray consisting of 92 melanoma patient tissue samples was analyzed via immunohistochemical staining for SIRT1 protein. Positive SIRT1 staining is indicated by the intensity of the red color from the Vector Red chromagen used in staining. Individual tissue cores were blindly scored as negative (0), weak (1+), moderate (2+), or strong (3+) staining in 50% of the cells examined. A) Representative core images for each level of staining. Large boxes are shown at 10× magnification, small boxes are shown at 40× magnification. B) Mean level of SIRT1 in melanoma samples and normal skin. Results are shown as mean +/− SEM of all samples analyzed. Statistical analysis was performed using a chi-squared test. C) Summary of SIRT1 staining results. Values shown are the number of samples receiving the specified score or staining localization for each tissue type. Percentages indicate the portion of samples within each tissue type receiving the score listed or localized within the area of the cell shown.
In addition to the change in overall SIRT1 staining intensity, there was a noticeable shift in the number of tissue samples exhibiting cytoplasmic localization of SIRT1. In the 43 normal skin samples analyzed, all positive staining for SIRT1 was localized in the nucleus, whereas in the melanoma samples, 12 (35%) contained nuclear staining, while 22 (65%) showed SIRT1 staining in the cytoplasm (Fig 2C). This shift to greater cytoplasmic localization in our study agrees with what Byles et al. found in lung, breast and prostate cancer cell lines, as well as in prostate cancer tissue [31]. Byles et al. further showed that in these cell lines, the cytoplasmic abundance of SIRT1 was independent of its nuclear-cytoplasmic translocation, and was instead the result of increased cytoplasmic protein stability mediated by PI3K/IGF-1R signaling. Thus, it is possible that increased levels of SIRT1 in cells could be the result of its increased stability in the cytoplasm, although an analysis of this hypothesis is outside of the scope of this study.
Overall, our results show that SIRT1 mRNA and protein are both significantly overexpressed in melanoma. In addition, cell lines which were isolated from primary tumors in the early stages of tumor development (WM35 and G361) showed lower levels of SIRT1 than the metastatic lines evaluated (A375, Hs294T, Sk-Mel-2). While the sample number is too small to determine what significance this might have, it suggests that an analysis of a potential correlation between SIRT1 levels and melanoma progression could be a viable area for future exploration.
Through observation of our cell culture in the laboratory, we also noticed that the cell lines that show the highest levels of SIRT1 are those which reach confluence first (data not shown), suggesting that there could be a correlation between melanoma growth rate and SIRT1 expression. Alternatively, a recent study involving the mouse melanoma lines B16F1 and B16F10 showed that in these cell lines, SIRT1 levels were affected by the cell culture density itself [32]. However, the significance of SIRT1 levels relative to the growth rate of melanoma cells remains to be determined.
Small molecule inhibitor Tenovin-1 significantly reduces growth and clonogenic survival of human melanoma cells
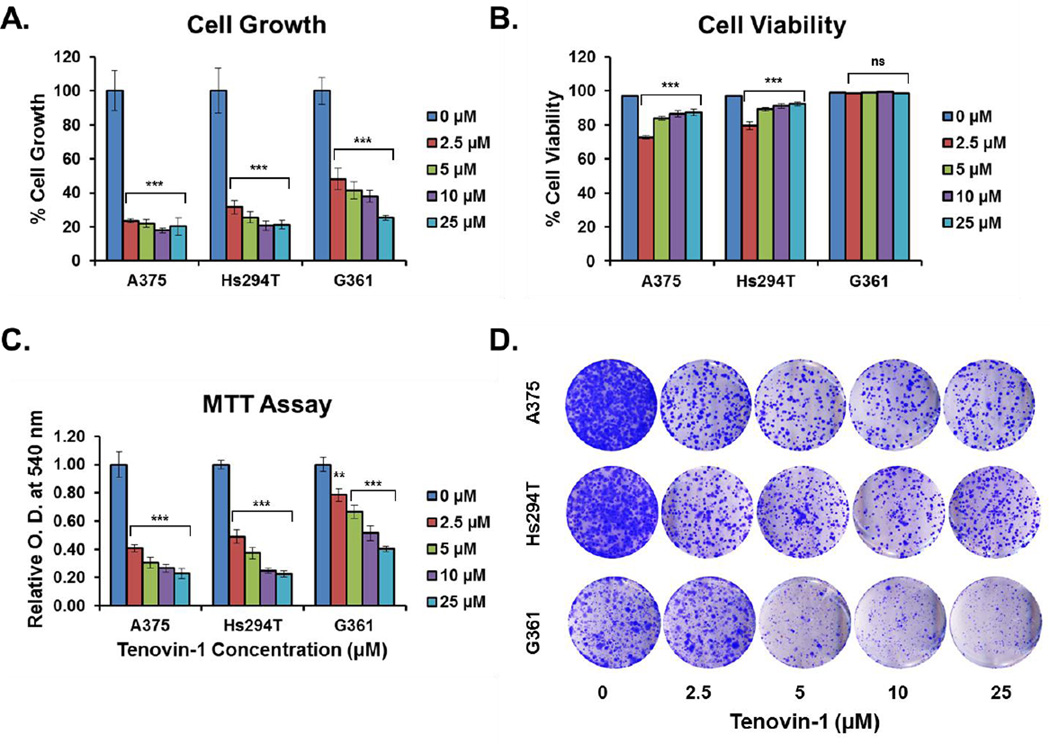
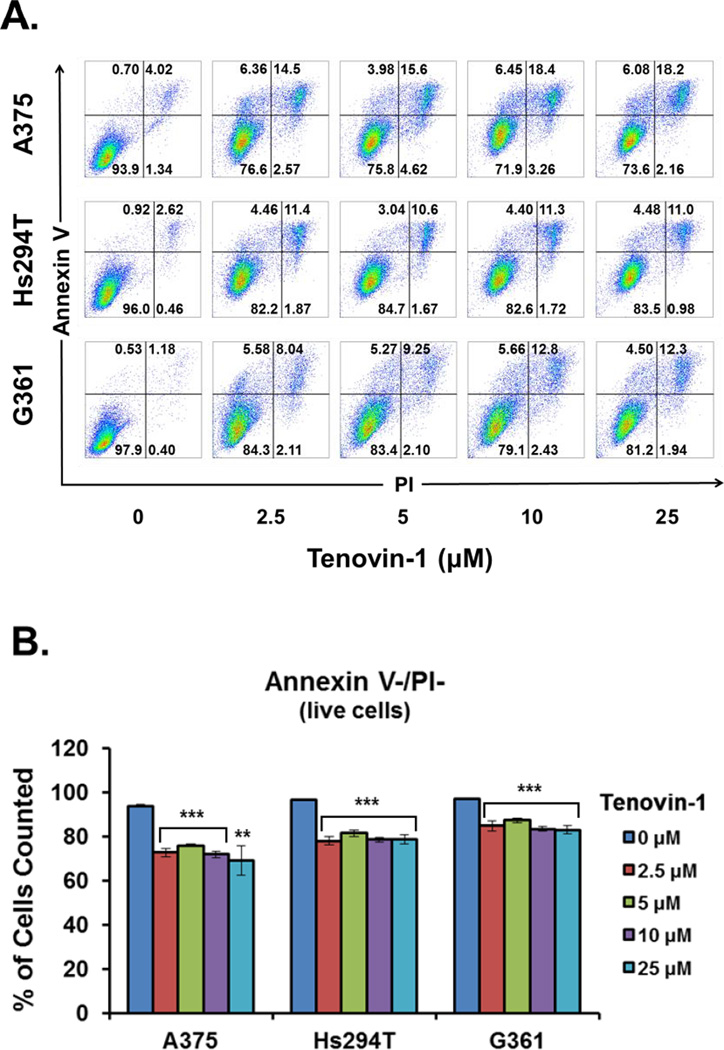
As an initial assessment of the effects of SIRT1 overexpression on tumor growth and survival, Tenovin-1, a small molecule inhibitor of SIRT1 was employed. Tenovin-1 is a Sirtuin specific inhibitor, although it inhibits both SIRT1 and SIRT2 at low levels, as well as SIRT3 at higher levels (SIRT3 IC50=67µM) [33]. While a more specific inhibitor of SIRT1 would be preferable, no adequate compound was commercially available at the time of the study initiation. In our initial experiments, we treated A375, Hs294T, and G361 cells with Tenovin-1 at 10, 25, or 50 µM Tenovin-1 (Supplementary Fig 1–2). However, due to a significant change in viability at the 50 µM level, we argued that this treatment level was high enough to involve inhibition of SIRT3. Therefore, we decided to lower the concentration of Tenovin-1 to minimize this effect. For all three cell lines, a 48 hour treatment with 2.5, 5, 10, or 25 µM Tenovin-1 resulted in a significant (p < 0.001) decrease in cell growth compared to untreated cells as determined by direct cell counting (Fig 3A). This decrease in growth was significant even at the lowest treatment level tested. Cell viability measured by trypan blue exclusion assay showed a mild, but statistically significant (p < 0.001) decrease in the viability of A375 and Hs294T cell lines, but not for G361 (Fig 3B). To confirm the results of the trypan blue analysis, Tenovin-1 treated cells were stained with Annexin V and propidium iodide (PI) and analyzed by flow cytometry. The number of cells staining positive for both Annexin V and PI showed a mild decrease across all treatment levels similar to that seen in the trypan blue staining for cell viability (Fig 4). However, the Annexin V/PI analysis showed a statistically significant (p < 0.01) decrease for all three cell lines, indicating that Tenovin-1 treatment does have a mild impact on melanoma cell viability.
Figure 3. Cell growth, viability, proliferation, and clonogenic survival of melanoma cells after Tenovin-1 treatment.
Melanoma cells were grown to 70% confluency, then treated for 48 hours with 0, 2.5, 5, 10, or 25 µM of Tenovin-1 prior to analysis. A) Cell growth was analyzed by Trypan Blue exclusion assay. Cell growth is expressed as percent growth relative to untreated control. The data are expressed as mean +/− SE of three experiments. Statistical analysis was performed using a two-tailed student’s t-test. (***p<0.001) B) Cell viability was analyzed by Trypan Blue exclusion assay. Cell viability is expressed as the percent of viable cells out of the total cells at each treatment level. The data are expressed as mean +/− SE of three experiments. Statistical analysis was performed using a two-tailed student’s t-test. (ns = no significance, ***p<0.001) C) Cell proliferation was analyzed by MTT assay. Results are displayed as a mean of the optical density at 540 nm at each treatment level normalized to the untreated control. The data are expressed as mean +/− SE of three experiments. Statistical analysis was performed using a two-tailed student’s t-test. (**p<0.01, ***p<0.001) D) After the 48 hr treatment with Tenovin-1, cells were collected and an equal number of viable cells were plated for each treatment level. After 10 days, cells were stained with crystal violet as an assessment of clonogenic survival. The results are representative of three separate experiments with similar results.
Figure 4. Cell viability of melanoma cells after Tenovin-1 treatment.
Melanoma cells were grown to 70% confluency, then treated for 48 hours with 0, 2.5, 5, 10, or 25 µM of Tenovin-1 prior to analysis. Cell viability was assessed by flow cytometric analysis of annexin V/propidium iodide (PI) staining. A) Two-dimensional dot plots of annexin V-FITC and PI fluorescence. Results are representative of two separate experiments with similar results. B) Quantification of annexin V/PI negative, viable cells. The data are expressed as mean +/− SE of six samples for each treatment level. Statistical analysis was performed using a two-tailed student’s t-test. (**p<0.01, ***p<0.001)
As further confirmation of the effects of Tenovin-1 on cell growth and viability, an MTT (3-[4, 5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazoliumbromide) assay was performed. We found significant (p < 0.01) decreases in the OD540 of Tenovin-1 treated cells relative to control cells at all levels tested (Fig 3C). This difference in OD540 levels between treated samples and controls is not as great as the observed differences in cell growth as measured via trypan blue exclusion assay. However, this is not unexpected since MTT assays rely largely on the activity of NAD(P)H-dependent oxidoreductases to reduce the MTT tetrazolium salt to a formazan detectable at 540 nm [34]. Therefore, the OD540 levels are impacted not only by cell growth and viability, but also by metabolic activity.
To show that the effects of this growth inhibition are long-lasting, cells were re-plated at low density after Tenovin-1 treatment and allowed to grow for 10–14 days under normal cell culture conditions. This clonogenic survival assay showed a visible decrease in the ability of previously treated cells to form new colonies for A375 and Hs294T starting at the 2.5 µM level, and for G361 at the 5 µM level (Fig 3D).
Our results show that Tenovin-1 treatment of human melanoma cell lines results in a significant decrease in cell growth, viability, and clonogenic survival.
Tenovin-1 inhibition of SIRT1 results in a significant increase in p53 protein and transcriptional activity in melanoma cells
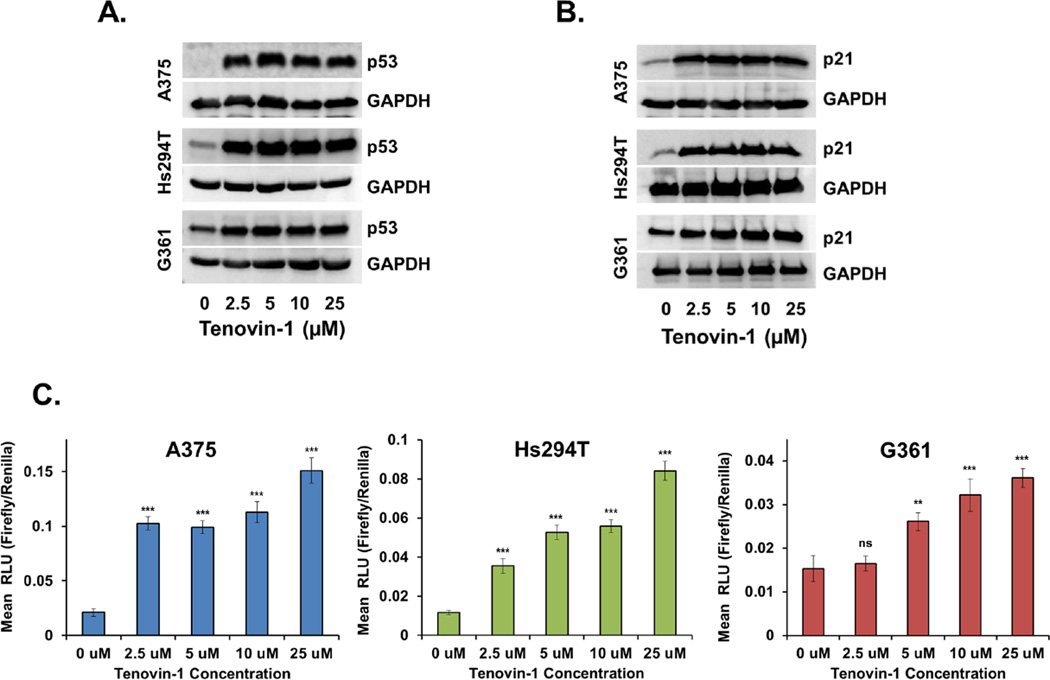
To assess whether the effects of Tenovin-1 treatment on cell growth are dependent on p53 expression, cells were treated for 48 hours, and p53 protein levels were evaluated via immunoblot analysis. As shown in Fig. 5A, Tenovin-1 treatment resulted in a marked increase in p53 protein levels starting at the lowest concentration tested in all three melanoma cell lines.
Figure 5. p53 protein and transcriptional activity in melanoma cells after Tenovin-1 treatment.
Melanoma cells were grown to 70% confluency, then treated for 24 hours with 0, 10, 25, or 50 µM of Tenovin-1 prior to analysis. A) p53 protein levels were determined by Western Blot analysis. Equal loading was confirmed by reprobing the blot for GAPDH. Data represent three separate experiments with similar results. B) p21 protein levels were determined by Western Blot analysis. Equal loading was confirmed by reprobing the blot for GAPDH. Data represent three separate experiments with similar results. C) p53 transcriptional activity was analyzed by transfecting cells with a p53 firefly luciferase reporter and a renilla luciferase control prior to Tenovin-1 treatment. Results are displayed as a ratio of firefly/renilla luminescence, or relative luminescence units (RLU). Data are expressed as mean +/− SE of three experiments. Statistical analysis was performed using a two-tailed student’s t-test. (ns = no significance, **p<0.01, ***p<0.001) There was a single outlier removed from the G361 5 µM treatment group as identified by a Grubbs test for statistical outliers with a significance level of α=0.0001.
To determine whether the increase in p53 protein corresponds with an increase in p53 activity, a p53 luciferase reporter assay was employed (Cignal reporter assay, SA Biosciences). This assay links tandem repeats of p53 transcriptional response elements to a firefly luciferase reporter, with a constitutively active renilla luciferase reporter as an internal control. 48 hours of Tenovin-1 treatment gave inconsistent results with this assay, presumably due to loss of the reporter plasmid at the collection time 72 hours after transfection (data not shown). Therefore, cells were collected and analyzed after 24 hours of treatment for this assay, and results show increasing p53 activity with increasing dose, with significance (p < 0.001) for A375 and Hs294T starting at the lowest concentration tested, and for G361 (p < 0.01) starting at the 5 µM concentration (Fig 5C). To confirm this increase in p53 activity, after 48 hours of Tenovin-1 treatment, protein levels of the p53 transcriptional target p21 were analyzed via immunoblot assay. Results show that p21 protein levels closely mirror those of p53, with a significant increase across all Tenovin-1 concentrations tested (Fig 5B).
From our data, it is clear that Tenovin-1 inhibition leads to an increase in p53 expression and activity, and an upregulation of the downstream effector p21. The increase in p53 expression is to be expected based on the deacetylation site of SIRT1 in the C-terminal domain of p53. Acetylation in this region protects p53 from degradation by preventing its ubiquitination by MDM2 [35, 36]. These changes in p53 activity and p21 expression occur concomitantly with a very mild decrease in cell viability and a drastic decrease in cell growth, which decreases the ability of cells to form new colonies for weeks after the treatment is removed. However, it is unclear whether this decrease in cell growth is indicative of a temporary cell cycle arrest or a permanent senescent state. P21 is known to play a role in both cellular senescence and cell cycle arrest, and distinguishing between the two will require further investigation.
The absence of a dramatic decrease in cell viability in response to Tenovin-1 treatment is not surprising based on a mechanism in which Tenovin-1 prevents SIRT1 deacetylation of p53. There are several known lysines which undergo acetylation in normally functioning p53. The target for SIRT1 deacetylation is lysine 382, which has been shown to initiate cell cycle arrest, but not apoptosis, via p21 when acetylated [15, 37]. Therefore, inhibition of SIRT1 in melanoma is predicted to cause cell cycle arrest through p21 transcriptional activation by p53 as observed here.
Conclusion
We have shown that SIRT1 is overexpressed in human melanoma cells and tissues, and that its inhibition by Tenovin-1 leads to decreased growth, viability, and clonogenic survivial of melanoma cells. These anti-proliferative effects are likely related to an observed increase in the expression and activity of the tumor suppressor p53 and its downstream effector p21. While we have shown these effects in an in vitro cell culture system, an in vivo model is still necessary to prove the therapeutic significance of SIRT1 inhibition in melanoma. In addition, further studies are needed to differentiate between the effects of SIRT1 and SIRT2 inhibition, as Tenovin-1 is an inhibitor of both of these Sirtuins.
Supplementary Material
Research Highlights.
The histone deacetylase SIRT1 is overexpressed in melanoma cells and clinical tissue samples.
SIRT1 localization shifts from entirely nuclear in skin to 65% cytoplasmic in melanoma tissues.
Tenovin-1 inhibition of SIRT1 causes an anti-proliferative response in melanoma cells.
The anti-proliferative response to Tenovin-1 correlates with increased p53 and p21 expression.
Acknowledgements
This work was partially supported by funding from the NIH (R01AR059130 and R01CA176748 and the Department of Veterans Affairs (VA Merit Review Award 1I01BX001008).
Abbreviations
- NHEM
normal human epidermal melanocyte
- NAD+
nicotinamide adenine dinucleotide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jemal A, Saraiya M, Patel P, Cherala SS, Barnholtz-Sloan J, Kim J, Wiggins CL, Wingo PA. Journal of the American Academy of Dermatology. 2011;65:S17.e1–S17.e11. doi: 10.1016/j.jaad.2011.04.032. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. CA: A Cancer Journal for Clinicians. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.van Leeuwen I, Lain S. In: Advances in Cancer Research. George FVW, George K, editors. Academic Press; 2009. pp. 171–195. [DOI] [PubMed] [Google Scholar]
- 4.Michan S, Sinclair D. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stunkel W, Campbell RM. J Biomol Screen. 2011;16:1153–1169. doi: 10.1177/1087057111422103. [DOI] [PubMed] [Google Scholar]
- 6.Rajendran R, Garva R, Krstic-Demonacos M, Demonacos C. J Biomed Biotechnol. 2011;2011:368276. doi: 10.1155/2011/368276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan H, Su L, Chen WY. Onco Targets Ther. 2013;6:1399–1416. doi: 10.2147/OTT.S37750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hida Y, Kubo Y, Murao K, Arase S. Arch Dermatol Res. 2007;299:103–106. doi: 10.1007/s00403-006-0725-6. [DOI] [PubMed] [Google Scholar]
- 9.Choi HN, Bae JS, Jamiyandorj U, Noh SJ, Park HS, Jang KY, Chung MJ, Kang MJ, Lee DG, Moon WS. Oncol Rep. 2011;26:503–510. doi: 10.3892/or.2011.1301. [DOI] [PubMed] [Google Scholar]
- 10.Jung-Hynes B, Huang W, Reiter RJ, Ahmad N. J Pineal Res. 2010;49:60–68. doi: 10.1111/j.1600-079X.2010.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalle AM, Mallika A, Badiger J, Alinakhi, Talukdar P, Sachchidanand Biochem Biophys Res Commun. 2010;401:13–19. doi: 10.1016/j.bbrc.2010.08.118. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Wang L, Wang Z, Ho Y, McDonald T, Holyoake TL, Chen W, Bhatia R. Cancer Cell. 2012;21:266–281. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma JX, Li H, Chen XM, Yang XH, Wang Q, Wu ML, Kong QY, Li ZX, Liu J. Neuropathology. 2013;33:7–16. doi: 10.1111/j.1440-1789.2012.01318.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhao G, Cui J, Zhang JG, Qin Q, Chen Q, Yin T, Deng SC, Liu Y, Liu L, Wang B, Tian K, Wang GB, Wang CY. Gene Ther. 2011;18:920–928. doi: 10.1038/gt.2011.81. [DOI] [PubMed] [Google Scholar]
- 15.Vaziri H, Dessain SK, Eaton EN, Imai S-I, Frye RA, Pandita TK, Guarente L, Weinberg RA. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 16.Beckerman R, Prives C. Cold Spring Harb Perspect Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuckerman V, Wolyniec K, Sionov RV, Haupt S, Haupt Y. J Pathol. 2009;219:3–15. doi: 10.1002/path.2584. [DOI] [PubMed] [Google Scholar]
- 18.Dahl C, Guldberg PER. APMIS. 2007;115:1161–1176. doi: 10.1111/j.1600-0463.2007.apm_855.xml.x. [DOI] [PubMed] [Google Scholar]
- 19.Soengas MS, Lowe SW. Oncogene. 2003;22:3138–3151. doi: 10.1038/sj.onc.1206454. [DOI] [PubMed] [Google Scholar]
- 20.Satyamoorthy K, Chehab NH, Waterman MJ, Lien MC, El-Deiry WS, Herlyn M, Halazonetis TD. Cell Growth Differ. 2000;11:467–474. [PubMed] [Google Scholar]
- 21.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herlyn M. Cancer Metastasis Rev. 1990;9:101–112. doi: 10.1007/BF00046337. [DOI] [PubMed] [Google Scholar]
- 23.Peebles PT, Trisch T, Papageorge AG. Pediatr Res. 1978;12:485–485. [Google Scholar]
- 24.Chen X, Lin J, Kanekura T, Su J, Lin W, Xie H, Wu Y, Li J, Chen M, Chang J. Cancer Res. 2006;66:11323–11330. doi: 10.1158/0008-5472.CAN-06-1536. [DOI] [PubMed] [Google Scholar]
- 25.Ahmad I, Muneer KM, Tamimi IA, Chang ME, Ata MO, Yusuf N. Toxicol Appl Pharmacol. 2013;270:70–76. doi: 10.1016/j.taap.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 26.Looi CY, Moharram B, Paydar M, Wong YL, Leong KH, Mohamad K, Arya A, Wong WF, Mustafa MR. BMC Complement Altern Med. 2013;13:166. doi: 10.1186/1472-6882-13-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fok JY, Ekmekcioglu S, Mehta K. Mol Cancer Ther. 2006;5:1493–1503. doi: 10.1158/1535-7163.MCT-06-0083. [DOI] [PubMed] [Google Scholar]
- 28.Giard DJ, Aaronson SA, Todaro GJ, Arnstein P, Kersey JH, Dosik H, Parks WP. J Natl Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 29.Creasey AA, Smith HS, Hackett AJ, Fukuyama K, Epstein WL, Madin SH. In Vitro. 1979;15:342–350. doi: 10.1007/BF02616140. [DOI] [PubMed] [Google Scholar]
- 30.Fogh J. Human tumor cells in vitro. New York: Plenum Press; 1975. pp. 120–121. [Google Scholar]
- 31.Byles V, Chmilewski LK, Wang J, Zhu L, Forman LW, Faller DV, Dai Y. Int J Biol Sci. 2010;6:599–612. doi: 10.7150/ijbs.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunimoto R, Jimbow K, Tanimura A, Sato M, Horimoto K, Hayashi T, Hisahara S, Sugino T, Hirobe T, Yamashita T, Horio Y. J Invest Dermatol. 2014 doi: 10.1038/jid.2014.50. [Epub Ahead of Print], http://dx.doi.org/10.1038/jid.2014.50. [DOI] [PubMed] [Google Scholar]
- 33.Lain S, Hollick JJ, Campbell J, Staples OD, Higgins M, Aoubala M, McCarthy A, Appleyard V, Murray KE, Baker L, Thompson A, Mathers J, Holland SJ, Stark MJR, Pass G, Woods J, Lane DP, Westwood NJ. Cancer Cell. 2008;13:454–463. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berridge MV, Herst PM, Tan AS. Biotechnol Annu Rev. 2005;11:127–152. doi: 10.1016/S1387-2656(05)11004-7. [DOI] [PubMed] [Google Scholar]
- 35.Li M, Luo J, Brooks CL, Gu W. J Biol Chem. 2002;277:50607–50611. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- 36.Poyurovsky MV, Katz C, Laptenko O, Beckerman R, Lokshin M, Ahn J, Byeon I-JL, Gabizon R, Mattia M, Zupnick A, Brown LM, Friedler A, Prives C. Nat Struct Mol Biol. 2010;17:982–989. doi: 10.1038/nsmb.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy S, Packman K, Jeffrey R, Tenniswood M. Cell Death Differ. 2005;12:482–491. doi: 10.1038/sj.cdd.4401581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.