Abstract
The role of the transcription factor nuclear factor of activated T cells (NFAT) was initially identified in T and B cell gene expression, but its role in regulating gene expression in macrophages during sepsis is not known. Our data show that NFATc3 regulates expression of inducible nitric oxide synthase (iNOS) in macrophages stimulated with lipopolysaccharide. Selective inhibition of NFAT by cyclosporine A and a competitive peptide inhibitor 11R-VIVIT inhibited endotoxin-induced expression of iNOS and nitric oxide (NO) release. Macrophages from NFATc3 knockout (KO) mice show reduced iNOS expression and NO release and attenuated bactericidal activity. Gel shift and chromatin immunoprecipitation assays show that endotoxin challenge increases NFATc3 binding to the iNOS promoter, resulting in transcriptional activation of iNOS. The binding of NFATc3 to the iNOS promoter is abolished by NFAT inhibitors. NFATc3 KO mice subjected to sepsis show that NFATc3 is necessary for bacterial clearance in mouse lungs during sepsis. Our study demonstrates for the first time that NFATc3 is necessary for macrophage iNOS expression during sepsis, which is essential for containment of bacterial infections.
Key Words: Inducible nitric oxide synthase, Macrophages, Nuclear factor of activated T cells, Sepsis, VIVIT
Introduction
Severe sepsis is a common medical condition characterized by a systemic inflammatory response that results from a harmful or damaging host response to infection that leads to multiple organ failure and death [1, 2]. It is imperative to improve the understanding of the dysregulation of the host response/interactions during sepsis, which would lead to new opportunities for therapeutic intervention [3, 4]. Macrophages have a major role in the maintenance of the tissue homeostasis, in part because of antimicrobial properties and through involvement in both the innate and adaptive immune responses during the course of inflammatory diseases. Macrophages are innate immune effector cells that are essential for generating beneficial inflammation that is necessary for pulmonary antibacterial host defense [2, 5]. Severe sepsis that is associated with Gram-negative bacteremia is characterized by neutrophilic lung inflammation and pulmonary vascular hyperpermeability, which are characteristic of the acute respiratory distress syndrome [3]. Macrophages express high levels of inducible nitric oxide synthase (iNOS) and are important sources of iNOS-derived nitric oxide (NO), which together with the respiratory burst of reactive oxygen species (ROS) leads to effective bactericidal activity [2, 6]. NO has a paradoxical role in severe sepsis, where it has a harmful effect by contributing to lung injury in animals and humans [7, 8] and a beneficial effect on the immune system and in the lungs through antimicrobial activity [9, 10, 11]. Recently it has been reported that monocyte/macrophage-derived iNOS plays a pivotal role in mediating resolution of lung injury by modulating lung immune responses, thus facilitating clearance of alveolar inflammation and promoting lung repair [12]. Activation of macrophages is a key component of the immune response, and several bacterial products and proinflammatory cytokines participate actively in triggering this process. Activation of macrophages is dependent on the Toll-like receptor (TLR) signaling pathway, which engages transcriptional regulators to induce proinflammatory genes for antimicrobial activity [13]. However, uncontrolled or excess proinflammatory mediators and ROS can exert deleterious effects such as those observed during severe sepsis and in persistent local inflammatory processes [14]. Thus it is important to understand the potential role of transcription factors that regulate the gene expression in macrophages and immune cells.
Nuclear factors of activated T cells (NFATs) are transcription factors that tightly control proinflammatory cytokine expression for adaptive immunity in T and B lymphocytes. However, little is known about the role of NFATs for innate immunity in macrophages. The NFAT gene family consists of 5 members; NFATc1-c4 are controlled by calcium levels, while NFAT5 is activated by osmotic stress [15, 16]. Calcium mobilization through Ca2+ release-activated Ca2+ channels has been observed in macrophages in response to bacterial infection [17, 18]. Recently it has been reported that the calcium entry controller channel transient receptor potential melastatin-4 mobilizes calcium entry and maintains Ca2+ homoeostasis, which specifically affects the functions of macrophages in sepsis [19]. An increase in intracellular calcium levels causes calcineurin, a calcium-dependent phosphatase, to dephosphorylate cytoplasmic NFAT, resulting in its activation and translocation to the nucleus and ensuing transcription of the target genes. Activated NFAT is negatively regulated by rephosphorylation activity of various nuclear NFAT kinases and exported back to the cytoplasm [15]. NFATc3 and NFATc4 are involved in the TLR-activated innate inflammatory response and cytokine induction [20]. In neurons, NFATc3 undergoes rapid dephosphorylation and nuclear translocation, whereas NFATc4 remains phosphorylated and localized to the cytosol [21]. Recently it has been reported that blocking NFAT function is beneficial in inflammatory diseases like inflammatory bowel disease and pulmonary arterial hypertension [22, 23], which suggests that NFAT may alter NO production. However, the molecular mechanisms and physiological relevance underlying the transcription regulation of iNOS in macrophages by NFAT have not been previously reported in the context of bactericidal activity.
In this study, we demonstrate that the transcription factor NFATc3 mediates the expression of iNOS in macrophages and that abrogation of NFAT results in unrestrained polymicrobial sepsis and inflammation in mice lungs.
Materials and Methods
Mouse Strains Used in This Study
NFATc3−/− [NFATc3 knockout (KO)] and wild-type (WT) C57BL/6J (NFATc3+/+) mice were purchased from The Jackson Laboratory. NFATc3−/− and WT mice 8-12 weeks of age were used in the study, and all the experiments were conducted with protocols approved by the Institutional Animal Care and Use Committee of The Ohio State University, Columbus, Ohio, and the University of Illinois, Chicago, Ill., USA.
Culture of Immortalized Mouse Bone Marrow-Derived Macrophages
Immortalized mouse bone marrow-derived macrophages generated from transgenic luciferase-expressing HLL mice (BMDM-Luc) were generated as described previously [24] and were cultured in complete medium [1× DMEM containing 10% FBS (HyClone), supplemented with penicillin (100 U/ml)/streptomycin (100 µg/ml; Invitrogen)] and maintained at 37°C in humidified air containing 5% CO2.
Isolation and Culture of BMDM
Femoral and tibia bone marrow was isolated from mice (WT, NFATc3 KO and HLL) as previously described [25]. Mouse bone marrow cells were flushed from femurs and tibias with Ca2+/Mg2+-free Hanks' balanced salt solution (Mediatech Inc.). Bone marrow progenitor cells were cultured in DMEM (Mediatech) containing 10% heat-inactivated FBS and 100 U of penicillin-streptomycin (Mediatech) and supplemented with 10% L929 Cell Culture Media. After 7 days, BMDM (approx. 99% macrophages based on flow cytometry using anti-F4/80) were collected for the experiments.
Expression of iNOS in Mouse Macrophages
BMDM-Luc were seeded in tissue culture dishes in complete medium, grown for 24 h and starved in 2% FBS-containing medium overnight. The cells were treated with 100 ng/ml lipopolysaccharide (LPS) Escherichia coli serotype R515, TLR grade (581-007-L002, Enzo Life Sciences), for various periods of time (0, 2, 4, 8, 16 and 24 h). The expression of iNOS was analyzed by reverse transcription quantitative real-time PCR (RT-qPCR) and Western blot.
Inhibition of iNOS Expression in Mouse Macrophages
Inhibition of iNOS expression in macrophage cells was examined by using the NFAT-specific inhibitors cyclosporine A (CsA; Enzo Life Sciences) and the cell-permeable peptide 11R-VIVIT (Calbiochem). 11R-VEET custom synthesized by GenScript was used as control peptide. The BMDM-Luc were seeded in culture dishes in complete medium for 24 h and then starved in 2% FBS-containing medium overnight. Subsequently, cells were treated with CsA (10 µM, 30 min) or 11R-VIVIT (5 µM, 1 h) and then challenged with LPS (100 ng/ml) for 12-16 h. The cells were lysed in radioimmunoprecipitation assay buffer and immunoblotted for iNOS.
Measurement of Nitrite in Macrophage Culture Media
BMDM-Luc and BMDM were treated either with LPS (100 ng/ml) alone or with CsA (10 μM), 11R-VEET (50 μM) or 11R-VIVIT (50 μM) for the indicated time periods. Culture supernatants were stored at −70°C for analysis of nitrite. Nitrite concentration was determined in culture supernatant with a Griess Reagent Kit (G-7921, Molecular Probes Inc.) in a 96-well microplate assay system according to the manufacturer's protocol. The nitrite concentration in the samples was calculated from a standard curve of sodium nitrite.
Reverse Transcription Quantitative Real-Time PCR
Total RNA was isolated using the RNeasyPlus Mini kit (Qiagen) according to the manufacturer's protocol, and the quality was assessed by agarose gel electrophoresis. Purified total RNA (2.5 μg) was reverse transcribed using the RevertAid First Strand cDNA Synthesis kit (Thermo Scientific) with random hexamer primers. The iNOS and β-actin mRNA levels were determined by RT-qPCR on a LightCycler 480 real-time PCR instrument using LightCycler 480 SYBR Green I Master reagent, and 2 μl of the cDNA was used with the following conditions: initial denaturation at 95°C for 10 min, and a loop of 45 cycles consisting of denaturation at 95°C for 15 s, annealing at 68°C for 30 s and extension at 68°C for 30 s. The results were analyzed with LightCycler 480 Software 1.5.0. by an advanced relative quantification algorithm (Roche Diagnostics). Sequences of murine iNOS primers are as follows: miNOS2F, 5′-CCG CCC TGG TGC AGG GAA TC-3′, and miNOS2R, 5′-ACC CAG TAG CTG CCG CTC TCA-3′. For murine β-actin, primers are mActinF (5′-GTC GAG TCG CGT CCA CCC GC-3′) and mActinR (5′-CGT CAT CCA TGG CGA ACT GGT GGC-3′).
Activation of NFATc3 during LPS Stimulation
The BMDM from WT mice were treated with LPS (100 ng/ml) for 0, 0.5 and 4 h and washed in cold 1× PBS. The cellular and nuclear protein fractions were separated using a Pierce NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (catalogue No. 78833), resolved on 10% SDS-polyacrylamide gels and immunoblotted with different NFAT antibodies. The antibodies used for immunoblotting were NFATc1 (8032S, Cell Signaling), NFATc2 (4389S, Cell Signaling), NFATc3 (4998S, Cell Signaling) and NFATc4 (sc-13036, Santa Cruz Biotechnology Inc.). Purity of cytoplasmic and nuclear preparations was assessed by β-actin and p53 (sc-6243, Santa Cruz Biotechnology).
Western Blot
After appropriate treatment, BMDM-Luc were lysed with 1× radioimmunoprecipitation assay buffer [50 mM Tris-HCl, pH 8.0, 150 mM sodium chloride, 1.0% Igepal CA-630 (NP-40), 0.5% sodium deoxycholate and 0.1% sodium dodecyl sulfate] containing 1× protease inhibitor cocktail (Roche Diagnostics). Cell lysates containing equal amounts of protein were fractionated by electrophoresis on a reducing polyacrylamide gel and electroblotted to a nitrocellulose membrane (BioRad). The nitrocellulose membranes were incubated with blocking buffer [50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% Tween-20 (TBST) containing 5% BSA] for 1 h at room temperature, followed by overnight incubation with primary antibody at 4°C in blocking buffer and then with a horseradish peroxidase-conjugated secondary antibody. After washing the membrane with TBST, the reactive bands were detected by enhanced chemiluminescent substrate (SuperSignal West Pico, Thermo Scientific). Antibodies used were anti-iNOS/NOS Type II (610332, BD Transduction Laboratories), anti-α-tubulin (sc-8035, Santa Cruz Biotechnology) and anti-β-actin (Sigma).
Analysis of iNOS Promoter
The genomic sequence containing the promoter region of mouse iNOS mRNA was downloaded from the National Center for Biotechnology Information database. Potential NFAT binding sites were identified by analysis of the promoter region using the online software Patch 1.0 (www.gene-regulation.com/pub/programs.html) and MatInspector (www.genomatix.de) [26].
Electrophoretic Mobility Shift Assay
BMDM-Luc were grown in complete medium, serum starved overnight, treated with CsA (10 µM) and then stimulated with LPS (0.1 µg/ml) for 30 and 60 min. The cells were washed with ice-cold 1× PBS and lysed with Buffer A [10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.1% Triton-X-100 and protease inhibitor cocktail (Roche)] for 15 min at 4°C. The content was centrifuged, and the pellet was washed with Buffer B [10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl and protease inhibitor cocktail (Roche)]. The supernatant was discarded, and the nuclear pellet was lysed with Buffer C [20 mM HEPES, pH 7.9, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, pH 8, 25% glycerol, 1 mM DTT and protease inhibitor cocktail (Roche)]. The NFAT consensus DNA probes were obtained from Santa Cruz Biotechnology (catalogue No. sc-2577) and labeled using the Biotin Labeling kit (catalogue No. 89818, Thermo Scientific). Equal amounts of protein from different treatments were incubated with a biotin-labeled NFAT consensus probe and resolved on 5% polyacrylamide native gel in 0.5× Tris-borate-EDTA buffer. The protein-DNA complexes were transferred to Biodyne A membranes (catalogue No. 77015, Thermo Scientific). For super shift assays, the nuclear extract was incubated with anti-NFATc3, and for controls, the respective IgGs were used. For the electrophoretic mobility shift (EMSA) competition assay, 5 µg of nuclear extract was incubated with 10 pmol of biotin-labeled NFAT consensus oligo using reaction buffer supplied in the Pierce LightShift EMSA kit (Thermo Scientific, No. 20148). In the cold competition assay, unlabeled NFAT consensus oligo was included in excess at a concentration of 10, 30 or 50 pmol separately in the above reaction mix and analyzed. The reaction products were resolved on 5% polyacrylamide native gel in 1× Tris-borate-EDTA buffer and transferred onto a Pierce Biodyne Nylon membrane. The biotinylated products were detected using the Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific) on the Gel Imaging System (BioRad).
Chromatin Immunoprecipitation
BMDM-Luc were treated as described above. The cells were washed twice with 1× PBS, and proteins were cross-linked with 1% formaldehyde (Electron Microscopy Sciences) for 10 min at room temperature. The reaction was quenched by adding 125 mM glycine for 5 min, and cells were washed twice with 1× PBS and stored at −80°C. Chromatin was extracted using the ChromaFlash Chromatin extraction kit (Epigentek Group Inc.), and the chromatin was sheared by using a sonicator (Virtis Virsonic Cell Disrupter) for 5 min, with a 10-second pulse on, and a gap of 20 s, and was repeated 3 times. Chromatin immunoprecipitation (ChIP) was carried out using the ChromaFlash One-Step ChIP kit (Epigentek) with the manufacturer's recommended protocol. The DNA-protein complex was immunoprecipitated using a mixture of anti-NFATc3 antibodies (sc-23814 and sc-1152, Santa Cruz Biotechnology). An unrelated goat polyclonal antibody (anti-GFP, sc-5385) and anti-RNA Polymerase II (sc-5943) were used as controls. The immunoprecipitated DNA fragments were analyzed by RT-qPCR using PerfeCTa SYBR Green FastMix (Quanta BioSciences Inc.) containing 250 nM of each primer (forward: 5′-CTG TTC CCC TTA GGT GGG-3′; reverse: 5′-ACA GGA GTG TGT GCA TCT G-3′) with initial denaturation at 95°C for 5 min and a loop of 45 cycles consisting of denaturation at 95°C for 15 s, annealing at 55°C for 30 s and extension at 72°C for 45 s, on a LightCycler 480 real-time PCR instrument (Roche Diagnostics). The crossing point values were analyzed with LightCycler 480 Software 1.5.0 SP4. The abundance of NFATc3 on the iNOS promoter or the fold enrichment was calculated using the equation mentioned in the ChromaFlash One-Step ChIP kit (Epigentek). The unprecipitated chromatin was used as input control for DNA and used for RT- qPCR. An unrelated DNA sequence (approx. 3 kb downstream of the start codon of iNOS) was analyzed for absence of NFAT binding sites by MatInspector, and primers were designed for control ChiP experiments (forward: 5′-AGG GTC ACT TCC CTC AGT AT-3′; reverse: 5′-CAC TCT GCC TGT GTC TTA TCT T-3′).
Synthesis of the Control Peptide 11R-VEET
The control peptide, 11R-VEET [23] (RRRRRRRRRRRGGGMAGPPHIVEETGPHVI; scrambled sequence of 11R-VIVIT), was custom synthesized and HPLC-purified (95.0% purity) by GenScript (Piscataway, N.J., USA).
In vitro Bactericidal Assay of Macrophages Using E. coli
The in vitro bactericidal assay was performed according to a previously published protocol with slight modifications [27]. BMDM were cultured from WT and NFATc3 KO mice in 12-well tissue culture plates starved of serum overnight in DMEM containing 2% FBS. Overnight culture of E. coli DH5α (Invitrogen Corporation) was used to assess the bactericidal activity of macrophages. Bacterial cell number was determined by measuring the optical density at 600 nm. In the first set of experiments, the WT BMDM were treated with the control peptide 11R-VEET or the NFAT inhibitor 11R-VIVIT and exposed to bacteria. In the second set, the BMDM from WT and NFATc3 KO mice were exposed to bacteria. E. coli DH5α was added to each well containing the macrophages, and after 90 min, cells were washed with DMEM to get rid of the nonphagocytized bacteria, and the medium was replenished with DMEM containing 2% FBS and incubated overnight. The macrophages were lysed with Milli-Q water, plated on Luria Bertani agar culture plates and incubated at 37°C. Colony-forming units were enumerated with the Colony Counter program using Quantity One Software (BioRad).
Polymicrobial Infection Assay in Lungs
The WT and NFATc3 KO mice were subjected to sepsis by the cecal ligation and puncture method (CLP) [28]. In this method, a midline incision of about 1 cm was made in the lower abdominal area, and the cecum was exposed, ligated with a sterile silk thread and punctured twice with a 21-gauge needle. After puncturing, a very small amount of fecal material was allowed to extrude into the abdominal cavity. The viscera and skin layers were sutured back in place with a 6.0 Nylon silk suture. In sham controls, the cecum was exposed by opening the visceral cavity but was neither ligated nor punctured. The animals were resuscitated with 0.5 ml of saline subcutaneously. Mice were maintained in aseptic conditions, and 24 h after surgery (sham and CLP), the mice were euthanized and lungs were surgically excised, minced in DMEM and ground using a handheld mechanical homogenizer. The contents were spun, and 100 µl of the tissue supernatant was spread on tryptic soy agar sheep blood agar culture plates (Remel Products, Thermo Fisher Scientific) and incubated at 37°C. Colony-forming units were enumerated with the Colony Counter program using Quantity One Software (BioRad).
Statistical Analysis
Statistical analyses were performed using a nonpaired Student's t test, and p ≤ 0.05 was considered significant.
Results
Transcription Factor NFATc3 Mediates Expression of iNOS in Macrophages
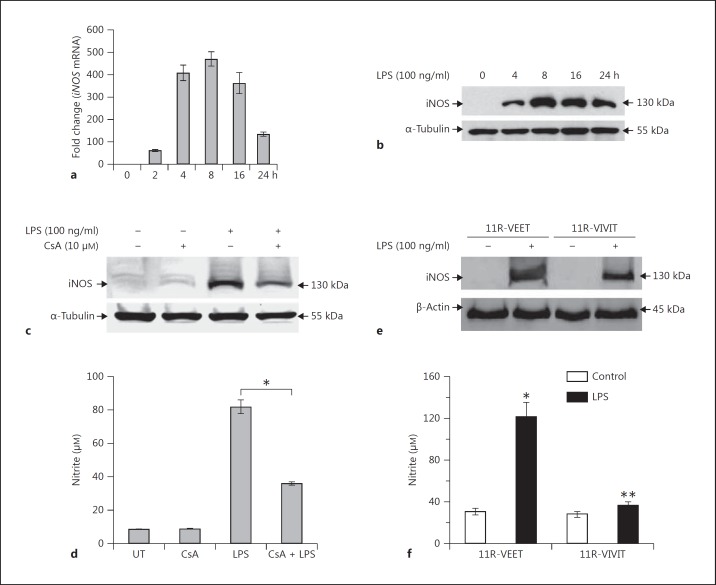
BMDM-Luc were used to analyze the time course for expression of iNOS in response to LPS (100 ng/ml). The expression of iNOS mRNA and protein peaked approximately 8-16 h after LPS treatment (fig. 1a, b), and this time point was used for subsequent experiments. The transcription factor NFAT has been shown to play an important role in the regulation of cytokine expression for adaptive immunity in T and B lymphocytes, and we hypothesized that NFAT plays an essential role in the expression of iNOS in macrophages. To address this hypothesis, we initially used 2 specific inhibitors of NFAT, CsA and a cell membrane-permeable peptide (11R-VIVIT), to determine the impact on expression of iNOS in endotoxin (LPS)-stimulated BMDM-Luc. CsA inhibits the activity of calcinuerin phosphatase of proteins including NFAT [29], whereas the cell-permeable 11R-VIVIT inhibits NFAT activation through interaction with the calcineurin binding site for NFAT and thus prevents nuclear translocation without affecting calcineurin phosphatase activity [30]. As shown, the expression of iNOS in the NFAT inhibitor-treated BMDM-Luc was decreased by more than 50% when compared with the cells treated with LPS alone (fig. 1c, e). We also assessed the accumulation of nitrite (NO2-) in macrophage culture media as an indicator of NO production, which is dependent on iNOS. Compared with the untreated macrophages, the accumulation of nitrite in NFAT inhibitor treatment was decreased by more than 50% (fig. 1d, f). Earlier literature indicated the role of NF-κB in modulating iNOS regulation [31, 32]. To specifically identify NFAT-dependent iNOS induction during sepsis, we used the NF-κB-responsive luciferase-harboring HLL BMDM cell line. The HLL BMDM were treated with NFAT and NF-κB inhibitors separately, stimulated with LPS and assayed for NFAT/ NF-κB-dependent iNOS expression/NO release and NF-κB luciferase reporter activities. Results of the inhibitor experiments indicate that inhibition of NFAT by 11R- VIVIT attenuates the NO release by 50–70% (online suppl. fig. 1; for all online suppl. material, see www.karger.com/doi/10.1159/000362647), whereas the NF-κB inhibitor decreases it by only about 30% (online suppl. fig. 1). Interestingly, NF-κB inhibition by QNZ decreased the NF-κB-dependent luciferase activity by more than 80%, whereas VIVIT decreased it by about 40%. This indicates that NO release is significantly regulated by NFAT. Together these data indicate that expression of iNOS in macrophages is significantly regulated by the transcription factor NFATc3.
Fig. 1.
Expression of iNOS in macrophages is regulated by NFAT. BMDM-Luc were treated with LPS (100 ng/ml), and expression of iNOS was determined at various time points. a, b iNOS mRNA levels were determined by RT-qPCR (a), and protein level was determined by Western blot (b). Expression peaked at approximately 8-16 h following treatment with LPS. c, e Macrophages pretreated with specific inhibitors of NFAT (CsA and 11R-VIVIT, respectively) and challenged with LPS showed significant inhibition of expression of iNOS (c, e, representative Western blots). d, f The concentration of nitrite in the cell culture media, which is an indicator of iNOS expression, was also measured. The concentration of nitrite in the presence of NFAT inhibitors CsA (d) and 11R-VIVIT (f) was analyzed. Treatment with specific NFAT inhibitors resulted in significant reduction of nitrite in cell culture media, indicating that the expression of iNOS was reduced. 11R-VEET was used as control. The error bars represent the SEM from 3 independent experiments. * p ≤ 0.05, ** p ≤ 0.005. UT = Untreated.
NFATc3 Modulates iNOS Expression at the Transcriptional Level
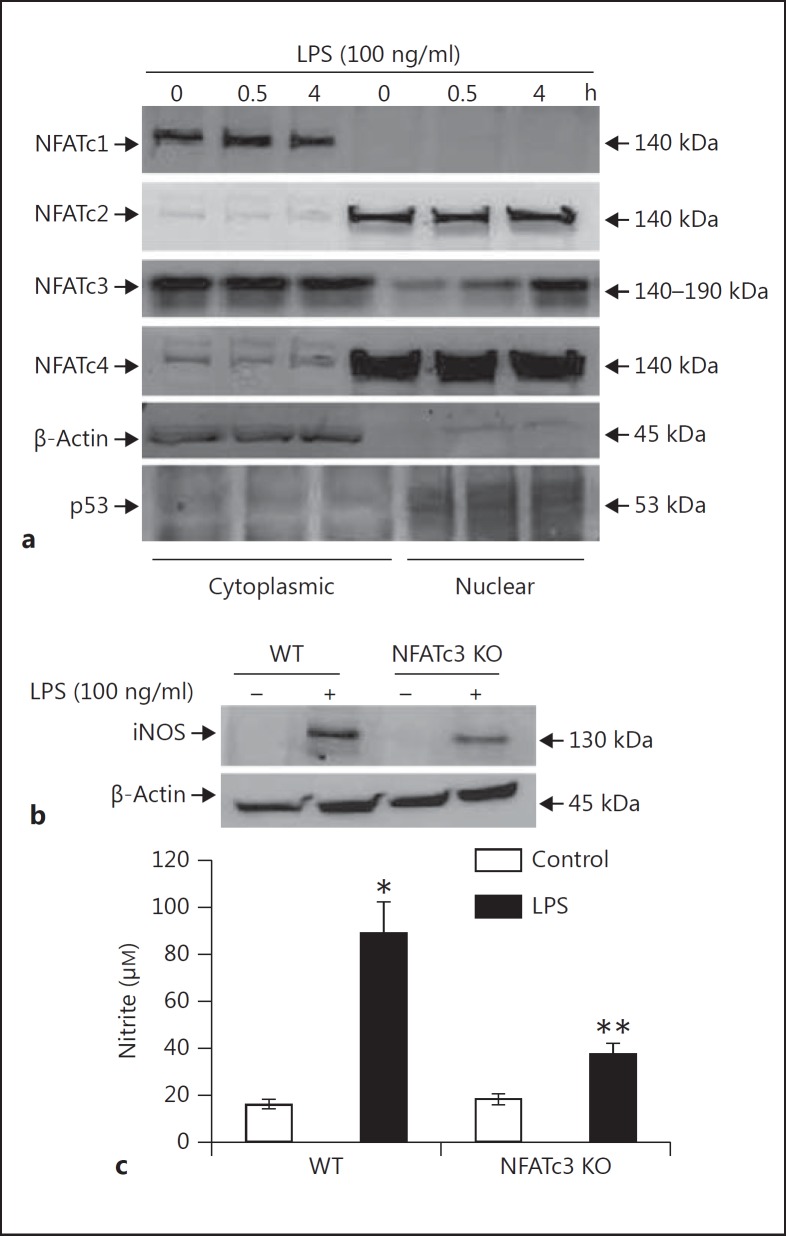
The transcription factor NFAT gene family consists of 5 members; NFATc1-c4 are controlled by calcium levels, while NFAT5 is activated by osmotic stress [15, 16]. So, to check which isoform contributed to the expression of iNOS in macrophages, we checked the nuclear translocation of the cytosolic NFAT isoforms. The WT BMDM were treated with LPS, and the nuclear and cytosolic proteins were isolated and probed for the translocation of different NFAT isoforms. Only NFATc3 translocated to the nucleus in response to LPS, compared to other members that were exclusively localized either in the cytosol or nucleus or both (fig. 2a). To further confirm that NFATc3 is actually involved in the regulation of iNOS, BMDM cultured from WT and NFATc3 KO mice were stimulated with LPS. The NFATc3 KO BMDM had significantly decreased expression of iNOS compared to WT BMDM, which also correlated with decreased NO release in culture medium (fig. 2b, c).
Fig. 2.
LPS causes NFATc3 nuclear translocation specifically, which regulates the iNOS expression. a BMDM were treated with 100 ng/ml LPS for the indicated time periods, and cells were harvested. Cytosolic and nuclear protein fractions were resolved and analyzed by immunoblotting with NFATc1, NFATc2, NFATc3 and NFATc4 antibodies. b WT (NFATc3+/+) and NFATc3−/− BMDM were stimulated with 100 ng/ml LPS and analyzed for iNOS expression as described in figure 1b. c WT (NFATc3+/+) and NFATc3−/− BMDM were stimulated with 100 ng/ml LPS and analyzed for NO release as described in figure 1d and f. * p ≤ 0.05, ** p ≤ 0.005.
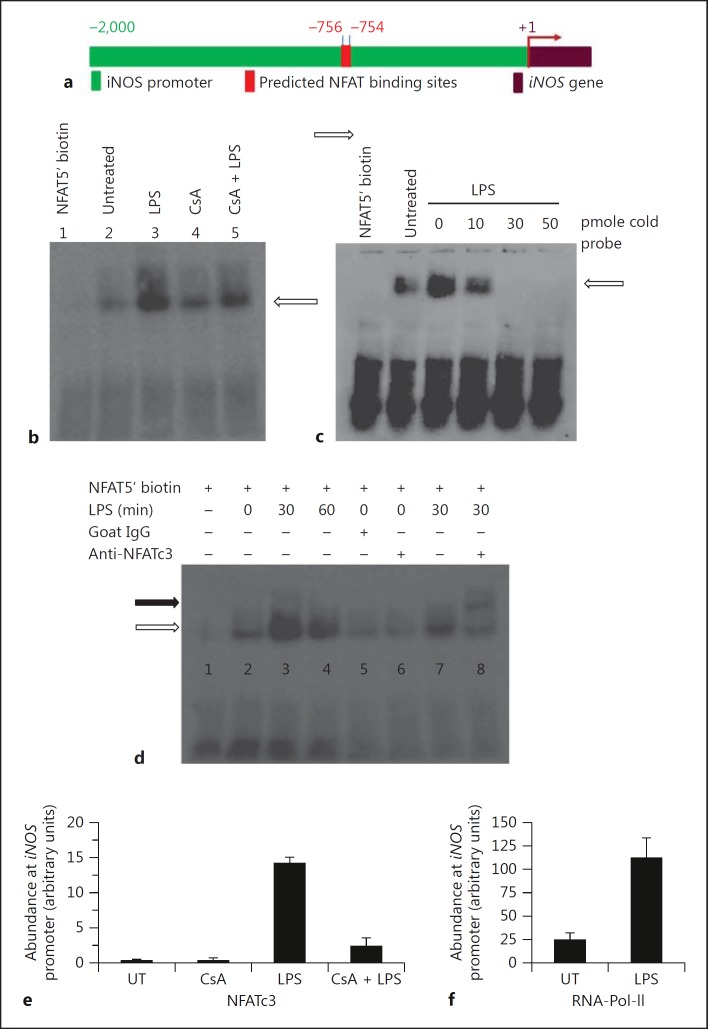
As LPS caused translocation of NFATc3 from the cytosol to the nucleus and associated upregulation of iNOS expression, we determined if the upregulation is mediated by binding of NFATc3 to the iNOS promoter using EMSA and ChiP assays on the iNOS promoter. In silico analysis of the genomic sequences (approx. 2 kb) flanking the mouse iNOS mRNA using the online software Patch 1.0. and MatInspector revealed two putative NFAT cognate binding sites upstream of the start codon (ATG) at around −756 and −754 (fig. 3a). In EMSA, binding of the biotin-labeled NFAT probe with the nuclear extract was significantly reduced in BMDM pretreated with CsA (NFAT inhibitor) in response to treatment with LPS compared to LPS alone (fig. 3b, lane 3 vs. lane 5). To further confirm the specificity of DNA-protein interactions, the unlabeled (cold) probe was used in a 10- to 50-pmol excess of labeled probe as competitor. The binding of the labeled probe was reduced and abolished in the presence of an increasing molar concentration of unlabeled probe (fig. 3c). The specificity of binding of the biotinylated probe with translocated NFATc3 was also confirmed using the anti-NFATc3 antibody, which caused a super shift of the DNA-protein complex (fig. 3d, lane 7 vs. lane 8). ChIP was performed to show the recruitment of NFATc3 on the iNOS promoter, by designing primers encompassing putative NFAT binding sites (-756 and −754). As shown, in BMDM treated with LPS, there was a high abundance of NFATc3 at the iNOS promoter, which was significantly reduced by CsA (fig. 3e); RNA Polymerase II was used as positive control for the ChIP assay (fig. 3f). We used the unprecipitated chromatin as input control for DNA, and used it for RT-qPCR, and all the reactions had similar crossing point values, indicating that an equal amount of DNA-protein complex was used for pull down (data not shown). To check the specificity of the ChIP, we designed primers from an unrelated downstream DNA sequence from the iNOS gene and subjected the immunoprecipitated DNA-protein complex to RT-qPCR, and there was no significant amplification (undetected signal by machine) with the control primers, indicating the specificity of ChIP.
Fig. 3.
DNA binding activity of NFATc3 is directly involved in transcriptional regulation of iNOS. a Schematic representation of the iNOS promoter (approx. 2-kb genomic region) representing cognate NFATc3 binding sites. b BMDM were treated with LPS and/or CsA for 30 min; nuclear extracts were prepared as described in Materials and Methods, and an equal amount of extract was incubated with biotin-labeled double-stranded DNA containing a consensus binding site for NFAT. The NFAT protein-DNA complex was resolved on a 5% native acrylamide gel, transferred onto a Nylon membrane and developed using a Pierce chemiluminescent detection kit. c Equal amounts of nuclear extract protein were incubated with biotin-labeled double-stranded DNA containing a consensus binding site for NFAT, competing against the unlabeled (cold) probe in increasing molar concentrations and processed as described in (b). d The specificity of DNA binding was determined using NFATc3 antibody, included in the oligo-protein reaction mixture. The black arrow represents the super shift band, and white arrows represent binding activity of the NFAT probe. e, f Recruitment of NFATc3 to iNOS promoter was assessed by ChIP. The eluted DNA was subjected to real-time PCR, and the abundance of NFATc3 (e) and RNA Polymerase II (RNA-Pol-II; f) at the iNOS promoter was calculated using the ratio of amplification efficiency (crossing point value) of the sample over nonimmune IgG. UT = Untreated.
Deficiency of NFATc3 Results in Attenuated Bactericidal Activity by Macrophages
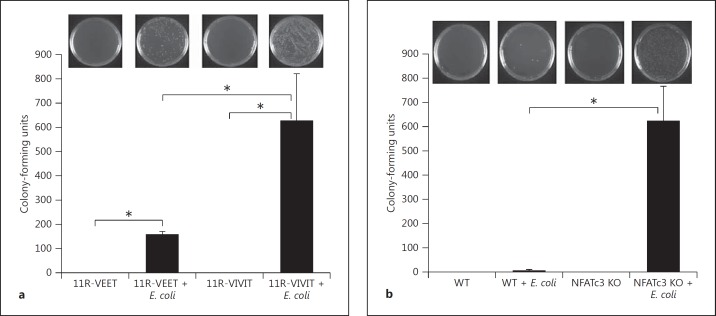
Macrophages are inflammatory/immune effector cells that are necessary for bactericidal activity in an event of microbial infection. This activity is largely attributable to iNOS and nicotinamide adenine dinucleotide phosphate (NADPH) oxidase, which together are the major source for reactive oxygen/nitrogen intermediates [6]. Thus, we examined the bactericidal activity of macrophages by pretreating them with a NFAT-specific inhibitor peptide (11R-VIVIT) and incubating with the bacterium E. coli. The macrophages that were treated with the inhibitor had attenuated bactericidal activity compared to control peptide (fig. 4a). In the second experiment, WT and NFATc3 KO BMDM were challenged with E. coli. The NFATc3 KO macrophages had attenuated bactericidal activity compared to WT macrophages (fig. 4b). These results clearly indicate that NFATc3 is required for bactericidal activity by macrophages.
Fig. 4.
Deficiency of NFATc3 results in attenuated bactericidal activity. a WT macrophages were treated with 11R-VIVIT (specific NFAT inhibitor) and incubated with E. coli, and bactericidal activity was analyzed by plating macrophage lysate on bacterial culture plates. The NFAT inhibitor-treated macrophages had attenuated bactericidal activity compared to the group treated with control peptide (11R-VEET). b Macrophages cultured from WT and NFATc3 KO mice were treated with E. coli, and bactericidal activity was analyzed by plating macrophage lysate on bacterial culture plates. The NFATc3-deficient macrophages had significantly attenuated bactericidal activity compared to WT macrophages. Error bars represent the standard deviation. * p < 0.01. n = 3.
Inhibition of NFATc3 Results in Compromised Bacterial Clearance
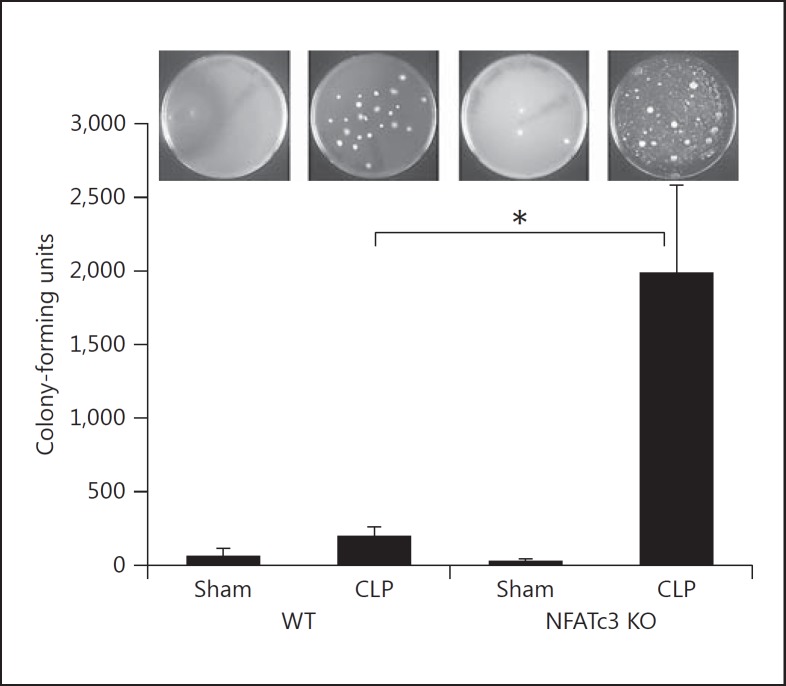
To further confirm the role of NFATc3 in mediating the pathogenesis of severe sepsis, we used WT and NFATc3 KO mice. The mice were subjected to abdominal sepsis by the CLP model [28]. The NFATc3 KO mice were more susceptible to sepsis and appeared moribund compared to WT mice. The lungs from these mice were excised and assessed for bacterial clearance. The NFATc3 KO mice subjected to CLP had significant polymicrobial infection in the lungs compared to the WT CLP mice (fig. 5). These data show a critical role of NFATc3 in controlling polymicrobial sepsis.
Fig. 5.
Inhibition of NFATc3 results in bacterial infection in lungs during sepsis. The WT and NFATc3 KO mice were subjected to CLP. After 24 h the mice were euthanized, and bacterial infection in the lungs was assessed by plating lung homogenate on sheep blood agar plates. The NFATc3 KO mice subjected to sepsis by CLP had severe bacterial infection compared to WT mice. Error bars represent the standard deviation. * p < 0.01. n = 3.
Discussion
Severe sepsis is a serious medical condition which often leads to an overwhelming systemic inflammatory response, multiple organ failure and death. The most common elements of sepsis are Gram-negative bacteria and the bacterial endotoxin (LPS), which activate the immune responses resulting in production of proinflammatory immune modulators [1]. The lung, abdominal cavity and primary infections of the bloodstream are the most common sites of infection in bacterial sepsis. There are currently no effective, specific treatments that are based on molecular pathogenesis; thus it is imperative to understand the molecular mechanism mediating the bacterial clearance during sepsis. Macrophages are innate immune effector cells that are involved in generating beneficial inflammation that is necessary for pulmonary host defense [33]. In contrast, dysregulated macrophages can contribute to harmful collateral lung damage due to either exuberant or persistent lung inflammation. Engagement of LPS with TLR4 on the surface of a pulmonary macrophage results in activation of NADPH oxidase by driving a protein-protein interaction among the cytoplasmic and membrane components of NADPH oxidase that results in production of ROS. It is well known that NO, which is a major contributing factor of cellular ROS in macrophages, is produced from iNOS, also known as type II NOS. NO plays important roles in many different aspects of mammalian biology and is an important component of host defense against intracellular pathogens; however, with subsequent formation of peroxynitrite and poly(adenosine diphosphate ribose) it is critically implicated in the pathophysiology of acute lung injury and sepsis [6]. Macrophages are intrinsically more responsive to endotoxin than neutrophils, basophils or eosinophils [34] due to increased expression of TLR4, which can be markedly upregulated by treatment with certain stimuli [35].
Until very recently, NF-κB proteins were the only Rel-like transcription factors known to be involved in the innate immune response in macrophages and other immune cells; however, recent publications have emphasized the role of the transcription factor NFAT in macrophages as well [20, 23]. NFATs play a key role in the development and function of immune cells [16] and were initially identified in modulating T and B cell gene expression. However, their role in the modulation of gene expression in macrophages during sepsis has not been explored. KO mice of each NFAT family member exhibit distinct phenotypic defects, possibly due to the different expression profiles of each NFAT among different cell types [36, 37, 38]. Recently it has been shown that expression of TLR-induced gene expression, including iNOS, in macrophages is regulated by NFAT5, and this finding is an intriguing phenomenon as it is independent of its osmoregulatory function [39].
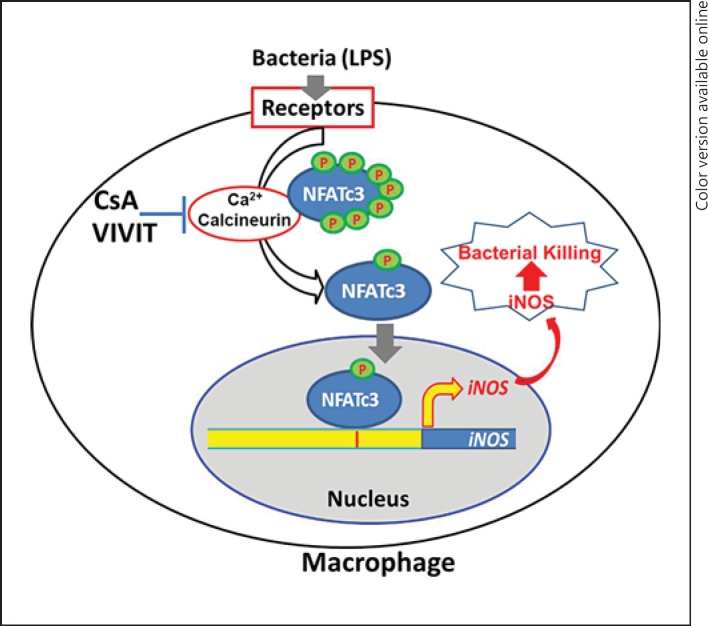
Here, we show that NFATc3 actively translocates from the cytosol to the nucleus of LPS-treated macrophages, expression of iNOS in macrophages is mediated by NFATc3 and, more importantly, blocking NFATc3 results in attenuated bactericidal activity. The NFATc3-deficient macrophages cultured from the KO mice had reduced iNOS expression/NO release and exhibited poor bactericidal activity. We tested the biological significance of these in vitro findings in a whole-animal model of severe sepsis due to CLP using NFATc3 KO mice. The KO mice showed decreased bacterial clearance in lungs compared to the control group, indicating that NFATc3 plays a crucial role in containment of bacterial sepsis. Our findings are summarized in the diagram shown in figure 6. During the onset of sepsis by bacterial infection, the transcription factor NFATc3 in macrophages is activated by the change in calcium levels, which results in its translocation to the nucleus, resulting in the transcriptional upregulation of iNOS. This results in robust generation of NO and other reactive oxygen/nitrogen intermediates, resulting in efficient bactericidal activity, thus contributing to the resolution of harmful inflammation.
Fig. 6.
Transcription factor NFATc3 regulates macrophage bactericidal activity. During the onset of sepsis by bacterial infection, the transcription factor NFATc3 in macrophages is activated by the change in the calcium levels, which results in its translocation to the nucleus, resulting in the transcription of iNOS. This results in robust generation of NO and other reactive oxygen/nitrogen intermediates, resulting in efficient bactericidal activity and thus contributing to the resolution of harmful bacterial infection and inflammation.
Disclosure Statement
The authors declare that there is no conflict of interests.
Supplementary Material
Supplementary data
Acknowledgements
This work was supported by National Institutes of Health Grants R01 HL075557, HL068610 and T32HL082547 and Department of Veterans Affairs Merit Review Grant 5I01BX000108.
References
- 1.Cohen J. The immunopathogenesis of sepsis. Nature. 2002;420:885–891. doi: 10.1038/nature01326. [DOI] [PubMed] [Google Scholar]
- 2.Fang FC, Vazquez-Torres A. Nitric oxide production by human macrophages: there's no doubt about it. Am J Physiol Lung Cell Mol Physiol. 2002;282:L941–L943. doi: 10.1152/ajplung.00017.2002. [DOI] [PubMed] [Google Scholar]
- 3.Sadowitz B, Roy S, Gatto LA, Habashi N, Nieman G. Lung injury induced by sepsis: lessons learned from large animal models and future directions for treatment. Expert Rev Anti Infect Ther. 2011;9:1169–1178. doi: 10.1586/eri.11.141. [DOI] [PubMed] [Google Scholar]
- 4.Toussaint S, Gerlach H. Immunoglobulins in adult sepsis and septic shock. Curr Infect Dis Rep. 2012;14:522–529. doi: 10.1007/s11908-012-0287-z. [DOI] [PubMed] [Google Scholar]
- 5.Pitt BR, St Croix., CM Complex regulation of iNOS in lung. Am J Respir Cell Mol Biol. 2002;26:6–9. doi: 10.1165/ajrcmb.26.1.f224. [DOI] [PubMed] [Google Scholar]
- 6.MacMicking J, Xie Q-W, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 7.Baron RM, Carvajal IM, Liu X, Okabe RO, Fredenburgh LE, Macias AA, Chen Y-H, Ejima K, Layne MD, Perrella MA. Reduction of nitric oxide synthase 2 expression by distamycin A improves survival from endotoxemia. J Immunol. 2004;173:4147–4153. doi: 10.4049/jimmunol.173.6.4147. [DOI] [PubMed] [Google Scholar]
- 8.Farley KS, Wang LF, Razavi HM, Law C, Rohan M, McCormack DG, Mehta S. Effects of macrophage inducible nitric oxide synthase in murine septic lung injury. Am J Physiol Lung Cell Mol Physiol. 2006;290:L1164–L1172. doi: 10.1152/ajplung.00248.2005. [DOI] [PubMed] [Google Scholar]
- 9.Diefenbach A, Schindler H, Donhauser N, Lorenz E, Laskay T, MacMicking J, Röllinghoff M, Gresser I, Bogdan C. Type 1 interferon (IFNα/β) and type 2 nitric oxide synthase regulate the innate immune response to a protozoan parasite. Immunity. 1998;8:77–87. doi: 10.1016/s1074-7613(00)80460-4. [DOI] [PubMed] [Google Scholar]
- 10.Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci USA. 2000;97:8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 12.D'Alessio FR, Tsushima K, Aggarwal NR, Mock JR, Eto Y, Garibaldi BT, Files DC, Avalos CR, Rodriguez JV, Waickman AT, Reddy SP, Pearse DB, Sidhaye VK, Hassoun PM, Crow MT, King LS. Resolution of experimental lung injury by monocyte-derived inducible nitric oxide synthase. J Immunol. 2012;189:2234–2245. doi: 10.4049/jimmunol.1102606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Fortin CF, McDonald PP, Fulop T, Lesur O. Sepsis, leukocytes, and nitric oxide (NO): an intricate affair. Shock. 2010;33:344–352. doi: 10.1097/SHK.0b013e3181c0f068. [DOI] [PubMed] [Google Scholar]
- 15.Feske S, Rao A, Hogan PG. The Ca2+-calcineurin-NFAT signalling pathway. New Compr Biochem. 2007;41:365–401. [Google Scholar]
- 16.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 17.Semenova SB, Kiselev KI, Mozhaeva GN. Low-conductivity calcium channels in the macrophage plasma membrane: activation by inositol-1,4,5-triphosphate. Neurosci Behav Physiol. 1999;29:339–345. doi: 10.1007/BF02465347. [DOI] [PubMed] [Google Scholar]
- 18.Lee C, Liu QH, Tomkowicz B, Yi Y, Freedman BD, Collman RG. Macrophage activation through CCR5- and CXCR4-mediated gp120-elicited signaling pathways. J Leukoc Biol. 2003;74:676–682. doi: 10.1189/jlb.0503206. [DOI] [PubMed] [Google Scholar]
- 19.Serafini N, Dahdah A, Barbet G, Demion M, Attout T, Gautier G, Arcos-Fajardo M, Souchet H, Jouvin MH, Vrtovsnik F, Kinet JP, Benhamou M, Monteiro RC, Launay P. The TRPM4 channel controls monocyte and macrophage, but not neutrophil, function for survival in sepsis. J Immunol. 2012;189:3689–3699. doi: 10.4049/jimmunol.1102969. [DOI] [PubMed] [Google Scholar]
- 20.Minematsu H, Shin MJ, Celil Aydemir AB, Kim KO, Nizami SA, Chung GJ, Lee FY. Nuclear presence of nuclear factor of activated T cells (NFAT) c3 and c4 is required for Toll-like receptor-activated innate inflammatory response of monocytes/macrophages. Cell Signal. 2011;23:1785–1793. doi: 10.1016/j.cellsig.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulrich JD, Kim MS, Houlihan PR, Shutov LP, Mohapatra DP, Strack S, Usachev YM. Distinct activation properties of the nuclear factor of activated T-cells (NFAT) isoforms NFAT3 and NFAT4 in neurons. J Biol Chem. 2012;287:37594–37609. doi: 10.1074/jbc.M112.365197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonnet S, Rochefort G, Sutendra G, Archer SL, Haromy A, Webster L, Hashimoto K, Bonnet S, Michelakis ED. The nuclear factor of activated T cells in pulmonary arterial hypertension can be therapeutically targeted. Proc Natl Acad Sci USA. 2007;104:11418–11423. doi: 10.1073/pnas.0610467104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elloumi HZ, Maharshak N, Rao KN, Kobayashi T, Ryu HS, Muhlbauer M, Li F, Jobin C, Plevy SE. A cell permeable peptide inhibitor of NFAT inhibits macrophage cytokine expression and ameliorates experimental colitis. PLoS One. 2012;7:e34172. doi: 10.1371/journal.pone.0034172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Musiek ES, Brooks JD, Joo M, Brunoldi E, Porta A, Zanoni G, Vidari G, Blackwell TS, Montine TJ, Milne GL, McLaughlin B, Morrow JD. Electrophilic cyclopentenone neuroprostanes are anti-inflammatory mediators formed from the peroxidation of the omega-3 polyunsaturated fatty acid docosahexaenoic acid. J Biol Chem. 2008;283:19927–19935. doi: 10.1074/jbc.M803625200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadikot RT, Zeng H, Yull FE, Li B, Cheng DS, Kernodle DS, Jansen ED, Contag CH, Segal BH, Holland SM, Blackwell TS, Christman JW. p47phox deficiency impairs NF-kappa B activation and host defense in Pseudomonas pneumonia. J Immunol. 2004;172:1801–1808. doi: 10.4049/jimmunol.172.3.1801. [DOI] [PubMed] [Google Scholar]
- 26.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 27.Campbell PA, Canono BP, Drevets DA. Measurement of bacterial ingestion and killing by macrophages. Curr Protoc Immunol. 2001 doi: 10.1002/0471142735.im1406s12. chapter 14:unit 14.6. [DOI] [PubMed] [Google Scholar]
- 28.Rittirsch D, Huber-Lang MS, Flierl MA, Ward PA. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat Protoc. 2009;4:31–36. doi: 10.1038/nprot.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Farmer JD, Jr, Lane WS, Friedman J, Weissman I, Schreiber SL. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 30.Noguchi H, Matsushita M, Okitsu T, Moriwaki A, Tomizawa K, Kang S, Li ST, Kobayashi N, Matsumoto S, Tanaka K, Tanaka N, Matsui H. A new cell-permeable peptide allows successful allogeneic islet transplantation in mice. Nat Med. 2004;10:305–309. doi: 10.1038/nm994. [DOI] [PubMed] [Google Scholar]
- 31.Xie QW, Kashiwabara Y, Nathan C. Role of transcription factor NF-kappa B/Rel in induction of nitric oxide synthase. J Biol Chem. 1994;269:4705–4708. [PubMed] [Google Scholar]
- 32.Xie Q, Nathan C. The high-output nitric oxide pathway: role and regulation. J Leukoc Biol. 1994;56:576–582. doi: 10.1002/jlb.56.5.576. [DOI] [PubMed] [Google Scholar]
- 33.Parihar A, Eubank TD, Doseff AI. Monocytes and macrophages regulate immunity through dynamic networks of survival and cell death. J Innate Immun. 2010;2:204–215. doi: 10.1159/000296507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabroe I, Jones EC, Usher LR, Whyte MK, Dower SK. Toll-like receptor (TLR)2 and TLR4 in human peripheral blood granulocytes: a critical role for monocytes in leukocyte lipopolysaccharide responses. J Immunol. 2002;168:4701–4710. doi: 10.4049/jimmunol.168.9.4701. [DOI] [PubMed] [Google Scholar]
- 35.Mita Y, Dobashi K, Shimizu Y, Nakazawa T, Mori M. Toll-like receptor 2 and 4 surface expressions on human monocytes are modulated by interferon-gamma and macrophage colony-stimulating factor. Immunol Lett. 2001;78:97–101. doi: 10.1016/s0165-2478(01)00241-3. [DOI] [PubMed] [Google Scholar]
- 36.Abbott KL, Friday BB, Thaloor D, Murphy TJ, Pavlath GK. Activation and cellular localization of the cyclosporine A-sensitive transcription factor NF-AT in skeletal muscle cells. Mol Biol Cell. 1998;9:2905–2916. doi: 10.1091/mbc.9.10.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amasaki Y, Adachi S, Ishida Y, Iwata M, Arai N, Arai K, Miyatake S. A constitutively nuclear form of NFATx shows efficient transactivation activity and induces differentiation of CD4(+)CD8(+) T cells. J Biol Chem. 2002;277:25640–25648. doi: 10.1074/jbc.M201860200. [DOI] [PubMed] [Google Scholar]
- 38.Obasanjo-Blackshire K, Mesquita R, Jabr RI, Molkentin JD, Hart SL, Marber MS, Xia Y, Heads RJ. Calcineurin regulates NFAT-dependent iNOS expression and protection of cardiomyocytes: co-operation with Src tyrosine kinase. Cardiovasc Res. 2006;71:672–683. doi: 10.1016/j.cardiores.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 39.Buxade M, Lunazzi G, Minguillon J, Iborra S, Berga-Bolanos R, Del Val M, Aramburu J, Lopez-Rodriguez C. Gene expression induced by toll-like receptors in macrophages requires the transcription factor NFAT5. J Exp Med. 2012;209:379–393. doi: 10.1084/jem.20111569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data