Abstract
Interleukin 22 (IL-22) is a cytokine that regulates tissue homeostasis at barrier surfaces. A variety of IL-22 producing cell types are known, but identification on the single cell level remains difficult. We therefore generated a fate reporter mouse that would allow the identification of IL-22 producing cells and their fate mapping in vivo. To trace IL-22 expressing cells, a sequence encoding Cre recombinase was cloned into the Il22 locus and Il22Cre mice were crossed with reporter mice expressing enhanced yellow fluorescence protein (eYFP) under control of the endogenous Rosa26 promoter. In IL22CreR26ReYFP mice, the fluorescent reporter permanently labels cells that have switched on Il22 expression irrespective of cytokine production. Despite a degree of underreporting, eYFP expression was detectable in non-immune mice and restricted to innate lymphoid cells (ILC3) in the gut and γδ T cells in skin or lung. Upon skin challenge with imiquimod, eYFP+ γδ and CD4 T cells expanded in the skin. Infection with Citrobacter rodentium was initially controlled by ILC3, followed by expansion of eYFP+ CD4 T cells, which were induced in innate lymphoid follicles (ILF) in the colon. No eYFP expression was detected in small intestinal Th17 cells and they did not expand in the immune response. Colonic eYFP+ CD4 T cells exhibited plasticity during infection with expression of additional cytokines in contrast to ILC3, which remained largely stable. Single cell qPCR analysis of eYFP+ CD4 T cells confirmed their heterogeneity, suggesting IL-22 expression is not strictly confined to particular subsets or a dedicated Th22 subset.
INTRODUCTION
Interleukin-22 (IL-22) is a cytokine expressed by immune cells but acting on non-haematopoetic cells. The receptor for IL-22 is expressed in barrier sites such as skin, intestine, lung as well as in liver, pancreas and kidney (1, 2). IL-22 production is attributed to many immune cells types such as CD4, CD8 and γδ T cells, NK cells and subsets of innate lymphoid cells (ILC) (3). Thus, the expression pattern of IL-22 and its receptor creates signaling directionality from the immune system to the tissues in line with the important function IL-22 has in maintaining tissue integrity. IL-22 plays an important role in the homeostasis of mucosal surfaces. During inflammation, IL-22 induces the expression of acute phase proteins, antimicrobial peptides and chemokines (4), which support resolution of the local inflammation, repair of injured tissue and re-establishment of homeostasis. IL-22 is required for protective immune responses against certain extracellular bacteria (5-9) and prevent the dissemination of intestinal microbiota (10). On the other hand, dysregulated production of IL-22 is associated with certain human auto-inflammatory diseases, including rheumatoid arthritis (RA), inflammatory bowel disease (IBD), and psoriasis (2, 11-13).
Despite the undisputed biological importance of IL-22 it remains difficult to follow its expression in vivo either in steady state or during inflammatory responses, due to technical problems of intracellular staining. An additional complication is the issue of effector cell plasticity, which makes it difficult to unequivocally assign IL-22 production to different subsets. We have previously addressed this issue for IL-17 producing cells by generating a fate reporter that marked cells that had initiated the IL-17 program with eYFP expression (14). This allowed easy identification of such cells ex vivo and further determination of their effector program irrespective of ongoing IL-17 production.
Here we have employed the same strategy to generate a knock-in mouse strain bearing a gene encoding Cre recombinase in the Il22 locus and breeding those mice with reporter mice expressing eYFP from the Rosa26 promoter to monitor expression of IL-22 in steady state and during infection with Citrobacter rodentium. Our data demonstrate a substantial expansion of eYPF+ CD4 T cells in the large intestinal lamina propria (LI LP) from day 5 after infection, whereas eYFP+ ILC are present in uninfected mice and do not substantially expand on infection. IL-22 expressing CD4 T cells predominantly associate with a Th17 profile, but show pronounced plasticity in the course of infection in contrast to ILC that remain committed to IL-22 production. Single cell qPCR analysis of gene expression for CD4 T cell subsets indicate substantial heterogeneity, which suggests that IL-22 expression is not strictly confined to particular subsets or a dedicated Th22 subset.
MATERIALS AND METHODS
Mice
Codon improved Cre recombinase (iCre)(15) was inserted into the first exon of the Il22 locus in by homologous recombination in B6/N mouse embryonic stem cells. The neo cassette was removed via FLPe-mediated recombination. To visualize Cre-mediated recombination, Il22Cre mice were intercrossed with R26ReYFP reporter mice (expressing eYFP from the Rosa26 promoter)(16), generating Il22CreR26ReYFP reporter mice.
C57Bl/6 (B6), B6.Rag2−/−CD45.1 and Il22CreR26ReYFP reporter mice were bred in the animal facility of the Medical Research Council National Institute for Medical Research. All mice were kept under specific pathogen-free conditions. All animal experiments were approved by the local Ethical Review panel at NIMR in accordance with the Institutional Committees on Animal Welfare of the UK Home Office (the Home Office Animals Scientific Procedures Act, 1986).
Generation of bone marrow chimeras
Bone marrow chimeras were generated by iv injection of 107 T cell depleted bone marrow cells from homozygous or heterozygous R26ReYFP reporter mice into B6.Rag2−/− CD45.1 hosts that were sublethally irradiated with 5Gy. Mice were used in experiment from 7 weeks after reconstitution.
Citrobacter rodentium infection
C. rodentium strain DBS100 (ATCC 51459; American Type Culture Collection) was cultured in LB broth overnight, diluted 1:400 with fresh LB and cultured additional 3-4h. Bacterial concentration was determined by measuring the optical density at 600 nm. The cultures were serially diluted and plated to confirm the inoculum. IL-22 fate reporter, Il22+/− or wild-type mice were gavaged with 2×109 CFU in 200μl phosphate-buffered saline (PBS). After infection, mice were weighed daily and weight loss was determined. Distal colon pieces were weighed and homogized in sterile PBS and serial dilutions were plated in duplicates onto Brilliance E Coli/ Coliform medium (Fisher Scientific) plates for measurement of colony-forming units. C. rodentium colonies were identified based on morphology and color after 18-24 h of incubation at 37°C.
Imiquimod induced psoriasiform-like skin inflammation
Mouse dorsal skin was shaved and 50 mg per mouse Aldara crème containing 5% Imiquimod (Meda AB) was applied daily for 5 consecutive days to induce psoriasiform skin inflammation. On day 5, a 8mm biopsy punch (Kai medical) was used to collect full-thickness skin biopsies of the treated area. Two skin biopsies were minced and then digested with a cocktail containing 400 μg/ml Liberase TL (Roche) and 1 mg/ml collagenase D (Roche) in IMDM medium for 2 hours at 37 °C with shaking, and mashed through a 70μl cell strainer to obtain single cell suspensions. Leukocytes were enriched using 36.5% Percoll (Amersham) density gradient centrifugation.
Flow cytometry analysis of tissue-resident cells
Lung, pancreas, kidney, liver and spleen were minced. Minced lung was incubated with a digestion cocktail containing 400 μg/ml Liberase TL (Roche) in IMDM medium for 30 minutes at 37°C with shaking. Lung, pancreas, kidney, liver, spleen, lymph nodes, thymus and Payer’s patches were mashed through a 70μl cell strainer to obtain single cell suspensions. Cells from skin were isolated as described above.
Gut-resident lymphocytes were prepared by cutting the small intestine or large intestine (colon and cecum) in small pieces and incubation in digestion buffer (IMDM supplemented with 3% (vol/vol) FCS, 5 mM EDTA, 25 mM HEPES, penicillin/streptomycin and 0.145mg/ml DTT for 20-40 min at 37°C, shaking. The supernatant containing epithelial and intraepithelial cells (IEL) was enriched for IEL using 36.5% Percoll (Amersham) density gradient centrifugation. The remaining tissue was further digested with a cocktail containing 400 μg/ml Liberase TL (Roche) and DNase I (10 U/ml; Sigma) for 30 min at 37 °C. For flow cytometry analysis cells were stained for surface molecules (see list of antibodies in Table 1) in the presence of 10% Fc block and acquired on a FACSCantoII (BD).
Table 1.
| Antigen | Clone | Fluorochrome | Supplier |
|---|---|---|---|
| CD3ε | 145-2C11 | APC | BioLegend |
| CD4 | GK1.5 | APC, V500 | BioLegend, BD Biosciences |
| CD8α | 53-6.7 | PerCP/Cy5.5, APC | BioLegend |
| CD11b | M1/70 | APC | BioLegend |
| CD11c | N418 | APC | BioLegend |
| CD19 | 6D5 | APC | BioLegend |
| CD45.2 | 104 | PE/Cy7, APC/Cy7 | BioLegend |
| cKit | 2B8 | PE | BioLegend |
| CD49b | DX5 | APC | BioLegend |
| Foxp3 | 150D | AlexaFluor 647 | BioLegend |
| γδTCR | UC7-13D5 | PE/Cy7, BV421, APC | BioLegend |
| Ly-6G/Ly-6C (Gr1) | RB6-8C5 | APC | BioLegend |
| IFNγ | XMG1.2 | PE | BD Biosciences |
| IL-17A | TC11-18H10.1 | AlexaFluor 647, Pacific Blue | BioLegend |
| IL-22 | AM-22 | APC | gift from JC Renauld (Brussels) |
| IL-4 | 11B11 | PE | BioLegend |
| IL-7R | A7R34 | PE, PE-Cy5 | BioLegend, eBioscience |
| IL-9 | RM9A4 | PE | BioLegend |
| NK1.1 | PK136 | PE, PerCP/Cy5.5 | BioLegend |
| NKp46 | 29A1.4 | Brilliant Violet 421 | BioLegend |
| Sca-1 | D7 | Pacific Blue | BioLegend |
| TCRβ | H57-597 | APC/Cy7, PerCP/Cy5.5, APC | BioLegend |
| Ter119 | TER-119 | APC | BioLegend |
| CD90.2 (Thy1.2) | 53-2.1 | PE/Cy7 | BioLegend |
| Rorγt | O31-378 | PE | BD Biosciences |
| GATA-3 | TWAJ | PerCP-eFluor710 | eBioscience |
In vitro T cell differentiation
Naïve T cells (CD4+ CD25− CD44low) were flow cytometry purified from spleen and lymph nodes of fate reporter mice and activated with plate-bound anti-CD3 (clone: 2C11 eBioscience) and anti-CD28 (clone: 37.51, Biolegend) (coated with 0.5 μg/ml and 5 μg/ml, respectively) in the presence of lineage polarizing cytokines as follows: IL-12 (5 ng/ml) for TH1, IL-4 (10 ng/ml) for TH2, IL-4 (10 ng/ml) and TGFβ (5 ng/ml) for TH9, TGFβ (10 ng/ml) for iTreg, IL-6 (20 ng/ml) and TGFβ (1 ng/ml) and IL1β (10 ng/ml), IL-23 (20 ng/ml) (all from R&D Systems) or FICZ (250 nM; Enzo) for TH17 cells. Alternatively, CD4+ cells were purified using EasySep Mouse CD4+ T Cell Enrichment Kit (STEMCELL Technologies) from spleen of fate reporter mice and activated as described above. Cells were cultured with anti-IFNγ (10 μg/ml, clone: XMG1.2, eBioscience), anti-IL-4 (10 μg/ml, clone: 11B11), IL-6, IL1β and FICZ, and with TGFβ (for TH17 cells) or with anti-TGFβ (10 μg/ml, clone: 1D11) for “TH22” cells. All cytokines were of mouse origin. Cells were cultured in Iscove’s modified Dulbecco medium (Sigma) supplemented with 5% FCS, 0.002M L-glutamine, 100U/ml penicillin, 100μg/ml streptomycin and 5×10−5M β-mercaptoethanol. Cells were analysed for intracellular cytokines on day 4.
Cytokine measurement
Cells were stimulated for 4 h with PdBU (500 ng/ml) and ionomycin (500 ng/ml) in the presence of brefeldin A (1 μg/ml), fixed with 4% PFA, permeabilized with 0.1% NP-40 or 0.1% saponin and stained for intracellular cytokines or transcription factors (antibodies in Table 1). Cells were acquired on a FACSCantoII (BD). The Th cell culture supernatants (Fig. 1D) were analysed for IL-22 using FlowCytomix according to the instructions of the manufacturer (Bender MedSystems, Austria).
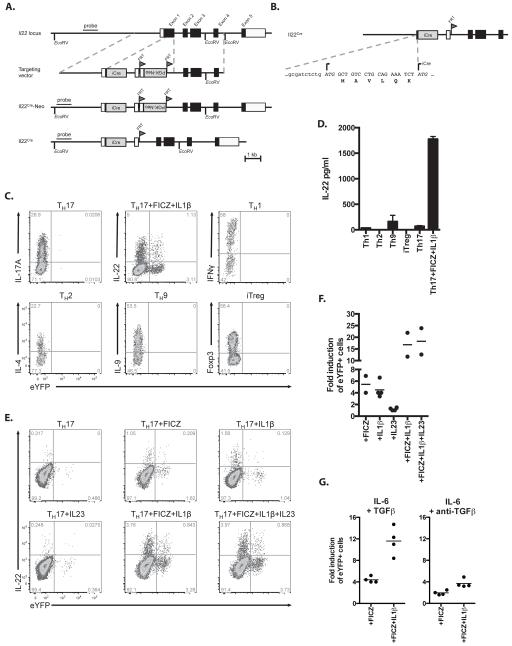
Figure 1. Fate-reporter eYFP+ cells are induced among IL22-producing cells.
A) Schematic presentation of the endogenous Il22 locus, the targeting vector, the targeted Il22 allele including the Neo resistance cassette (Il22Cre-Neo) and the final targeted allele (Il22Cre) after FLPe-mediated recombination. The iCre component is a minigene composed of a codon improved Cre followed by an intron and polyadenylation site derived from SV40 B) Sequence of Il22 exon 1 after targeting showing the original 6 amino acids after Il22 ATG-site and the respective start of the iCre minigene. C) FACS plots showing the expression of eYFP and intracellular cytokines or Foxp3 in TH1, TH2, TH9, TH17 or inducible regulatory T (iTreg) cells after 4 days of in vitro differentiation D) Concentration of IL22 in day 4 supernatants of in vitro differentiated TH1, TH2, TH9, TH17 or iTreg cells. Data shown as mean ± SEM are representative of 3 independent experiments. E) FACS plots showing the expression of eYFP and intracellular IL22 in naïve CD4+CD44loCD25− T cells cultured for 4 days in TH17 differentiating conditions (TGFβ+IL6) supplemented with FICZ, IL1β or IL23 or their combination. F,G) The fold induction in the percentage of eYFP+ cells cultured for 4 days in modified TH17 conditions (as indicated on the x axis) compared to TH17 cells cultured in the presence of TGFβ and IL6 only F) or in the presence of IL-6 and anti-TGFβ (G). Data shown are representative of 3 independent experiments (C, E).
eYFP+ and eYFP− CD4 T cells (CD45+CD4+TCRβ+) were FACS sorted from LI LP of C. rodentium infected fate reporter mice d14 p.i., and 25 000 cells were cultured on anti-CD3 coated wells for 18 hours. Medium only wells were included as controls. The culture supernatants were analysed for IL-22, IL-17A and IFNγ as described above.
For serum IL-22 measurement, blood was collected from both Citrobacter infected animals and steady-state control mice and processed by centrifugation to isolated the serum. The concentration of IL-22 in the serum was then determined by capture ELISA according to the manufacturer’s instructions (eBioscience).
Single cell gene expression analysis
eYFP+ CD4 T cells from large intestine lamina propria were FACS sorted and subsequently deposited into 96-well PCR plates using an automated cell deposition unit on a MoFlo sorter (Becton Dickinson, UK) containing 10 μl lysis buffer. Sorter provided single cells in >99% of the wells and no wells with more than one cell as assessed by routinely sorting fluorescent beads or cells prior to and after single cell sorting. Cells were processed for qRT-PCR analysis using Single Cell-to-CT™ qRT-PCR Kit (Ambion) according to the manufacturer’s protocol. A total of 175 single cells were analysed (d0; 75, d5; 44, d25; 56).
The resultant cDNA served as a template for the amplification of the genes of interest by real-time PCR with TaqMan Gene Expression assays and universal PCR Master Mix on an ABI-PRISM 7900 Sequence Analyzer (all from Applied Biosystems).
Immunohistology
Frozen sections of large intestine were prepared at 8μm thickness and fixed in 4% paraformaldehyde. Sections were counterstained with 4′6′-diamidino-2-phenylindole dihydrochloride (DAPI) and mounted using Fluoromount-G (Southern Biotechnology). Monoclonal antibodies were as listed in Table 1. Confocal images were obtained using a Leica TCS-SP2 microscope equipped with 405, 488, 594 and 647 nm lasers and image analysis was done in Adobe Photoshop.
Skin was fixed in neutral buffered formalin (Sigma) and tissue sections were stained with H&E for histopathology analysis. Epidermal and scale thickness was quantified as previously described (17).
Statistical Analysis
Statistical analysis was performed using Prism version 6.0 (GraphPad Software). If two groups were compared, significance was calculated using unpaired t test. If more than two groups were assessed, significance was calculated using one-way analysis of variance and Dunnett’s or Sidak’s corrected p value for multiple comparisons. For weight loss data, significance was calculated using two-way analysis of variance and Tukey’s multiple comparisons test.
RESULTS
Generating an IL-22 fate reporter mouse
To generate a reporter system which would allow not only the identification of IL-22 cytokine producing cells, but also to map the fate of these cells in vivo, we cloned a sequence encoding Cre recombinase into the Il22 locus (Il22Cre). To visualize Cre activity, the IL22Cre mice were then crossed with reporter mice expressing enhanced yellow fluorescence protein (eYFP) under the control of the endogenous Rosa26 promoter (R26ReYFP). In IL22CreR26ReYFP mice, the fluorescent reporter permanently labels cells that have switched on Il22 expression regardless of the present production status of this cytokine. The endogenous Il22 locus, the gene targeting vector, the targeted Il22 allele including the Neo resistance gene cassette (Il22cre-Neo) and the final targeted allele (IL22Cre) after FLPe-mediated recombination are schematically depicted (Fig.1A). Fig.1B shows the sequence of IL22 exon 1 with a linker consisting of six amino acids, placed between the original Il22 ATG-site and the respective start of the iCre mini gene.
We first tested reporter activity in vitro following stimulation of FACS selected naïve CD4 T cells for 3 days under Th17, Th1, Th2, Th9 or iTreg conditions. IL-22 had previously been identified as a Th17 cytokine (4). In accordance we detected eYFP expression in Th17 cells, but not in any other in vitro differentiated CD4 T cell subset (Fig.1C). This was confirmed by measuring the secreted IL-22 in the culture supernatants (Fig.1D). IL-22 expression by Th17 cells differentiated in the presence of IL-6 and TGFβ is normally low to absent, depending on the culture medium (18), whereas stimulation in the presence of agonists of the aryl hydrocarbon receptor (AhR) induces IL-22 expression (19). Culture under Th17 conditions with IL-1β had a similar effect as culture with FICZ, whereas IL-23 had no enhancing effect on IL-22/eYFP expression. Optimal expression was achieved by combining FICZ and IL-1β (Fig.1E,F). There was only very marginal induction of eYFP or IL-22 under “Th22” conditions using IL-6 and anti-TGFβ (Fig.1G).
However, there was only a small proportion of cells stained for intracellular IL-22 that co-expressed eYFP and in addition a fraction of eYFP+ cells that did not stain for IL-22, suggesting that there is a disconnection between the production of IL-22 cytokine and induction of the reporter. We had seen a degree-albeit less pronounced-of ‘underreporting’ also in the IL-17Cre fate reporter model. A likely explanation for incomplete reporting is the delay between cytokine induction and production of Cre, which is necessary to reveal the fluorescent reporter. In the case of IL-22 cytokine production may be of shorter duration, thus compromising effective Cre induction. Furthermore, IL-22 expression, like that of the closely related cytokine IL-10, may be monoallelic and therefore more severely compromised combined with the disruption of one allele by Cre.
Detecting IL22-eYFP+ cells in vivo under steady state conditions
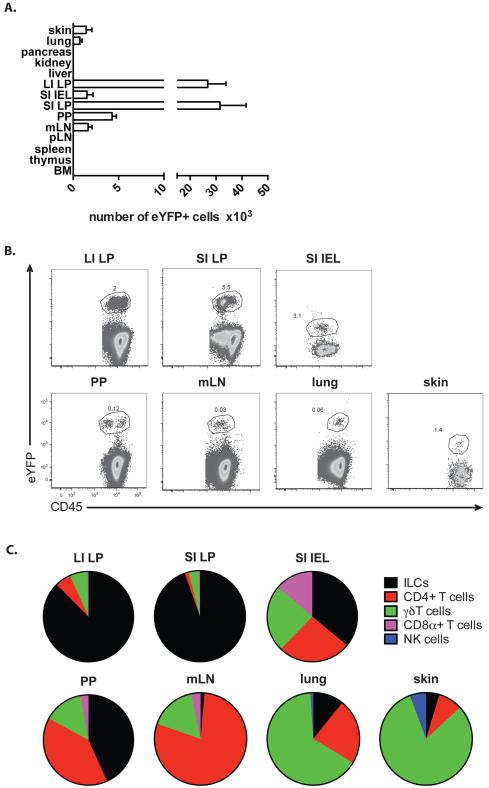
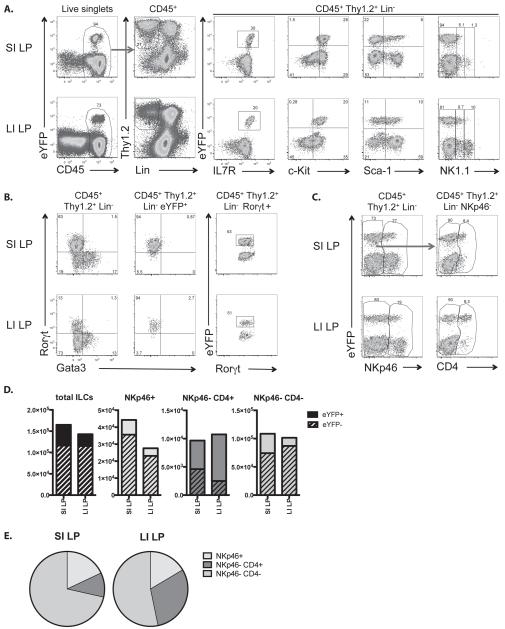
To assess which cells express IL-22 in unchallenged mice, different organs, including gut, lung, skin, lymph nodes, spleen, thymus, liver, pancreas, kidney and bone marrow were analysed for the presence of eYFP+ cells in. In concordance with the predominant function of IL-22 in maintaining epithelial barriers, eYFP+ cells were found only in tissues involved in barrier function, namely gut, mesenteric LN (mLN), lung and skin (Fig.2A,B). However, the cell type expressing eYFP was tissue-dependent (Fig.2C): in the gut the majority of eYFP+ cells were ILC, whereas in skin or lung the majority of eYFP+ cells were γδT cells. We did not detect eYFP+ cells in the resident Th17 population of the lamina propria (LP) in the small intestine (SI) with the caveat that a small number of such cells might have escaped detection due to underreporting. However, there were eYPF+ CD4 T cells in the intraepithelial lymphocyte (IEL) population and Peyer’s patches as well as CD8αα and γδ T cells and ILC (Fig.2B,C). Amongst the eYFP+ ILC fraction in the small intestinal lamina propria (SI LP) and large intestinal lamina propria (LI LP)- see gating strategy in Fig.3A) the majority was NKp46−CD4−, but also CD4+ (LTi) and NKp46+ ILC were present (Fig.3C-E). All eYFP+ ILC expressed RORγt and therefore can be allocated to the ILC3 subset (Fig.3B).
Figure 2. Prevalence of eYFP+ cells in untreated fate reporter mouse.
A) Bar graph showing the number of eYFP+ cells in various organs of untreated fate reporter mice. LI LP; large intestine lamina propria, SI IEL; small intestine intraepithelial lymphocytes, SI LP; small intestine lamina propria, PP; Peyer’s patches, mLN; mesenteric lymph nodes, pLN; peripheral lymph nodes, BM; bone marrow. Data shown as mean ± SEM are representative of 2-9 independent experiments. B) FACS plots showing the expression of eYFP and CD45 in various organs of untreated fate reporter mice. Data are representative of 2-9 independent experiments. C) Pie charts showing the relative number of eYFP+ cell types in intestinal tissues, skin and lung. Gating strategy to obtain different cell populations: Cells were first gated for lymphocytes (FSC-A vs. SSC-A) and singlets (FSC-A vs. FSC-W). These cells were analysed for their CD45 expression. CD45+ cells were analysed for their surface markers as follows: CD4+ T cells: TCRβ+ CD4+, CD8α+ T cells: TCRβ+ CD8α+, γδT cells: γδTCR+, NK cells: NK1.1+, ILCs: Lineage- Thy1.2+ (Lineage cocktail: CD11c, CD11b, DX5, Gr1, Ter119, γδTCR, TCRβ, CD3, CD8α and CD19). Data shown as mean are representative of 2-9 independent experiments.
Figure 3. eYFP+ ILC3 cells in untreated fate reporter mice.
A) Gating strategy for flow cytometry analysis. Cells were first gated for lymphocytes (FSC-A vs. SSC-A) and singlets (FSC-A vs. FSC-W). These cells were analysed for their CD45 expression. CD45+ cells were analysed for their Thy1.2 and lineage marker (CD11c, CD11b, DX5, Gr1, Ter119, γδTCR, TCRβ, CD3, CD8α and CD19) expression. Thy1.2+ lineage− cells were further analysed for eYFP, IL-7R, c-Kit, Sca-1 and NK1.1 expression. B) Thy1.2+ lineage− cells (left panels) and eYFP+ Thy1.2+ lineage− cells (middle panels) were further analysed for Rorγt and Gata3 expression. Right panel: eYFP expression on gated Thy1.2+ RORγt+ lineage− cells. C) Thy1.2+ lineage− cells analysed for eYFP and NKp46 expression (left panels). NKp46− cells analysed for eYFP and CD4 expression (right panels). D) Bar graphs showing the numbers of eYFP+ and eYFP− total ILCs and ILC3 subpopulations. E) Pie charts showing the relative number of eYFP+ ILC3 subpopulations in SI LP and LI LP. Data shown as mean are representative of at least 4 independent experiments (C, D).
Induction of eYFP expression during inflammatory responses
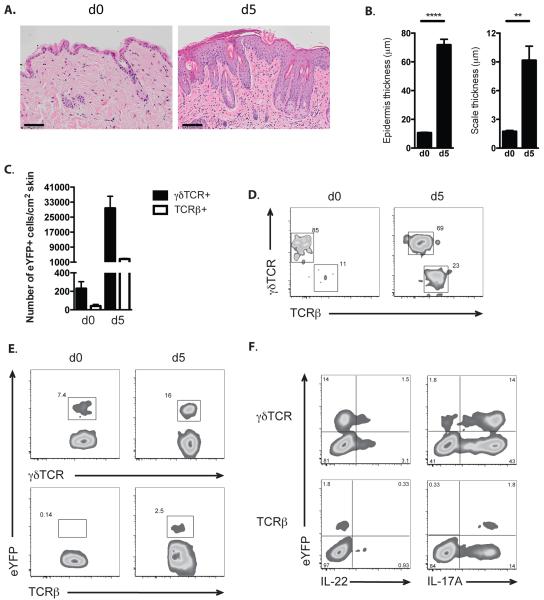
IL-22 expression has been documented in inflammatory skin conditions, such as psoriasis and the mouse model of psoriasiform inflammation induced by the TLR7 ligand imiquimod (20, 21). We therefore treated IL22CreR26ReYFP mice with imiquimod containing Aldara cream for 5 consecutive days and analysed eYFP expression on day 5. This treatment resulted in increased epidermal thickness and scaling (Fig.4AB) and expansion of primarily γδ T cells that expressed eYFP and a considerably smaller number of eYFP+ CD4 T cells, (Fig.4C-E) consistent with reports for a predominant role for γδ T cells in this model (20, 22) As recently reported (17) we did not detect any CD8 T cells or innate lymphoid cells expressing eYFP in this model. The concordance of IL-22 and eYFP expression in γδ T cells was about 30% (Fig.4F), which is considerably higher than what was seen under in vitro culture conditions. Importantly, there was a considerably larger fraction of IL-17 producers in the eYFP+ fraction, indicating that IL-22 production may be a specialization of the Th17 program, rather than an independent program.
Figure 4. Fate reporter eYFP+ cells in skin inflammation.
A) Representative images of H&E staining of skin sections from untreated (d0) and IMQ-treated (d5) IL-22 fate reporter mice at day 5. Scale bar represents 100 μm. B) Quantification of epidermal (left) and scale (right) thickness of untreated (d0) fate reporter mice and at day 5 after IMQ treatment. Bars show mean ± SEM, n=4 mice per group. ** P < 0.01 and *** P < 0.001. C) Bar graph showing the number of eYFP+ γδTCR+ and eYFP+ TCRβ+ cells in dorsal skin of untreated fate reporter mouse (d0) and after five days of imiquimod treatment (d5).Data shown as mean ± SEM are representative of 5 independent experiments. D) FACS plot of eYFP+ CD45+ cells from the dorsal skin of untreated (d0) and imiquimod treated (d5) fate reporter mouse analysed for their γδTCR and TCRβ expression. E) FACS plot of γδTCR+ and TCRβ+ cells analysed for their eYFP expression in dorsal skin of untreated (d0) and imiquimod treated (d5) fate reporter mouse. F) FACS plot of CD45+ γδTCR+ and TCRβ+ cells from the dorsal skin of imiquimod treated fate reporter mouse analysed for their eYFP, IL-22 and IL-17A expression. Data are representative of 5 independent experiments (C-F).
We next investigated the induction of eYFP during the course of infection with Citrobacter rodentium, an effacing attaching bacterial pathogen that causes severe inflammation of the large intestine and requires IL-22 for restoration of the intestinal barrier following infection (7-9). It was noticeable that IL22CreR26ReYFP mice showed a more pronounced transient weight loss following infection than wildtype B6 mice and even heterozygous IL-22+/− mice (Fig.5A). This is indicative of stochastic monoallelic expression of IL-22, which in combination with only one functional allele will result in lower IL-22 expression than that of heterozygous IL-22+/− mice. However, at the time points these mice were analysed (from d28) they had all cleared the infection and the kinetics of Citrobacter clearance in IL22CreR26ReYFP followed those reported for wildtype mice (23) (data not shown).
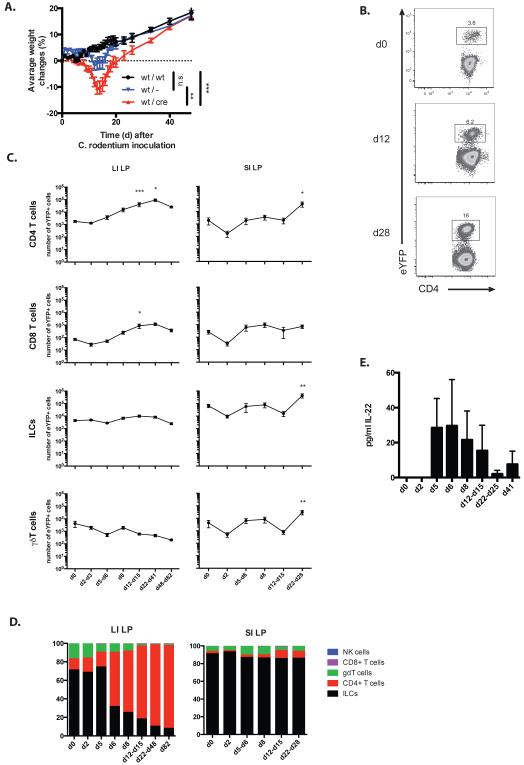
Figure 5. Response of fate reporter mice to infection with Citrobacter.
A) Weight curve of C57BL/6 (wt/wt), IL-22+/− (wt/−) and IL-22 fate reporter (wt/cre) mice infected with C. rodentium. N=6 per group. Data shown as mean ± SEM, n.s. non-significant, ** P < 0.01 and *** P < 0.001. B) FACS plot of CD45+ cells from LI LP of untreated (d0) or C. rodentium infected fate reporter mice at d12 and d28 analysed for their eYFP and CD4 expression. Data are representative of 2-8 independent experiments. C) Kinetics of expansion of eYFP+ CD4 T cells, CD8 T cells, ILC and γδT cells in LI LP and SI LP of fate reporter mice during the course of infection with C. rodentium. Data shown as mean ± SEM are representative of 2-12 independent experiments. For the statistical analysis, all the time points were compared to untreated sample * P < 0.05, ** P < 0.01 and *** P < 0.001. D) Relative number of eYFP+ NK cells, CD8 T cells, γδT cells, CD4 T cells and ILC in LI LP and SI LP of C. rodentium infected fate reporter mice at different time points post inoculation. Data shown as mean are representative of 2-12 independent experiments. E) Serum levels of IL-22 protein in IL-22 fate reporter mice during the course of infection with C. rodentium. Data shown as mean+/− SEM are representative of 2-4 independent experiments.
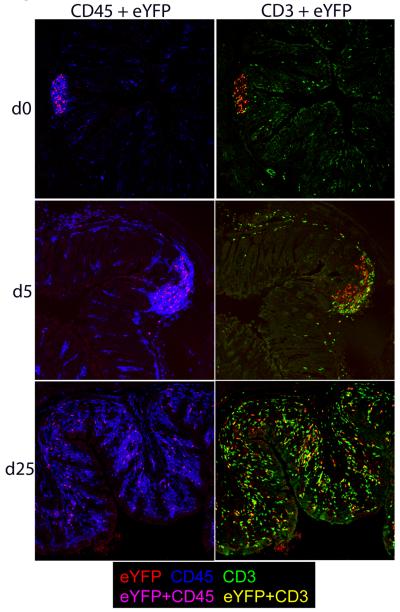
Kinetic analysis between days 2-82 after infection for appearance and expansion of eYFP+ expressing CD4 T cells (Fig.5B) and other cell types in LI LP and SI LP showed that cellular expansion was prominent in the CD4 cell population in the LI LP, whereas eYFP+ cells in the SI LP increased only in the later stages, presumably due to overspill from the LI LP (Fig.4C,D). eYFP+ ILC or γδ T cells did not expand significantly throughout the course of infection. IL-22 protein was detectable in serum from day 5 after infection (Fig.5E). Histological analysis of colon from IL22CreR26ReYFP mice showed eYFP+ cells in structures resembling isolated lymphoid follicles (ILF) and their expansion throughout the colon following infection. Initially eYFP+ cells were CD3-ve, indicative of ILC, whereas after infection eYFP expression was extended to CD3+ cells (Fig.6).
Figure 6. Immune histology of colon from fate reporter mice.
Sections from the large intestine (distal) of untreated (d0) and C. rodentium infected fate reporter mice at two time points (d5 and d25) post inoculation were stained with anti-GFP (green), anti-CD45 (blue) and anti-CD3 (red). Magnification: 20×. Representative results from one of at least three experiments per time point are shown.
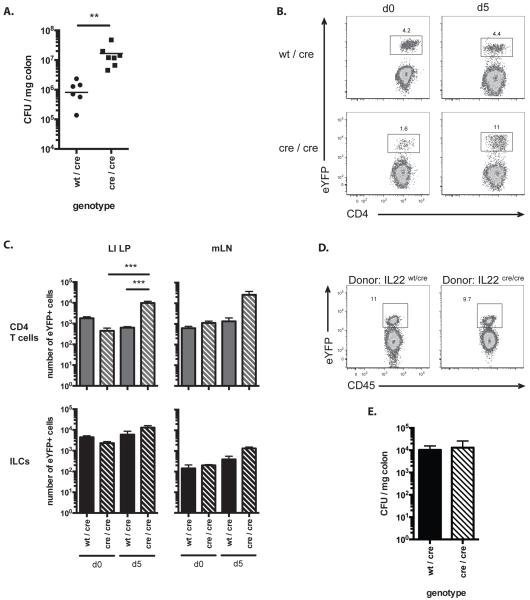
Homozygous IL22CreR26ReYFP cells expand in infection, but fail to control Citrobacter
We next investigated the fate of eYFP+ CD4 T cells in IL22CreR26ReYFP mice with homozygous expression of Cre. Such mice are IL-22 deficient, but still allow identification of the cell type via eYFP reporting (Fig.7B). IL-22cre/cre reporter mice were unable to clear Citrobacter as indicated by a log higher CFU count on day 5 after infection (the mice succumbed to the infection by day 8) (Fig.7A). Interestingly, the number of eYFP+ CD4 T cells, but not ILC in LI LP and mLN was substantially higher (Fig.7C). This could either indicate continuous stimulation of CD4 T cells to curb bacterial expansion or could indicate that homozygous Cre expression improved the reporter intensity. In order to avoid complications due to the differential outgrowth of Citrobacter, we generated chimeric mice in which we co-transferred wildtype B6 bonemarrow with bonemarrow from either IL22CreR26ReYFP (heterozygous for Cre, IL-22wt/cre) or from Cre homozygous reporter mice (IL-22cre/cre) into B6. Rag2−/− CD45.1 hosts. This allowed us to monitor the extent of eYFP induction in CD4 T cells from heterozygous or homozygous donors in mice with comparable bacterial burden. As shown in Fig.7D, there was no difference in eYFP+ CD4 T cells in recipients of homozygous vs heterozygous Cre reporter bonemarrow and the bacterial burden was similar (Fig.7E). This makes it less likely that the hyper-expansion of eYFP+ cells in homozygous reporter mice shown in Fig.7B was simply due to a higher reporting efficiency due to the presence of two Cre alleles, but rather suggests an ongoing stimulatory drive to CD4 T cells to combat the outgrowth of Citrobacter, which is frustrated due to the lack of IL-22.
Figure 7. Homozygous and heterozygous fate reporter mice infected with C. rodentium.
A) Log10 CFU of C. rodentium in colon of fate reporter mice on day 5 post inoculation. Horizonal lines represent mean CFU values ** P < 0.01. B) FACS plot of CD45+ cells from LI LP of untreated (d0) or C. rodentium infected (d5) fate reporter mouse analysed for eYFP and CD4 expression. C) Bar graphs showing the number of eYFP+ CD4 T cells (top panel) and ILC (lower panel) in LI LP (left) and mLN (right) of untreated (d0) or C. rodentium infected (d5) fate reporter mice. Data shown as mean ± SEM are representative of 3-8 independent experiments (B, C). Wt/cre; heterozygous IL-22 fate reporter mouse, cre/cre; homozygous IL-22 fate reporter mouse. *** P < 0.001. D) FACS plots showing eYFP expression within donor CD45.2+ CD4 T cells from LI LP of C. rodentium infected chimeric mice 12 days post infection. Donors: CD45.2 heterozygous (IL-22Cwt/cre) or homozygous (IL22cre/cre) fate reporter mice. Recipients: CD45.1 Rag2−/− mice. Data are representative of 3 independent experiments. E) Log10 CFU of C. rodentium in colon of chimeric mice on day 12 post inoculation. Data shown as mean ± SEM are representative of 3 independent experiments.
Functional plasticity of cytokine expression in eYFP+ cells is prominent in T cells, but limited in ILC
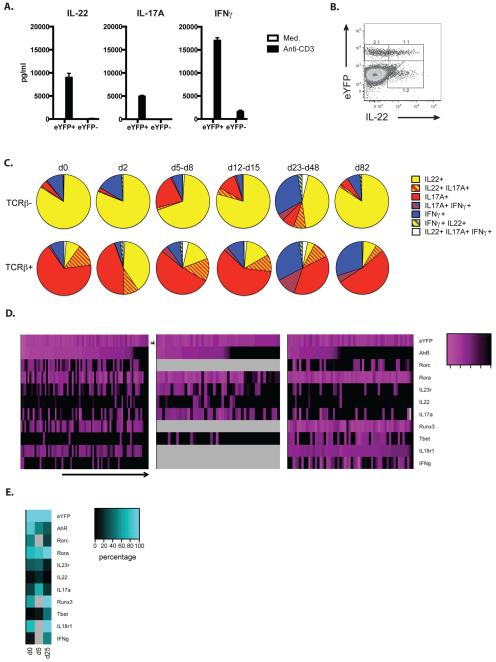
In order to investigate the cytokine profile of eYFP+ cells, T cells (gated on TCRβ expression) and ILC (TCRβ-ve) were analysed throughout the course of infection for the single as well as co-expression of IL-22, IL-17 and IFNγ by intracellular staining. Firstly, we investigated cytokine production of FACS purified eYFP+ and eYFP- CD4 T cells from Citrobacter infected mice (d14), restimulated with anti-CD3 in vitro. Interestingly, IL-22, IL-17 and IFNγ were all predominantly produced by eYPF+ CD4 T cells, indicating the substantial plasticity of these cells during the course of infection (Fig.8A). Fig.8B shows the concordance of eYPF and IL-22 expression in CD45+ cells, which is higher than what was observed for the in vitro culture (see Fig1).
Figure 8. Cytokine and transcription factor expression profile of eYFP+ fate reporter mice.
A) eYFP+ and eYFP- CD4 T cells purified from LI LP of C. rodentium infected mice d14 p.i. were stimulated with anti-CD3 or medium for 18h. Bar graphs show the amount of IL-22 and IL-17A produced after stimulation. B) FACS plot of CD45+ cells from LI LP of C. rodentium infected fate reporter mice at d2 analysed for their eYFP and IL-22 expression. Data are representative of 4 independent experiments C) Pie charts showing the relative numbers of eYFP+ TCRβ+ and TCRβ− cytokine expressing cells based on intracellular cytokine staining in LI LP of untreated (d0) fate reporter mice and at different timepoints after infection with C.rodentium. LI LP; large intestine lamina propria. Data shown as mean are representative of 2-7 independent experiments. D) Heat map presentation for single cell gene expression of selected transcription factors, cytokines or cytokine receptors as well as eYFP in FACS purified eYFP+ CD4 T cells from LI LP of untreated (d0) and C. rodentium infected fate reporter mice at two time points (d5 and d25) post infection. Expression levels are expressed as Ct values. E) Summary of data in Fig 8B: Heat map indicating quantitative values of gene expression (Ct<35) at different time points. Light grey areas indicate missing values.
eYFP+ ILC remained predominantly IL-22 positive throughout infection with only a proportion expressing IL-17 and/or IFNγ. In contrast, the predominant cytokine expressed in eYFP+ T cells was IL-17 alone or together with IL-22 in the early phase of infection with a gradual increase in IFNγ expression alone or together with the other cytokines (Fig.8C). This confirmed the predominant association of IL-22 with the Th17 subset on the one hand and indicated substantial plasticity on the other hand as previously seen with Th17 cells in intestinal inflammation (24). In an attempt to identify the developmental origin and fate of eYFP expressing CD4 T cells in Citrobacter infection, we performed single cell qPCR for a range of markers associated with different T cell subsets, including transcription factors and cytokines.
The expression changes for these markers were determined from the uninfected basal state to day 5 the onset of CD4 T cell expansion to day 25 when Citrobacter was cleared. They indicate a gradual shift towards induction of Tbet and IFNγ (not determined for day 5) in the later stages whilst retaining the expression of Rora, with less prominent expression of Rorc (not determined at day 5) and reduced AhR expression. The co-expression of IL-22 with eYFP was low in steady state as well as in late stages of infection, but more pronounced on day 5 (Fig.8 D). The magnitude of gene expression during the three time points is given in the format of a heat map (Fig.8E grey fields indicate absence of data). Overall these data indicate substantial heterogeneity of eYFP+ CD4 T cells suggesting that IL-22 induction may not be strictly associated with a particular CD4 T cell subset.
DISCUSSION
Generation of an IL-22 fate reporter mouse was driven by the technical difficulties encountered in identifying such cells without resorting to in vitro culture. Although antibodies for intracellular staining are now available it is difficult to predict the kinetics of IL-22 production and expression of this cytokine appears to be transient and tightly controlled. For this reason the fate reporter approach offers the advantage of allowing identification of cells that had initiated this pathway at any time point without having to rely on active production of the cytokine. We used the same approach for the generation of the Cre knock-in construct that we had successfully used for the generation of IL17CreR26ReYFP mice. However, we noticed immediately that the penetrance of reporting as well as the co-expression of eYFP and IL-22 cytokine was considerably lower in IL22CreR26ReYFP mice. IL-22 belongs to the IL-10 family of cytokines and it has been shown that expression of IL-10 is largely monoallelic and stochastically regulated via control of expression frequency (25). It is likely that IL-22 producing CD4 T cells – similar to IL-10 producing cells- have a low probability for activation of the IL-22 allele so that expression is predominantly monoallelic. As our construct disables one allele by Cre insertion, IL-22 expression is further reduced, which is particularly evident in the single cell qPCR analysis. With hindsight it would have been better to avoid destruction of one allele by Cre insertion.
Nevertheless, despite these technical shortcomings, the reporter appears faithful and under in vitro conditions reveals IL-22 production exclusively under Th17 conditions with important contribution by IL-1β and the AhR ligand FICZ. AhR stimulation promotes the expression of IL-1R (26) and the induction of IL-22 in the presence of Th17 conditions with IL-1β without addition of the AhR ligand reflects the presence of AhR agonists in culture medium (18).
IL-23, which is important for IL-22 induction in vivo (8, 27) does not play a noticeable role in vitro under these culture conditions, which are too short to promote expression of IL-23 receptor (28). We have not been able to detect substantial eYFP expression under IL-6 stimulation and in our hands TGFβ is essential in vitro. The in vitro conditions for induction of IL-22 production remain controversial and could be influenced also by ambient TGFβ concentrations in medium constituents. We strongly believe that in vitro culture conditions for both IL-17 and IL-22 producers do not accurately reflect the stimuli these cells obtain in vivo, where multiple cell types, including stromal cells, might contribute to optimize their polarization.
In vivo the fate reporter revealed IL-22 producers in steady state and under inflammatory conditions consistent with previous reports that emphasised the importance of this cytokine at barrier sites of intestine, lung and skin. The expression of eYFP was restricted to tissue resident cell types at these locations, such as γδ T cells in skin and lung and ILC3 in the intestine, indicating their important role in homeostatic maintenance of epithelial barrier integrity. Interestingly, the resident Th17 population present in the small intestinal lamina propria of non-immune mice did not show eYFP expression, which instead was restricted to ILC with the characteristics of the ILC3 subset (29) in this location with the caveat that low numbers of eYFP+ cells might have been missed due to underreporting.
In line with earlier reports, IL-22 producing intestinal ILC3 control Citrobacter rodentium infection in the early phase for about 5 days until the adaptive T cell response ensues (30). This could involve the action of retinoic acid which has been shown to attenuate colon inflammation via increased IL-22 production by ILC3(31). The induction of IL-22/eYFP expression in CD4 T cells was noticeable first in the colon lamina propria and to a lesser degree in the mesenteric lymph node, suggesting that the adaptive CD4 response was induced locally. Histologically it appeared that structures resembling colonic patches (CLP) and isolated lymphoid follicles (ILF), were the main sites of eYFP expression. These structures are to be induced by the lymphotoxin pathway upstream of IL-22 and both are cooperating in the organization and maintenance of CLP and ILF during infection with Citrobacter rodentium (32).
CD4 T cells from the small intestine did not seem to be involved in the immune response to Citrobacter and eYFP+ cells in this location could only be detected at later stages when they might have spilled over from the colon. To some extent this may be due to the fact that Citrobacter infection targets the distal colon (33) and thus does not directly involve the small intestinal Th17 cell population. In addition, however, it may indicate that SI LP Th17 cells are not participating in inflammatory immune responses.
The ILC3 response to Citrobacter infection remained largely focused on IL-22 with only a minor contribution of IFNγ and IL-17 production. In contrast, the T cell response was highly plastic with a large fraction of IL-17 producers and a gradual shift towards IFNγ as well as dual cytokine production. Cytokine analysis of FACS purified CD4 cell populations confirmed that production of IL-22, IL-17 and IFNγ was restricted to eYFP+ cells, emphasising their plasticity as well as the fidelity of this reporter mouse.
IL-22 was detected maximally around day 2 of infection prior to the main expansion of CD4 T cells and appeared transient compared with IL-17 that remained dominant throughout the infection period. It is conceivable that tight control of IL-22 secretion is necessary to avoid negative side effects associated with this cytokine, such as the promotion of colonization by pathogens such as Salmonella through suppression of related commensal bacteria (34), or the perpetuation of bacteria-induced colon cancer (35).
Citrobacter rodentium was cleared around 21 days after infection (data not shown), but elevated numbers of CD4 T cells persisted in the colon (see Fig.6 for d25) up to at least 82 days albeit the majority of these were not producing any cytokines anymore (data not shown). Detection of cells that had initiated the IL-22 program via expression of eYFP allowed the analysis of single cells via qPCR (36) and confirmed the considerable heterogeneity of eYFP+ CD4 T cells throughout the course of infection.
Single cell data indicated a gradual shift in profile from a more Th17-like phenotype towards a Th1-like phenotype at later stages. Expression of Rora was quite consistent throughout, whereas Rorc expression was not prominent and further reduced at the expense of Tbet on day 25 and also Ahr expression was reduced at the later time points. Runx3 was uniformly expressed in eYFP+ CD4 T cells at day 25 after infection and this together with Tbet promotes the generation of pathogenic IFNγ producing Th17 cells(37). Overall the emerging picture from the IL-22 fate reporter suggests that in CD4 T cells IL-22 is transiently associated with a Th17 population that is highly plastic and prone to adopt a more Th1-like profile in the course of infection. The shortcomings of this reporter, resulting in underreporting of IL-22 make it impossible to unequivocally state that there is no Th22 subset dedicated to producing this cytokine, but the substantial plasticity of inflammatory CD4 T cells in this setting make such a scenario less likely.
Acknowledgements
We thank J.-C. Renauld (Ludwig Institute for Cancer Research Brussels) for the AM.3 anti-IL-22 antibody. We gratefully acknowledge the contributions of our Biological Services facility, the Flow Cytometry Lab and the Histology Lab.
This work was supported by the Medical Research Council UK (U117512792).
References
- 1.Tachiiri A, Imamura R, Wang Y, Fukui M, Umemura M, Suda T. Genomic structure and inducible expression of the IL-22 receptor alpha chain in mice. Genes and immunity. 2003;4:153–159. doi: 10.1038/sj.gene.6363934. [DOI] [PubMed] [Google Scholar]
- 2.Wolk K, Kunz S, Witte E, Friedrich M, Asadullah K, Sabat R. IL-22 increases the innate immunity of tissues. Immunity. 2004;21:241–254. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol. 2011;29:71–109. doi: 10.1146/annurev-immunol-031210-101312. [DOI] [PubMed] [Google Scholar]
- 4.Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, Diefenbach A. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- 6.Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, Fu YX, Zhou L. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, Reinhart TA, McAllister F, Edeal J, Gaus K, Husain S, Kreindler JL, Dubin PJ, Pilewski JM, Myerburg MM, Mason CA, Iwakura Y, Kolls JK. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nature medicine. 2008;14:275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nature medicine. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 9.Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 2011;34:122–134. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonnenberg GF, Artis D. Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity. 2012;37:601–610. doi: 10.1016/j.immuni.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brand S, Beigel F, Olszak T, Zitzmann K, Eichhorst ST, Otte JM, Diepolder H, Marquardt A, Jagla W, Popp A, Leclair S, Herrmann K, Seiderer J, Ochsenkuhn T, Goke B, Auernhammer CJ, Dambacher J. IL-22 is increased in active Crohn’s disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. American journal of physiology. Gastrointestinal and liver physiology. 2006;290:G827–838. doi: 10.1152/ajpgi.00513.2005. [DOI] [PubMed] [Google Scholar]
- 12.Andoh A, Zhang Z, Inatomi O, Fujino S, Deguchi Y, Araki Y, Tsujikawa T, Kitoh K, Kim-Mitsuyama S, Takayanagi A, Shimizu N, Fujiyama Y. Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology. 2005;129:969–984. doi: 10.1053/j.gastro.2005.06.071. [DOI] [PubMed] [Google Scholar]
- 13.Lo YH, Torii K, Saito C, Furuhashi T, Maeda A, Morita A. Serum IL-22 correlates with psoriatic severity and serum IL-6 correlates with susceptibility to phototherapy. Journal of dermatological science. 2010;58:225–227. doi: 10.1016/j.jdermsci.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 14.Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, Ahlfors H, Wilhelm C, Tolaini M, Menzel U, Garefalaki A, Potocnik AJ, Stockinger B. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol. 2011;12:255–263. doi: 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimshek DR, Kim J, Hubner MR, Spergel DJ, Buchholz F, Casanova E, Stewart AF, Seeburg PH, Sprengel R. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 2002;32:19–26. doi: 10.1002/gene.10023. [DOI] [PubMed] [Google Scholar]
- 16.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Meglio P, Duarte JH, Ahlfors H, Owens ND, Li Y, Villanova F, Tosi I, Hirota K, Nestle FO, Mrowietz U, Gilchrist MJ, Stockinger B. Activation of the aryl hydrocarbon receptor dampens the severity of inflammatory skin conditions. Immunity. 2014;40:989–1001. doi: 10.1016/j.immuni.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veldhoen M, Hirota K, Christensen J, O’Garra A, Stockinger B. Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J Exp Med. 2009;206:43–49. doi: 10.1084/jem.20081438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 20.Van Belle AB, de Heusch M, Lemaire MM, Hendrickx E, Warnier G, Dunussi-Joannopoulos K, Fouser LA, Renauld JC, Dumoutier L. IL-22 is required for imiquimod-induced psoriasiform skin inflammation in mice. J Immunol. 2012;188:462–469. doi: 10.4049/jimmunol.1102224. [DOI] [PubMed] [Google Scholar]
- 21.Boniface K, Guignouard E, Pedretti N, Garcia M, Delwail A, Bernard FX, Nau F, Guillet G, Dagregorio G, Yssel H, Lecron JC, Morel F. A role for T cell derived interleukin 22 in psoriatic skin inflammation. Clin Exp Immunol. 2007;150:407–415. doi: 10.1111/j.1365-2249.2007.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pantelyushin S, Haak S, Ingold B, Kulig P, Heppner FL, Navarini AA, Becher B. Rorgammat+ innate lymphocytes and gammadelta T cells initiate psoriasiform plaque formation in mice. J Clin Invest. 2012;122:2252–2256. doi: 10.1172/JCI61862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann C, Hill DA, Minkah N, Kirn T, Troy A, Artis D, Bushman F. Community-wide response of the gut microbiota to enteropathogenic Citrobacter rodentium infection revealed by deep sequencing. Infection and immunity. 2009;77:4668–4678. doi: 10.1128/IAI.00493-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrison PJ, Bending D, Fouser LA, Wright JF, Stockinger B, Cooke A, Kullberg MC. Th17-cell plasticity in Helicobacter hepaticus-induced intestinal inflammation. Mucosal Immunol. 2013;6:1143–1156. doi: 10.1038/mi.2013.11. [DOI] [PubMed] [Google Scholar]
- 25.Calado DP, Paixao T, Holmberg D, Haury M. Stochastic monoallelic expression of IL-10 in T cells. J Immunol. 2006;177:5358–5364. doi: 10.4049/jimmunol.177.8.5358. [DOI] [PubMed] [Google Scholar]
- 26.Duarte JH, Di Meglio P, Hirota K, Ahlfors H, Stockinger B. Differential influences of the aryl hydrocarbon receptor on Th17 mediated responses in vitro and in vivo. PLoS One. 2013;8:e79819. doi: 10.1371/journal.pone.0079819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Y, Danilenko DM, Valdez P, Kasman I, Eastham-Anderson J, Wu J, Ouyang W. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445:648–651. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- 28.Veldhoen M, Hocking RJ, Flavell RA, Stockinger B. Signals mediated by transforming growth factor-beta initiate autoimmune encephalomyelitis, but chronic inflammation is needed to sustain disease. Nat Immunol. 2006;7:1151–1156. doi: 10.1038/ni1391. [DOI] [PubMed] [Google Scholar]
- 29.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, Powrie F, Vivier E. Innate lymphoid cells--a proposal for uniform nomenclature. Nature reviews. Immunology. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 30.Basu R, O’Quinn DB, Silberger DJ, Schoeb TR, Fouser L, Ouyang W, Hatton RD, Weaver CT. Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity. 2012;37:1061–1075. doi: 10.1016/j.immuni.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mielke LA, Jones SA, Raverdeau M, Higgs R, Stefanska A, Groom JR, Misiak A, Dungan LS, Sutton CE, Streubel G, Bracken AP, Mills KH. Retinoic acid expression associates with enhanced IL-22 production by gammadelta T cells and innate lymphoid cells and attenuation of intestinal inflammation. J Exp Med. 2013;210:1117–1124. doi: 10.1084/jem.20121588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ota N, Wong K, Valdez PA, Zheng Y, Crellin NK, Diehl L, Ouyang W. IL-22 bridges the lymphotoxin pathway with the maintenance of colonic lymphoid structures during infection with Citrobacter rodentium. Nat Immunol. 2011;12:941–948. doi: 10.1038/ni.2089. [DOI] [PubMed] [Google Scholar]
- 33.MacDonald TT, Frankel G, Dougan G, Goncalves NS, Simmons C. Host defences to Citrobacter rodentium. Int J Med Microbiol. 2003;293:87–93. doi: 10.1078/1438-4221-00247. [DOI] [PubMed] [Google Scholar]
- 34.Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W, Raffatellu M. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity. 2014;40:262–273. doi: 10.1016/j.immuni.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirchberger S, Royston DJ, Boulard O, Thornton E, Franchini F, Szabady RL, Harrison O, Powrie F. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J Exp Med. 2013;210:917–931. doi: 10.1084/jem.20122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chattopadhyay PK, Gierahn TM, Roederer M, Love JC. Single-cell technologies for monitoring immune systems. Nat Immunol. 2014;15:128–135. doi: 10.1038/ni.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Godec J, Ben-Aissa K, Cui K, Zhao K, Pucsek AB, Lee YK, Weaver CT, Yagi R, Lazarevic V. The Transcription Factors T-bet and Runx Are Required for the Ontogeny of Pathogenic Interferon-gamma-Producing T Helper 17 Cells. Immunity. 2014;40:355–366. doi: 10.1016/j.immuni.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]