Abstract
To determine the breadth and underpinning of changes in immunocyte gene expression due to genetic variation in mice we performed, as part of the Immunological Genome Project, gene expression profiling for CD4+ T cells and neutrophils purified from 39 inbred strains of the Mouse Phenome Database. Considering both cell types, a large number of transcripts showed significant variation across the inbred strains, 22% of the transcriptome varying by two-fold or more. These included 119 loci with apparently complete loss-of-function, where the corresponding transcript was not expressed in some of the strains, representing a useful resource of “natural knockouts”. We identified 1,222 cis- expression quantitative trait loci (cis-eQTL) that control some of this variation. Most (60%) cis-eQTLs were shared between T cells and neutrophils, but a significant portion uniquely impacted one of the cell types, suggesting cell-type specific regulatory mechanisms. Using a conditional regression algorithm we predicted regulatory interactions between transcription factors and potential targets, and demonstrated that these predictions overlap with regulatory interactions inferred from transcriptional changes during immunocyte differentiation. Finally, comparison of these and parallel data from CD4+ T cells of healthy humans demonstrated intriguing similarities in variability of a gene's expression: the most variable genes tended to be the same in both species, and there was an overlap in genes subject to strong cis-acting genetic variants. We speculate that this “conservation of variation” reflects a differential constraint on intra-species variation in expression levels of different genes, either through lower pressure for some genes, or by favoring variability for others.
Introduction
For more than a century, inbred mice have played a unique role in biomedical research. Their group homogeneity, phenotypic reproducibility, and genetic stability over time haveled tokey discoveries in essentially every area of biomedical research (1), including the discovery of fundamental concepts of immunology such as histocompatibility, MHC restriction, or genetic susceptibility to autoimmune diseases. The near-homogeneous nature of an inbred strain's genome underlies the extraordinary power of targeted germline modifications, and has supported mapping of loci associated with disease or phenotypic traits. The genomes of laboratory strains have been molded by strong selective pressures linked to their domestication by mouse fanciers in China and Europe, then to inbreeding and allele fixation in biomedical research colonies. These genomes incorporate segments from several origins (2), as now clearly established by the decoding of the complete genome of the reference C57BL/6J, followed by a number of other inbred strains (3,4). Efforts to standardize and integrate phenotypic and genetic information, as exemplified by the Mouse Phenome Database project (MPD) (5), are also helping to exploit the full potential of inbred strains in biomedical research.
The Immunological Genome Project (ImmGen) is an international collaboration of laboratories that collectively perform a thorough dissection of gene expression and its regulation in the immune system of the mouse. Genome-wide gene expression data have been collected for ∼250 immunological cell types of the mouse, yielding insights into genomic correlates of immunocyte differentiation and lineages (6). The assembled data also enabled predictions about regulatory networks that underlie mouse hematopoiesis (7). The first phase of the ImmGen project mainly used the reference C57BL/6J strain, and thus focused on identifying changes in gene expression during differentiation and activation in the context of a unique genome. Yet there is much value in analyzing the impact of functional genetic variation on gene expression levels. Variants influencing gene expression are pervasive in mammalian species, and comprise a large majority of the disease-related variants identified in genome wide association studies (GWAS)(8). Combined analysis of gene expression and genotype data across a genetically diverse population is a powerful means to understand the impact of genotypic variation on cellular processes, and ultimately to build mechanistic models that link genetic variation to detailed cellular processes in a context-specific manner (8,9). Several comparative analyses of gene expression have been performed across inbred mouse strains(10-14), but were of limited breadth and/or performed in celltypes not directly relevant to ImmGen.
In terms of understanding human disease, while the mouse models have been invaluable in establishing fundamental paradigms of immunologic function, caution has been suggested in translating findings from the mouse to the human immune system (15). Similarities and differences have been reported in the genomic underpinning of immune lineages of human and mouse, whether at steady state or after cell activation (16-19). A direct comparison of the genetic underpinning of these differences would also be valuable in ascertaining what mouse models can be usefully applied to understand human diseases and their genetics.
To better understand the effect of genetic variation on the mouse immune system, we generated RNA expression data for 39 of the main inbred strains in the MPD “Priority Strain Panel”. Using rigorous ImmGen standard operating procedures, genome-wide expression data were generated for two immunological cell types, CD4+ T cells (T4) and polymorphonuclear neutrophils (granulocytes, GN). These were chosen to represent the main lymphoid and myeloid branches of the immune system, and its adaptive and innate facets. This effort paralleled a study of similar design in an ethnically diverse population of healthy humans, the “ImmVar” study, where genotype and gene expression data were collected for CD4+ T cell and CD14+CD16- monocytes [(20,21) and Ye et al, submitted]. This matching study design allowed us to compare transcriptional variability and its roots in the two species. Here, we first report on the impact of genetic background on gene expression levels in mouse T4and GN, identify cis expression quantitative trait loci (eQTL), and chart regulatory interactions that can be inferred from the perturbation of the regulatory network by genetic variation. Second, we compare the impact of functional variation in human and mouse, by exploring the overlap between expression variability and its genetics in the two species.
Materials and Methods
Gene expression and genotype data
Inbred mouse strains from the MDP Priority Strain Panel, representing 39 strains, were obtained from the Jackson Laboratory production facility in Bar Harbor, Maine, at five weeks of age. All mice were bred in the Jackson Laboratory production facility under SPF conditions. CD3+CD4+CD62L+ naïve T splenocytes and CD11b+Ly6G+ bone marrow granulocytes were sorted from pools of two to three mice. Two biological replicates were generated for each strain using the ImmGen standard operating protocol (SOP; www.immgen.org). Gene expression data was generated for bone marrow granulocytes (GN) and CD4+ T splenocytes (T4) using Affymetrix ST1.0 microarrays, the platform used for the main ImmGen compendium, resulting in the quantification of expression levels for 25,134 probes corresponding to 21,951 unique genes. Data were processed and normalized using the ImmGen standard operating protocol (www.immgen.org). When indicated, data were filtered to only include genes with >0.95 probability of expression (or a mean of >120 expression on the intensity scale; see SOP). This filtering criteria resulted in 11,598 and 11,285 expressed transcripts in T4, and GN, respectively, with 131,85 transcripts expressed in one or the other, and 9,698 transcripts expressed in both cell types. A threshold for absence of expression was also set at <0.05 probability of expression (or a <42 expression level on intensity scale). Genotype data was obtained from the mouse HapMap genotype resource (http://mouse.cs.ucla.edu/mousehapmap) (22). Only genotyped SNPs with minor allele frequency (MAF) greater than 0.05 and no more than 10% missing rate (resulting in a total of 96,779 SNPs) were used in this study.
Defining the true variability (TV) metric, bimodality in gene expression, and complete loss of function loci
All analyses were performed in the MATLAB computing environment (R2013a, v 8.1.0.604).
At least two biological replicates were available for each mouse and each cell type (for the strains for which there were more than two replicates, we randomly chose two of the replicates for this analysis). For the TV metric, two quantities were computed for each gene and each cell type using the log transformed data: (1) the between-strains mean absolute deviation, and then divided by the mean gene expression level for that gene; (2) the average of within-strains mean absolute deviation, where mean absolute deviation (MAD) for each strain was computed using the two replicates for that strain, and then divided by the mean gene expression level for that gene. The TV score for each gene was defined as the difference between the first quantity, representing both meaningful and unwanted variability, and the second quantity, representing the unwanted variability. We note that there are two main differences between the TV metric proposed here and a standard ANOVA approach: first, we chose to quantify variability using MAD as opposed to variance because the latter gives more weight to extreme values. Second, as opposed to an associated F-statistic in ANOVA, where the test statistics (interpreted as the true variability score) is the ratio of two variances, here we use the difference of the two MADs as the score. We chose to use the difference so to emphasize the magnitude of the variability, in addition to the relative variability of the within-strains and between-strains MAD.
Bimodal genes were identified using two criteria: (1) based on the assessment of the fit of a mixture of Gaussian with two components to expression levels across the strains, and (2) a threshold on the fold difference between high and low expressing strains. The mixture of Gaussians were fit using MATLAB's gm distribution function (MATLAB R2013a, v 8.1.0.604). A likelihood ratio test was used to assign a bimodality p-value to each gene by comparing the likelihood of mixture of Gaussian with two components with simply the fit of a single Gaussian. Genes with bimodality p-value < 10-6 and at least a two-fold difference in top two high expressing and bottom two low expressing strains were identified as bimodal. Complete loss-of-function loci were identified as those bimodal genes that additionally satisfied a strict threshold on expression levels: an expression of less than 42 (corresponding to <0.05 probability of expression) for at least two strains and expression greater than 120 (corresponding to >0.95 probability of expression) for at least two strains.
Expression quantitative trait loci (eQTL) association mapping for mouse
It is well appreciated that genetic association studies in inbred strains are impacted by population stratification, which violates the assumptions of standard statistical tests and lead to an abundance of false positive associations (and therefore an inflation of association p-values)(23). To account for population stratification, we used linear regression, regressing out the effect of the top two genotype PCs from log gene expression data. We chose two PCs by quantifying the inflation of observed p-values using the λ statistic (24) as we varied the number of removed genotype PCs from one to five. A cis window of 1Mb centered on transcription start site (TSS) was used to identify all cis SNPs for each gene.
Joint analysis
To increase statistical power, for the joint analysis, residual expression data (after removing genotype PCs, see above) from both cell types were concatenated, resulting in a dataset with 2×39 samples and 13,185 expressed transcripts (expressed in at least one cell type). For each SNP-gene pair, the Wilcoxon rank sum statistic (as implemented in MATLAB R2013a, v 8.1.0.604) was used to test whether the expression of the gene was significantly different between strains with the reference or the alternative allele at the given SNP. 10,000 permutations were performed for each SNP-gene pair, permuting the assignment of SNP values to strains while keeping intact the correspondence between genotype assigned to the T4 and GN sample for the same strain (thus accounting for “repeated” samples). A gene-level p-value was assigned that accounted for the number of tested SNPs per gene by using the minimum permutation p-value across all tested SNPs for that gene as the null distribution (25,26). Final set of cis-eQTLs were defined by setting a 5% FDR threshold on the gene-level p-values.
Cell-specific eQTL analysis
Cell-specific eQTLs were identified by testing the significance of an interaction term between genotype and cell-type indicator in a linear regression setting, where the fit of the baseline model (no interaction) with one that additionally included a cell-type-indicator by genotype interaction term was assessed using an F-test. In particular, we model the expression level of gene gin tissue t for strain i as xg,t,i = αg,t + βgsi + γg,tsi where αg,t is genotype-independent tissue-specific effect for tissue t and gene g, βg is the tissue-shared genotype effect, and γg,t represents the cell-specific genotype effect for tissue t. As above, gene-level p-values were computed using 10,000 permutations (permuting the assignment of genotype values to the strains).
Constructing regulatory networks in mouse and validation using Ontogenet links
For constructing regulatory networks, genes expressed in both cell types and identified to have non-negligible TV scores (as per Figure S1A) were used, which resulted in 3675 analyzed genes. Among these 164 are TFs, as defined in (7). Two networks (one for each cell type) were constructed using stepwise regression, where a sparse set of TFs (regulators) were identified for each target gene (set of targets includes both TFs and non-regulatory genes). More specifically, for each target gene, stepwise regression was performed using all regulators (excluding auto-regulation), and inferred regulators were identified using 5% FDR to correct for the number of TFs tested for each target. A “joint” network was also constructed using the same approach but applied on concatenated expression data form both cell types (after removing mean gene expression from each cell type). Networks were constructed on genotype-PC corrected data.
We used the joint network constructed from T4 and GN data to compare the co-expression-based links derived here with those derived from the ImmGen data (using the Ontogenet algorithm (7)). We decided to use the joint network as we observed a high degree of overlap between networks constructed individually from each cell type (see Results), and in order to identify persistent, and thus more likely true positive, relationships. Regulatory interactions and modules defined by Ontogenet were downloaded from the ImmGen web server (www.immgen.org). Note that in (7) two types of modules were defined: initially 81 larger “coarse-grained modules” were defined, and subsequently some of these modules were refined into smaller modules with more coherent expression, resulting in 334 “fine modules”. Coarse modules were constructed to capture the mechanisms that co-regulate a larger set of genes in one cell-lineage, whereas fine modules were constructed to capture distinct regulatory mechanism controlling only a smaller subset of these genes in the sub-lineage(s). Only “fine modules” and their “top regulators”, representing more functionally specific gene groups and links, were used in the present analyses. Based on this data, a list of 4,083 testable links, connecting the top regulators to all genes in their assigned module was generated. First, the replication rate for this list in the current study was computed by assigning a p-value to each link in this study based on the co-expression of the corresponding regulator-target pair, and then assessing the proportion of true-positive p-values using Storey's π1 (27). To correct for the overall inflation of p-values between all pairs of genes, as is often observed in co-expression data, we used the distribution of p-values for co-expression of all gene-gene pairs as the null distribution to assign a p-value to each of the 4,083 links. Second, the links identified in this study were tested for consistency with those identified by the Ontogenet algorithm on the ImmGen data using a hypergeomtric test---this test identified regulators whose inferred targets were also co-regulated (i.e., assigned to the same module) according to Ontogenet. Third, we computed the proportion of links identified here that were also reported by Ontogenet, and used the hypergeometric test to compute a p-value for the overlap.
Gene expression, genotype, and eQTL discovery in human
Genotype and gene expression for T4 and neutrophils were obtained from the ImmVar study. As done for the mouse data, cis-eQTLs were defined using a 1Mb window centered on the TSS. Gene expression data was corrected for three genotype PCs and 30 expression PCs (to increase statistical power by removing variability due to environmental or non-local genetic factors). The number of removed expression PCs was set by evaluating the improvement in number of cis-eQTLs that were detected based on data from one (“training”) chromosome (chromosome 18). In particular, to select the number of PCs that are removed, the number of cis-eQTL discoveries in raw data was compared to PC-corrected data where we varied the number of removed PCs from one to 50. In order to avoid overfitting, we optimized the number of removed PCs based on cis-eQTL discovery on just one chromosome (and not the whole dataset). As previously observed (28), the improvement in cis-eQTL discovery greatly increased with removal of PCs, and there was a stable plateauing effect when we removed 20-40 PCs (see for example (21)). As described for the mouse data above, in the joint eQTL analysis, gene expression data from both cell types were combined and a gene-level p-value was computed for each gene using permutation analysis (1000 permutations per gene). Here, the Spearman rank correlation was used as the test statistic.
Constructing regulatory networks for human/mouse comparison
Stepwise regression was used to construct a regulatory network for T4 data. For this analysis, we used the set of genes expressed in both human and mouse (in T4s) and were considered to have non-negligible TV scores for T4 data in mouse (as defined by Figure S1A), which resulted in a set of 3,407, of which 183 are TFs. For constructing the network, human data was corrected for batch, population structure (three genotype PCs), gender, and age whereas mouse data was corrected for two genotype PCs (mouse data was done in one batch, and the mice had identical gender and age). Significant links were identified at 5% FDR.
The replication rate of links identified in one species onto the other was computed using the π1 statistic to quantify the proportion of true-positives among the co-expression p-values for the relevant links (links being replicated). As above, co-expression p-values were adjusted using the distribution of all co-expression p-values as the null.
The stepwise regression approach above identifies regulatory links in a target-centric manner, identifying “top” regulators for each target. In addition, in a TF-centric manner, top targets for each TF were identified based on the ranking of their co-expression value (Pearson correlation coefficient) with the given TF. In particular, two analyses were conducted. First, for each TF in mouse (human), top ten targets were defined based on co-expression values, and the overlap of these targets were assessed in the top N=[10, 20, 30, 50] targets for the same TF in human (mouse). The significant of the overlaps were determined using the hypergeometric test, and corrected for the number of TFs tested. Second, the evidence for conservation of the top N=[10, 20, 30, 50] targets of each TF in mouse (human) was assessed in human by using the Wilcoxon rank sum test to compare the distribution of the co-expression values for the top N targets compared to the distribution of co-expression values between that TF and all genes.
Results
The mice tested here included 35 classic laboratory inbred strains (M.m.domesticus) that represent all the major branches of the inbred tree (1) and four “wild-derived” strains (CAST/EiJ, PWD/PhJ, JF1/Ms and MSM/Ms, which are representative of the M.m.castaenus, M.m.musculus, M.m.molossinus species, respectively). Gene expression data for bone marrow granulocytes (GN) and CD4+ T splenocytes (T4) were quantified using Affymetrix ST1.0 microarrays (Methods). Matching genotype data were obtained from the Mouse HapMap Genotype Imputation Resource (29), and included 132,285 genotyped SNPs (Methods). As we did not attempt here to identify causal variants, because of the limitations imposed by the relatively large size of LD blocks in inbred mice, the analyses only used genotyped SNPs for computational efficiency. All expression datacan be browsed or accessed on the ImmGen website (www.immgen.org).
Extent and distribution of expression variation across strains
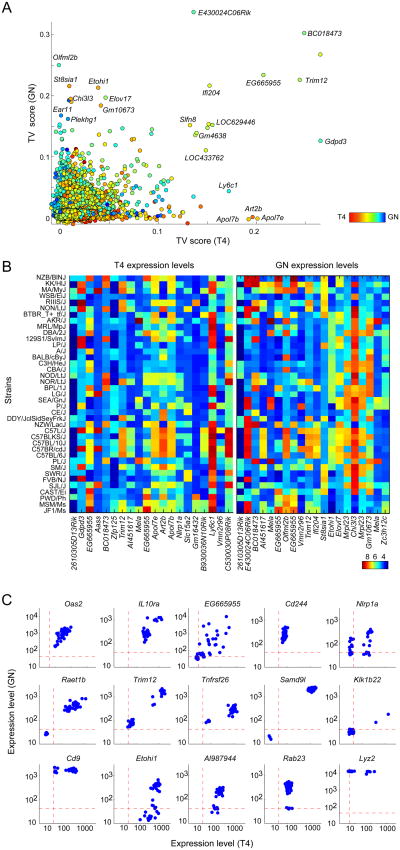
We first investigated the nature and extent of the transcript variability across the inbred strain panel. Overall we observed some variability in expression levels for the majority of genes (58% of tested genes, or 8,544 genes in T4—or 39% of genes—, and 10,006 genes in GN—46% of genes— at 5% FDR; Figure S1A). Of these, 2,508 genes in T4, and 3,711 genes in GN, had a greater than two-fold difference between the highest two and lowest two expressing strains. Some of the most variable genes correspond to retroviral elements (Mela, EG665955), and some to loci with known copy number variation (e.g. Cd244, Trim12, Glo1 (30)). A “true variability” (TV) score was computed for each gene (and per cell type) to identify transcripts whose variance across the strains could be attributed to meaningful differences, by factoring out technical factors and unwanted variability(Figure 1A; Figure S1A). In practice we computed a TV score for each gene by contrasting a measure of within-strain variability (computed from biological replicates) to between-strain variability (Methods). We validated the reproducibility of these TV scores by (a) comparing them to TV quantified from a previous gene expression dataset from macrophages for the Hybrid Mouse Diversity Panel, which included 22 of the strains tested here(11), and (b) assessing the correspondence with reported variability in DNA as ehypersensivity sites in eight inbred strains (31). Reassuringly, we found a significant correlation between the TV scores in GN and T4 with that computed from macrophage data (Spearman rho=0.26 for GN and rho=0.2 for T4, p-value<10-100) (Figure S1B). We also observed significantly higher TV scores for genes previously identified to have variable DNase sites nearby, compared to the background TV scores (p-value<10-3; Figure S1C).
Figure 1. The extent and patterns of gene expression variation between inbred mice.
A TV score was quantified for each transcript by contrasting a measure of between strain variability, computed using biological replicates for each strain, with that of within strain variability. (A) TV scores plotted per transcript based on T4 and GN data; each point represents a transcript. Colors depict preferential expression in T4 (red) or GN (blue) as quantified by the difference between mean expression levels. (B) Heatmap of expression levels for the top 20 most variable transcripts based on T4 and GN data. (C) Examples of three types of “variation patterns”. For each example transcript, each point represents a (mouse) strain, x-axis shows expression in T4 and y-axis shows expression in GN.
The distribution of expression across the strains for variable genes covered a wide range, with varying patterns (Fig. 1B,C). In most cases, a continuous spectrum was observed, hinting at a complex genetic determinism (Figure 1C, top row). In others, bimodal patterns were observed, which we quantified by assessing the fit of a Gaussian mixture model to expression pattern of each gene (433 and 567 such bimodal genes for T4 and GN expression, respectively, were identified at a Bonferroni-corrected p-value<0.05; Figures 1C; Methods). We also searched for instances of complete loss of function by using a combination of the bimodality test and expression below the 0.05 probability of expression in at least two strains(Methods). Overall, we identified 67 and 53complete loss-of-function loci in T4 and GN, respectively, of which 10 lost expression in both cells (Figure 1C, middle row; a complete list of loss-of-function loci is available from www.immgen.org). An example gene displaying such an on/off pattern was Raet1b, which encodes a natural killer cell lectin-like receptor ligand; it was silent in five of the strains but highly expressed in all others. This pattern was consistent for T4 and GN, likely reflecting the variation in composition of the Rae1α–ε family, and more generally the multiplicity of targets of NKG2D (32). There were also several instances of “conditional loss-of-function”, loci whose expression was sometimes absent in one cell type but present in all strains in the other cell type (Figure 1C, bottom row); for example, Rab23 transcripts were absent in GNs for some of strains, but present in all T4s. Several of these strains can thus serve as “natural knockouts” or “natural knockdowns”, either directly or by backcrossing the segments involved.
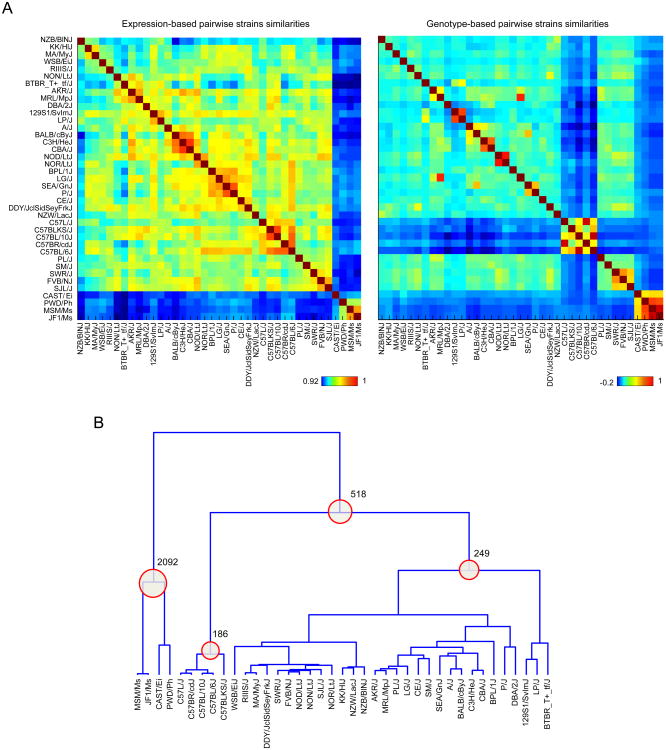
We assessed the impact of genetic variation on gene expression at a global level by comparing the relationships between the strains inferred from gene expression data with known genealogies and with genotype-derived relationships (Figure 2A). Simple examination of the parallel correlation maps of Figure 2A showed a significant correspondence between strain relationships as derived from the gene expression data and strain genotypes (1,33). Differences are sharper on the genotype than on the expression matrix, most trivially because the former inherently focuses on differences (SNPs) rather than on transcripts that are largely shared, and/or because most SNPs have no transcriptional consequence. As expected, the wild-derived strains (CAST/EiJ, PWD/PhJ, JF1/Ms, MSM/Ms) were more similar to each other than the classical inbred strains;the CAST/Ei strain, derived from M. m. castaneus species, was the most distant outlier, while the two M. m. molossinus derived strains (JF1/Ms and MSM/Ms) were more closely related to each other. Other relationships expected from strain histories (34) include the “C57 black” group of strains, the high pairwise similarity between CBA and C3H, or between NOD and NOR, both of which derive from the same stock through selection for susceptibility or resistance to diabetes (35).
Figure 2. Expression-based and genotype-based strain similarities.
(A) The similarity between each pair of strains was computed from genotype data or gene expression data using Spearman correlation. For expression data, all expressed genes (expressed in either cell type) were used in the computation. For genotype data, all variants satisfying the initial criteria (MAF > 0.05 and missing rate < 10%) were used. Each element in the heatmap (matrix) represents the strength of the similarity between a pair of strains. Expression-based and genotype-based similarity heatmaps follow the same row and column order. (B) The dendrogram was derived from a genotype-based similarity matrix using hierarchical agglomerative clustering. To account for strain-based scale differences between the distribution of similarities, pairwise strain distances for constructing the dendrogram were derived from ranked pairwise similarities for each strain. Each internal node in the dendrogram was used to define two groups (clusters) of strains—represented by descending leaves, and non-descending leaves. Numbers of differentially expressed genes (DEGs) (defined using t-test and a 5% FDR threshold) for groupings that yielded > 100 DEGs at 5% FDR are shown.
For a better handle on the number and identity of differentially-expressed transcripts that underlie these relationships, we created a genotype-based dendrogram depicting the relationship between the strains, and identified differentially expressed genes that characterized each group (Figure 2B). The wild-derived group was associated with 2,092 differentially expressed genes (5% FDR, of which 204 differ by a fold-change>2). These “wild-specific” genes have a range of functionalities, as evidenced by the absence of enrichment for any particular functional category based on GO analysis. Manual exploration of the top associations identified several suggestive differences: the marked under-expression of some toll-like-receptors (Tlr1 and Tlr7) in T4 cells from wild-derived strains; several members of the NK family (Klrd1, Klrb1f) or of the interferon-response pathway (Ifitm1, Ifitm2) were uniquely expressed in wild-derived T4;transcriptsencoding cell-surface molecules whose distribution is normally restricted to myeloid cells (Atp1a3, CD163, and Anxa3) but were present in T4 from wild-derived strains.
We also noted an intriguing differential expression of Eps8l1 in the C57 black group. Mutations in Eps8 family members lead to diverse auditory phenotypes, and the C57 strains are known to develop age-related hearing loss(36). At its inception, this project aimed to find, in the genetic and gene expression data, correlates to the phenotypic traits of these mouse strains, as assembled in the MPD database. Unfortunately, a systematic test for association between gene expression levels and an extensive set of behavioral and physiological traits (∼1500 traits from the Mouse Phenome Database) (37) did not yield significant findings, when corrected for random association. Reasons for this may include the limited number of strains for which complete phenotypes were available, buffering of gene expression by regulatory networks, or that the two cell types examined here are not relevant to the traits currently in the Phenome database.
Identifying cis-eQTLs for Neutrophils and CD4+ T cells
By correlating local genotype and expression data for the mice, we next identified specific cis genetic variants that impact gene expression levels in T4 and/or GN (our study did not have the statistical power to detect trans-eQTLs). To eliminate broad population-based trends that can result in the inflation of association p-values(29), we removed the effect of the top two Principal Components (PC) of the genotype, which represent population structure, from the gene expression data using linear regression. We chose two principal components by assessing the inflation factor\lambda (24) (see Methods). We performed a cis-eQTL analysis with the residuals of this fit, defining cis SNPs as mapping in a 1Mb window from the transcription start site. To increase our power to detect eQTLs that are shared by the two cell types while also detecting cell-specific eQTLs, we performed two analyses: (1) in a “joint” analysis, we combined data from the two cell types, and evaluated the significance of each SNP-to-gene association using permutation analysis; (2) in a “cell-specific” analysis, we explicitly tested the significance of a cell-specific SNP effect (Methods). In both cases, using permutation analysis, we obtained a gene-level p-value that took into account the number of tested cis variants(25,26,38), and defined significant cis-eQTLs at 5% FDR based on these gene-level p-values.
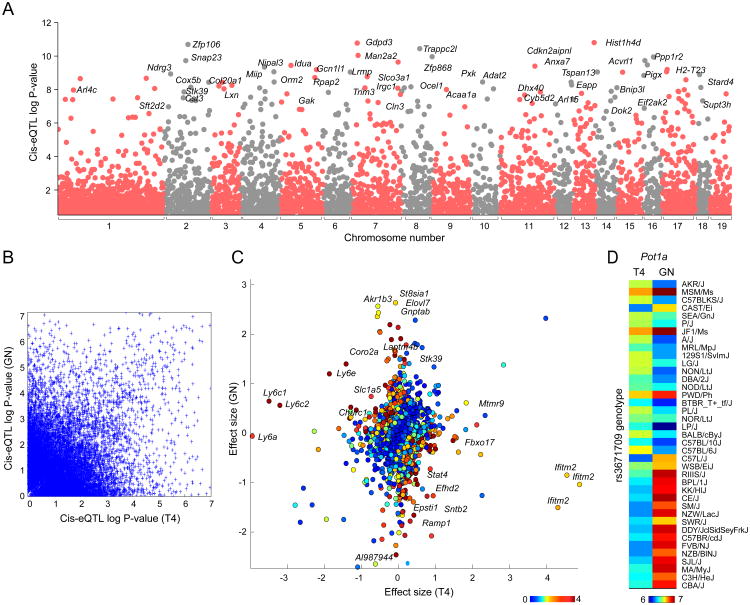
Using the joint analysis, we identified 1,047 genes with cis-eQTLs (Figure 3A, listed in TableS1 and available for browsing on the ImmGen server). The joint analysis increased our discovery power: we identified 262 eQTLs that were not detected in separate analyses of GN and T4 data (774 and 958 eQTLs in separate analysis of T4 and GN, respectively). We observed a significant correlation between cis-eQTL association strengths and TV (Spearman rho=0.29, p-value<10-100).
Figure 3. GN and T4 joint-discovered and cell-specific cis-eQTLs.
(A) Association p-values for each transcript and all of its cis SNPs (1Mb from the TSS) were computed using the Wilcoxon rank sum test. Association statistics were computed from both T4 and GN data (“joint analysis”). Each point represents a transcript; only the best association for each transcript is shown. (B) For each cell type, association p-values for SNP-transcript pairs were computed separately. The association p-values are shown for the best GN SNP and best T4 SNP for each transcript (i.e., each transcript is represented twice). (C) Cell-specific SNP-transcript association p-values were computed by testing the significance of an interaction term (see Methods). Effect sizes for the best SNP for each transcript and each cell type were computed as the mean difference between strains with alternative and reference alleles. Colors depict the strength (negative log10 p-value) of cell-specificity at gene-level. Figure shows effect size and association strengths for the best SNP for all expressed genes. (D) Heatmap shows the expression levels for the gene Pot1a in GN and T4. Strains are sorted based on the genotype of the best SNP for Pot1a.
Previous studies have identified cis-eQTLs for inbred mouse in various tissues including liver (10,12-14), and immunocytes(10,11). We compared our set of cis-eQTLs with those identified in macrophages(11), which was the most relevant and comparably sized. Orozsco et al. identified 1,937 genes (corresponding to 4,897 SNP-gene pairs) with cis-eQTLs controlling transcripts in primary macrophages that were testable in this study. To robustly compare results, we used Storey's π1 statistic(27), and observed a replication rate of 55% (p-value<0.001 under permutation testing). This estimate of overlap is similar to those previously reported in the literature for studies involving different cell types or conditions(28,39-41).
To identify cell-specific cis-eQTLs, which should denote genetic impact on cell-specific regulatory pathways, we considered 9,698 genes that are expressed in both cell types. We identified 234 significant cell-specific cis-eQTL, which indicates that ∼30% of discovered cis-eQTLs are cell-specific (Figure 3B)—an estimate consistent with recent reports of tissue- and cell-type specificity of eQTLs in human studies(40,41). For many genes with a cell specific eQTLs signal, we found major differences between effect sizes for the associated SNP in the two cell types (Figure 3C). This analysis also identified 17 eQTL where expression values correlate in an opposite manner in the two cell types. For ten of the 17 genes, the same top SNP was identified from both GN and T4 data. One of the strongest eQTL with opposite directionality of effect was observed for Pot1a (Figure 3D). The proportion of directional cis-eQTL discovered here is similar to those previously detected using primary immunocytes in human(21,42). This divergence may reflect the fact that a factor recruited to the same motif acts in an opposite manner in the two cells, but it is also possible that the SNP identified is in linkage disequilibrium with two different causal SNPs, each active in one cell type only.
Identifying regulatory links by co-expression analysis
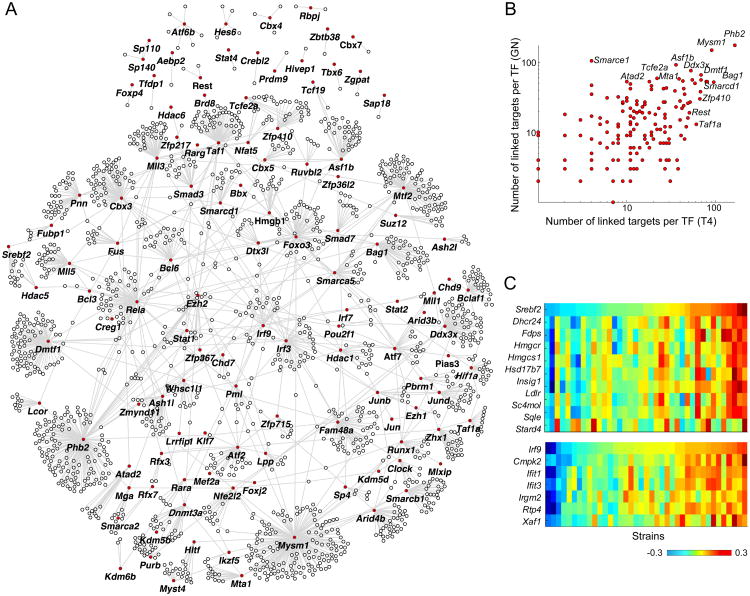
Gene expression datasets that carry small “perturbations” such as those resulting from genetic variation can be fruitfully exploited to reverse-engineer the structure of genetic regulatory networks (43-45), with the caveat that relationships based solely on baseline co-expression cannot resolve causal from merely correlative associations. We constructed regulatory networks where we inferred interactions (links) between a set of 164 transcription factors (TFs) and 3,675 candidate downstream targets using stepwise regression —this analysis included only genes that were expressed in both cell types and had a non-negligible TV score (as per Figure S1A). As above, to avoid artifacts from broad population structure, we used the genotype PC-corrected data. We identified 3,462 and 3,321 significant (5% FDR) links in T4 data and GN data, respectively, and 4,927 links in a joint network constructed using both T4 and GN data. For these networks, few regulatory hubs correlated with expression levels of a large number of targets (>100) and most TFs were linked to 15 or less targets (Figure 4A,B). The major hubs mostly include chromatin modifiers and generic transcriptional activators such as Smarcd1 and Smarce1 (SWI/SNF related chromatin regulators), Asf1b (a histone chaperone), Phf21a (a histone deacetylase), and the histone de-ubiquitinase Mysm1 (Figure 4B).
Figure 4. Analysis of gene co-expression in mouse.
(A) Overall network showing TF-target links discovered from T4 and GN data (“joint network”; for visualization purpose, figure only shows a limited set of the strongest links). TFs are marked in red. (B) Figure shows the node degree for each TF in T4 (x-axis) and GN (y-axis) networks. (C) Expression heatmaps for selected regulators and their inferred targets; top heatmap shows data for Srebf2 and nine of its target, bottom heatmap shows data for Irf9 and six of its targets (only targets that overlap with the Ontogenet predictions based on ImmGen data are shown).
We evaluated the overlap between GN and T4 inferred regulatory links using Storey's π1 statistic(27). Considering only the interactions passing the statistical significance threshold in the discovery sample (5% FDR), we estimated replication rates of π1=53% and 49%, for T4 links in GN and vice versa, respectively, indicating that a large fraction of these associations are shared among the two cell types. Conversely, by directly testing the significance of a cell-type-specific effect (Methods), we estimated that 17% of total interactions are truly cell specific (at 5% FDR). With the interaction test, Lmo2 was one of the most differential regulatory hubs, with 51 inferred links in GN, but only four potential target genes in T4, which likely denotes a very specific role in GN (its targets in GN do not correspond to a distinct functional category in GO analysis).
For an independent validation of co-expression relationships identifiable from this data, we compared a joint set of links identified from analysis of both cell types (“joint network”; Methods) with a previous network constructed from the ImmGen compendium using the Ontogenet algorithm. Ontogenet exploits variation in expression through differentiation cascades to identify regulatory relationships(7). We hypothesized that true TF-target pairs identified by Ontogenet would also show evidence of co-expression when natural genetic variation was the network perturbant. First, we evaluated the strength of co-expression between pairs of TF-targets previously identified by Ontogenet, and, using the π1 statistic on adjusted p-values for co-expression correlation coefficients(Methods), we found that 27% of these links show evidence of co-expression here. Conversely, we checked whether the targets of each TF are also more likely to belong to the same Ontogenet module by testing for significantly enriched Ontogenet modules among the predicted targets of each TF using the hypergeometric test. For 11 of the 127 TFs with at least 10 inferred targets, the targets were significantly enriched in an Ontogenet (fine) module at 5% FDR (Table S2). For example, Srebf2, which encodes asterol regulatory TF, was associated with 33 genes here, 9 of which were part of the same module and predicted by Ontogenet to be regulated by Srebf2(p-value<10-15; Figure 4C). Another well-known set of replicated links was between Irf9 and six of its known targets within the interferon response signature (p-value<10-8; Figure 4C). Although less robust to differences in inference method and sample sizes, we also directly evaluated the overlap between the inferred regulatory links here and that of Ontogenet, where we observed a modest (4%) but significant overlap (hypergeometricp-value<10-10).
Co-expression relationships that underlie the regulatory links here are not conclusive of directionality. To disentangle causal from simply correlative associations in the present network, we examined the propagated influence of cis variants associated with the inferred TFs (46). In practice, we asked whether a cis-eQTL SNP for a TF was also correlated with the expression levels of the TF's inferred targets. Within the set of links identified in the joint network (4,927 links) 230 links were testable as they were incident to one of the 15 TFs for which a cis-eQTL had been identified above; 50 links(22%) were “causally” supported, in that the genotype at the cis-eQTL was significantly associated with the expression of the TF's targets at 5% FDR.
Comparison of variation in gene expression in human and mouse
Comparative studies of gene expression patterns across species have mainly focused on comparing similarities and differences in expression across tissues, cell types, or responses to triggers. In these studies, conserved cell-type specificity or response to similar triggers across species is taken to indicate conserved functionality(19,47-52). The impact of genetic variation in each species is averaged, smoothed or factored out in such analyses. Instead, we sought to exploit the diversity of genetic background across inbred mice and across the human population sampled in the ImmVar study (which includes 360 healthy individuals from Asian, Africa, and European backgrounds with available expression data for CD4 and CD14 cells - the derivation and analysis of ImmVar datasets are detailed elsewhere (21)). ImmVar was designed to match the present analysis in several respects (parallel profiling of CD4+ T in both human and mouse). We took advantage of these congruent datasets to explore the similarities and differences in expression variability, impact of cis regulatory variation, and inferred regulatory interactions in mouse and human. For this analysis, we considered 14,130 genes with one-to-one human/mouse orthology (MGI HMD_Human5 set), and restricted the analysis to 5964 genes expressed in T4 (we only analyzed the T4 data, because of the exact correspondence of this cell type in our data from the two species).
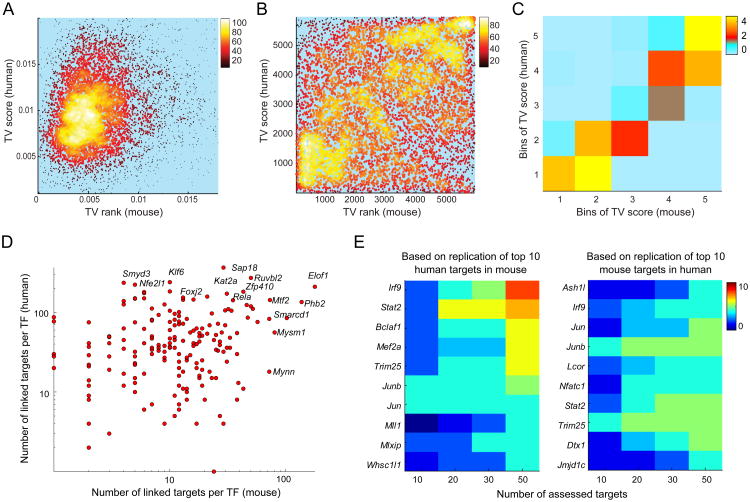
First, we applied the same TV metric of variation discussed above to compare the variability in genes' expression in human and mouse. The TV scores were calculated for human genes by using replicate samples prepared from the same donor (collected at intervals ranging from 3 to 25 weeks) after accounting for batch, age, and gender (using linear regression). The TV metric allowed us to eliminate gene-wise technical variability and only capture biological variability (responding to environmental and/or genetic cues). Human versus mouse comparison of the TV scores showed interesting patterns (Figure 5A-C); some genes were variable in one species or the other, but in general there was a correlation between TV in mouse and human (Spearman rho=0.16, p-value<10-10). We categorized genes into five equally-sized bins in each species based on TV scores and found significant predictability of TV scores in the second species based on the assigned bin in the first species (Figure 5C; Wilcoxon rank sum test p-values 10-4 to 10-8 for bins created in mouse and human, respectively): for example 26% of the genes in the top bin (most variable) in one species are also categorized in the top bin the other species.
Figure 5. Sharing of variability and co-expression in mice and humans.
(A) Figure shows a density map of the scatter plot for TV scores (expressed genes only) in T4s of human and mouse (Spearman correlation rho=0.16). For visualization purpose, the figure is zoomed in on the high density region. (B) Figure shows the density map of the scatter plot for TV ranks (TV scores were transformed to ranks; in each species, highest TV score is assigned a rank of 5965). (C) To quantify the significance of the overlap of variable genes in the two species, mouse and human TV scores were each binned into five equally-sized bins, and for each pair of TV bins (a square), the density of genes observed in that square was compared to density of genes in the same square under random mapping of human genes to mouse genes. 50,000 permutations (permuting the mapping between human and mouse genes) were performed to quantify the significance of the observed density in each square. Colors depict the strength of these p-values (negative log10 p-value). (D) Figure shows the network degree (number of inferred links) for each TF in the human and mouse networks. (E) Heatmap summarizes the number of shared top targets for each TF in human and mouse. The top ten targets for each TF in human (mouse) were first identified, and then the overlap with the top N=[10, 20, 30, 40, 50] mouse (human) targets for the same TF was computed. Only regulators with at least 4 overlapping targets in the list of top 50 targets (p-value<10-6) are shown. The second plot shows the result where the top 10 targets for each regulator in mouse were evaluated in human data.
The variability captured by the TV metric encompasses environmental and other factors beyond the impact of genetic variation. To compare the extent of genetically determined variation in gene expression in both species, we evaluated the overlap of cis-eQTLs in human and mouse. Using the same methodology as above, we identified 2,285 cis-eQTL genes in the human ImmVar datasets among the set of 7,098 expressed genes in both species (either cell type)(eQTLs identified using the joint analysis; Methods). Of the 674 genes associated with an eQTL discovered in mice for this set of expressed genes, 275 were also associated with an eQTL based on human data (hypergeometirc p-value<10-6), implying that genes which show a significant impact of local genetic variation tend to overlap in mice and humans, even though the variants themselves are certainly unrelated.
Next we compared gene regulatory networks constructed from the T4 dataset for both human and mouse. The motivation was to analyze the evolutionary conservation of these regulatory links, and from a practical standpoint to validate the inferences by confirmation in another species. For each species, we first constructed a network using stepwise regression as above (Methods). At a global level, we observed a correlation between TFs' out-degree (the number of targets connected to each TF; Figure 5D), with 38% of the top 20% hubs in one species shared with the second species (p-values <0.01). Per above, chromatin modifiers tend to be strong regulatory hubs in both species. We used the π1 statistic to estimate the fraction of TF-to-target links identified in one species that are replicated in the second species. A 47% replication rate was observed for mouse links in the human T4 dataset, and a 19% replication rate for human links in the mouse dataset (permutation analysis p-value<0.001) (Figure S2A,B). Finally, in a regulator-centric analysis, we also assessed the correspondence between top co-expressed links for each TF in the two species. To do so, we assessed the overlap and the distribution of co-expression values (correlation coefficients) for the top N=[10,20,30, 50] targets of each TF in the second species (Methods). Of the 189 TFs that were analyzed, we identified 17 TFs whose top 10 targets were highly conserved (hypergeometic test p-value<10-6; Figure 5E), and the top targets of an additional set of 42 TFs showed significant evidence of high co-expression values in the second species (using the Wilcoxon rank sum test; Figure S2C,D). Among these highly conserved co-expression links, we identified well-known relationships, including co-expression between Irf9 and interferon response genes Dh×58, Ifi35, Irf1, Pml, Trafd1, Stat2 and strong co-expression between Jun and Fos and known early response genes (Ier2, Gadd45b). We did not attempt to interpret the divergent regulatory links within these datasets: these are not conclusive of true differences, since multiple confounding factors can underlie such differences (different environmental influences, much smaller sample size for the mouse data, imperfect mapping of human to mouse probes). Overall, these comparisons show that many of the regulatory connections that can be inferred from the inter-individual variation in expression profiles are conserved between these two mammalian species.
Discussion
Our motivation, in the context of the ImmGen and MDP programs, was three-fold: to serve as a reference of genomic and genetic information relevant to the immune function in mouse; to provide additional material for the dissection of genetic regulatory networks; to provide a documented basis for comparison of the mouse and human immune systems.
In terms of resource, the present data provide useful information at several levels, and are all available interactively from the ImmGen and MDP web browsers (www.immgen.org,phenome.jax.org). We detected a number of genes with greater than a two-fold change in expression across the strains (the empirical rule-of-thumb for functional significance). It will be interesting to see how these traits segregate in settings such as the Collaborative Cross strains, where the chromosomal segments can be traced in the recombinant chromosomes, allowing refinement of the genetic control and/or discovery of epistatic modifiers. Variation followed both bimodal and continuous expression patterns across mouse strains, including a few loci with complete loss of expression in some of the mouse strains. As such, these can serve as are source of “natural knockdowns” or “natural knockouts” (some affecting both cell types, others cell-specific). The 1,222 cis-eQTLs detected in the two immunological cell types are also available through the dedicated ImmGen interface. However, the relatively large size of the LD blocks in these inbred mouse strains, relative to outbred humans or mice(22), make it impossible to pinpoint with precision the causal variant, and the SNPs listed should only be considered as likely proxies of the truly relevant variant. Never the less, the patterns of variation and the eQTLs described here, and their conservation across species, may help to interpret differences in susceptibility to infection or autoimmune diseases, in a manner than translates to genetic in risk human populations.
The patterns of inter-strain variability followed, as expected, the patterns of genetic distance and genealogical history between the strains. Wild-derived strains were predictably more distant from the classic inbred lines. Some of this genetic distance may be directly related to selective events during mouse domestication or to the input from non-domesticus subspecies. We previously reported that a variant at the Il1b locus, which leads to a 5- to 10-fold greater Interleukin-1 response to stimulation through innate receptor pathways, is frequent in wild-derived strains but quite rare among classical inbred strains (53), and some of the expression variations uncovered here may be of the same nature (e.g. Tlr1 and Tlr7, although in this instance it is the wild-derived strains that show low or absent expression in T4). Some genes whose expression is normally confined to myeloid cells were expressed in CD4+ T cells of the wild-derived strains. Some of these conditionally expressed genes are surprising, such as the expression of CD163, a scavenger receptor on macrophages whose function in CD4+ T cells is not immediately obvious. We might speculate that this reflects a mode of innate sensing by CD4+ T cells that was lost during domestication (interestingly, however, human T cells do not express these monocyte genes).
The distribution of cis-acting genetic variation was significantly correlated with the variation in expression for the most variable genes, although many of the genes with a high TV score did not show an active cis-eQTL. Recent literature indicates a larger impact for local sequence variation, which may have been detectable with larger sample sizes(28,54), perhaps attainable with a larger study of outbred or Collaborative Cross mice. We note that the number of cis-eQTLs detected here is more than what would be expected from an equally sized human dataset (28) where the effect of environment cannot be as effectively controlled.
The co-expression-based network estimated here extended the analysis of the regulatory networks of immunocytes initiated in ImmGen(7), and we observed a comforting degree of overlap between the two analyses. Although co-expression cannot formally identify causal directionality in a correlated pair (i.e. who controls whom), the selection of transcriptional regulators provides a functional prior for directionality. Indeed, when we searched for causal chains of associations, by correlating a cis variant impacting the expression of a TF with the TF's downstream effects on its inferred targets, a significant portion (22%) of the testable links turned out to be causally driven. Interestingly, connections identified from inter-strain variation more frequently involved generic regulators such as chromatin modifiers, which showed up here as major hubs, than classic sequence-specific DNA-binding TFs and lineage determination factors (which were predictably more prevalent in the Ontogenet analysis). This difference is in line with the paucity of cis-eQTLs for classic transcription factors regulators involved in differentiation or lineage determination, as previously shown in human cells (28,55). One might speculate that a degree of “noise” in transcript level resulting from variations in redundant and pleiotropic factors is better tolerated (or even favored) than variation in more specific factors that form the blueprint of cell differentiation and lineage determination. This dominance of broad transcriptional regulators as major co-expression hubs was strikingly reproduced in the human datasets.
Finally, we observed sharing of the patterns of expression variability between human and mouse. Both genetic and non-genetic factors can result in expression variability, and we also observed significantly non-random overlaps in genes that are associated with cis-eQTLs in both species. From an evolutionary standpoint, this “conservation of variability” can be explained by species-shared strength of selection pressure on gene expression levels (56): variation in more redundant and/or less essential genes is better tolerated, and these characteristics would tend to be conserved. It is also possible that some of this species-shared variability is in genes whose intra-species variation is favorable. The extraordinary diversification of coding sequence in MHC genes favors heterozygosity in individuals and diversity at the level of the species to best meet variable pathogen challenges (57). Similarly, it may be advantageous to diversify the levels of expression, and hence of response potential, in pathways of the immune system. Genes controlling activating and inhibitory NK receptors would plausibly fall in that category. From a mechanistic standpoint, one might also imagine different scenarios for the roots of this reproducible variability: some regions of the genome may be inherently noisier, a characteristic preserved during the evolutionary shuffling of syntenic chromosomal regions; regulatory feedback loops that control individual genes or sets of genes may be more or less robust; miRNAs or other non-coding RNAs might make for a variable degree of control. Any of these mechanisms may have been, to an extent, preserved through 200 million years of evolution, to conserve immunologically relevant variation.
Supplementary Material
Acknowledgments
We thank ImmGen participants for comments and suggestions, J. Ericson, S. Davis for help with RNA preparation and profiling, S. Davis for data processing, and Jonathan Flint for helpful discussions.
Abbreviations used
- T4
CD4+ T cell
- GN
granulocyte (polymorphonuclear neutrophil)
- LD
linkage disequilibrium
- eQTL
expression quantitative trait locus
- MPD
Mouse Phenome Database
Footnotes
This work was supported by a resource grant from the NIH to the Immunological Genome Project (R24 AI072073); DA028420 and AG038070 to M.B.
References
- 1.Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, Festing MF, Fisher EM. Genealogies of mouse inbred strains. Nat Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- 2.Wade CM, Daly MJ. Genetic variation in laboratory mice. Nat Genet. 2005;37:1175–1180. doi: 10.1038/ng1666. [DOI] [PubMed] [Google Scholar]
- 3.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O'Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 4.Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellaker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assuncao JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogue MA, Grubb SC. The Mouse Phenome Project. Genetica. 2004;122:71–74. doi: 10.1007/s10709-004-1438-4. [DOI] [PubMed] [Google Scholar]
- 6.Heng TS, Painter MW Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 7.Jojic V, Shay T, Sylvia K, Zuk O, Sun X, Kang J, Regev A, Koller D, Best AJ, Knell J, Goldrath A, Jojic V, Koller D, Shay T, Regev A, Cohen N, Brennan P, Brenner M, Kim F, Rao TN, Wagers A, Heng T, Ericson J, Rothamel K, Ortiz-Lopez A, Mathis D, Benoist C, Bezman NA, Sun JC, Min-Oo G, Kim CC, Lanier LL, Miller J, Brown B, Merad M, Gautier EL, Jakubzick C, Randolph GJ, Monach P, Blair DA, Dustin ML, Shinton SA, Hardy RR, Laidlaw D, Collins J, Gazit R, Rossi DJ, Malhotra N, Sylvia K, Kang J, Kreslavsky T, Fletcher A, Elpek K, Bellemare-Pelletier A, Malhotra D, Turley S. Identification of transcriptional regulators in the mouse immune system. Nat Immunol. 2013;14:633–643. doi: 10.1038/ni.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montgomery SB, Dermitzakis ET. From expression QTLs to personalized transcriptomics. Nat Rev Genet. 2011;12:277–282. doi: 10.1038/nrg2969. [DOI] [PubMed] [Google Scholar]
- 9.Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014;15:34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerrits A, Li Y, Tesson BM, Bystrykh LV, Weersing E, Ausema A, Dontje B, Wang X, Breitling R, Jansen RC, de HG. Expression quantitative trait loci are highly sensitive to cellular differentiation state. PLoS Genet. 2009;5:e1000692. doi: 10.1371/journal.pgen.1000692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orozco LD, Bennett BJ, Farber CR, Ghazalpour A, Pan C, Che N, Wen P, Qi HX, Mutukulu A, Siemers N, Neuhaus I, Yordanova R, Gargalovic P, Pellegrini M, Kirchgessner T, Lusis AJ. Unraveling inflammatory responses using systems genetics and gene-environment interactions in macrophages. Cell. 2012;151:658–670. doi: 10.1016/j.cell.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, Neubauer M, Neuhaus I, Yordanova R, Guan B, Truong A, Yang WP, He A, Kayne P, Gargalovic P, Kirchgessner T, Pan C, Castellani LW, Kostem E, Furlotte N, Drake TA, Eskin E, Lusis AJ. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS, Ferris MT, Frelinger JA, Heise M, Frieman MB, Gralinski LE, Bell TA, Didion JD, Hua K, Nehrenberg DL, Powell CL, Steigerwalt J, Xie Y, Kelada SN, Collins FS, Yang IV, Schwartz DA, Branstetter LA, Chesler EJ, Miller DR, Spence J, Liu EY, McMillan L, Sarkar A, Wang J, Wang W, Zhang Q, Broman KW, Korstanje R, Durrant C, Mott R, Iraqi FA, Pomp D, Threadgill D, de Villena FP, Churchill GA. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 2011;21:1213–1222. doi: 10.1101/gr.111310.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClurg P, Janes J, Wu C, Delano DL, Walker JR, Batalov S, Takahashi JS, Shimomura K, Kohsaka A, Bass J, Wiltshire T, Su AI. Genomewide association analysis in diverse inbred mice: power and population structure. Genetics. 2007;176:675–683. doi: 10.1534/genetics.106.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis MM. A prescription for human immunology. Immunity. 2008;29:835–838. doi: 10.1016/j.immuni.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Payne KJ, Crooks GM. Immune-cell lineage commitment: translation from mice to humans. Immunity. 2007;26:674–677. doi: 10.1016/j.immuni.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, Carninci P, Daub CO, Forrest AR, Gough J, Grimmond S, Han JH, Hashimoto T, Hide W, Hofmann O, Kamburov A, Kaur M, Kawaji H, Kubosaki A, Lassmann T, van NE, MacPherson CR, Ogawa C, Radovanovic A, Schwartz A, Teasdale RD, Tegner J, Lenhard B, Teichmann SA, Arakawa T, Ninomiya N, Murakami K, Tagami M, Fukuda S, Imamura K, Kai C, Ishihara R, Kitazume Y, Kawai J, Hume DA, Ideker T, Hayashizaki Y. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shay T, Jojic V, Zuk O, Rothamel K, Puyraimond-Zemmour D, Feng T, Wakamatsu E, Benoist C, Koller D, Regev A. Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc Natl Acad Sci U S A. 2013;110:2946–2951. doi: 10.1073/pnas.1222738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee MN, Ye C, Villani AC, Raj T, Li W, Eisenhaure TM, Imboywa SH, Chipendo PI, Ran FA, Slowikowski K, Ward LD, Raddassi K, McCabe C, Lee MH, Frohlich IY, Hafler DA, Kellis M, Raychaudhuri S, Zhang F, Stranger BE, Benoist CO, De Jager PL, Regev A, Hacohen N. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343:1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raj T, Rothamel K, Mostafavi S, Ye C, Lee MN, Replogle JM, Feng T, Lee M, Asinovski N, Frohlich I, Imboywa S, Von KA, Okada Y, Patsopoulos NA, Davis S, McCabe C, Paik HI, Srivastava GP, Raychaudhuri S, Hafler DA, Koller D, Regev A, Hacohen N, Mathis D, Benoist C, Stranger BE, De Jager PL. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirby A, Kang HM, Wade CM, Cotsapas C, Kostem E, Han B, Furlotte N, Kang EY, Rivas M, Bogue MA, Frazer KA, Johnson FM, Beilharz EJ, Cox DR, Eskin E, Daly MJ. Fine mapping in 94 inbred mouse strains using a high-density haplotype resource. Genetics. 2010;185:1081–1095. doi: 10.1534/genetics.110.115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 25.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, Ingle CE, Sekowska M, Smith GD, Evans D, Gutierrez-Arcelus M, Price A, Raj T, Nisbett J, Nica AC, Beazley C, Durbin R, Deloukas P, Dermitzakis ET. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proc Natl Acad Sci U S A. 2005;102:12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Battle A, Mostafavi S, Zhu X, Potash JB, Weissman MM, McCormick C, Haudenschild CD, Beckman KB, Shi J, Mei R, Urban AE, Montgomery SB, Levinson DF, Koller D. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang HM, Ye C, Eskin E. Accurate discovery of expression quantitative trait loci under confounding from spurious and genuine regulatory hotspots. Genetics. 2008;180:1909–1925. doi: 10.1534/genetics.108.094201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orozco LD, Cokus SJ, Ghazalpour A, Ingram-Drake L, Wang S, van NA, Che N, Araujo JA, Pellegrini M, Lusis AJ. Copy number variation influences gene expression and metabolic traits in mice. Hum Mol Genet. 2009;18:4118–4129. doi: 10.1093/hmg/ddp360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hosseini M, Goodstadt L, Hughes JR, Kowalczyk MS, De GM, Otto GW, Copley RR, Mott R, Higgs DR, Flint J. Causes and consequences of chromatin variation between inbred mice. PLoS Genet. 2013;9:e1003570. doi: 10.1371/journal.pgen.1003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunol Rev. 2010;235:267–285. doi: 10.1111/j.0105-2896.2010.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petkov PM, Ding Y, Cassell MA, Zhang W, Wagner G, Sargent EE, Asquith S, Crew V, Johnson KA, Robinson P, Scott VE, Wiles MV. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;14:1806–1811. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morse H. Origins of inbred mice: proceedings of a workshop; Bethesda, Maryland. February 14-16, 1978; New York: Academic Press; 1978. [Google Scholar]
- 35.Kikutani H, Makino S. The murine autoimmune diabetes model: NOD and related strains. Adv Immunol. 1992;51:285–322. doi: 10.1016/s0065-2776(08)60490-3. [DOI] [PubMed] [Google Scholar]
- 36.Zheng QY, Johnson KR, Erway LC. Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hear Res. 1999;130:94–107. doi: 10.1016/s0378-5955(99)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grubb SC, Bult CJ, Bogue MA. Mouse phenome database. Nucleic Acids Res. 2014;42:D825–D834. doi: 10.1093/nar/gkt1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nica AC, Parts L, Glass D, Nisbet J, Barrett A, Sekowska M, Travers M, Potter S, Grundberg E, Small K, Hedman AK, Bataille V, Tzenova BJ, Surdulescu G, Dimas AS, Ingle C, Nestle FO, Di MP, Min JL, Wilk A, Hammond CJ, Hassanali N, Yang TP, Montgomery SB, O'Rahilly S, Lindgren CM, Zondervan KT, Soranzo N, Barroso I, Durbin R, Ahmadi K, Deloukas P, McCarthy MI, Dermitzakis ET, Spector TD. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grundberg E, Small KS, Hedman AK, Nica AC, Buil A, Keildson S, Bell JT, Yang TP, Meduri E, Barrett A, Nisbett J, Sekowska M, Wilk A, Shin SY, Glass D, Travers M, Min JL, Ring S, Ho K, Thorleifsson G, Kong A, Thorsteindottir U, Ainali C, Dimas AS, Hassanali N, Ingle C, Knowles D, Krestyaninova M, Lowe CE, Di MP, Montgomery SB, Parts L, Potter S, Surdulescu G, Tsaprouni L, Tsoka S, Bataille V, Durbin R, Nestle FO, O'Rahilly S, Soranzo N, Lindgren CM, Zondervan KT, Ahmadi KR, Schadt EE, Stefansson K, Smith GD, McCarthy MI, Deloukas P, Dermitzakis ET, Spector TD. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flutre T, Wen X, Pritchard J, Stephens M. A statistical framework for joint eQTL analysis in multiple tissues. PLoS Genet. 2013;9:e1003486. doi: 10.1371/journal.pgen.1003486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fairfax BP, Makino S, Radhakrishnan J, Plant K, Leslie S, Dilthey A, Ellis P, Langford C, Vannberg FO, Knight JC. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012;44:502–510. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 44.Gardner TS, Bernardo D, Lorenz D, Collins JJ. Inferring genetic networks and identifying compound mode of action via expression profiling. Science. 2003;301:102–105. doi: 10.1126/science.1081900. [DOI] [PubMed] [Google Scholar]
- 45.Bar-Joseph Z, Gerber GK, Lee TI, Rinaldi NJ, Yoo JY, Robert F, Gordon DB, Fraenkel E, Jaakkola TS, Young RA, Gifford DK. Computational discovery of gene modules and regulatory networks. Nat Biotechnol. 2003;21:1337–1342. doi: 10.1038/nbt890. [DOI] [PubMed] [Google Scholar]
- 46.Zhu J, Zhang B, Smith EN, Drees B, Brem RB, Kruglyak L, Bumgarner RE, Schadt EE. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat Genet. 2008;40:854–861. doi: 10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller JA, Horvath S, Geschwind DH. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci U S A. 2010;107:12698–12703. doi: 10.1073/pnas.0914257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan ET, Quon GT, Chua G, Babak T, Trochesset M, Zirngibl RA, Aubin J, Ratcliffe MJ, Wilde A, Brudno M, Morris QD, Hughes TR. Conservation of core gene expression in vertebrate tissues. J Biol. 2009;8:33. doi: 10.1186/jbiol130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng-Bradley X, Rung J, Parkinson H, Brazma A. Large scale comparison of global gene expression patterns in human and mouse. Genome Biol. 2010;11:R124. doi: 10.1186/gb-2010-11-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strand AD, Aragaki AK, Baquet ZC, Hodges A, Cunningham P, Holmans P, Jones KR, Jones L, Kooperberg C, Olson JM. Conservation of regional gene expression in mouse and human brain. PLoS Genet. 2007;3:e59. doi: 10.1371/journal.pgen.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Enard W, Khaitovich P, Klose J, Zollner S, Heissig F, Giavalisco P, Nieselt-Struwe K, Muchmore E, Varki A, Ravid R, Doxiadis GM, Bontrop RE, Paabo S. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- 53.Ohmura K, Johnsen A, Ortiz-Lopez A, Desany P, Roy M, Besse W, Rogus J, Bogue M, Puech A, Lathrop M, Mathis D, Benoist C. Variation in IL-1β gene expression is a major determinant of genetic differences in arthritis aggressivity in mice. Proc Natl Acad Sci U S A. 2005;102:12489–12494. doi: 10.1073/pnas.0504325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lappalainen T, Sammeth M, Friedlander MR, 't Hoen PA, Monlong J, Rivas MA, Gonzalez-Porta M, Kurbatova N, Griebel T, Ferreira PG, Barann M, Wieland T, Greger L, van IM, Almlof J, Ribeca P, Pulyakhina I, Esser D, Giger T, Tikhonov A, Sultan M, Bertier G, MacArthur DG, Lek M, Lizano E, Buermans HP, Padioleau I, Schwarzmayr T, Karlberg O, Ongen H, Kilpinen H, Beltran S, Gut M, Kahlem K, Amstislavskiy V, Stegle O, Pirinen M, Montgomery SB, Donnelly P, McCarthy MI, Flicek P, Strom TM, Lehrach H, Schreiber S, Sudbrak R, Carracedo A, Antonarakis SE, Hasler R, Syvanen AC, van Ommen GJ, Brazma A, Meitinger T, Rosenstiel P, Guigo R, Gut IG, Estivill X, Dermitzakis ET. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferraro A, D'Alise AM, Raj T, Asinovski N, Phillips R, Ergun A, Replogle JM, Bernier A, Laffel L, Stranger BE, De Jager PL, Mathis D, Benoist C. Interindividual variation in human T regulatory cells. Proc Natl Acad Sci U S A. 2014;111:E1111–E1120. doi: 10.1073/pnas.1401343111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Georgi B, Voight BF, Bucan M. From mouse to human: evolutionary genomics analysis of human orthologs of essential genes. PLoS Genet. 2013;9:e1003484. doi: 10.1371/journal.pgen.1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parham P, Ohta T. Population biology of antigen presentation by MHC class I molecules. Science. 1996;272:67–74. doi: 10.1126/science.272.5258.67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.