Abstract
Invariant natural killer T (iNKT) cells have been recently classified into NKT1, NKT2 and NKT17 lineages with distinct transcription factor and cytokine profiles, but mechanisms underlying such fate decisions remain elusive. Here, we report crucial roles of mTOR signaling especially mTORC2 in iNKT cell development and fate determination of NKT17 cells. Loss of Rictor, an obligatory component of mTORC2, decreased thymic and peripheral iNKT cells, associated with defective survival. Strikingly, Rictor deficiency selectively abolished the NKT17 lineage, as indicated by marked reduction of RORγt and IL-17 expression. Moreover, deletion of Pten upregulated mTORC2 activity and enhanced NKT17 generation, but concomitant loss of Rictor completely reversed the NKT17 dysregulation. In contrast, mTORC1 regulators Raptor and Rheb are dispensable for NKT17 differentiation, despite their importance in iNKT cell thymic development. Our findings establish pivotal and unique roles of mTORC2 signaling, which is reciprocally regulated by Rictor and Pten, in NKT17 lineage determination.
Keywords: iNKT, mTORC2, NKT17, Pten
Introduction
The invariant NKT (iNKT) cells are a unique group of αβ T cells characterized by their expression of a semi-invariant TCR-α (Vα14-Jα18) chain and a TCR-β chain of limited repertoire (1, 2). iNKT cells play an important role in bridging innate and adaptive immunity. In response to self- and bacteria-derived lipid antigens presented by CD1d molecules, iNKT cells rapidly produce a broad range of cytokines, including IFNυ, TNFα, IL-4 and IL-17, and mediate key immune functions (1, 2). The development of iNKT cells has been traditionally divided into four distinct stages based on unique surface molecules CD24, CD44 and NK1.1, namely immature Stage 0 (CD24+CD44−NK1.1−), transitional Stage 1 (CD24−CD44−NK1.1−) and Stage 2 (CD24−CD44+NK1.1−), and mature Stage 3 cells (CD24−CD44+NK1.1+) (3). A number of transcription factors have been implicated in iNKT cell development, with the most notable example being PLZF, a lineage-specific factor crucial for iNKT cell early development and functional differentiation (4, 5). In view of distinct transcription factor and cytokine profiles, iNKT cells have been recently classified into three effector lineages: NKT1, NKT2 and NKT17, as indicated by their expression of transcriptional factors T-bet, GATA3 and RORγt, respectively (6, 7). Despite the characterization of multiple regulators of iNKT cell development, mechanisms underlying the diversity of iNKT effector lineages remain elusive. Moreover, little is understood about the relationship between the developmental maturation and lineage diversification.
The mechanistic target of rapamycin (mTOR) signaling integrates immune signals and metabolic cues in orchestrating T cell responses (8, 9). mTOR signaling is comprised of two complexes mTORC1 and mTORC2, defined by the respective signature components Raptor and Rictor. mTORC1 and mTORC2 exhibit distinct functions in peripheral T cell responses, especially T cell activation and differentiation (10-14), but their developmental roles are considerably less understood (15). Of note, NKT thymocytes, unlike conventional T cells, undergo blasting and cell division during thymic development (1, 2). Consistent with this notion, loss of mTORC1 activity by deletion of Raptor has been shown in very recent studies to block development of iNKT cells at early stage and impair their functionality (16, 17), while permitting the normal development of conventional T cells. However, the role of mTOR in lineage diversification of iNKT cells is less clear (18). In particular, there has been no evidence linking mTORC2 to iNKT cell development or effector differentiation.
In this study, we have genetically defined the functions of various mTOR components in iNKT cell development, and established the critical role of mTORC2 signaling in NKT17 lineage differentiation. Loss of Rictor decreases iNKT cells in the thymus and periphery, associated with defective NKT cell survival but largely normal maturation. Strikingly, deficiency of Rictor, but not Raptor or Rheb (a crucial upstream regulator of mTORC1 (8, 9), abrogates the NKT17 lineage in the thymus and periphery. Moreover, NKT17 development is markedly enhanced by the deletion of the PI3K inhibitory molecule Pten, while concomitant loss of Rictor rescues this defect. Our studies have therefore identified crucial and unique roles for mTORC2 signaling, which is reciprocally regulated by Rictor and Pten, in establishing the NKT17 lineage.
Materials and Methods
Mice
C57BL/6J and Ptenfl mice were purchased from the Jackson Laboratory. Rptorfl, Rictorfl, Rhebfl and CD4-Cre mice have been described previously (13, 14). All mice have been backcrossed onto the C57BL/6J background, and were kept in specific pathogen-free conditions in Animal Resource Center at St. Jude. Animal protocols were approved by Institutional Animal Care and Use Committee of St. Jude.
Cellular assays
Cell isolation and flow cytometry of iNKT cells were as described (18, 19). For ex vivo stimulation, iNKT cells were enriched from total thymocytes by depleting CD8α+ cells (20), and stimulated with PMA and ionomycin in the presence of monensin for 5 h. For in vivo α-GalCer challenge, mice were injected intraperitoneally with 200 μg brefeldin A and intravenously with 2 μg α-GalCer 30 min later; splenic iNKT cells were analyzed for cytokine production at 3 h after α-GalCer treatment. For α-GalCer stimulation in vitro, enriched thymocytes were stimulated with 125 ng/ml α-GalCer for 72 h.
Statistical analysis
One-way ANOVA with Tukey’s test was performed to analyze statistical significance (Graphpad Prism 5.01). * p<0.05, ** p<0.01 and *** p<0.001 were used to show statistical significance throughout the article.
Results and Discussion
Loss of Rictor impairs the development of iNKT cells
To investigate the roles of Rictor in iNKT cell development, we crossed mice with loxP-flanked Rictor with CD4-Cre mice to delete the Rictor conditional alleles specifically in T cells (termed Rictor−/− mice). Rictor−/− mice had significantly decreased frequencies and numbers of iNKT cells (CD1d-PBS57+TCR-β+) in the thymus, spleen and liver, which were approximately one third of wild-type levels (Fig. 1A, 1B and 1C). To evaluate the role of Rictor in the developmental maturation of iNKT cells, we examined the expression of CD24, CD44 and NK1.1 that can be used to distinguish different stages of iNKT cells (3). Rictor−/− iNKT cells showed a modestly reduced frequency of Stage 2 (CD24−CD44+NK1.1−) but a normal percentage of Stage 3 cells (CD24−CD44+NK1.1+) (Fig. 1D and 1E). Therefore, Rictor contributes to normal development of iNKT cells but is largely dispensable for their terminal maturation.
Figure 1.
mTORC2 promotes iNKT cell development. Cells were from WT, Rictor−/−, Rptor−/−and Rictor−/−Rptor−/− mice. (A) Flow cytometry of iNKT cells (CD1d-PBS57+TCR-β+) in the thymus (Thy), spleen (Spl) and liver. Percentages (B) and numbers (C) of iNKT cells in the thymus, spleen and liver are shown. (D and E) Analysis of iNKT cell developmental stages in the thymus. Results are representative of 2-5 independent experiments. *, p<0.05; **, p<0.01; ***, p<0.001.
To explore the underlying basis for the reduced cellularity of iNKT cells in Rictor−/− mice, we first examined the proliferation of thymic iNKT cells by measuring BrdU incorporation and Ki67 expression. WT and Rictor-deficient iNKT cells had comparable BrdU incorporation and Ki67 expression (Supplemental Fig. 1A and 1B), indicating a largely dispensable role of Rictor in the proliferation of iNKT cells. However, Rictor−/− iNKT cells exhibited elevated Caspase activity, indicative of a survival defect (Supplemental Fig. 1C). Moreover, when stimulated with α-GalCer in vitro, Rictor−/− iNKT cells had more profound cell death than WT cells, as indicated by increased 7-AAD staining and Caspase activity (Supplemental Fig. 1D and 1E). Collectively, these data indicate that Rictor deficiency impairs survival of iNKT cells that likely contribute to the defective development.
Selective roles of mTOR signaling components in iNKT cell development
We next compared the effects of Rictor deficiency with the disruption of mTORC1 activity on iNKT cell development. To this end, we used the CD4-Cre system to delete Raptor and Rheb (Rptor−/− and Rheb−/−), key molecules associated with mTORC1 activity (8, 9). In contrast to the modest reduction of iNKT cells in Rictor−/− mice, Raptor deficiency caused a more profound impairment of iNKT cellularity in the thymus, spleen and liver (Fig. 1A, 1B and 1C). These defects were largely recapitulated in mice lacking both Rictor and Raptor in T cells (Rictorfl/flRptorfl/flCD4-Cre; Rictor−/−Rptor−/−) (Fig. 1A, 1B and 1C). Additionally, thymic development of Rptor−/− iNKT cells was blocked at an early stage, as indicated by the accumulation of Stage 0 and Stage 1 cells and a marked loss of Stage 3 cells (Fig. 1D and 1E). Severe defects were also observed in Rictor−/−Rptor−/− iNKT cells (Fig. 1D and 1E). These results indicate an essential role of Raptor in iNKT cell development.
In contrast, deficiency of Rheb resulted in a small reduction of iNKT cells in the thymus (Supplemental Fig. 2A-C), with marginal effects on the distribution of iNKT cells at different developmental stages (Supplemental Fig. 2D). These results indicate that Rheb-independent mTORC1 signaling plays a dominant role in promoting terminal maturation of iNKT cells, and highlight the discrete requirement of mTOR signaling in iNKT cell development.
Rictor but not Raptor or Rheb promotes NKT17 lineage differentiation
iNKT cells have been recently classified into three functional lineages according to the expression of the signature transcription factors (6, 7). We therefore used intracellular staining of PLZF and lineage-specific transcription factors as described in (6), to determine the roles of Rictor, Raptor and Rheb in iNKT effector lineages. Rictor−/− iNKT cells showed a marked reduction of the NKT17 lineage (PLZFintRORγt+), but contained normal percentages of NKT1 (PLZFloRORγt− or PLZFloT-bet+) and NKT2 (PLZFhiRORγt− or PLZFhiGATA3+) lineages (Fig. 2A). In contrast, Rptor−/− thymocytes had drastically reduced NKT1 cells, a corresponding increase of NKT2 cells, and a normal frequency of NKT17 cells (Fig. 2A). Finally, mice deficient in Rheb contained largely normal percentages of NKT1 and NKT17 cells, and a moderately increased NKT2 cells (Supplemental Fig. 2E). These results point to the distinct effects of mTOR components at impinging upon the lineage diversification of iNKT cells. Because of the selective requirement of Rictor in NKT17 differentiation, we focused on this control mechanism in our following analyses.
Figure 2.
mTORC2 is required for NKT17 cell development. Cells were from WT, Rictor−/− and Rptor−/− mice. (A) Intracellular staining of PLZF, RORγt, T-bet and GATA3 in iNKT cells from the thymus. NKT1, NKT2 and NKT17 were labeled as described (6). NKT1: WT vs Rictor−/−, p>0.05; WT vs Rptor−/−, p<0.001. NKT2: WT vs Rictor−/−, p>0.05; WT vs Rptor−/−, p<0.001. NKT17: WT vs Rictor−/−, p<0.05; WT vs Rptor−/−, p>0.05 (n=4-7 mice per group). (B) Intracellular staining of PLZF and RORγt in iNKT cells from the spleen (Spl), peripheral lymph nodes (PLN) and liver. Spl, p<0.001; PLN, p<0.001; Liver, p<0.001 (n=4 mice per group). (C and D) Cytokine production by iNKT cells (gated on CD1d-PBS57+TCR-β+ cells) from the thymus (C), spleen, peripheral lymph nodes and liver (D), after stimulation with PMA and ionomycin in the presence of monensin for 5 h. n=4-6 mice per group. (E) Cytokine production of splenic iNKT cells in response to in vivo-GalCer challenge. n=3-5 mice per group. (F) IL-17 production by thymic iNKT cells stimulated with α-GalCer in vitro. Thymocytes were cultured with 125 ng/ml α-GalCer for 72 h, and treated with PMA, ionomycin and monensin in the last 5 h. n=4-5 mice per group. Results are representative of 2-4 independent experiments. Data shown are Mean ± SEM.
Consistent with the reduction of NKT17 cells in the thymus, Rictor−/− mice also contained diminished NKT17 cells in peripheral lymphoid organs including spleen, peripheral lymph nodes and liver (Fig. 2B). NKT1, NKT2 and NKT17 are enriched at different developmental stages, that is, NKT1 in Stage 3, NKT2 in Stages 1 and 2, and NKT17 in Stage 2 (6). Rictor−/− iNKT cells showed a marked reduction of NKT17 cells at all stages examined (data not shown), indicating an intrinsic effect of mTORC2 at promoting NKT17 differentiation. We next examined cytokine production of NKT cells in vitro and in vivo. Consistent with the transcription factor analysis, thymic Rictor−/− iNKT cells had substantially reduced IL-17 production but normal IFNγ and TNFα production (Fig. 2C). Reduced IL-17 production was also manifested in Rictor−/− iNKT cells in spleen, peripheral lymph nodes and liver (Fig. 2D). Moreover, in response to α-GalCer in vivo challenge or in vitro stimulation, Rictor−/− iNKT cells had a severe impairment in IL-17 production (Fig. 2E and Fig. 2F). Collectively, these results indicate that Rictor is selectively required for RORγt expression, IL-17 production, and NKT17 cell generation.
Pten deficiency potentiates NKT17 differentiation in a Rictor-dependent manner
To determine whether NKT17 development is controlled by regulators of mTOR activity, we analyzed mice deficient in Pten, a negative regulator of mTOR signaling (8), using the CD4-Cre system (Ptenfl/flCD4-Cre; Pten−/−). As expected, Pten−/− mice had increased AKT S473 phosphorylation, a signature activity of mTORC2 activity in mature thymocytes (Fig. 3A). iNKT cells were increased in the thymus of Pten−/− mice, but this was largely rescued after concomitant loss of Rictor in Ptenfl/flRictorfl/flCD4-Cre mice (Pten−/−Rictor−/−) (Fig. 3B). The development of Pten−/− iNKT cells was blocked at the transition from Stage 2 to Stage 3, as shown by the marked accumulation of Stage 2 cells and the loss of Stage 3 cells (Fig. 3C), similar as the phenotype observed following Lck-Cre-mediated deletion of Pten (21). Notably, this phenotype persisted in Pten−/−Rictor−/−mice (Fig. 3C), indicating the involvement of an mTORC2-independent activity. Strikingly, Pten−/− iNKT cells contained markedly increased RORγt+ NKT17 cells, and a corresponding reduction of NKT1 cells, and these defects were completely reversed upon the additional loss of Rictor (Fig. 3D). Consistent with these observations, IL-17 production from iNKT cells was greatly upregulated in Pten−/− but not in Pten−/−Rictor−/− mice, while IFNγ production from iNKT cells was reduced in Pten−/− but not in Pten−/−Rictor−/− mice (Fig. 3E). Therefore, Rictor-mediated signaling drives the expansion of NKT17 cells in Pten−/− mice, but does not contribute to the defective terminal maturation in these mice.
Figure 3.
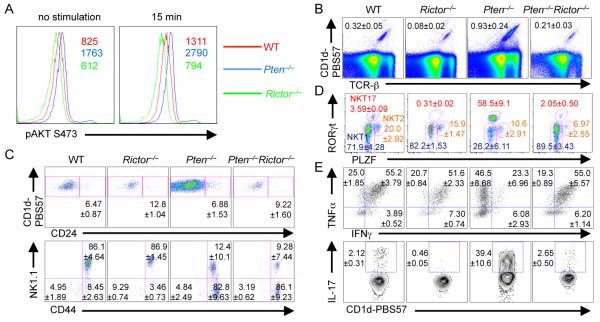
Pten limits NKT17 cell differentiation by inhibiting mTORC2 activity. Thymocytes were from WT, Rictor−/−, Pten−/− and Pten−/−Rictor−/− mice. (A) AKT S473 phosphorylation levels in CD4 single-positive thymocytes without or with stimulation with PMA and ionomycin for 15 min. Mean fluorescent intensity is indicated. (B) Flow cytometry of iNKT cells (CD1d-PBS-57+TCR-β+) in the thymus. WT vs Pten−/−, p<0.05; WT vs Pten−/−Rictor−/−, p>0.05 (n=3-7 mice per group). (C) Analysis of iNKT cell developmental stages in the thymus. Stage 2: WT vs Pten−/−, p<0.01; WT vs Pten−/−Rictor−/−, p<0.001; Pten−/− vs Pten−/−Rictor−/−, p>0.05 (n=3 mice per group). (D) Intracellular staining of PLZF and ROR t in thymic iNKT cells. NKT17: WT vs Pten−/−, p<0.001; WT vs Pten−/−Rictor−/−, p>0.05; Pten−/− vs Pten−/−Rictor−/−, p<0.001 (n=3-5 mice per group). (E) Cytokine production by thymic iNKT cells (gated on CD1d-PBS57+TCR-β+ cells), after stimulation with PMA and ionomycin in the presence of monensin for 5 h. n=3-7 mice per group. Results are representative of 2-5 independent experiments. Data shown are Mean ± SEM.
Much progress has been made on the mechanisms of peripheral CD4 T cell differentiation as well as the control of thymic iNKT cell development. Emerging evidence also suggests that iNKT cells diverge into distinct effector lineages, similar as helper T cell differentiation (6, 7), but how iNKT lineage determination is orchestrated remains obscure. Here, we have compared and contrasted the roles of mTORC1 and mTORC2 signaling in the coordination of iNKT cell development and lineage determination. Raptor-mediated mTORC1 signaling plays a dominant role in iNKT cell development, terminal maturation and NKT1 differentiation, and these effects are largely independent of Rheb functions. Rictor-mediated mTORC2 signaling also contributes to the development and peripheral maintenance of iNKT cells, at least partially, by facilitating their survival. Moreover, mTORC2 is selectively required for NKT17 lineage determination, as loss of Rictor and Pten reciprocally affects NKT17 cell generation, RORγt expression, and IL-17 production. Interestingly, while deletion of Rictor reversed NKT17 dysregulation in Pten-deficient cells, it did not affect the blocked maturation of Stage 2 into Stage 3 iNKT cells. These results provide crucial genetic evidence that developmental maturation and lineage differentiation of iNKT can be uncoupled, lending additional support for the recently proposed model of lineage diversification of iNKT cells (6, 7). Furthermore, despite the analogy and shared transcription factors between effector differentiation of iNKT cells in the thymus and antigen-stimulated helper CD4 T cells in the periphery (6, 7), these processes employ distinct mTOR signaling. For instance, whereas Raptor and Rheb, but not Rictor, have been implicated in TH17 cell generation (10-12), we show here that Rictor, but not Raptor or Rheb, is crucial for NKT17 cell differentiation. Altogether, the new insight into mTOR-dependent programming of iNKT cell differentiation contributes to our understanding of fundamental mechanisms of lineage commitment and fate determination in adaptive immunity.
Supplementary Material
Acknowledgments
We acknowledge C. Cloer and B. Rohde for help with animal colony management, and the NIH Tetramer Facility for the CD1d-PBS57 tetramer.
This work was funded by NIH CA176624, AI101407 and AI105887, American Cancer Society, and Crohn’s & Colitis Foundation of America (to H.C.).
Abbreviations used in this paper
- α-GalCer
α-galactosylceramide
- iNKT
invariant NKT
- mTOR
mechanistic target of rapamycin
- PLZF
the promyelocytic leukaemia zinc finger
- Pten
phosphatase and tensin homolog
- WT
wild-type
Footnotes
Disclosures Authors have no financial conflict of interest.
References
- 1.Salio M, Silk JD, Jones EY, Cerundolo V. Biology of CD1- and MR1-restricted T cells. Annu Rev Immunol. 2014;32:323–366. doi: 10.1146/annurev-immunol-032713-120243. [DOI] [PubMed] [Google Scholar]
- 2.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol. 2013;13:101–117. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- 3.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 4.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, Pandolfi PP, Sant’Angelo DB. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14:1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Constantinides MG, Bendelac A. Transcriptional regulation of the NKT cell lineage. Curr Opin Immunol. 2013;25:161–167. doi: 10.1016/j.coi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annu Rev Immunol. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, Magnuson MA, Boothby M. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, Powell JD. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurebayashi Y, Nagai S, Ikejiri A, Ohtani M, Ichiyama K, Baba Y, Yamada T, Egami S, Hoshii T, Hirao A, Matsuda S, Koyasu S. PI3K-Akt-mTORC1-S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORgamma. Cell Rep. 2012;1:360–373. doi: 10.1016/j.celrep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Yang K, Shrestha S, Zeng H, Karmaus PW, Neale G, Vogel P, Guertin DA, Lamb RF, Chi H. T Cell Exit from Quiescence and Differentiation into Th2 Cells Depend on Raptor-mTORC1-Mediated Metabolic Reprogramming. Immunity. 2013;39:1043–1056. doi: 10.1016/j.immuni.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499:485–490. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee K, Nam KT, Cho SH, Gudapati P, Hwang Y, Park DS, Potter R, Chen J, Volanakis E, Boothby M. Vital roles of mTOR complex 2 in Notch-driven thymocyte differentiation and leukemia. J Exp Med. 2012;209:713–728. doi: 10.1084/jem.20111470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin J, Wang S, Deng W, Wu J, Gao J, Zhong XP. Mechanistic target of rapamycin complex 1 is critical for invariant natural killer T-cell development and effector function. Proc Natl Acad Sci U S A. 2014;111:E776–783. doi: 10.1073/pnas.1315435111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Tschumi BO, Corgnac S, Ruegg MA, Hall MN, Mach JP, Romero P, Donda A. Mammalian Target of Rapamycin Complex 1 Orchestrates Invariant NKT Cell Differentiation and Effector Function. J Immunol. 2014 doi: 10.4049/jimmunol.1400769. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Yang J, Yang K, Wang H, Gorentla B, Shin J, Qiu Y, Que LG, Foster WM, Xia Z, Chi H, Zhong XP. iNKT cells require TSC1 for terminal maturation and effector lineage fate decisions. J Clin Invest. 2014;124:1685–1698. doi: 10.1172/JCI69780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang K, Neale G, Green DR, He W, Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat Immunol. 2011;12:888–897. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finlay DK, Kelly AP, Clarke R, Sinclair LV, Deak M, Alessi DR, Cantrell DA. Temporal Differences in the Dependency on Phosphoinositide-Dependent Kinase 1 Distinguish the Development of Invariant V{alpha}14 NKT Cells and Conventional T Cells. J Immunol. 2010;185:5973–5982. doi: 10.4049/jimmunol.1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kishimoto H, Ohteki T, Yajima N, Kawahara K, Natsui M, Kawarasaki S, Hamada K, Horie Y, Kubo Y, Arase S, Taniguchi M, Vanhaesebroeck B, Mak TW, Nakano T, Koyasu S, Sasaki T, Suzuki A. The Pten/PI3K pathway governs the homeostasis of Valpha14iNKT cells. Blood. 2007;109:3316–3324. doi: 10.1182/blood-2006-07-038059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




