Abstract
Transcriptional mechanisms governing hematopoietic stem cell (HSC) quiescence, self-renewal, and differentiation are not fully understood. Sequence-specific single-stranded DNA-binding protein 2 (SSBP2) is a candidate acute myelogenous leukemia (AML) suppressor gene located at chromosome 5q14. SSBP2 binds the transcriptional adaptor protein Lim-domain binding protein 1 (LDB1) and enhances LDB1 stability to regulate gene expression. Notably, Ldb1 is essential for HSC specification during early development and maintenance in adults. We previously reported shortened lifespan and greater susceptibility to B cell lymphomas and carcinomas in Ssbp2 −/− mice. However, whether Ssbp2 plays a regulatory role in normal HSC function and leukemogenesis is unknown. Here, we provide several lines of evidence to demonstrate a requirement for Ssbp2 in the function and transcriptional program of hematopoietic stem and progenitor cells (HSPCs) in vivo. We found that hematopoietic tissues were hypoplastic in Ssbp2−/− mice and the frequency of lymphoid-primed multipotent progenitor cells in bone marrow was reduced. Other significant features of these mice were delayed recovery from 5-fluorouracil treatment and diminished multilineage reconstitution in lethally irradiated bone marrow recipients. Dramatic reduction of Notch1 transcripts and increased expression of transcripts encoding the transcription factor E2a and its downstream target Cdkn1a also distinguished Ssbp2−/− HSPCs from wild-type HSPCs. Finally, a tendency towards coordinated expression of SSBP2 and the AML suppressor NOTCH1 in a subset of The Cancer Genome Atlas AML cases suggested a role for SSBP2 in AML pathogenesis. Collectively, our results uncovered a critical regulatory function for SSBP2 in HSPC gene expression and function.
Introduction
A small pool of pluripotent stem cells capable of self-renewal maintains normal cellular hematopoiesis and recovery from cytotoxic stress (1, 2). The delicate balance between hematopoietic stem cell (HSC) self-renewal and differentiation during these processes requires precise coordination of gene expression with the abundance of their protein products. The core multiprotein transcriptional complex that regulates hematopoietic and progenitor stem cell (HSPC) activity is comprised of the b-HLH protein TAL1/SCL, the Lim-only protein 2 (LMO2), LIM domain-binding protein 1 (LDB1), and the zinc finger protein GATA2. Initial gene targeting studies demonstrated an absolute requirement for the HSC-expressed and lineage restricted factor, TAL1/SCL1, in embryonic HSC specification (3). In contrast, TAL1/SCL appears to be dispensable for hematopoietic maintenance in adults due to functional redundancy with LYL1 (4). In erythroid and megakaryocytic progenitor cells, the non–DNA-binding transcriptional adaptor LMO2 is thought to bridge TAL1/SCL1 and the self-dimerizing cofactor LDB1 to nucleate the assembly of b-HLH-GATA factor- and LDB1-containing multisubunit complexes (5, 6).This pathway raises the question of whether a similar mechanism occurs in HSCs. Furthermore, unlike TAL1/SCL1, LMO2 and LDB1 are continuously required for post-natal HSC maintenance (7–9). Elegant global chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-Seq) studies have localized LDB1, TAL1/SCL1 and GATA2 to conserved promoter elements on the TAL1/SCL1, Gata2, Runx1, Lmo2, E2a and Myb genes. Furthermore, decreased expression of Tal1/Scl1, Gata2, Runx1, Lmo2 and Myb in Ldb1−/− mouse HSPCs has unequivocally demonstrated these encoded proteins to be positive regulators of HSPC-specific gene expression (9).
LDB1 is a member of a multi-protein transcription complex. Human single-stranded DNA-binding protein 2 (SSBP2) was positionally cloned as a myeloid leukemia suppressor from a chromosomal disruption within a critical region of loss in 5q13-14 in an acute myelogenous leukemia (AML) cell line (10). SSBP2 and the related SSBP3 and SSBP4 were so designated due to the in vitro single-stranded DNA-binding activity of the founding member CSDP, the chicken ortholog of SSBP3 (11); however, the significance of this putative single-stranded DNA binding activity in vivo is unknown. All three proteins can bind LDB1 through a highly conserved amino terminal domain; in turn, LDB1 binds the LIM domains of LMO or LIM homeodomain proteins (LHX) through an evolutionarily conserved carboxy-terminal LIM interacting domain.
We and others have established that SSBPs enhance promoter occupancy and transcriptional activity of LMO2- and LHX-containing complexes by preventing the ubiquitylation and ultimately the proteasomal degradation of LDB1 by the E3 ubiquitin ligase RNF12/RLIM (12–15). Clearly, SSBPs have been selected for and evolutionarily maintained as key modulators of LDB1 activity. In Caenorrhabditis elegans, for example, the sole SSBP ortholog SAM-10 coordinates with the LDB1 ortholog in a cell-autonomous fashion to regulate synaptic differentiation (16). Similarly, the expression of the Drosophila ortholog, SSDP, is a rate-limiting cofactor that regulates combinatorial transcriptional signals from CHIP (LDB1)-APTEROUS (LHX) or CHIP (LDB1)-PANNIER (GATA) complexes (17, 18). Finally, in Zebra fish, SSDPs regulate neural patterning and sensory neuronal growth in part through LDB1 stabilization (19). Although SSBPs from all of these species binds and stabilizes LDB1, consistent with extensive evolutionary conservation of these members of this gene family (>99% identity), there may also be distinct function for each member. Consistent with this notion, Ssbp2−/− mice are viable (16) whereas Ssbp3−/− mice die in utero (20, 21). In addition, a genome wide in vivo screen by genetic cross of two strains of mice which differ in hippocampal neurogenesis suggested Ssbp2 to be a quantitative trait locus regulating neuronal survival and regeneration (22). Finally, the highly penetrant autoimmune defects and enhanced predisposition to B cell lymphomas and carcinomas of Ssbp2−/− mice suggest a unique role of Ssbp2 in lymphoid differentiation and tumor suppression (15).
The pivotal role of Ldb1 in HSPC maintenance suggests that Ssbp2 plays a similarly important role. In this report, we report here that Ssbp2 exerts a non-overlapping regulatory function to maintain murine HSPCs. In addition, reestablishment of homeostasis after elimination of cycling HSPCs by myeloablative treatment is impaired upon Ssbp2 ablation. Moreover, Ssbp2−/− bone marrow (BM) competes poorly in multilineage reconstitution of lethally irradiated recipients. Further, decreased expression of Notch1 and elevated expression of E2a and its transcriptional target Cdkn1a in Ssbp2−/− HSPCs identify a role for Ssbp2 in the HSPC-specific gene expression program. Finally, since accumulating evidence suggests a myeloid suppressor role for NOTCH1 (23–25), we evaluated the expression of NOTCH1 and SSBP2 from data obtained from The Cancer Genome Atlas (TCGA). Interestingly, NOTCH1 expression was modestly correlated with SSBP2 expression in primary AML samples. Together, these initial biological findings lay the foundation for future mechanistic investigations on SSBP2 regulated transcriptional networks in HSPCs.
Materials and Methods
Mice
Ssbp2−− mice were described previously (15). For the competitive transplantation experiments, the mice were backcrossed to a C57BL/6 (CD45.2) background for 10 generations. Mouse bleeding and tissue harvests were according to Institutional Animal Care and User Committee (IACUC) approved protocols following standard operating procedures.
Immunofluorescence analysis
BM mononuclear cells enriched by centrifugation or depleted of lineage-committed cells through magnetic-activated cell separation were stained for SSBP2. Lin−Sca1+c-Kit+ (LSK) cells were purified by staining the Lin− cells with anti-Sca1–PE and anti-c-Kit– FITC antibodies (BD Pharmingen) followed by cell sorting using a BD FACS Aria IIu sorter. Cells were centrifuged onto slides and fixed in 4% paraformaldehyde in PBS for 20 min, which was followed by permeabilization and then staining with rabbit polyclonal anti-SSBP2 antibody or mouse anti-LDB1 antibody (Molecular Probes) as detailed previously (26).
Phenotypic analysis of HSPCs by flow cytometry
Seven-color FACS analysis (Pacific Blue, Alexa 488, APC-Cy7, Alexa 647, PE, PerCP- Cy5.5) was performed using a Fortessa cell analyzer (BD Bioscience) and data were analyzed with FlowJo software (Tree Star, Inc.). The antibodies used against mouse antigens, including Pacific Blue lineage cocktail [Pacific Blue anti-mCD3, -mLy-6G(Ly-6C), -mCD11b, -mCD45R(B220), and -mTer-119], Sca1–Alexa 488, c-Kit–APC-Cy7, CD34–Alexa 647, Flt3–PE, and CD48–PerCP-Cy5.5 were from BD Pharmingen, eBioscience or BioLegend. BM mononuclear cells were stained with Pacific Blue-conjugated lineage cocktail along with Sca1–Alexa 488, c-Kit–APC-Cy7, and Sca1+, c-Kit+ cells were defined as LSK cells. Similar procedures were used to analyze Flt3−CD34− LSK cells (LT-HSCs), Flt3−CD34+ LSK cells (ST-HSCs), Flt3loCD34+ LSK cells (multipotent progenitor cells; MPPs), Flt3hiCD34+ LSK cells (lymphoid-primed multipotent progenitor cells; LMPPs), and CD48+ or CD48− LSK cells.
Hematopoietic recovery from myelotoxic agents
Mice were administered a single intraperitoneal dose of 5-fluorouracil (5-FU) at 150 mg/kg of body weight. 50ul aliquots of peripheral blood samples collected from 5-FU–treated and control (untreated) mice were analyzed at 6, 12, and 18 days after 5-FU injection in a Siemens Advia 120 auto analyzer for total and differential counts.
Two complementary approaches were taken to examine the role of Ssbp2 in HSPC recovery from 5 FU. In the first, wild type bone marrow was transplanted into lethally irradiated Ssbp2 null mice and the mice challenged with 5 FU 4 months after transplantation. In the second set, donor BM cells (CD45.2) were prepared from Ssbp2−/− or Ssbp2+/+ control mice and mixed with congenic competitor BM cells (CD45.1) at a 10:1 ratio. The cell mixture (5x106) was transplanted into the irradiated recipients (CD45.1+) by tail vein injection and the mice challenged with 5 FU as detailed above.
Competitive BM transplantation
Age-matched female congenic B6.SJL-PtrcaPep3b/BoyJ (B6.BoyJ;CD45.1+) mice (Jackson Laboratories) were used as transplant recipients. These mice were lethally irradiated (9.5 Gy). Donor BM cells (CD45.2) were prepared from Ssbp2−/− or Ssbp2+/+ control mice and were mixed with congenic competitor BM cells (CD45.1) at a 1:1 or 10:1 ratio. The mixed cells (5x106) were transplanted into the irradiated recipients (CD45.1+) by tail vein injection. Donor-derived engraftment and multilineage reconstitution in BM and peripheral blood from recipients were assessed by flow cytometry using CD45.1–APC or CD45.2–PE-Cy7 at 4 and 16 weeks post-transplantation to evaluate for short- and long-term engraftment, respectively.
Real-time quantitative PCR
RNA was isolated using the RNeasy mini kit (Qiagen). cDNA was generated using SuperScript II reverse transcriptase (Invitrogen). Samples were normalized using 18S rRNA (4319413E) (Applied Biosystems), and gene expression levels were determined using the relevant primers and probes (Applied Biosystems) on a Taqman ABI7900HT instrument (Applied Biosystems). Each experiment was performed five times in triplicate. Data are expressed as mean ± SD.
Analysis and data mining
TCGA AML database (27) was accessed and the pertinent data analyzed with cBioPortal for AML cases with mutations, copy number alterations, or an at least twofold change in expression level by RNA-sequence analysis (28, 29).
Statistical analysis
Data were analyzed for significance using two-tailed Student's t-test. A p<0.05 was considered statistically significant.
Results
Expression of Ssbp2 in HSPCs and hypoplasia of hematopoietic organs in Ssbp2−/− mice
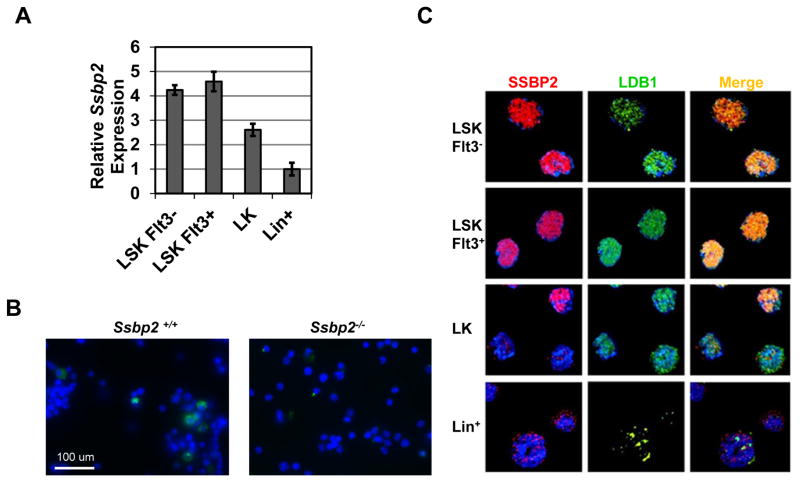
To determine whether Ssbp2 expression was regulated in a differentiation-specific manner, we examined BM cells enriched for HSPC subpopulations for transcript levels. Overall, Ssbp2 expression was higher in the long-term [Flt3−Lin−Sca+c-Kit+ (Flt3− LSK)] and short-term (Flt3+ LSK) HSPC populations than in committed progenitor cells (LKs), expression was lower in the lineage-positive (Lin+) cells than in the committed progenitor cells (Fig. 1A). These findings suggested that Ssbp2 expression was repressed as the cells differentiated. Immunofluorescence staining of whole BM confirmed restricted expression, with expression in approximately 5% of all cells (Fig. 1B). Consistent with our previous characterization of the high specificity of anti-SSBP2 antibodies (30), no specific signal was detected in Ssbp2−/− BM. Flow cytometry-sorted populations revealed abundant levels of nuclear expression and colocalization with LDB1 in both Flt3+ LSKs and Flt3− LSKs (Fig. 1C). These images reflected the relative change in Ssbp2 transcripts shown in Fig. 1A. The localization of SSBP2 to nuclear speckles in LK and Lin− cells was reminiscent of our previous findings with epithelial cells and fibroblasts in which SSBP2 localized to punctate nuclear structures (26, 31). Although the nature of these structures is unknown, our work in progress suggests that non–LDB1-interacting elements may also be present.
Fig 1. Abundant Ssbp2 expression and colocalization with LDB1 in murine HSPCs.
A. Ssbp2 RNA expression pattern in subpopulations of mouse BM cells as detected by real-time PCR. Cells were sorted by flow cytometry based on their surface markers, including LT-HSCs (Lin−Sca1+c-Kit+Flt3− [Flt3− LSKs]), ST-HSCs (Lin−Sca1+c-Kit+Flt3+ [Flt3+ LSKs]), progenitor cells (Lin−c-Kit+ [LKs]), and Lin+ cells. Data represent mean ± S.D. from three independent experiments.
B. SSBP2 expression is restricted to a few cells in normal BM. Mononuclear cells from whole BM were immunostained with anti-SSBP2 antibodies. Nuclear DNA was stained with DAPI (blue). Representative fields illustrate antibody specificity. A few cells in WT BM were immunoreactive. No signal was detected in null mice.
C. SSBP2 and LDB1 colocalize in the nucleus. FACS-sorted subpopulations of HSPCs were fixed, permeabilized, and stained with anti-SSBP2 and -LDB1 antibodies. Nuclear DNA was stained with DAPI.
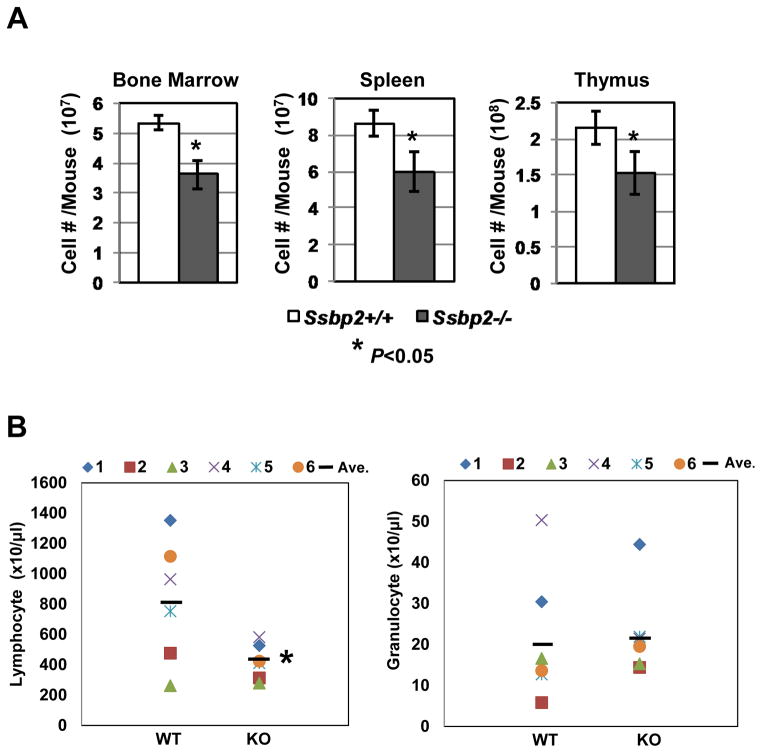
To evaluate whether hematopoiesis was affected in the absence of Ssbp2, we first assessed mice 6-8 weeks of age for the absolute number of total nucleated cells in the BM, spleen and thymus. In all three tissues, the mononuclear population was reduced by 30–40% (p<0.05 for each tissue type), suggesting an overall reduction in hematopoietic activity (Fig. 2A). The absolute number of peripheral blood lymphocytes was twice as high in wild-type (WT) than in null mice (Fig. 2B, left). Although there was a tendency for an increase in granulocytes, the values did not reach statistical significance (Fig. 2B, right). Likewise, the absolute number of red blood cells and platelets in WT and Ssbp2-null mice were not significantly different(data not shown).
Fig 2. Hypoplasia of hematopoietic tissues in Ssbp2−/− mice.
A. Reduced cellularity of BM, spleen and thymus in Ssbp2-null mice. Cells from single-cell suspensions prepared from BM, spleen and thymus from Ssbp2−/− mice and their WT littermates were counted.
B. Peripheral blood lymphocytes were decreased in the absence of Ssbp2. Absolute numbers of peripheral blood lymphocytes and granulocytes are shown.
Data represent mean ± S.D. from 6 age- and gender-matched pairs of WT and null mice. *p<0.05 vs. WT as determined with unpaired two-tailed t-test.
HSPC homeostasis is impaired in Ssbp2−/− mice
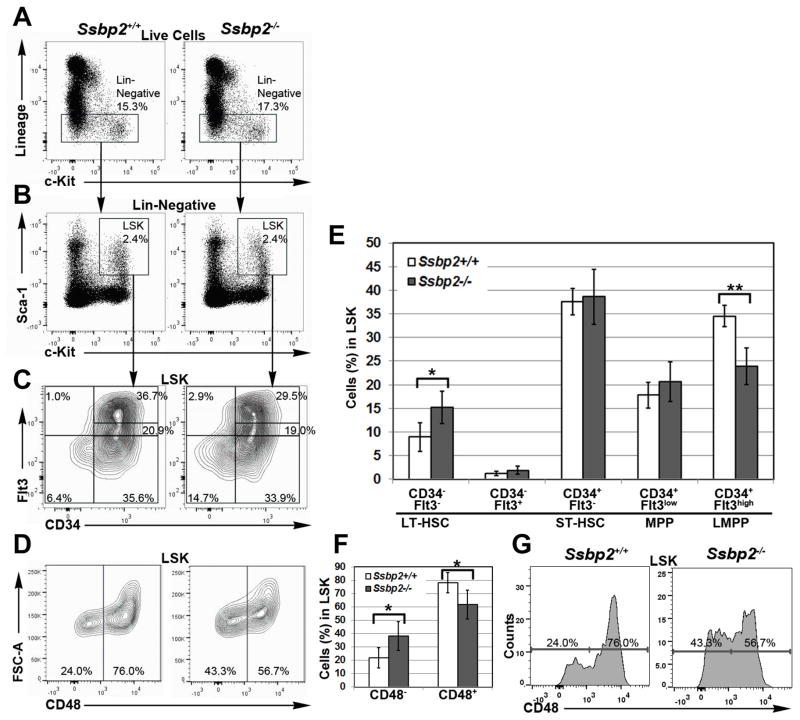
The abundant expression of SSBP2 in HSPCs prompted us to determine whether normal HSPC homeostasis was perturbed in the absence of Ssbp2. Accordingly, flow cytometric analyses for stem and early progenitor cell subpopulations were performed with null mice and their WT littermates. Characterization of the Lin Sca1+c-Kit+ (LSK) population, which is comprised of stem and early progenitor cells, revealed no significant difference in frequencies (Fig. 3B). Within the LSK compartment, the frequency of CD34− cells was significantly lower in WT than null mice, suggesting an increase in the ratio of early immature HSPCs (Supplemental Fig. 1B). CD34 and FLT3 expression were used to further subdivide the LSK subpopulation (Fig. 3C). The CD34−Flt3− fraction which corresponded to LT-HSCs was twice as high in null than in control mice, whereas there was no significant difference in the ST-HSC or CD34+FLT3− population (Fig. 3E). FLT3 expression is associated with lymphoid priming and increased lymphoid gene expression program. The frequency of CD34+FLT3hi LMPPs was reduced by a third. Although there was a trend towards fewer LMPPs, the differences were not statistically significant in this small group of mice (WT: n = 4, null: n = 5). Furthermore, there was no compensatory increase in the frequency of CD34+FLT3lo population, suggesting the decrease in FLT3hi expression in flow cytometry was not due to absence of Flt3 uprgulation by SSBP2 in the pre-LMPP cells. Likewise, the signaling lymphocyte activation molecule (SLAM) marker CD150 distinguishes LT-HSC in the larger the LSK population whereas CD48 expression detects an uncommitted, early progenitor population(32). Once again, the frequency of CD48− cells representing LT-HSCs was increased in the absence of Ssbp2, in contrast to the decreased frequency of CD48+ uncommitted progenitor cells with restricted amplifying potential (Fig. 3D, 3F).
Fig 3. Ssbp2 deletion affects the frequency of HSPC subpopulations.
A. Frequency of Lin− cells is unaffected by Ssbp2−/− BM. A representative image of gating within live population for lineage-negative cells B. Frequency of LSK cells was unaffected by Ssbp2−/− BM. A representative image of c-Kit, Sca1 gating in Lin− cells. C and E. Frequency of LMPPs (Flt3hiCD34+ LSKs) is decreased in Ssbp2−/− BM. C. Representative flow cytometry analysis showing LT-HSCs (Flt3−CD34− LSKs), ST-HSCs (Flt3−CD34+ LSKs), MPPs (Flt3loCD34+ LSKs), and LMPPs (Flt3hiCD34+ LSKs) within LSKs in Ssbp2-null mice and their WT littermates. E. Frequency of LT-HSCs, ST-HSCs, MPPs and LMPPs within the LSK population in WT (n = 5) and null mice (n = 5). D, F and G. Frequency of CD48 cells is increased in Ssbp2−/− BM. D. Representative flow cytometry analysis showing CD48+ cells within LSKs in Ssbp2-null mice and their WT littermates. F. Frequency of CD48+ and CD48− population within the LSK compartment in WT (n = 5) and null mice (n = 5). G. Representative histogram for anti-CD48 staining. Note the increase in frequency in CD48− cells in null mice. Data in E and F represent mean ± S.D from age-matched WT (n = 5) and null mice (n = 5)*p<0.05) as determined by unpaired, two tailed t test between WT and Ssbp2−/− mice.
In summary, the frequency of both CD34− cells and FLT3− cells within the LSK compartment increased and that of LSK cells expressing CD34 or FLT3 decreased (Suppl. Fig. 1 and 2). That these results would reflect a change in absolute numbers if a larger cohort of mice were used cannot be ruled out. Collectively, these data suggested a seminal role for Ssbp2 in maintaining early HSPC homeostasis.
Delayed hematopoietic recovery of Ssbp2−/− mice after myeloablative treatment
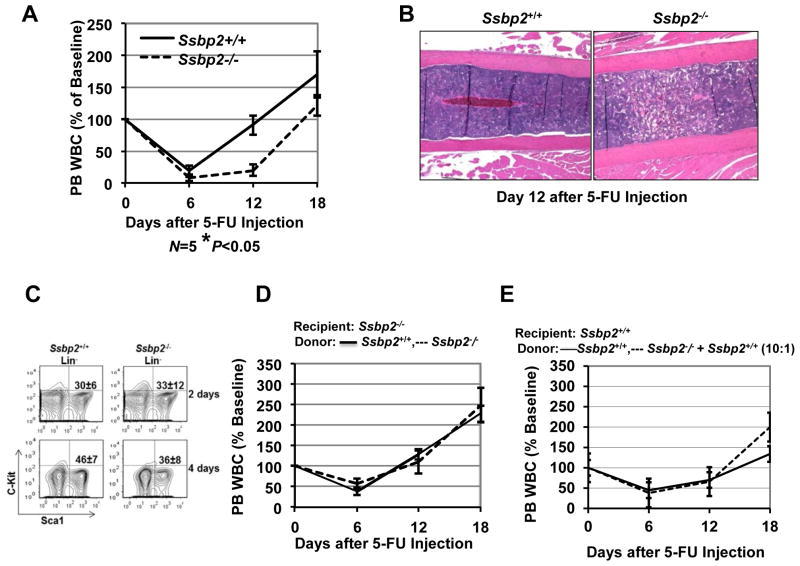
To assess whether loss of Ssbp2 affected the hematopoietic response to the cytotoxic stress we observed, we challenged mice with a single dose of the myeloablative drug 5-FU, which eliminates cycling HSPCs. Ssbp2−/− mice displayed a markedly delayed recovery as reflected in serial peripheral blood leukocyte counts post-drug injection. On day 12, the total percentage of peripheral blood leukocytes was 75±10% in WT mice but only 20±5% in null mice (p<0.05) (Fig. 4A). The recovery of both myeloid cells and lymphoid cells was impaired; likewise, platelet and red blood cell recovery was delayed (data not shown). The normal BM cellularity of a WT mouse on day 12 was noticeably altered, with decreased BM cellularity observed in null mice (Fig. 4B). Hypoplasia of both myeloid and erythroid lineages were evident in Ssbp2−/− BM (data not shown).
Fig 4. Delayed hematopoietic recovery from cytotoxic stress in Ssbp2−/− mice.
A. Peripheral blood hematopoietic recovery is delayed in Ssbp2−/− mice. Hematopoietic reconstitution was monitored by serial peripheral blood counts in mice injected with a single dose of 5-FU (150 mg/kg, i.p.). Total WBC counts are shown as a percentage of the initial values for each group of mice. B. Hypoplastic BM on day 12 post 5-FU treatment in Ssbp2−/− mice. Hematoxylin and eosin staining of BM from Ssbp2-null mice and their WT littermates at day 12 post 5-FU injection. C. Early post 5-FU HSPC recovery is compromised in Ssbp2−/− BM. A representative flow cytometric analysis showing Lin Sca1+ cells in BM from Ssbp2−/− mice and their WT littermates 2 days and 4 days after 5-FU injection. D. WT BM rescues Ssbp2−/− mice from delayed post–5-FU recovery. Lethally irradiated WT and Ssbp2−/− cells were reconstituted with WT BM. Four months after transplantation, recipients were challenged with a single dose of 5-FU and hematopoietic recovery was followed with serial peripheral blood counts. WBC counts are shown as a percentage of the initial values for each group of mice. *p<0.05) as determined by unpaired two-tailed t-test between WT and Ssbp2−/− mice. Each experiment was conducted twice with a group of six age-matched mice per genotype. E. WT mice reconstituted with Ssbp2−/− enriched marrow show normal hematopoietic recovery after 5-FU treatment.
Weissman et al. (33) demonstrated that early stages of recovery from 5-FU treatment are characterized by a transient expansion of Lin−Sca+ HSPCs. Therefore, we examined the BM of null and WT mice by flow cytometry at 48 and 96 hours after 5-FU treatment. Unlike the 50% expansion of Lin−Sca+ cells seen in the WT mice (from 30% to 46%) between days 2 and 4, Ssbp2−/− mice showed a more modest 12% expansion (from 33% to 36%) (Fig. 4C). These findings suggested a diminished potential for stress-mediated HSPC expansion in the absence of Ssbp2.
These mice were homozygously deleted for Ssbp2 in all tissues, so it was important to examine cell intrinsic vs microenvironmental mechanisms producing these results. To determine whether the defects were intrinsic or extrinsic to HSCs, we transplanted isogenic ( CD45.2) WT BM cells into lethally irradiated congenic Ssbp2−/− mice which had been lethally irradiated to ablate autologous hematopoiesis. The recipients were also challenged with 5-FU after they had been fully engrafted (4 months after the transplantation) and we found no significant difference in the recovery rate between transplanted null mice and the control mice at that point. Regardless, the poor reconstituting potential of Ssbp2−/− BM prevented us from conducting the reverse experiments (i.e., testing the 5-FU response in WT mice reconstituted with null mouse BM). In a complementary approach Ssbp2−/− or WT BM cells ( CD45.2 ) were mixed with WT CD45.1 donor BM cells at a ratio of 10:1 and transplanted into lethally irradiated WT CD45.1 mice (Fig.4E) and the recipients challenged with 5-FU after they had been fully engrafted (4 months after the transplantation). Surprisingly, we found no significant difference in the recovery rate between mice transplanted with Ssbp2 −/− or WT enriched bone marrow (Fig. 4E). However, due to technical limitations in monitoring CD45.1 or CD45.2 alleles during recovery, these findings should be interpreted with caution. Overall these findings are consistent with a contribution from non-hematopoietic cells of the bone marrow niche to the recovery Ssbp2−/− HSPCs from cytotoxic stress.
Multilineage repopulating activity of Ssbp2−/− BM is severely impaired
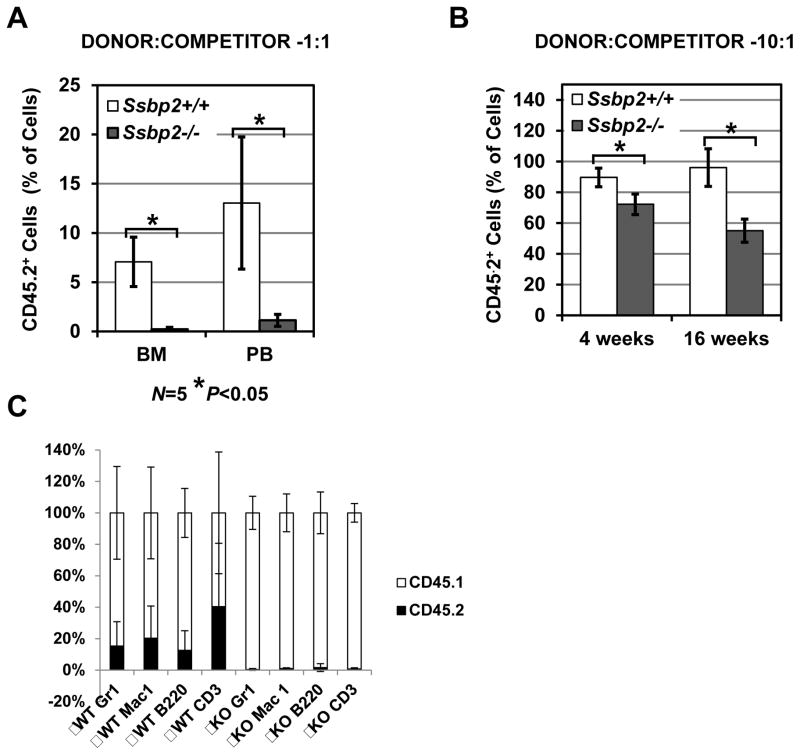
Next, we tested the ability of Ssbp2−/− BM cells to competitively reconstitute hematopoiesis in lethally irradiated mice. We assessed their relative short- and long-term repopulating potential at 4 and 16 weeks by transplanting BM cell mixtures containing different ratios of Ssbp2−/− donor to WT competitor cells into lethally irradiated recipients. Initial transplantations at a ratio of 1:1 donor to competitor revealed that WT cells predominated in both BM (7% vs. 1%; p<0.05) and peripheral blood (13% vs. 2%; p<0.05) at 4 weeks post-transplantation. However, when the ratio was increased to 10:1, Ssbp2−/− cells could be detected in ~50% to 70% of the recipients (Fig.5A). Despite the transplantation of a much higher number of Ssbp2−/− than WT cells, the frequency of donor cells in the Ssbp2−/− recipients was still significantly lower than that of WT recipients at 4 weeks (72.2±6.7% vs. 89.6±6.1%; p<0.05) and at 16 weeks (55±7.5% vs. 96 ±12.2%; p<0.05), suggesting a diminished potential for short- or long-term reconstitution (Fig.5B).
Fig 5. Long-term and short-term repopulating ability is decreased in the absence of Ssbp2.
A.Ssbp2−/− BM cells do not reconstitute at a 1:1 donor-to-competitor ratio. Short-term engraftment (4 weeks) results as determined with CD45.2-expressing cells in BM and peripheral blood are depicted. Data represent mean ± S.D. from 5 recipient mice. B. Reduced frequency of Ssbp2−/− BM donor cells in peripheral blood of recipients transplanted at a donor-to-competitor ratio of 10:1. Engraftment analyses were performed at 4 weeks (short term) and 16 weeks (long-term) after transplantation. Data represent mean ± S.D. from one of two experiments each with 5 mice per genotype.*p<0.05) as determined by unpaired two-tailed t-test between WT and Ssbp2−/− mice. C. Contribution to all the peripheral blood lineages is compromised in the absence of Ssbp2. Representative analysis showing % CD45.1 and CD45.2 populations within granulocyte(Gr1+), macrophage (Mac1+), B lymphocyte (B220+) and T lymphocyte (CD3+), in the peripheral blood from Ssbp2−/− BM recipients 16 weeks after transplantation.
We next examined the peripheral blood contribution of Ssbp2−/− cells to specific hematopoietic lineages. Consistent with the decreased bone contribution to bone marrow homeostasis, the peripheral blood cell types including granulocytes, macrophages, B and T lymphocytes was severely compromised ( Fig. 5C).
To rule out the possibility that the reduced repopulating capacity of Ssbp2−/− HSCs was due to impaired homing of these cells to BM, we compared the ability of Ssbp2−/− cells (CD45.2) and their WT counterparts to move into the BM of irradiated recipients (CD45.1). At 24 hours and 48 hours post-transplantation, there was no significant difference between the percentage of CD45.2−-expressing WT and Ssbp2−/− cells in homing to the BM (24 hours: 1.2% vs. 1.0%); 48 hours: 31% vs. 29%) (Fig. 2A). In addition, we sorted the transplanted cells for Flt3 expression 18 hours after transplantation and found no significant difference between WT and null donors (32% vs. 29%). Taken together, these data demonstrated reduced multilineage repopulating activity and a pronounced loss of B lymphoid potential in Ssbp2−/− HSPCs.
Abundant expression of b-HLH factor E2a and loss of Notch1 expression in the absence of Ssbp2
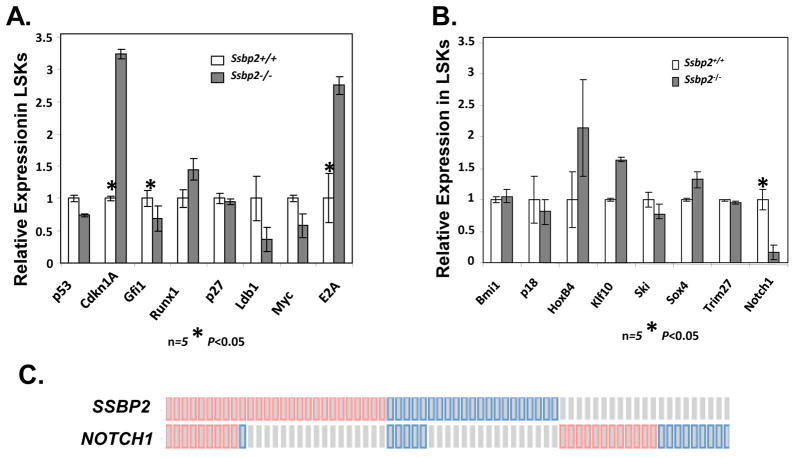
To assess whether the impaired HSC activity in Ssbp2−/− mice correlates with an altered gene expression pattern, we evaluated transcript levels of 17 genes encoding known regulators of quiescence and self-renewal in purified HSPCs: Trp53, Myc, p27Kip1, Cdkn1a (p21), Gfi1, Ldb1, Runx1, E2a, HoxB4, Bmi1, Klf10, Ski, Sox4, Trim27, p18Ink4c, and Notch1). Of these, E2a and Cdkn1A (p21) were expressed at a significantly higher level and Gfi1 at a significantly lower level in null HSPCs than in WT HSPCs (p<0.05 for each gene). Although high expression levels of E2a and Cdkn1a positive regulators of quiescence and a modest decrease in Gfi1 expression a negative regulator of HSC quiescence appear contradictory, the net outcome reflected impaired stress recovery and engraftment defects and suggested perturbation in the transcriptional network overall.
Accumulating evidence has suggested a myeloid leukemia suppressor role for NOTCH1 (23, 25). Therefore, we queried TCGA AML dataset for SSBP2 and NOTCH1 expression as determined by RNA-Seq studies. As shown in Fig. 6C, 21 out of 166 identified cases (12.6%) had loss of SSBP2 expression and 27 cases (16.2%) had increased expression. Twenty four percent of the patients with low SSBP2 expression also had decreased NOTCH1 expression, whereas 9 of the 27 cases (37% of total) with higher SSBP2 expression also had elevated NOTCH1 expression. The coordinated expression of both genes was moderately significant (p=0.046). Considering the genetic and phenotypic heterogeneity in AML (34), these results, taken together with the absence of Notch1 expression in Ssbp2−/− HSPCs, suggest that this axis is preserved in a subset of AMLs.
Fig 6. Gene expression alterations in Ssbp2−/− HSPCs.
A and B. E2a and Cdkn1a transcript levels are increased and Notch1 transcript levels are decreased in the absence of Ssbp2. Quantitative RT-PCR results illustrate differences between Ssbp2−/− and WT counterparts in the expression of HSC quiescence (A) and self-renewal regulators (B) normalized to 18sRNA control. Representative results of triplicates from one of two separate cDNA pools from a group of 5 mice. *p<0.05 between paired samples. C. Tendency towards coordinated SSBP2 and NOTCH1 expression in primary AMLs. The cBIOPortal database was queried for at least >2-fold alteration in expression as determined by RNA-Seq; 166 AML cases were identified. Each column represents a patient. p-value at 95% confidence interval is 0.047.
Discussion
Our studies revealed Ssbp2 as a novel positive regulator of HSC homeostasis and recovery from cytotoxic stress. These findings address the consequences of disrupting a critical transcriptional pathway in HSPC homeostasis, as SSBPs directly interacting with LDB1 and are present in all of the same transcriptional complexes examined to date. Earlier ChIP-Seq studies established the colocalization of LDB1, SCL1, and GATA2 on the Tal1/SCL1, Gata2, Runx1, Lmo2, E2a and Myb gene promoters in HSPCs. More important, a feed-forward autoregulatory loop may exist, as the expression all these genes except E2A was decreased in the absence of LDB1 (9).
Abundant SSBP2 expression and colocalization with LDB1 in HSPCs
High expression of Ssbp2 in undifferentiated stem and progenitor cells, mirroring Ldb1 expression (9), is indicative of a role for Ssbp2 in normal hematopoiesis (Fig. 1A). These findings are in agreement with global expression profiling studies. SSBP2 is frequently on the list of genes preferentially expressed in HSPCs (35–38). Because nuclear localization of Drosophila orthologs (SSDPs) depends on the nuclear localization signal of LDB1 (39), the colocalization with LDB1 raises the possibility of an LDB1-dependent role for SSBP2 in HSPCs (Fig. 1C). Such an interaction suggests that some of the phenotypic consequences of Ssbp2 loss may be Ldb1 dependent. Moreover, the unexpected localization of SSBP2 to nuclear speckles in differentiated cells suggests that SSBP2-LDB1 interactions may be less crucial in mature cells. As we reported previously, in most epithelial cells (MCF7, HeLa, and A549) and in fibroblasts (IMR90), SSBP2 localizes to punctate structures in both the nucleus and nucleolus (26, 31). In some of these cells SSBP2 co-localized with the DNA/RNA-binding protein families of ILF2 and EWS (Liang et al. manuscript in preparation). Future studies should shed light on the role of LDB1-dependent and -independent SSBP2 pathways in hematopoiesis.
Impaired HSPC homeostasis and stress response in Ssbp2−/− mice
The biological consequences of Ssbp2 ablation, reduced cellularity in multiple hematopoietic tissues (Fig. 2), increased frequency of LT-HSCs, and reduced frequency of LMPPs, underscore the significance of high Ssbp2 expression in HSPCs (Fig. 3). While the absolute number of HSPC subpopulations was not significantly altered, the increased frequency of LT-HSCs and decreased frequency of LMPPs pointed to impediments to normal HSPC differentiation and stress response in the absence of Ssbp2. A larger cohort of mice may better detect a small but biologically significant change in the absolute number of cells.
When cycling HSPCs are eliminated by 5-FU treatment or other hematopoietic stress, quiescent HSCs capable of self-renewal enter the cell cycle rapidly and re-establish the stem cell pool before returning to dormancy (33, 40, 41). The initial proliferative burst of HSCs, which expands the Lin−Sca+ population, was severely compromised in the absence of Ssbp2 (Fig. 4). Whether the HSPC defects are HSC intrinsic or due to a niche contribution or some combination of the two cannot be readily answered by the present studies. These findings regardless, raise the possibility of a role for Ssbp2 in an interplay between HSC and bone marrow niche, as reconstitution with either WT marrow or the microenvironment rescued the delayed recovery from cytotoxic stress. Finally, the possibility that Ssbp2 may be a strain specific quantitative trait locus in hematopoiesis analogous to neuronal survival (22) cannot be ruled out.
The overall decrease in the short- and long-term reconstituting potential of BM from null mice likewise implicates Ssbp2 in HSPC expansion (Fig. 5). The subtle but significant differences in the LT-HSC and LMPP frequency and pronounced defects in myelo-regenerative response, and competitive repopulating activity of knockout BM cells underscore a requirement for Ssbp2 in maintaining normal HSPC integrity.
Aberrant expression of regulatory genes and potential role in AML
Our characterization of aberrant E2a and Notch1 expression in the absence of Ssbp2 raises a number of questions. Both E2a and Notch1 have critical regulatory roles and potentially important roles in HSPC maintenance, and our findings implicated SSBP2 directly or indirectly in modulating E2a and Notch1 transcription. E2a expression, which is first detectable in LT-HSCs, increases steadily as those cells develop into MPPs and LMPPs, then rises dramatically in common lymphoid progenitor cells (42). The enhanced LT-HSC cycling and stem cell exhaustion seen in E2a−/− mice has uncovered a role for E2A in HSC cycling through transcriptional up-regulation of CDK inhibitors (43, 44). Elegant serial adoptive transfer studies with compound heterozygotes for E2a and Cdkn1a suggest an absolute requirement for this pathway in maintaining LT-HSCs (28). Furthermore, a global genetic screen identified E2a promoted Cdkn1a expression as a regulator of cell cycle arrest in the absence of cell death in several tumor cell lines (45). Although high expression levels of E2a and Cdkn1a potential positive regulators of quiescence and the modest decrease in Gfi1 expression a negative regulator of HSC quiescence, appear contradictory, the net outcome of Ssbp2 loss was impaired stress recovery and engraftment defects.
In normal BM, Notch1 expression increases modestly from LT-HSC to ST-HSCs. Similar to E2a expression, Notch1 expression increases progressively from LT-HSC to MPPs to LMPPs before rising dramatically in common lymphoid progenitor cells (46). The reduced frequency of LMPPs in Ssbp2−/− BMcould potentially be a direct consequence of increased E2a expression, as HSPCs and downstream progeny are exquisitely sensitive to E2a dosage (43) and to loss of Notch1 expression, which is critical for LMPP expansion.
Clearly, the resolution of HSPC analyses and gene expression changes in subpopulations presented in this report can be enhanced considerably with the use of larger cohorts of mice and more sensitive signaling lymphocyte activation molecule markers (47). ChIP studies will further elucidate the mechanistic underpinnings of SSBP2-regulated transcriptional network. Regardless, the preliminary findings shown in Fig. 7 identify Ssbp2−/− mice as a novel tool with which to examine the complex E2A-NOTCH1 axis in regulating HSPC integrity.
Human SSBP2 was positionally cloned as a candidate myelodysplasia/AML suppressor gene from chromosome 5q14 (10). Although most patients harbor large deletions, the smallest region of overlap is thought to map to 5q31. However, a recent study with high-density single-nucleotide polymorphism arrays demonstrated that larger deletions are associated with poorer outcome and shortened disease-free survival (48). In addition, SSBP2 expression is decreased in AML stem cells and cell lines, in contrast to normal HSCs (30, 49). Unlike murine HSCs, which acquire self-renewal abilities without transformation, with HoxB4 overexpression in two primates induced AML in both (50). One of these monkeys harbored retroviral integration in the SSBP2 gene and an associated reduction in expression by 70%. Moreover, inducible expression of SSBP2 in the myelomonocytic cell line U937 resulted in growth arrest and differentiation, although the mechanisms are unknown (30). While these findings suggest a role for AML suppressor, direct evidence for SSBP2-mediated myeloid leukemia suppression is lacking (50).
AML is genetically diverse (51, 52). The role of NOTCH1 in myeloid differentiation and transformation is controversial and may reflect loss and gain offunction roles. Overexpression of HES1, a NOTCH1 downstream target, exacerbates chronic myelogenous leukemia blast crisis in BCR-ABL–induced murine models (53). Further, inactivating NOTCH1 mutations are found in chronic myelomonocytic leukemia. Together, loss of SSBP2 and altered NOTCH1 expression may identify a distinct subset of Leukemia initiating cells ( LICs) originating HSCs or MPPs, while enhanced SSBP2 and NOTCH1 expression may define LMPPs as a target of transformation. The present report is an early step in connecting these intricate networks with HSPC activity and, potentially, AML pathogenesis.
Supplementary Material
Acknowledgments
We thank Dr. Margaret Goodell for helpful discussions and critical review of the results. Verlene Henry and Adrienne Ferguson are acknowledged for assistance with tail vein injections and bleeding. Our special thanks are to Hank Adams for assistance with microscopy.
These studies were supported by HL744409, a developmental research project award from SPORE grant P50-CA100632, and the Loper family research funds to L.N. Flow cytometry, genotyping and veterinary services were supported by NCI Core Grant CA16672 to The University of Texas MD Anderson Cancer Center.
References
- 1.Wilson A, Laurenti E, Trumpp A. Balancing dormant and self-renewing hematopoietic stem cells. Current opinion in genetics & development. 2009;19:461–468. doi: 10.1016/j.gde.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Trumpp A, Essers M, Wilson A. Awakening dormant haematopoietic stem cells. Nature reviews. Immunology. 2010;10:201–209. doi: 10.1038/nri2726. [DOI] [PubMed] [Google Scholar]
- 3.Shivdasani RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 4.Souroullas GP, Salmon JM, Sablitzky F, Curtis DJ, Goodell MA. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell Stem Cell. 2009;4:180–186. doi: 10.1016/j.stem.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meier N, Krpic S, Rodriguez P, Strouboulis J, Monti M, Krijgsveld J, Gering M, Patient R, Hostert A, Grosveld F. Novel binding partners of Ldb1 are required for haematopoietic development. Development. 2006;133:4913–4923. doi: 10.1242/dev.02656. [DOI] [PubMed] [Google Scholar]
- 6.Lecuyer E, Lariviere S, Sincennes MC, Haman A, Lahlil R, Todorova M, Tremblay M, Wilkes BC, Hoang T. Protein stability and transcription factor complex assembly determined by the SCL-LMO2 interaction. The Journal of biological chemistry. 2007;282:33649–33658. doi: 10.1074/jbc.M703939200. [DOI] [PubMed] [Google Scholar]
- 7.Warren AJ, Colledge WH, Carlton MB, Evans MJ, Smith AJ, Rabbitts TH. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 8.Mukhopadhyay M, Teufel A, Yamashita T, Agulnick AD, Chen L, Downs KM, Schindler A, Grinberg A, Huang SP, Dorward D, Westphal H. Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development. 2003;130:495–505. doi: 10.1242/dev.00225. [DOI] [PubMed] [Google Scholar]
- 9.Li L, Jothi R, Cui K, Lee JY, Cohen T, Gorivodsky M, Tzchori I, Zhao Y, Hayes SM, Bresnick EH, Zhao K, Westphal H, Love PE. Nuclear adaptor Ldb1 regulates a transcriptional program essential for the maintenance of hematopoietic stem cells. Nature immunology. 2011;12:129–136. doi: 10.1038/ni.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castro P, Liang H, Liang JC, Nagarajan L. A novel, evolutionarily conserved gene family with putative sequence-specific single-stranded DNA-binding activity. Genomics. 2002;80:78–85. doi: 10.1006/geno.2002.6805. [DOI] [PubMed] [Google Scholar]
- 11.Bayarsaihan D, Lukens LN. Single-strand-DNA-binding factors specifically recognize the pyrimidine element in the chick alpha2(I) collagen gene promoter. Biochem J. 1996;314(Pt 1):293–296. doi: 10.1042/bj3140293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai Y, Xu Z, Nagarajan L, Brandt SJ. Single-stranded DNA-binding proteins regulate the abundance and function of the LIM-homeodomain transcription factor LHX2 in pituitary cells. Biochem Biophys Res Commun. 2008;373:303–308. doi: 10.1016/j.bbrc.2008.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gungor C, Taniguchi-Ishigaki N, Ma H, Drung A, Tursun B, Ostendorff HP, Bossenz M, Becker CG, Becker T, Bach I. Proteasomal selection of multiprotein complexes recruited by LIM homeodomain transcription factors. Proc Natl Acad Sci U S A. 2007;104:15000–15005. doi: 10.1073/pnas.0703738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Z, Meng X, Cai Y, Liang H, Nagarajan L, Brandt SJ. Single-stranded DNA-binding proteins regulate the abundance of LIM domain and LIM domain-binding proteins. Genes Dev. 2007;21:942–955. doi: 10.1101/gad.1528507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Klumpp S, Amin HM, Liang H, Li J, Estrov Z, Zweidler-McKay P, Brandt SJ, Agulnick A, Nagarajan L. SSBP2 is an in vivo tumor suppressor and regulator of LDB1 stability. Oncogene. 2010;29:3044–3053. doi: 10.1038/onc.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Q, Schaefer AM, Nonet ML. Regulation of C. elegans presynaptic differentiation and neurite branching via a novel signaling pathway initiated by SAM-10. Development. 2011;138:87–96. doi: 10.1242/dev.055350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bronstein R, Levkovitz L, Yosef N, Yanku M, Ruppin E, Sharan R, Westphal H, Oliver B, Segal D. Transcriptional regulation by CHIP/LDB complexes. PLoS genetics. 2010;6:e1001063. doi: 10.1371/journal.pgen.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bronstein R, Segal D. Modularity of CHIP/LDB transcription complexes regulates cell differentiation. Fly (Austin) 2011;5:200–205. doi: 10.4161/fly.5.3.14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong Z, Ma H, Taniguchi-Ishigaki N, Nagarajan L, Becker CG, Bach I, Becker T. SSDP cofactors regulate neural patterning and differentiation of specific axonal projections. Developmental biology. 2011;349:213–224. doi: 10.1016/j.ydbio.2010.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishioka N, Nagano S, Nakayama R, Kiyonari H, Ijiri T, Taniguchi K, Shawlot W, Hayashizaki Y, Westphal H, Behringer RR, Matsuda Y, Sakoda S, Kondoh H, Sasaki H. Ssdp1 regulates head morphogenesis of mouse embryos by activating the Lim1-Ldb1 complex. Development. 2005;132:2535–2546. doi: 10.1242/dev.01844. [DOI] [PubMed] [Google Scholar]
- 21.Enkhmandakh B, Makeyev AV, Bayarsaihan D. The role of the proline-rich domain of Ssdp1 in the modular architecture of the vertebrate head organizer. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11631–11636. doi: 10.1073/pnas.0605209103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kempermann G, Chesler EJ, Lu L, Williams RW, Gage FH. Natural variation and genetic covariance in adult hippocampal neurogenesis. Proc Natl Acad Sci U S A. 2006;103:780–785. doi: 10.1073/pnas.0510291103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S, van De Walle I, Cathelin S, Trimarchi T, Araldi E, Liu C, Ibrahim S, Beran M, Zavadil J, Efstratiadis A, Taghon T, Michor F, Levine RL, Aifantis I. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473:230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Freudenberg J, Cui K, Dale R, Song SH, Dean A, Zhao K, Jothi R, Love PE. Ldb1-nucleated transcription complexes function as primary mediators of global erythroid gene activation. Blood. 2013;121:4575–4585. doi: 10.1182/blood-2013-01-479451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong Y, Castro E, Yagi R, Zhu J, Lesourne R, Love PE, Feigenbaum L, Bosselut R. Thpok-independent repression of Runx3 by Gata3 during CD4+ T-cell differentiation in the thymus. Eur J Immunol. 2013;43:918–928. doi: 10.1002/eji.201242944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fleisig HB, Orazio NI, Liang H, Tyler AF, Adams HP, Weitzman MD, Nagarajan L. Adenoviral E1B55K oncoprotein sequesters candidate leukemia suppressor sequence-specific single-stranded DNA-binding protein 2 into aggresomes. Oncogene. 2007;26:4797–4805. doi: 10.1038/sj.onc.1210281. [DOI] [PubMed] [Google Scholar]
- 27.Network CGAR. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santos PM, Ding Y, Borghesi L. Cell-intrinsic in vivo requirement for the E47-p21 pathway in long-term hematopoietic stem cells. J Immunol. 2014;192:160–168. doi: 10.4049/jimmunol.1302502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang H, Samanta S, Nagarajan L. SSBP2, a candidate tumor suppressor gene, induces growth arrest and differentiation of myeloid leukemia cells. Oncogene. 2005;24:2625–2634. doi: 10.1038/sj.onc.1208167. [DOI] [PubMed] [Google Scholar]
- 31.Fleisig HB, Liang H, Nagarajan L. Adenoviral oncoprotein E1B55K mediates colocalization of SSBP2 and PML in response to stress. Journal of molecular signaling. 2010;5:6. doi: 10.1186/1750-2187-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 33.Randall TD, Weissman IL. Phenotypic and functional changes induced at the clonal level in hematopoietic stem cells after 5-fluorouracil treatment. Blood. 1997;89:3596–3606. [PubMed] [Google Scholar]
- 34.Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. The New England journal of medicine. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eckfeldt CE, Mendenhall EM, Flynn CM, Wang TF, Pickart MA, Grindle SM, Ekker SC, Verfaillie CM. Functional analysis of human hematopoietic stem cell gene expression using zebrafish. PLoS Biol. 2005;3:e254. doi: 10.1371/journal.pbio.0030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He X, Gonzalez V, Tsang A, Thompson J, Tsang TC, Harris DT. Differential gene expression profiling of CD34+ CD133+ umbilical cord blood hematopoietic stem progenitor cells. Stem Cells Dev. 2005;14:188–198. doi: 10.1089/scd.2005.14.188. [DOI] [PubMed] [Google Scholar]
- 37.Wagner W, Ansorge A, Wirkner U, Eckstein V, Schwager C, Blake J, Miesala K, Selig J, Saffrich R, Ansorge W, Ho AD. Molecular evidence for stem cell function of the slow-dividing fraction among human hematopoietic progenitor cells by genome-wide analysis. Blood. 2004;104:675–686. doi: 10.1182/blood-2003-10-3423. [DOI] [PubMed] [Google Scholar]
- 38.Laurenti E, Doulatov S, Zandi S, Plumb I, Chen J, April C, Fan JB, Dick JE. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nature immunology. 2013;14:756–763. doi: 10.1038/ni.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Meyel DJ, Thomas JB, Agulnick AD. Ssdp proteins bind to LIM-interacting co-factors and regulate the activity of LIM-homeodomain protein complexes in vivo. Development. 2003;130:1915–1925. doi: 10.1242/dev.00389. [DOI] [PubMed] [Google Scholar]
- 40.Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, Lio P, Macdonald HR, Trumpp A. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 41.Venezia TA, Merchant AA, Ramos CA, Whitehouse NL, Young AS, Shaw CA, Goodell MA. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS biology. 2004;2:e301. doi: 10.1371/journal.pbio.0020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1930–1935. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Q, Kardava L, St Leger A, Martincic K, Varnum-Finney B, Bernstein ID, Milcarek C, Borghesi L. E47 controls the developmental integrity and cell cycle quiescence of multipotential hematopoietic progenitors. J Immunol. 2008;181:5885–5894. doi: 10.4049/jimmunol.181.9.5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andrysik Z, Kim J, Tan AC, Espinosa JM. A genetic screen identifies TCF3/E2A and TRIAP1 as pathway-specific regulators of the cellular response to p53 activation. Cell reports. 2013;3:1346–1354. doi: 10.1016/j.celrep.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oh P, Lobry C, Gao J, Tikhonova A, Loizou E, Manent J, van Handel B, Ibrahim S, Greve J, Mikkola H, Artavanis-Tsakonas S, Aifantis I. In vivo mapping of notch pathway activity in normal and stress hematopoiesis. Cell Stem Cell. 2013;13:190–204. doi: 10.1016/j.stem.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jerez A, Gondek LP, Jankowska AM, Makishima H, Przychodzen B, Tiu RV, O'Keefe CL, Mohamedali AM, Batista D, Sekeres MA, McDevitt MA, Mufti GJ, Maciejewski JP. Topography, clinical, and genomic correlates of 5q myeloid malignancies revisited. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:1343–1349. doi: 10.1200/JCO.2011.36.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qian Z, Fernald AA, Godley LA, Larson RA, Le Beau MM. Expression profiling of CD34+ hematopoietic stem/ progenitor cells reveals distinct subtypes of therapy-related acute myeloid leukemia. Proc Natl Acad Sci U S A. 2002;99:14925–14930. doi: 10.1073/pnas.222491799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang XB, Beard BC, Trobridge GD, Wood BL, Sale GE, Sud R, Humphries RK, Kiem HP. High incidence of leukemia in large animals after stem cell gene therapy with a HOXB4-expressing retroviral vector. J Clin Invest. 2008;118:1502–1510. doi: 10.1172/JCI34371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Majeti R, Becker MW, Tian Q, Lee TL, Yan X, Liu R, Chiang JH, Hood L, Clarke MF, Weissman IL. Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc Natl Acad Sci U S A. 2009;106:3396–3401. doi: 10.1073/pnas.0900089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen SJ, Shen Y, Chen Z. A panoramic view of acute myeloid leukemia. Nature genetics. 2013;45:586–587. doi: 10.1038/ng.2651. [DOI] [PubMed] [Google Scholar]
- 53.Nakahara F, Sakata-Yanagimoto M, Komeno Y, Kato N, Uchida T, Haraguchi K, Kumano K, Harada Y, Harada H, Kitaura J, Ogawa S, Kurokawa M, Kitamura T, Chiba S. Hes1 immortalizes committed progenitors and plays a role in blast crisis transition in chronic myelogenous leukemia. Blood. 2010;115:2872–2881. doi: 10.1182/blood-2009-05-222836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.