Abstract
The high molecular weight glycosaminoglycan, hyaluronic acid (HA), makes up a significant portion of the brain extracellular matrix (ECM). Glioblastoma multiforme (GBM), a highly invasive brain tumor, is associated with aberrant HA secretion, tissue stiffening, and overexpression of the HA receptor CD44. Here, transcriptomic analysis, engineered materials, and measurements of adhesion, migration, and invasion were used to investigate how HA/CD44 ligation contributes to the mechanosensing and invasive motility of GBM tumor cells, both intrinsically and in the context of RGD/integrin adhesion. Analysis of transcriptomic data from The Cancer Genome Atlas (TCGA) reveals up-regulation of transcripts associated with HA/CD44 adhesion. CD44 suppression in culture reduces cell adhesion to HA on short time scales (0.5h post-incubation) even if RGD is present, whereas maximal adhesion on longer time scales (3h) requires both CD44 and integrins. Moreover, time-lapse imaging demonstrates that cell adhesive structures formed during migration on bare HA matrices are more short-lived than cellular protrusions formed on surfaces containing RGD. Interestingly, adhesion and migration speed were dependent on HA hydrogel stiffness, implying that CD44-based signaling is intrinsically mechanosensitive. Finally, CD44 expression paired with an HA-rich microenvironment maximized three-dimensional invasion, whereas CD44 suppression or abundant integrin-based adhesion limited it. These findings demonstrate that CD44 transduces HA-based stiffness cues, temporally precedes integrin-based adhesion maturation, and facilitates invasion.
Keywords: CD44, Cell adhesion, Hyaluronic acid/hyaluronan, Tumor invasion
Introduction
The prognosis of glioblastoma multiforme (GBM), a highly invasive and rapidly lethal brain tumor, has improved only incrementally in the past several decades. This dire prognosis is in large part attributed to the aggressively invasive nature of glioma cells, which in turn has fueled interest in exploring new strategies for slowing invasion, including identifying and limiting interactions between tumor cells and pro-invasive components of the tumor extracellular matrix (ECM) (1). In addition to invading brain tissue along vascular structures, GBM tumors are characterized by a diffuse single-cell invasion pattern of glioma cells into brain parenchyma (2, 3), which contains ECM characteristically devoid of the fibrillar adhesive proteins found in connective tissue, but rich in HA (3, 4). This chemical signature of brain ECM is significant because an emerging body of literature has revealed that the HA/CD44 interaction can act as a powerful regulator of cell proliferation, survival, and anti-apoptotic pathways (5–7). In the ECM of gliomas, HA is much more abundant than in normal brain (8), suggesting that its oversecretion may contribute to the aggressive invasion pattern of glioma cells in brain parenchyma. Finally, CD44 expression directly contributes to the survival of glioma stem-like cells hypothesized to be the driver of tumor recurrence (9).
While changes in ECM biochemistry are key features of GBM, we and others have shown that ECM biophysical properties also strongly influence glioma cell invasion in an in vitro setting (10, 11). This is consistent with the observation that GBM tumors are stiffer than normal brain tissue, to the extent that ultrasound imaging can be used to delineate tumor margins intraoperatively (12, 13). This has led to the hypothesis that part of the aggressive nature of GBM may be regulated by biophysical interactions between glioma cells and the brain ECM. Matrix stiffness cues encoded in the ECM are traditionally thought to be transduced by integrins, and this signaling is altered in cells derived from a variety of cancer cell types. While the importance of integrin-mediated signaling in these scenarios has been well-characterized, the significance of non-integrin ECM adhesion receptors to tumor cell mechanobiology remains largely unexplored. The abundant presence of HA in brain ECM and the established role of CD44-mediated signaling in tumor progression beg the question of how HA/CD44 interactions contribute to glioma adhesion and invasion in ECMs composed of HA, integrin-adhesive domains, or both.
Despite the acknowledged association of GBM with altered HA deposition, CD44-based signaling, and tissue mechanics, little is known about the causal relationships between these phenomena in the pathogenesis of the disease, particularly with respect to tumor invasion. We therefore sought to investigate these connections by combining transcriptomic analysis of human GBM tumors to explore correlations in the expression of HA/CD44-related genes, engineered hydrogel materials to recapitulate the compliant, HA-rich nature of brain ECM (14–16), and biophysical studies of tumor cell adhesion, migration, and invasion. We find that GBM tumors preferentially express certain HA/CD44-related genes relative to normal brain tissue and that HA/CD44 interactions strongly contribute to tumor cell adhesion, mechanosensing, and invasive motility. These effects are both experimentally separable and functionally distinct from contributions of integrin-based adhesion.
Materials and Methods
The Cancer Genome Atlas (TCGA) gene expression analysis
Data from the publicly available data browser was queried for expression of CD44 and related genes. The cBioPortal analysis tool from Memorial Sloan-Kettering was used to retrieve mRNA expression data for genes of interest from all available GBM tumor samples. Correlations with CD44 expression were analyzed by Pearson’s product-moment correlation coefficient.
HA hydrogel synthesis
HA hydrogels were synthesized as previously described (14, 17). Briefly, methacrylic anhydride was used to functionalize HA with methacrylate groups (Me-HA). The degree of methacrylation was characterized by 1H NMR as detailed previously (14), and the Me-HA used for the experiments discussed here was characterized to have 50% of disaccharides methacrylated. Me-HA could then be conjugated via Michael Addition reactions with molecules containing free thiol groups. In some cases, Me-HA was conjugated with the cysteine-containing RGD peptide (Ac-GCGYGRGDSPG-NH2, Anaspec) to add integrin-adhesive functionality at a concentration of 0.5 mM. Finally, hydrogels were formed by crosslinking 5 wt% Me-HA in DMEM (Invitrogen) with varying concentrations of the bifunctional thiol dithiothreitol (DTT, Sigma-Aldrich), ranging from 2.79 mM (to yield 0.15 kPa) to 22.3 mM (to yield 6.9 kPa). After 1 h crosslinking time, the hydrogels were rinsed thoroughly with PBS prior to cell seeding.
Rheological measurements
The shear modulus of various hydrogel formulations was measured using oscillatory rheometry as described previously (14). Briefly, hydrogels were first crosslinked by incubation for 1 h in a humidified 37°C chamber. Rheological testing consisted of frequency sweeps ranging from 100 Hz to 0.1 Hz at 0.5% amplitude, also in a humidified 37°C chamber. Shear modulus was reported as the storage modulus at an oscillation frequency of 0.1 Hz.
Functionalization of HA hydrogels with full-length proteins
After crosslinking, some HA hydrogels were functionalized with adhesive proteins in a method adapted from a previous study (18). Since the hydrogels are resistant to passive protein adsorption, crosslinked hydrogels were conjugated with poly-L-lysine by carbodiimide chemistry using 0.5 M EDC (Pierce) and 0.5 M NHS (Sigma) in 0.1 M MES buffer at pH 5.8. After rinsing, a solution of 0.5% poly-L-lysine (Sigma) in PBS was added for 1.5 h, then a 0.1 mg/mL solution of human plasma fibronectin (Millipore) or rat laminin (Life Technologies) was adsorbed for 1 h at room temperature. Before cell seeding, the hydrogels were rinsed with PBS.
Cell culture
U373-MG and U87-MG human glioblastoma cells were obtained from the University of California, Berkeley Tissue Culture Facility, which sources its cultures directly from the ATCC. We note that ATCC U373-MG cells have been discovered to share common origins with SNB-19 and U251-MG cell lines, although these three lines appear to have since evolved to exhibit distinct karyotypes and drug sensitivities (19). Whatever their origins, all of these lines are understood to be fully distinct from U87-MG cells and thus serve as an appropriate culture model for comparison. Cells were cultured as previously described (10) in DMEM (Invitrogen) supplemented with 10% calf serum (JR Scientific), 1% penicillin-streptomycin, MEM nonessential amino acids, and sodium pyruvate (Invitrogen). Accutase (Innovative Cell Technologies, Inc.) was used to harvest cell cultures from tissue culture polystyrene.
Lentiviral shRNA transfection
To create CD44 knockdown cells, two distinct CD44-specific shRNA constructs (V2LHS_111682 and V2LHS_111684, Thermo Scientific) were screened in addition to a non-targeting scramble sequence (plasmid 1864, Addgene) as a negative control. Viral particles were packaged using human embryonic kidney 293T cells, then transfected into U373-MG human glioblastoma cells with a multiplicity of infection of 1 IU/cell. Cells were selected using 1 μg/mL puromycin, and expression of the GFP-containing vectors was confirmed using flow cytometry.
Western Blots
The efficiency of CD44 knockdown was confirmed using Western blots. RIPA buffer with protease and phosphatase inhibitors were used to lyse cells. Lysates were heated to 70°C, run through a 4–12% bis-tris gel, and then blotted onto a PVDF membrane. Membranes were probed with rat anti-pan-CD44 primary antibody (Hermes-1, Pierce) or goat anti-RHAMM primary antibody (E-19, Santa Cruz Biotechnology), and mouse anti-GAPDH (Sigma-Aldrich). HRP-conjugated secondary antibodies against rat, mouse (Invitrogen), or goat (Zymed), and SuperSignal West Dura reagent (Thermo Scientific) were used for chemiluminescent detection. Membranes were imaged using the ChemiDoc XRS+ system (Biorad), and bands were quantified using ImageLab software (Biorad) and normalized to GAPDH content.
Centrifugal adhesion assay
In some cases, cells were pre-treated by incubation in serum-free media with blockers of adhesion for 15 min, including pan-CD44-neutralizating antibody (Hermes-1, Pierce), rat IgG2a control antibody (eBioscience), and soluble RGD peptide (Ac-GCGYGRGDSPG-NH2, Anaspec). Cells were seeded either on crosslinked HA hydrogels with or without RGD functionality, or on tissue culture polystyrene surfaces coated overnight with high molecular weight HA from Streptococcus equi (Sigma), low molecular weight HA (66–90 kDa, Lifecore Biomedical), BSA (Sigma), and/or human plasma fibronectin (Millipore). After a specified adhesion time, wells were completely filled with fresh serum-free media, and the cell culture plate was sealed with an adhesive plate sealer. The plate was then inverted and centrifuged for 5 min at 100 g. Cells remaining on the hydrogels were then fixed and stained with 4′,6-diamidino-2-phenylindole (DAPI, Invitrogen). Automated Otsu thresholding analysis of the DAPI images were performed on ImageJ (NIH) to determine a total count of the number of cells on each hydrogel. Each condition was tested in at least 3 wells, over at least 3 experiments.
Integrin characterization assay
To characterize the integrin expression pattern of control and CD44 knockdown cells, an α/β Integrin-mediated Cell Adhesion Array Combo Kit (ECM532, EMD Millipore) was used, in which antibodies specific for a particular integrin subunit or heterodimer are immobilized onto microtiter wells. Cells were non-enzymatically harvested by incubation in 5 mM EDTA for 20 m, then incubated on microtiter plates for 2 h in serum-free medium. Non-adherent cells were gently washed off, and the number of cells was quantified with a colorimetric readout per manufacturer’s instructions. Each integrin subunit or heterodimer antibody was assayed in duplicate wells, and the experiment was repeated three times. The specificity of each antibody was verified, and published accounts of the antibody clones (20, 21) used differed slightly from the kit description; these literature-validated antibody specificities are reported here.
Live cell time-lapse microscopy
U373-MG cells were seeded at least 4 h prior to the beginning of each time-lapse experiment. Every 15 m, phase contrast images were obtained with a 10x objective. Migration speed was measured using manual tracking plugin in Image J. For each cell, the displacement of the nucleus for each 15 m time interval was averaged over 6 h to obtain the mean cell speed. At least 50 cells were analyzed per condition, over at least 3 separate experiments. Persistence parameter was calculated as the end-to-end path distance divided by the total distance travelled during a 6 h time span.
Immunofluorescence staining and cell area measurements
Mouse anti-vinculin primary antibody (Sigma) and AlexaFluor 546 goat anti-mouse secondary antibody (Molecular Probes) were used to visualize vinculin. Rat anti-CD44 primary antibody (Hermes-1, Pierce) and AlexaFluor 647 chicken anti-rat secondary antibody (Molecular Probes) were used to visualize CD44. Filamentous actin was stained using AlexaFluor 488 phalloidin (Invitrogen), and nuclei were labeled with DAPI. Confocal images were obtained with a swept-field confocal microscope (Prairie Technologies). Cell area was measured by manually tracing the cell edges of live cell phase contrast images taken 24 h after seeding using ImageJ software.
Transwell invasion assay
Transwell membrane inserts with 8 μm pores, the smallest pore size we found to be permissible for cell invasion in U373-MG cells, were coated overnight with 2 mg/mL high molecular weight HA from Streptococcus equi, 3.7 or 9.3 μg/mL human plasma fibronectin, or a combination of both HA and fibronectin. Membranes not adsorbed with fibronectin were blocked with BSA (Sigma). Membranes were then thoroughly rinsed with PBS, and cells were seeded at a density of 28,000 cells/cm2 in cell medium supplemented with 1% calf serum. In some cases, soluble HA was added to the cell medium in both the upper and lower chambers at a concentration of 0.125 or 0.625 mg/mL. Phase contrast microscopy was used to visualize cells on either side of clear membranes. To visualize only cells on the underside of the membrane, which had therefore invaded through pores, GFP-expressing cells were seeded on Fluoroblok membranes and imaged with fluorescence microscopy. Cells were fixed 4 h after seeding, and cells remaining on tops of membranes were removed by scraping with a pipette tip. To quantify cells that had invaded through the membrane, fluorescence images of DAPI-stained cells were thresholded, and the total number of nuclei in each membrane was counted with particle analysis software (ImageJ). To confirm specificity of CD44 with adsorbed HA, control cells were pre-incubated with CD44-neutralizing antibody and seeded on HA-coated transwells. To confirm specificity of RGD-ligating integrins with adsorbed fibronectin, cells were pre-incubated with soluble RGD peptide and seeded on fibronectin-coated transwells.
Statistics
Analysis of percentage data for adhesion and invasion assays was preceded by an arcsin transformation. For normally distributed data, statistical significance was tested using ANOVA followed by Tukey-Kramer multiple comparison, and represented by bar plots with error bars representing standard error. Non-normally distributed data was tested by Kruskal-Wallis followed by Dunn’s multiple comparison, and represented by box-and-whiskers plots. Boxes represent 25th and 75th percentiles, whiskers represent 10th and 90th percentiles, and squares represent the mean. All error bars represent standard error.
Results
Expression of proteins associated with HA/CD44-based adhesion is frequently aberrant in GBM tumors
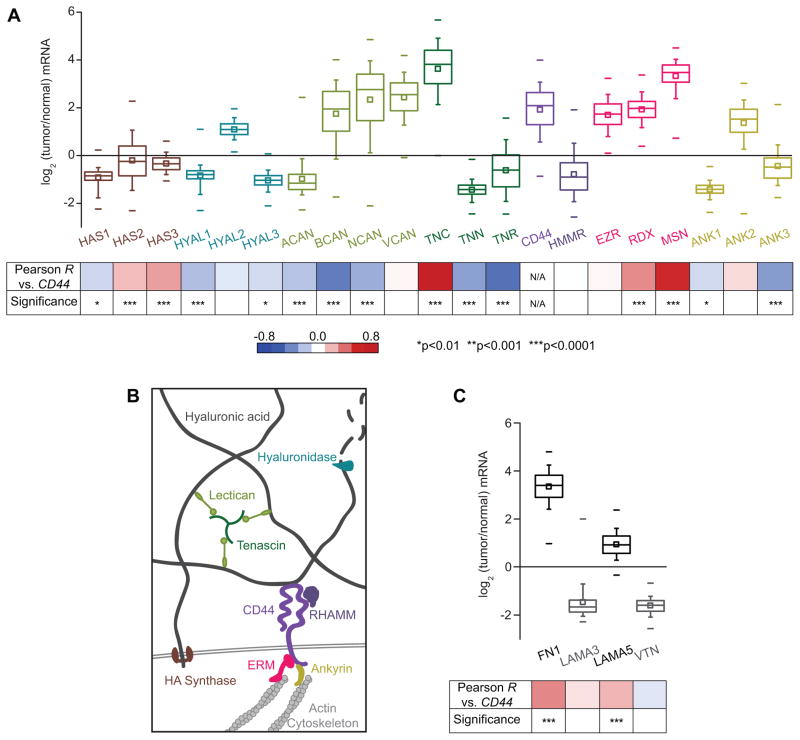
Aberrant expression of CD44 is a common feature of many cancers, so we began by investigating to what extent CD44 itself, and related proteins that link it to the cytoskeleton or ECM, are misregulated in GBM tumors (22) (Fig. 1). On the mRNA level, correlation analysis of transcriptomic data from The Cancer Genome Atlas (TCGA) reveals that CD44 is indeed frequently overexpressed in GBM tumors, with a median gene expression 4.25-fold higher than that of normal brain tissue (Fig. 1B), in agreement with many previous smaller-scale histological studies (8, 23–26). Further analysis reveals that many other CD44-related genes that are also commonly overexpressed in GBM, particularly those involved in HA synthesis and degradation. The genes HAS1, HAS2, and HAS3, which encode the HA synthases, are aberrantly expressed. Importantly, induced HAS2 expression has been shown to promote anchorage-independent growth and increases tumorigenicity (27). However, the level of HA ultimately found in the matrix is affected both by rates of HA synthesis and several post-transcriptional levels of regulation, so the median decrease in HA synthase transcription is not necessarily inconsistent with the elevated levels of HA in the tumor microenvironment. For instance, the expression of hyaluronidases or hyals, which are responsible for HA matrix catabolism, are also misregulated, with hyaluronidase-1 and hyaluronidase-3 generally poorly expressed relative to normal brain tissue. On the other hand, hyaluronidase-2 is expressed at high levels during development but at low levels in the adult brain (28), and its induced overexpression promotes tumor invasion in a mouse model (29).
Figure 1. CD44 is frequently overexpressed in glioblastoma multiforme tumors, and correlated with misregulation of other proteins involved in CD44-mediated pathways.
(A) Transcriptomic analysis of The Cancer Genome Atlas (TCGA) microarray data: HA synthases (HAS); hyaluronidases (HYAL); the lecticans aggrecan (ACAN), brevican (BCAN), neurocan (NCAN), and versican (VCAN); the tenascins tenascin-C (TNC), tenascin-N (TNN), and tenascin-R (TNR); CD44; RHAMM (HMMR); the ERM proteins (EZR, RDX, MSN), and the ankyrins (ANK). Percentiles are represented by boxes (25, 50, 75), whiskers (10, 90), and dashes (1, 99). Bottom panel: analysis of gene expression correlation with CD44. Red-blue heat map indicates value of Pearson product-moment correlation coefficient between CD44 expression and each gene, and p values indicate the statistical significance of the correlation. (B) Expression of proteins relevant to hyaluronic acid (HA)-based brain ECM and associated downstream signaling. HA synthase proteins synthesize HA, while hyaluronidase enzymes degrade HA. Other ECM components that interact with HA include lecticans, which bind to both tenascins and HA to form a mesh-like matrix. CD44 and RHAMM are the main HA receptors. Intracellularly, CD44 links to the actin cytoskeleton through the ERM (ezrin-radixin-moesin) family proteins, and ankyrins. (C) Analysis of expression of genes encoding other extracellular matrix components previously determined to be key for glioblastoma multiforme progression: fibronectin (FN1), laminin α-5 (LAMA5), laminin α-3 (LAMA3), and vitronectin (VTN).
In addition to proteins that regulate turnover of the HA network, other proteins that physically crosslink linear HA chains together also exhibit altered transcription patterns (Fig. 1A). Among these include the family of lecticans and tenascins, which, with HA, form the basic structural components of brain ECM (30). The expression of genes that encode proteins key to CD44-mediated intracellular signaling are also misregulated. A notable example is the ERM family proteins (ezrin, ridixin, and moesin), which mediates interactions between the cytoplasmic domain of CD44 and the actin cytoskeleton, and have known pro-oncogenic function (31). All three ERM proteins are overexpressed in tumor tissue, with expression of MSN correlating strongly and positively with CD44 expression. Interestingly, while RHAMM (receptor for hyaluronic acid-mediated motility) is generally described as a pro-invasive HA receptor and has been found to be overexpressed in smaller GBM studies (32), TCGA analysis revealed that the median mRNA expression of the encoding gene HMMR in tumors is only 0.53-fold the expression level of normal brain tissues. This suggests that post-transcriptional regulation may have significant effects on HMMR, or that RHAMM overexpression may be a feature of a specific subset of GBM tumors. Finally, since HA functions as a key organizational scaffold for brain ECM proteins, we examined the gene expression of other ECM proteins (Fig. 1C). Indeed, GBM tumors overexpress several proteins that ligate integrins through RGD peptides, such as fibronectin. Together, these findings motivated us to more deeply investigate contributions of CD44 and integrin binding to GBM tumor cell adhesion, migration, and invasion.
CD44-mediated adhesion is important even when RGD is abundant
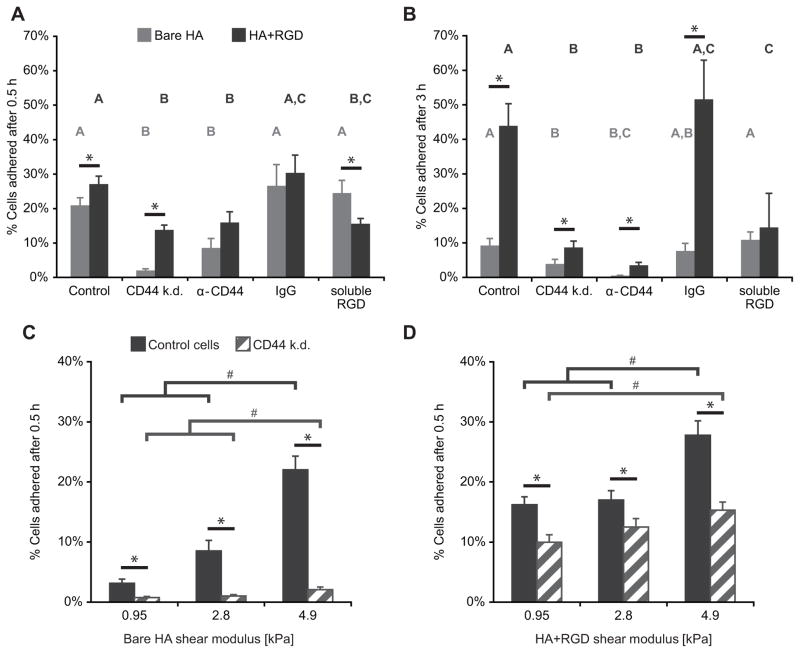
Given the wealth of data supporting the importance of integrins in glioma cell-ECM adhesion, we first compared the biophysical contributions of CD44 and integrin engagement to total cell-ECM adhesion. We initially chose to work with HA hydrogels with stiffnesses similar to that of brain tissue, which we fabricated according to our previous description (14). Using a centrifugal detachment assay, we measured cell adhesion strength to bare HA hydrogels and HA hydrogels functionalized with RGD peptide, both with a shear modulus of 4.6 kPa. Our use of the RGD sequence was motivated by the fact that this is the key integrin-binding sequence in fibronectin, which is enriched in the ECM of GBM tumors when compared to normal brain tissue. RGD is also ubiquitously used within synthetic biomaterials as a modular integrin-binding sequence (33–35), which allowed us to more readily compare our findings with work in these other materials systems. We seeded U373-MG human glioblastoma cells transduced virally with either a CD44-targeting shRNA (yielding 64% protein expression knockdown, Supplementary Fig. S1A–B) or a non-targeting control sequence, and allowed the cells to adhere for 0.5 h or 3 h in serum-free culture medium. We then measured adhesion strength by quantifying the percentage of cells that remained adherent after exerting a detachment force of 100 g to remove weakly adhered cells.
When cells were centrifuged 0.5 h after initial seeding, 21% of control cells remained adherent to the bare HA hydrogel (Fig. 2A). Incorporation of an RGD peptide into the hydrogel yielded a modest but statistically significant increase in cell adhesion. When the same surfaces were presented to the CD44 knockdown cells, bare HA hydrogels yielded negligible adhesion, while adhesion to HA-RGD hydrogels was reduced twofold relative to control cells. To confirm the molecular specificity of this effect with an alternative approach, we incubated control cells with a CD44-neutralizing antibody, which yielded qualitatively identical results. We also confirmed that CD44 knockdown altered neither integrin subtype expression nor cell viability (Supplementary Fig. S2). Notably, and as reported previously (36), CD44 suppression increased expression of the HA receptor RHAMM (Supplementary Fig. S1C–D), indicating that the effects of depleting CD44 could not be fully rescued by enhancement of other HA adhesion proteins.
Figure 2. U373-MG cell adhesion to hyaluronic acid (HA) hydrogels is CD44- and stiffness-dependent.
(A) Adhesion of cells on 4.6 kPa hydrogels at short (0.5 h) time scales. Control cells or CD44 knockdown (k.d.) cells were allowed to attach to substrates in the absence or presence of adhesion blockers, including α-CD44 neutralizing antibody, IgG isotype control antibody, or soluble RGD peptide; then centrifuged to induce detachment of weakly adhered cells. (B) Adhesion of cells on 4.6 kPa hydrogels at longer (3 h) time scales. (C) Adhesion of cells on bare HA hydrogels as a function of stiffness after 0.5 h adhesion time. (D) Adhesion of cells on RGD-functionalized HA hydrogels as a function of stiffness after 0.5 h adhesion time. #p<0.05 differences between stiffnesses by Tukey-Kramer, *p<0.05 by Student’s t-test.
As expected, pre-incubation of both cell types with an isotype-matched IgG antibody led to similar attachment as the untreated control condition. Pre-treatment of cells with a soluble RGD peptide reduced adhesion to HA-RGD but not to bare HA hydrogels, confirming that the cells adhered to the crosslinked HA through an RGD receptor-independent mechanism. When we repeated this assay for a longer adhesion time of 3 h, the only conditions that resulted in increased adhesion were those with both CD44- and integrin-based adhesions (Fig. 2B). Together, these data suggest that CD44 is largely responsible for adhesion on early time scales (0.5 h), and that these initial adhesions are later reinforced by integrin-mediated adhesions, if available, on longer time scales (3 h).
CD44-mediated adhesion, spreading, and motility is dependent on HA stiffness
It is well established that integrin-based adhesion and signaling are strongly sensitive to ECM stiffness (37–40). Given that GBM invasion is accompanied by HA deposition (8) and tissue stiffening (12), and that glioma invasion requires the ability to sense and transduce mechanical force, we wondered whether this integrin-independent HA/CD44-mediated adhesion might also be stiffness-sensitive. We therefore systematically varied the stiffness of our bare HA hydrogels while keeping HA concentration constant at 5 wt%, and used the same centrifugation assay to ask whether HA matrix stiffness altered cell adhesion (Fig. 2C–D). Our studies revealed that on bare HA hydrogels, adhesion of CD44-positive cells increased with stiffness, whereas adhesion of CD44 knockdown cells, while still sensitive to stiffness, was vastly reduced (Fig. 3D). Thus, CD44-HA adhesion is intrinsically mechanosensitive to matrix stiffness in a manner that does not require integrin ligation. Incorporation of RGD into the hydrogels increased adhesion at all stiffnesses for both cell types, but still resulted in a stiffness-dependent trend. Even with integrin ligation present, CD44 knockdown reduced cell adhesion at all stiffnesses tested (Fig. 2D).
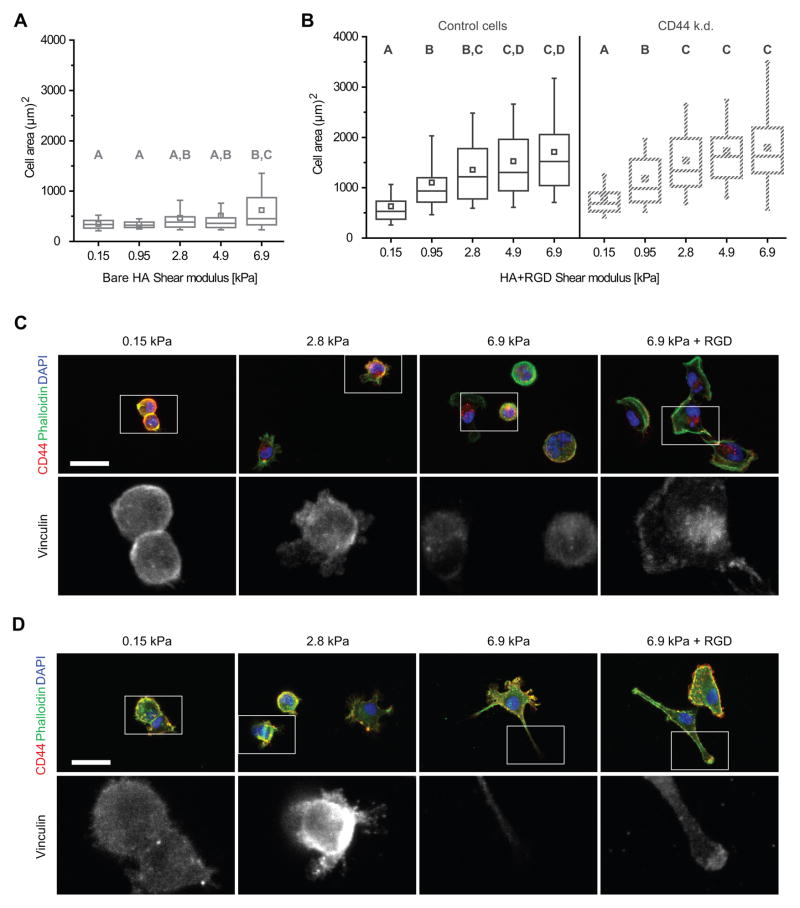
Figure 3. U373-MG glioma cell spreading and morphology on HA-based hydrogel is stiffness-dependent.
(A–B) Cell spread area after 24 h of adhesion on bare HA hydrogels (A), or HA hydrogels functionalized with RGD (B). Hydrogel shear modulus is displayed on the x-axis. A,B,C statistical families show p<0.05 from Dunn’s test for multiple comparison of non-normally distributed data. Boxes represent 25th and 75th percentiles, whiskers represent 10th and 90th percentiles. (C) Cell morphology of U373-MG human glioblastoma cells on HA hydrogel in the absence of integrin-based binding differs from cells spread on HA functionalized with RGD. Colors in top row represent localization of CD44 (red), F-actin (phalloidin, green) and nuclear DNA (DAPI, blue). Bottom row depicts localization of vinculin. Scale bar represents 50 μm. (D) Cell morphology of U87-MG human glioblastoma cells with same conditions described in (C).
To determine whether this CD44-mediated mechanosensing had functional consequences for cell behaviors relevant to tumor invasion, we next explored the relative roles of CD44 and integrins in stiffness-dependent spreading and motility on HA-based hydrogels. When allowed to adhere over a period of 24 h in medium containing 10% serum, control U373-MG cells exhibited differing morphologies depending on stiffness and presence of RGD (Fig. 3). Cell area analysis performed on images obtained 24 h after seeding shows that, consistent with our past observations (14), HA hydrogels functionalized with RGD induced stiffness-dependent spreading (Fig. 3B). Surprisingly, cells seeded on HA substrates devoid of RGD still exhibited stiffness-dependent spreading, although the stiffness dependence was less steep than that of cells on HA-RGD hydrogels (Fig. 3A). On compliant bare HA hydrogels of 0.95 kPa, all cells remained rounded and formed sparse multicellular aggregates, as expected for a low-adhesion environment (41). On stiffer 4.9 kPa and 6.9 kPa bare HA hydrogels, cells predominantly adhered as single cells, with CD44-positive processes and without vinculin-positive adhesive plaques. Cell spreading was dramatically reduced compared to cells on RGD-functionalized surfaces (Fig. 3C). In contrast, cells on 6.9 kPa HA-RGD hydrogels exhibited stress fibers, which were absent in all stiffnesses of bare HA hydrogels. Similar results were seen with U87- MG human glioblastoma cells, albeit with expected variations in morphology between cell lines (Fig. 3D). In both cases, addition of RGD to the substrate resulted in localization of vinculin staining to the tips of protrusions, while vinculin staining remained diffuse on bare HA hydrogels.
Consistent with the CD44-specificity of this adhesion, cell spreading measurements on bare HA hydrogels could not be obtained for CD44 knockdown cells due to insufficient adhesion to the hydrogel. With the addition of RGD to the matrix, however, these cells exhibited similar spreading as control cells on the same matrix (Fig. 3B). This implies that over timescales of roughly 24 h and with the addition of 10% serum to the culture medium, integrin-based adhesions are able to compensate for the lack of CD44 adhesions on two-dimensional cell spreading and motility.
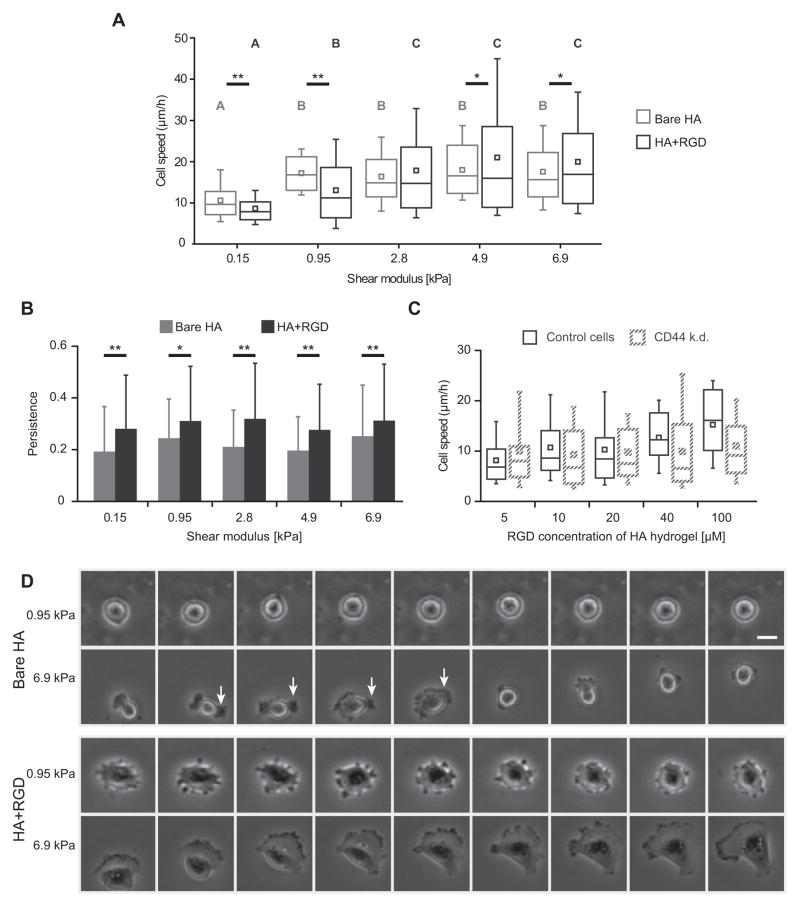
To gain further insight into CD44-dependent cell motility, we used time lapse microscopy to record cell migration on HA and HA-RGD matrices 12–18 h after seeding. Cells were able to productively migrate at comparable speeds with or without RGD functionalization. The speed of this migration was sensitive to HA matrix stiffness, both in the absence and presence of RGD peptide (Movie 1, Fig. 4A). Interestingly, the speed-stiffness relationships for the two materials crossed over, such that bare HA hydrogels supported faster migration for the lowest stiffnesses (<1 kPa) whereas HA-RGD hydrogels supported faster migration for the higher stiffnesses (>4 kPa). On all but the softest substrates tested, cells on bare HA surfaces at times exhibited protrusions that permitted productive motility, but were generally shorter-lived than the larger lamellipodial structures seen on cells with RGD-integrin binding (Fig. 4D). Bare HA substrates supported less directionally persistent migration compared to hydrogels with RGD at all stiffnesses (Fig. 4B). Thus, despite the lack of classically “spread” 2D morphology, glioma cells are still able to productively migrate on two-dimensional bare HA hydrogels using only CD44-based adhesion and without integrin-based adhesion. As with spread area and morphology, CD44 knockdown cells exhibited RGD concentration-dependent migration equivalent to that of control cells (Fig. 4C). In other words, knockdown of CD44 does not affect cell spreading or motility when abundant integrin-based signaling is available. To demonstrate that these results generally apply to a range of integrin ligands and not just RGD peptides, we functionalized HA hydrogels with full-length fibronectin or laminin and obtained similar cell spread area and cell speed dependence on hydrogel stiffness (Supplementary Fig. S3).
Figure 4. U373-MG glioma cell motility on HA-based hydrogel is CD44- and stiffness-dependent.
(A) Cell migration speed 12–18 h after seeding on bare HA hydrogels and on RGD-functionalized HA hydrogels. Hydrogel shear modulus is displayed on the x-axis. A,B,C statistical families show p<0.05 Dunn’s test for multiple comparison of non-normally distributed data. Boxes represent 25th and 75th percentiles, whiskers represent 10th and 90th percentiles. For pairwise comparisons between cell speeds on HA and HA-RGD hydrogels at a given shear modulus, *p<0.1, **p<0.05 by Student’s t-test. (B) Comparison of cell migration speed of control U373-MG cells with CD44 knockdown counterparts on HA hydrogels of varying RGD concetration. Comparison could not be made on bare HA hydrogels due to insufficient adhesion of CD44 knockdown cells in the absence of RGD functionalization. With HA functionalization of 1 μM RGD, very few CD44 knockdown cells attached. At all other RGD concentrations tested, no statistical difference in the migration speed between the two cell lines was found. Under all conditions tested, differences between control and CD44 knockdown cell speeds were not statistically significant. (C) Persistence parameter of random cell motility paths on bare HA and RGD-functionalized hydrogels. *p<0.1, **p<0.05 by Student’s t-test. (D) Still images taken from time lapse microscopy. Each column represents a 0.5 h interval. On bare HA of 0.95 kPa, cells consistently remain rounded. On bare HA of 6.9 kPa, cells extend small protrusions that rupture quickly, such as the one indicated by white arrows, which lead to productive migration. In contrast, cells on stiff RGD+HA hydrogels exhibit broad, stable lamellipodia. Scale bar represents 25 μm.
CD44-mediated adhesion promotes glioma invasiveness
The above studies reveal that CD44 can mediate mechanotransductive signaling and support matrix adhesion and two-dimensional motility without giving rise to mature, vinculin-positive focal adhesions. To test whether this mode of adhesion might also support the highly aggressive invasion of glioma cells through the narrow interstices in brain parenchyma, we employed a three-dimensional transwell invasion model.
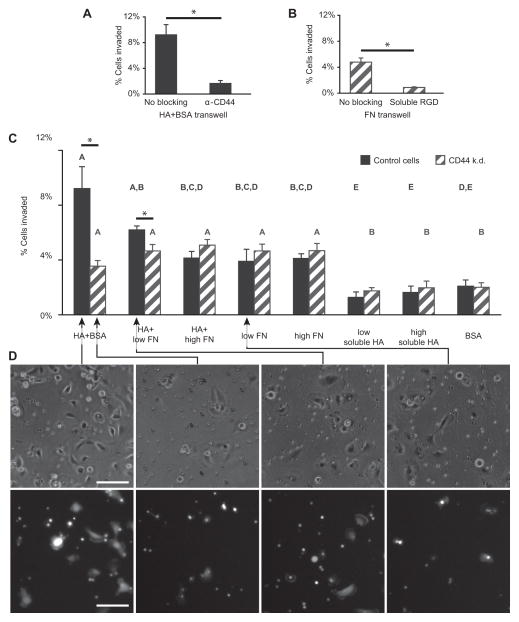
To confirm that adsorbed HA and fibronectin would still engage CD44 and integrins in a manner similar to that of the crosslinked HA hydrogels (as seen in Fig. 2), we first performed a series of adhesion control experiments with full-length fibronectin, which revealed similar ligand density-dependent spreading behavior as with RGD (Supplementary Figure S3). To confirm that CD44- and integrin-speicific adhesions are capable of inducing transwell invasion, we next performed receptor blocking experiments (Fig. 5A–B). First, control cells were pre-incubated with CD44-neutralizing antibody in suspension, then seeded on transwell membranes coated with HA and blocked with BSA. The resulting drop in cell invasion compared to cells not blocked with CD44-neutralizing antibody indicated that an appreciable component of the adhesion to adsorbed HA could be attributed to CD44 (Fig. 5A). Second, CD44 knockdown cells were pre-incubated with soluble RGD peptide, and the resulting abrogation of cell invasion on fibronectin-coated surfaces compared to unblocked cells indicated that the same receptors, presumably integrins, engage both RGD and fibronectin (Fig. 5B).
Figure 5. U373-MG glioma cell invasion through transwell membranes is promoted by HA-CD44 adhesion, but not by fibronectin-integrin adhesion.
(A) Incubating control cells plated on HA+BSA coated transwells with CD44-neutralizing antibody drastically reduces invasion, demonstrating that interaction with HA is CD44-dependent. (B) Incubating CD44 knockdown cells with soluble RGD peptide on membranes coated with 3.7 μg/mL fibronectin strongly attenuates invasion, demonstrating that adhesion to fibronectin is integrin-dependent. #p<0.05 multiple comparison by Tukey-Kramer, *p<0.05 by Student’s t-test. (C) Percent of control or CD44 knockdown U373-MG glioma cells that invaded through 8 μm pores in membranes coated with combinations of HA, varying concentrations of fibronectin (FN) and BSA, or with HA dissolved in the cell media (soluble HA). Statistically significant differences of p<0.05 from multiple comparisons within cell line groups by Tukey-Kramer are grouped by A,B,C statistical families. *p<0.05 t-test pairwise comparisons of cell lines on the same substrate. (D) Representative phase contrast images of cells either above or below clear transwell membranes (top row), and GFP-expressing cells below Fluoroblok membranes (bottom row). Both types of membranes have 8 μm pores, and images were taken 2.5 h after cell seeding. Scale bars represent 100 μm.
To test how CD44 and integrin ligation contribute to cell invasion when both cognate matrix elements were present, we then coated transwell membrane inserts containing 8 μm pores with HA, various concentrations of fibronectin, or both HA and fibronectin. The transwell invasion model has previously been shown to be predictive of glioma cell invasion through tissue (42). Membranes coated only with HA were also blocked with BSA, and membranes coated only with BSA were used as a negative control to measure baseline invasion with minimal receptor-mediated adhesion. Interestingly, CD44-expressing control cells were most invasive across membranes coated with HA with BSA, but not fibronectin (Fig. 5C). This phenomenon is CD44-specific, because the invasive motility of CD44 knockdown cells across these same membranes was significantly lower and indistinguishable from results obtained with BSA-coated membranes. Moreover, inclusion of fibronectin on HA-coated membranes reduced invasive motility in a concentration-dependent fashion; adsorption from a solution of 9.3 μg/mL fibronectin with HA reduced invasion of control cells 2.2-fold relative to HA-coated membranes, whereas 3.7 μg/mL fibronectin with HA yielded intermediate levels of invasion. To compare the effect of soluble HA to adsorbed HA, we added soluble HA in the media of both the top and bottom chambers of the transwell membrane. This did not lead to a significant increase in cell invasion compared to the BSA control.
To gain mechanistic insight into these results, we obtained both phase contrast images of cells adhered to either side of each membrane and fluorescence images of GFP-expressing cells that had invaded through the membrane to the undersides of Fluoroblok membranes (Fig. 5D). Consistent with adhesion results on HA and HA-RGD gels (Fig. 2A), control cells attached readily to membranes coated with HA and/or fibronectin. However, control cells invaded membranes coated in HA much more than membranes coated with either fibronectin alone or both HA and fibronectin. This implies that fibronectin enhanced ECM engagement and cell spreading, but hindered invasion. When neither type of adhesion was available (CD44 knockdown cells on HA+BSA matrix, or any cell population on BSA matrix), cells rarely attached or spread, which also precluded invasion. Only in an intermediate range of adhesion strength, in which HA-CD44 adhesion was available but integrin adhesion was not, did invasion proceed appreciably higher than baseline levels.
Discussion
Given the biochemical and biophysical changes in brain tissue associated with GBM progression, we were motivated to investigate how glioma invasion is affected by the nature of cell-matrix adhesions and the mechanical properties of the ECM. In addition to exposing previously underappreciated correlations between expression of CD44, pro-oncogenic signals, and HA-synthesizing proteins, we have also found evidence that CD44-HA adhesion is intrinsically mechanosensitive and occurs on a faster time scale than integrin-based binding. Most importantly, we find that while both adhesion systems support robust two-dimensional motility, CD44-based adhesion is accompanied by extension of small, short-lived processes, whereas integrin-based adhesion is associated with broad lamellipodia and more directionally persistent migration. These differences extend to three-dimensional migration through constricted pores, as CD44-HA adhesion drives optimal invasion in a transwell paradigm.
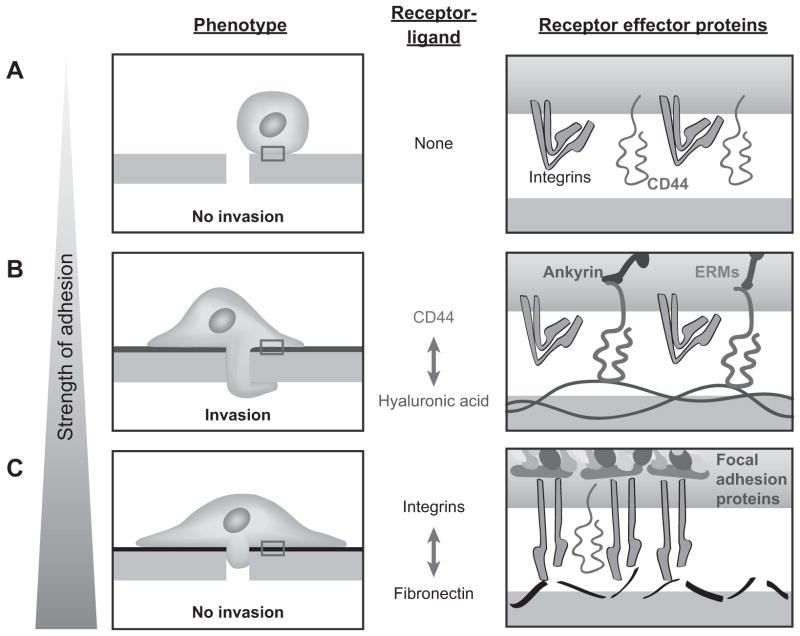
To explain our transwell results, we assembled a model based on previously described models of glioma invasion through constricted spaces (42) and adhesion strength on cell migration (43). Based on these and our observations, we propose a model in which optimal cell invasion requires a balance between cell-matrix adhesion and turnover (Fig. 6). In other words, we postulate that while CD44 alone can support cell adhesion, these adhesions are less mechanically reinforced than those formed by integrins. Consistent with this idea, integrin ligation is accompanied by formation of actomyosin bundles and focal adhesions (Fig. 3C–D), which slows migration through narrow pores. This model is supported by our measurements of adhesion, spreading, and migration on two-dimensional bare HA surfaces, where the finding that CD44 can support adhesion without lamellapodial spreading suggests that the latter is not required for robust adhesion or migration. Thus, invasion through three-dimensional pores is not directly predictable from two-dimensional motility alone, which is broadly consistent with the recent observation that two-dimensional protrusive propensity predicts three-dimensional migration speed much more accurately than does two-dimensional migration speed (44). Therefore, previously described causal relationships between CD44 expression, tumor size, and survival time in vivo (7) may not only be due to CD44-mediated stimulation of canonical pro-oncogenic signaling, but also through promotion of glioma invasion by biophysical mechanisms.
Figure 6. Proposed model for influence of adhesion strength on cell invasion.
We propose a model for cell invasion that involves a balance between formation and turnover of cell adhesions. (A) With little to no cell adhesion to the substrate, extrusion of the cell body through a confined space is not possible. (B) With an optimal balance between adhesion and detachment, as is the case with the relatively labile CD44-HA interaction, cells have the highest invasion potential. (C) When abundant integrin-based adhesion is present, the cells spread extensively and adhere robustly on the substrate but do not invade.
Using transwell assays and engineered HA hydrogels, we have demonstrated that in a range of timescales from 0.5–24 h, CD44 is capable of supporting glioma cell adhesion in the absence of integrin engagement. Ultimately, our results indicate that CD44 may serve a directly pro-invasive function through the formation of short-lived matrix adhesions. Previous studies support the first observation that HA promotes glioma cell invasion, either by addition of HA into a Matrigel coating (45), addition of soluble HA in the cell culture medium, (46) or overexpression of the HA synthases HAS1 (47) or HAS2 (48). Consistent with these earlier studies, we have directly shown that adsorption of HA onto transwell membranes strongly enhances invasion in a CD44-dependent manner. While integrins have been extensively demonstrated to contribute to GBM tumor growth and invasion (49, 50), our data suggest a complex interplay between integrins and CD44 that remains to be fully elucidated.
Remarkably, glioma cell adhesion, spreading, and two-dimensional motility are all sensitive to HA stiffness via CD44. We note that cell spreading is less sensitive to stiffness when CD44, rather than integrins, mediate adhesion; whether this is due to differences in the receptors themselves, the signals they transduce, or integrin-CD44 interactions remains to be investigated. This CD44-mediated mechanosensing has functional consequences for cell behaviors relevant to tumor invasion. It has long been appreciated that the molecular weight of HA can vary widely from oligomers of 4 disaccharides to macromolecules composed of up to 30,000 disaccharides, and these can have strikingly different effects on the activation of downstream signals (51). Full length HA is anti-angiogenic, whereas oligosaccharides of HA trigger angiogenesis (52) by a variety of mechanisms, including activation of PKCα and Src (53). However, the biophysical basis of HA molecular weight effects remain unexplored. In particular, the recent discovery that exceptionally high molecular weight HA strongly promotes cancer resistance and longevity in the naked mole rat (54) adds further relevance to our observations. Our data suggest a mechanism in which cells are able to differentiate HA molecular weight in part by detecting the mechanical rigidity of CD44-bound ligands. To date, one previous study to date has provided evidence that CD44 may play a role in sensing these rigidity differences in fibroblast cells (55), and another recent study indicates that CD44 can support or alter integrin-mediated mechanical signals in cardiac myocytes (56). In GBM, the extreme overexpression of HYAL-2 (Fig. 1A), typically seen only in developing brain, suggests that tumor cells may resurrect the pro-migratory and angiogenic processes associated with development (28) by generating low molecular weight HA fragments.
An important innovation of our study is the use of solid-state HA substrates, which are a more physiologically-mimetic form of the high-molecular weight HA that composes much of brain ECM. This also added another level of control by enabling us to vary HA stiffness. HA must be chemically methacrylated prior to crosslinking, which raises the potential concern that this functionalization may alter the binding affinity of HA for CD44. However, our adhesion assay results (Fig. 2A–B) demonstrate that HA methacrylation preserves CD44 binding function. Furthermore, the main binding site of CD44 for HA does not include the hydroxyl group of the N-acetyl glucosamine monomer, which is most likely to become methacrylated, and this hydroxyl group points away from the N-acetyl group that is essential for CD44 binding (57). Ultimately, additional layers of complexity are needed to capture key aspects of perivascular invasion, in which GBM cells infiltrate tissue by following vascular structures.
According to the “motor-clutch” model for cellular mechanosensing (58, 59), the comparatively weaker CD44-HA bond would be more sensitive to lower substrate stiffness than stronger integrin-based adhesions. This is supported by previously reported binding affinities for CD44 to HA10, 50 μM (57), which are much lower than that of activated α5β1 for fibronectin, reported as 90 nM (60). Given that brain matrix is a relatively compliant tissue that presumably limits cell-induced tissue stresses, it stands to reason that under these conditions, the weaker CD44-HA adhesion pair might serve as an important mechanotransducer. Indeed, our two-dimensional motility data agrees with this model; addition of RGD to the HA matrix expands the range over which cell speed is sensitive to matrix stiffness, whereas cell speed on bare HA hydrogels plateaus at lower stiffness values. Finally, our data also show that there is a time-dependent cooperation between the two adhesion receptors, in which CD44-HA adhesions form relatively quickly within 0.5 h, and integrin-RGD adhesions take longer to mature. If CD44 is not available, then over a period of 0.5–3 h, adhesion is drastically decreased if serum is not present, but over longer timescales of 24 h in the presence of serum, integrin-based adhesion is able to compensate. This lag time between the maturation of the two types of adhesions may be due to the large ensemble of proteins that are required to form mature focal adhesions, while CD44 adhesions require the association of relatively few components. The initiation of adhesion by cell membrane-bound HA to substrates temporally followed by integrin-based interaction has previously been observed with chondrocytes on protein-coated glass (61), but our results demonstrate definitively that CD44 is responsible for this early adhesion, and that the extracellular matrix can be engineered to modulate these interactions.
Our study raises a number of new questions in both cellular mechanobiology and the pathophysiology of GBM: What is the molecular basis of CD44-based mechanosensing, and to what extent do these mechanisms crosstalk with integrin-based mechanosensing? How do these CD44-based mechanotransductive signals ultimately influence gene expression? And finally, can interruption of these mechanotransductive signals limit tumor growth and invasion in vivo? Orthogonal control of these two adhesive systems with the use of engineered materials, systems biological tools, and animal models will help clarify these important issues, and potentially yield unexpected new insights into the biophysical basis of tumor invasion.
Supplementary Material
Implications.
This study reveals that the CD44 receptor, which is commonly overexpressed in glioblastoma multiforme tumors, is critical for cell adhesion, invasion, and mechanosensing of a hyaluronic acid-based matrix.
Acknowledgments
The authors thank Joanna Phillips and Anna Wade for guidance in conducting TCGA gene expression data analysis. Y. K. gratefully acknowledges the support of Graduate Fellowships from the NSF and U.S. Department of Defense (NDSEG). S.K. gratefully acknowledges grant support from the Arnold and Mabel Beckman Foundation, the NSF (CMMI 0727420), and the NIH (1DP2OD004213, Director’s New Innovator Award, part of the NIH Roadmap for Medical Research; 1U54CA143836, Physical Sciences Oncology Center Grant).
Footnotes
The authors do not disclose any conflicts of interest.
References
- 1.Silver DJ, Siebzehnrubl FA, Schildts MJ, Yachnis AT, Smith GM, Smith AA, et al. Chondroitin sulfate proteoglycans potently inhibit invasion and serve as a central organizer of the brain tumor Microenvironment. J Neurosci. 2013;33:15603–17. doi: 10.1523/JNEUROSCI.3004-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 3.Bellail AC, Hunter SB, Brat DJ, Tan C, Van Meir EG. Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int J Biochem Cell Biol. 2004;36:1046–69. doi: 10.1016/j.biocel.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Baier C, Baader SL, Jankowski J, Gieselmann V, Schilling K, Rauch U, et al. Hyaluronan is organized into fiber-like structures along migratory pathways in the developing mouse cerebellum. Matrix Biol. 2007;26:348–58. doi: 10.1016/j.matbio.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Bourguignon LYW, Zhu H, Shao L, Chen YW. CD44 interaction with c-Src kinase promotes cortactin-mediated cytoskeleton function and hyaluronic acid-dependent ovarian tumor cell migration. J Biol Chem. 2001;276:7327–36. doi: 10.1074/jbc.M006498200. [DOI] [PubMed] [Google Scholar]
- 6.Lin Y-H, Yang-Yen H-F. The Osteopontin-CD44 Survival Signal Involves Activation of the Phosphatidylinositol 3-Kinase/Akt Signaling Pathway. J Biol Chem. 2001;276:46024–30. doi: 10.1074/jbc.M105132200. [DOI] [PubMed] [Google Scholar]
- 7.Xu Y, Stamenkovic I, Yu Q. CD44 attenuates activation of the hippo signaling pathway and is a prime therapeutic target for glioblastoma. Cancer Res. 2010;70:2455–64. doi: 10.1158/0008-5472.CAN-09-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delpech B, Maingonnat C, Girard N, Chauzy C, Olivier A, Maunoury R, et al. Hyaluronan and hyaluronectin in the extracellular matrix of human brain tumour stroma. Eur J Cancer. 1993;29:1012–7. doi: 10.1016/s0959-8049(05)80214-x. [DOI] [PubMed] [Google Scholar]
- 9.Pietras A, Katz Amanda M, Ekström Elin J, Wee B, Halliday John J, Pitter Kenneth L, et al. Osteopontin-CD44 Signaling in the Glioma Perivascular Niche Enhances Cancer Stem Cell Phenotypes and Promotes Aggressive Tumor Growth. Cell Stem Cell. 2014;14:357–69. doi: 10.1016/j.stem.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulrich TA, de Juan Pardo EM, Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69:4167–74. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y-l, Motte S, Kaufman LJ. Pore size variable type I collagen gels and their interaction with glioma cells. Biomaterials. 2010;31:5678–88. doi: 10.1016/j.biomaterials.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 12.Unsgaard G, Rygh OM, Selbekk T, Müller TB, Kolstad F, Lindseth F, et al. Intra-operative 3D ultrasound in neurosurgery. Acta Neurochir. 2006;148:235–53. doi: 10.1007/s00701-005-0688-y. [DOI] [PubMed] [Google Scholar]
- 13.Chakraborty A, Bamber JC, Dorward NL. Preliminary investigation into the use of ultrasound elastography during brain tumour resection. Ultrasound. 2012;20:33–40. [Google Scholar]
- 14.Ananthanarayanan B, Kim Y, Kumar S. Elucidating the mechanobiology of malignant brain tumors using a brain matrix-mimetic hyaluronic acid hydrogel platform. Biomaterials. 2011;32:7913–23. doi: 10.1016/j.biomaterials.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bian L, Guvendiren M, Mauck RL, Burdick JA. Hydrogels that mimic developmentally relevant matrix and N-cadherin interactions enhance MSC chondrogenesis. Proc Natl Acad Sci U S A. 2013;110:10117–22. doi: 10.1073/pnas.1214100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seidlits SK, Khaing ZZ, Petersen RR, Nickels JD, Vanscoy JE, Shear JB, et al. The effects of hyaluronic acid hydrogels with tunable mechanical properties on neural progenitor cell differentiation. Biomaterials. 2010;31:3930–40. doi: 10.1016/j.biomaterials.2010.01.125. [DOI] [PubMed] [Google Scholar]
- 17.Marklein RA, Burdick JA. Spatially controlled hydrogel mechanics to modulate stem cell interactions. Soft Matter. 2010;6:136–43. [Google Scholar]
- 18.Flanagan LA, Ju Y-E, Marg B, Osterfield M, Janmey PA. Neurite branching on deformable substrates. Neuroreport. 2002;13:2411–5. doi: 10.1097/01.wnr.0000048003.96487.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stepanenko AA, Kavsan VM. Karyotypically distinct U251, U373, and SNB19 glioma cell lines are of the same origin but have different drug treatment sensitivities. Gene. 2014;540:263–5. doi: 10.1016/j.gene.2014.02.053. [DOI] [PubMed] [Google Scholar]
- 20.Burns GF, Cosgrove L, Triglia T, Beall JA, López AF, Werkmeister JA, et al. The IIb-IIIa glycoprotein complex that mediates platelet aggregation is directly implicated in leukocyte adhesion. Cell. 1986;45:269–80. doi: 10.1016/0092-8674(86)90391-0. [DOI] [PubMed] [Google Scholar]
- 21.Huang X, Wu J, Spong S, Sheppard D. The integrin alphavbeta6 is critical for keratinocyte migration on both its known ligand, fibronectin, and on vitronectin. J Cell Sci. 1998;111:2189–95. doi: 10.1242/jcs.111.15.2189. [DOI] [PubMed] [Google Scholar]
- 22.Mammoto T, Jiang A, Jiang E, Panigrahy D, Kieran MW, Mammoto A. Role of Collagen Matrix in Tumor Angiogenesis and Glioblastoma Multiforme Progression. The American Journal of Pathology. 2013;183:1293–305. doi: 10.1016/j.ajpath.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ariza A, López D, Mate JL, Isamat M, Musulén E, Pujol M, et al. Role of CD44 in the invasiveness of glioblastoma multiforme and the noninvasiveness of meningioma: an immunohistochemistry study. Hum Pathol. 1995;26:1144–7. doi: 10.1016/0046-8177(95)90278-3. [DOI] [PubMed] [Google Scholar]
- 24.Oz B, Karayel FA, Gazio NL, Ozlen F, Balci K. The distribution of extracellular matrix proteins and CD44S expression in human astrocytomas. Pathology oncology research : POR. 2000;6:118–24. doi: 10.1007/BF03032361. [DOI] [PubMed] [Google Scholar]
- 25.Ranuncolo SM, Ladeda V, Specterman S, Varela M, Lastiri J, Morandi A, et al. CD44 expression in human gliomas. J Surg Oncol. 2002;79:30–6. doi: 10.1002/jso.10045. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida T, Matsuda Y, Naito Z, Ishiwata T. CD44 in human glioma correlates with histopathological grade and cell migration. Pathol Int. 2012;62:463–70. doi: 10.1111/j.1440-1827.2012.02823.x. [DOI] [PubMed] [Google Scholar]
- 27.Kosaki R, Watanabe K, Yamaguchi Y. Overproduction of Hyaluronan by Expression of the Hyaluronan Synthase Has2 Enhances Anchorage-independent Growth and Tumorigenicity. Cancer Res. 1999;59:1141–5. [PubMed] [Google Scholar]
- 28.Strobl B, Wechselberger C, Beier DR, Lepperdinger G. Structural Organization and Chromosomal Localization of Hyal2, a Gene Encoding a Lysosomal Hyaluronidase. Genomics. 1998;53:214–9. doi: 10.1006/geno.1998.5472. [DOI] [PubMed] [Google Scholar]
- 29.Novak U, Stylli SS, Kaye AH, Lepperdingerm G. Hyaluronidase-2 overexpression accelerates intracerebral but not subcutaneous tumor formation of murine astrocytoma cells. Cancer Res. 1999;59:6246–50. [PubMed] [Google Scholar]
- 30.Ruoslahti E. Brain extracellular matrix. Glycobiology. 1996;6:489–92. doi: 10.1093/glycob/6.5.489. [DOI] [PubMed] [Google Scholar]
- 31.Zhu X, Morales FC, Agarwal NK, Dogruluk T, Gagea M, Georgescu M-M. Moesin Is a Glioma Progression Marker That Induces Proliferation and Wnt/β-Catenin Pathway Activation via Interaction with CD44. Cancer Res. 2013;73:1142–55. doi: 10.1158/0008-5472.CAN-12-1040. [DOI] [PubMed] [Google Scholar]
- 32.Zhou R, Wu X, Skalli O. The hyaluronan receptor RHAMM/IHABP in astrocytoma cells: expression of a tumor-specific variant and association with microtubules. J Neurooncol. 2002;59:15–26. doi: 10.1023/a:1016373015569. [DOI] [PubMed] [Google Scholar]
- 33.Massia SP, Hubbell JA. An RGD spacing of 440 nm is sufficient for integrin alpha V beta 3-mediated fibroblast spreading and 140 nm for focal contact and stress fiber formation. The Journal of Cell Biology. 1991;114:1089–100. doi: 10.1083/jcb.114.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. Cell adhesion and motility depend on nanoscale RGD clustering. J Cell Sci. 2000;113:1677–86. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 35.Reinhart-King CA, Dembo M, Hammer DA. Endothelial Cell Traction Forces on RGD-Derivatized Polyacrylamide Substrata. Langmuir. 2002;19:1573–9. [Google Scholar]
- 36.Nedvetzki S, Gonen E, Assayag N, Reich R, Williams RO, Thurmond RL, et al. RHAMM, a receptor for hyaluronan-mediated motility, compensates for CD44 in inflamed CD44-knockout mice: a different interpretation of redundancy. Proc Natl Acad Sci U S A. 2004;101:18081. doi: 10.1073/pnas.0407378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaman MH, Trapani LM, Sieminski AL, Siemeski A, Mackellar D, Gong H, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci U S A. 2006;103:10889–94. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. Substrate Compliance versus Ligand Density in Cell on Gel Responses. Biophys J. 2004;86:617–28. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, et al. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 40.Wang N, Ingber DE. Control of cytoskeletal mechanics by extracellular matrix, cell shape, and mechanical tension. Biophys J. 1994;66:2181–9. doi: 10.1016/S0006-3495(94)81014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinhart-King CA, Dembo M, Hammer DA. Cell-Cell Mechanical Communication through Compliant Substrates. Biophys J. 2008;95:6044–51. doi: 10.1529/biophysj.107.127662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beadle C, Assanah MC, Monzo P, Vallee R, Rosenfeld SS, Canoll P. The Role of Myosin II in Glioma Invasion of the Brain. Mol Biol Cell. 2008;19:3357–68. doi: 10.1091/mbc.E08-03-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature. 1997;385:537–40. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 44.Meyer AS, Hughes-Alford SK, Kay JE, Castillo A, Wells A, Gertler FB, et al. 2D protrusion but not motility predicts growth factor–induced cancer cell migration in 3D collagen. J Cell Biol. 2012;197:721–9. doi: 10.1083/jcb.201201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Radotra B, McCormick D. Glioma invasion in vitro is mediated by CD44–hyaluronan interactions. The Journal of pathology. 1997;181:434–8. doi: 10.1002/(SICI)1096-9896(199704)181:4<434::AID-PATH797>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 46.Hanagiri T, Shinohara S, Takenaka M, Shigematsu Y, Yasuda M, Shimokawa H, et al. Effects of hyaluronic acid and CD44 interaction on the proliferation and invasiveness of malignant pleural mesothelioma. Tumor Biol. 2012;33:2135–41. doi: 10.1007/s13277-012-0473-5. [DOI] [PubMed] [Google Scholar]
- 47.Golshani R, Lopez L, Estrella V, Kramer M, Iida N, Lokeshwar VB. Hyaluronic Acid Synthase-1 Expression Regulates Bladder Cancer Growth, Invasion, and Angiogenesis through CD44. Cancer Res. 2008;68:483–91. doi: 10.1158/0008-5472.CAN-07-2140. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, Li L, Brown TJ, Heldin P. Silencing of hyaluronan synthase 2 suppresses the malignant phenotype of invasive breast cancer cells. Int J Cancer. 2007;120:2557–67. doi: 10.1002/ijc.22550. [DOI] [PubMed] [Google Scholar]
- 49.Schnell O, Krebs B, Wagner E, Romagna A, Beer AJ, Grau SJ, et al. Expression of Integrin αvβ3 in Gliomas Correlates with Tumor Grade and Is not Restricted to Tumor Vasculature. Brain Pathol. 2008;18:378–86. doi: 10.1111/j.1750-3639.2008.00137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tabatabai G, Weller M, Nabors B, Picard M, Reardon D, Mikkelsen T, et al. Targeting integrins in malignant glioma. Targeted Oncology. 2010;5:175–81. doi: 10.1007/s11523-010-0156-3. [DOI] [PubMed] [Google Scholar]
- 51.Noble PW. Hyaluronan and its catabolic products in tissue injury and repair. Matrix Biol. 2002;21:25–9. doi: 10.1016/s0945-053x(01)00184-6. [DOI] [PubMed] [Google Scholar]
- 52.West D, Hampson I, Arnold F, Kumar S. Angiogenesis induced by degradation products of hyaluronic acid. Science. 1985;228:1324–6. doi: 10.1126/science.2408340. [DOI] [PubMed] [Google Scholar]
- 53.Slevin M, Kumar S, Gaffney J. Angiogenic Oligosaccharides of Hyaluronan Induce Multiple Signaling Pathways Affecting Vascular Endothelial Cell Mitogenic and Wound Healing Responses. J Biol Chem. 2002;277:41046–59. doi: 10.1074/jbc.M109443200. [DOI] [PubMed] [Google Scholar]
- 54.Tian X, Azpurua J, Hine C, Vaidya A, Myakishev-Rempel M, Ablaeva J, et al. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature. 2013;499:346–9. doi: 10.1038/nature12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hachet E, Van Den Berghe H, Bayma E, Block MR, Auzély-Velty R. Design of Biomimetic Cell-Interactive Substrates Using Hyaluronic Acid Hydrogels with Tunable Mechanical Properties. Biomacromolecules. 2012;13:1818–27. doi: 10.1021/bm300324m. [DOI] [PubMed] [Google Scholar]
- 56.Chopra A, Murray ME, Byfield FJ, Mendez MG, Halleluyan R, Restle DJ, et al. Augmentation of integrin-mediated mechanotransduction by hyaluronic acid. Biomaterials. 2014;35:71–82. doi: 10.1016/j.biomaterials.2013.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Banerji S, Wright AJ, Noble M, Mahoney DJ, Campbell ID, Day AJ, et al. Structures of the Cd44–hyaluronan complex provide insight into a fundamental carbohydrate-protein interaction. Nat Struct Mol Biol. 2007;14:234–9. doi: 10.1038/nsmb1201. [DOI] [PubMed] [Google Scholar]
- 58.Chan CE, Odde DJ. Traction Dynamics of Filopodia on Compliant Substrates. Science. 2008;322:1687–91. doi: 10.1126/science.1163595. [DOI] [PubMed] [Google Scholar]
- 59.Bangasser Benjamin L, Rosenfeld Steven S, Odde David J. Determinants of Maximal Force Transmission in a Motor-Clutch Model of Cell Traction in a Compliant Microenvironment. Biophys J. 2013;105:581–92. doi: 10.1016/j.bpj.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anido J, Sáez-Borderías A, Gonzàlez-Juncà A, Rodón L, Folch G, Carmona MA, et al. TGF-β Receptor Inhibitors Target the CD44high/Id1high Glioma-Initiating Cell Population in Human Glioblastoma. Cancer Cell. 2010;18:655–68. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 61.Cohen M, Kam Z, Addadi L, Geiger B. Dynamic study of the transition from hyaluronan- to integrin-mediated adhesion in chondrocytes. EMBO J. 2006;25:302–11. doi: 10.1038/sj.emboj.7600960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.