Abstract
Malignant pleural mesothelioma (MPM) is associated with asbestos exposure and is a cancer that has not been significantly impacted by small molecule-based targeted therapeutics. Previously, we demonstrated the existence of functional subsets of lung cancer and head and neck squamous cell carcinoma (HNSCC) cell lines in which fibroblast growth factor receptor (FGFR) autocrine signaling functions as a non-mutated growth pathway. In a panel of pleural mesothelioma cell lines, FGFR1 and FGF2 were co-expressed in 3 of 7 cell lines and were significantly associated with sensitivity to the FGFR-active tyrosine kinase inhibitor (TKI), ponatinib, both in vitro and in vivo using orthotopically propagated xenografts. Furthermore, RNAi-mediated silencing confirmed the requirement for FGFR1 in specific mesothelioma cells and sensitivity to the FGF ligand trap, FP-1039, validated the requirement for autocrine FGFs. None of the FGFR1-dependent mesothelioma cells exhibited increased FGFR1 gene copy number, based on a FISH assay, indicating that increased FGFR1 transcript and protein expression were not mediated by gene amplification. Elevated FGFR1 mRNA was detected in a subset of primary MPM clinical specimens and like MPM cells, none harbored increased FGFR1 gene copy number. These results indicate that autocrine signaling through FGFR1 represents a targetable therapeutic pathway in MPM and that biomarkers distinct from increased FGFR1 gene copy number such as FGFR1 mRNA would be required to identify MPM patients bearing tumors driven by FGFR1 activity.
Implications
FGFR1 is a viable therapeutic target in a subset of malignant pleural mesotheliomas, but FGFR TKI-responsive tumors will need to be selected by a biomarker distinct from increased FGFR1 gene copy number, possibly FGFR1 mRNA or protein levels.
INTRODUCTION
Malignant pleural mesothelioma (MPM) arises from the mesothelial cells lining the pleural cavity surrounding the lungs, and less frequently from the peritoneum (1). The incidence of MPM rose over the second half of the 20th century, coinciding with an increased industrial use of asbestos (1) and is associated with an extremely long latency of ~50 years (2). While the predicted incidence of MPM (2,000 to 3,000 cases per year) may have peaked in the United States, incidence is expected to continue to increase worldwide due to variable asbestos regulation (1, 3). Three histological subtypes of MPM have been defined, epithelioid, sarcomatoid and biphasic (mixed). Median survival ranges from 6 to 13 months with epithelioid tumors exhibiting the longest survival time (11–13 months) and sarcomatoid tumors the shortest (6–7.5 months) (4, 5). Combined cisplatin and pemetrexed is currently the only approved therapy with a median survival of 12.1 months (6).
Self-sufficiency in growth signaling, frequently through deregulation and activation of receptor tyrosine kinase (RTK) pathways, is a hallmark of cancer (7). Based on success in identifying and targeting specific mutated oncogene drivers in lung adenocarcinomas (8–10), similar investigations have proceeded in MPM. While gain-of-function mutations in EGFR are exceedingly rare in MPM (11, 12), epidermal growth factor receptor (EGFR) is highly expressed in as many as 97% of tumors. However, no clinical benefit was observed in MPM patients treated with EGFR-specific TKIs, gefitinib and erlotinib (13, 14). Besides EGFR, evidence supports activity of other RTKs in MPM including MET (15, 16), platelet-derived growth factor receptors (PDGFRs) (17, 18), vascular-endothelial growth factor receptors (VEGFRs) (19), AXL (20, 21) and insulin-like growth factor receptor (IGF1R) (22, 23). In fact, a growth network comprised of multiple RTKs has been proposed such that combined inhibition of multiple pathways yields greater efficacy (15, 24).
Deregulation of the FGFR signaling pathways is observed in various tumor types through multiple mechanisms (25). Activating mutations occur in FGFR2 and FGFR3 leading to constitutive dimerization in bladder, endometrial and squamous cell lung cancers (26–28). Chromosomal translocations in FGFR genes have also been observed in various malignancies (29, 30). Amplification of FGFR2 is commonly seen in gastric cancers and FGFR1 in breast cancer, squamous cell lung cancers and HNSCC (FGFR1) (31–34). The increase in FGFR1 gene copy number is especially relevant to lung cancer and serves as the key biomarker for patient recruitment to two open trials of FGFR-specific TKIs in solid tumors (NCT01004224 and NCT00979134). By contrast, ligand-dependent autocrine and paracrine signaling provides a mechanism that is independent of somatic mutation and has been demonstrated by our group in both non-small cell lung cancer (NSCLC) and HNSCC (35, 36). Herein, we demonstrate that FGFR1 is co-expressed with FGF2 in a subset of MPM cell lines and is strongly associated with sensitivity to the TKI, ponatinib, both in vitro and in vivo. Finally, FGFR1 dependency is demonstrated by RNAi-mediated silencing. The studies thereby identify a novel and therapeutically relevant target in malignant pleural mesothelioma.
MATERIALS AND METHODS
Cell Lines
H28, H513, H2052, H2452, MSTO211H, and Met5A cells were obtained from ATCC (Manassas, VA) and passaged less than 6 months after receipt for completion of the studies. H226 and H290 cells were obtained from and authenticated by (finger print analysis) the University of Colorado Cancer Center Tissue Culture shared resource. The cell lines were cultured in RPMI-1640 growth medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Luciferase expressing H226 cells were generated by stable transduction with a lentiviral vector containing the firefly luciferase gene (gift of Dr. Robert Doebele, UC Anschutz Medical Campus, Aurora, CO). Following selection with 1 µg/mL puromycin, individual colonies were screened for luciferase activity and a subclone expressing high activity was used for the orthotopic experiments in immune deficient mice. Mouse embryo fibroblasts were derived from day E13 embryos and immortalized using the 3T3 culture technique.
Quantitative Real-Time PCR (RT-PCR)
Total RNA was purified from cells using RNeasy mini prep kit (Qiagen, Valencia, CA) and aliquots (5 µg) were reverse transcribed in a volume of 20 µL using Maxima First Strand cDNA Synthesis Kit (Fermentas, Glen Burnie, MD). Aliquots (5 µL) of a 1:25 dilution of the reverse transcription reactions were submitted to PCR in 25µL reactions with SYBR green Jumpstart Taq Readymix (Sigma, St. Louis, MO) with FGFR1 and FGF2 primers previously described (35, 36) using a My iQ real time-PCR detection system (BioRad, Hercules, CA). GAPDH mRNA levels were measured as a housekeeper gene for normalization and data are presented as “relative expression.”
Immunoblot Analysis
For analysis of the phosphorylation status of signaling proteins in response to drug treatment, cells were seeded in 6-well dishes at 400,000 cells per well. After 24 hrs, cells were placed in HITES medium for 2 hrs and subsequently treated with ponatinib (0 – 300nM) for 2 hrs. Cells were rinsed in phosphate-buffered saline, lysed in 500 µL MAP Kinase Lysis Buffer (MKLB; 1× Protease Inhibitor Cocktail #8340 (Sigma), 0.5% Triton X-100, 50 mM β-glycerophosphate (pH 7.2), 0.1 mM Na3VO4, 2 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.3 M NaCl, and 4 mg/ml aprotinin) and centrifuged (5 min at 13,000 rpm). The supernatants were mixed with SDS sample buffer and submitted to SDS-PAGE. Following electrophoretic transfer onto nitrocellulose filters, the filters were blocked in 3% bovine serum albumin (Cohn Fraction V, ICN Biomedicals, Inc., Aurora, OH) in Tris-buffered saline with 0.1% Tween 20 (TTBS) and then incubated with antibodies to phospho-ERK-T202/Y204 (Cell Signaling Technology, Danvers, MA) for 16 hrs at 4°C. The filters were washed thoroughly in TTBS, then incubated with alkaline phosphatase-coupled goat anti-rabbit or mouse antibodies and developed with LumiPhos reagent (Pierce, Rockford, IL) according to the manufacturer’s instructions. The filters were subsequently stripped and reprobed for total ERK1 (sc–93) and ERK2 (sc–154) (Santa Cruz Biotechnology, Inc., Dallas, Texas).
For Immunoblot analysis of FGFR1, FGFR2, FGFR3 and the α-subunit of the NaK-ATPase, cells were collected in phosphate-buffered saline, centrifuged and suspended in MKLB. Aliquots of the cell lysates containing 150 µg of protein were submitted to SDS-PAGE and immunoblotted for FGFR1 (Origene, Rockville, MD, #TA301021) or FGFR2 (sc–122), FGFR3 (sc-13121) and NaK-ATPase α-subunit (sc-21712) (Santa Cruz Biotechnology).
Measurement of FGFR1 gene copy number in cell lines by fluorescence in situ hybridization (FISH)
Genomic FGFR1 status of all cell lines was analyzed using a FISH assay. Cells (60–80% confluent when harvested) were incubated with 10 ml of 0.075M KCl at 37°C and then fixative solution (Acetic acid/Methanol) was added. Slides were pretreated with 2× SSC at 37°C for 60 min and digested with Digest-All III (dilution 1:25) at 37°C for 6 min. FGFR1 target probe and a commercially available centromeric reference probe of chromosome 8 (CEP8) (Metasystems, Altussheim, Germany) were denatured at 73°C for 5 min and immediately placed on the slides. Co-denaturation of the cells and the FGFR1 probes (commercial green reference probe, Metasystems, Altussheim, Germany; BAC RP11-148D21 was used as a target probe spanning the FGFR1 gene locus 8p11.23 to 8p11.22; Invitrogen, Darmstadt, Germany) was performed at 85°C for 4 min and then hybridization proceeded at 37°C overnight. The slides were extensively washed with 0.5× SCC and then signals were visualized with a fluorescence microscope.
Anchorage Independent Growth and Cell Proliferation Assays
For measurement of anchorage-independent cell growth in soft agar, 15,000 cells (H2052, MSTO211H, H513, H226, Met5A, H2452) were suspended in 1.5 mL RPMI-1640 containing 10% fetal bovine serum and 0.35% agar noble and overlaid on base layers containing 1.5 mL RPMI-1640 containing 10% fetal bovine serum and 0.5% agar noble in six-well plates. The wells were overlayed with 2 mL growth media containing ponatinib or FP-1039 and replaced once per week. The plates were incubated for 14 days and viable colonies were stained for 24 hours with 200 µL of 1 mg/mL nitroblue tetrazolium. The wells were photographed and submitted to quantification using the MetaMorph (Molecular Devices, Downingtown, PA) imaging software program. Data are presented as percentage of the DMSO-treated control. For measurement of proliferation, 10,000 cells (H28) or 15,000 cells (H290) were plated in 2 mL RPMI-1640 containing 10% fetal bovine serum in 6-well plates. The cells were treated with ponatinib for 7 days and following tryspinization, viable cells were counted with a Cellometer (Nexcelom, Lawrence, MA) and compared with the DMSO untreated cell number for each cell line and presented as percent control.
FGFR1 Silencing with shRNAs
Distinct FGFR1-targeting shRNAs (clone IDs TRCN0000121185 and TRCN0000121186) in the pLKO.1 lentiviral vector were obtained from University of Colorado Cancer Center Functional Genomics shared resource. The pLKO.1 constructs encoding the FGFR1 shRNAs as well as pLKO.1 encoding a control shRNA targeting a non-expressed gene, green fluorescent protein (GFP), were packaged in 293T cells with component vectors pCMV-VSV-G and pΔ8.9. The lentiviruses released into the medium were filtered through a 0.45 µm filter and incubated with cell lines seeded in 6-well plates at 200,000 cells/well. Transfected cells were selected with puromycin (1mg/ml) for 8 days after which the plates were rinsed with phosphate-buffered saline (PBS) and fixed and stained with 0.5% (wt/vol) crystal violet in 6.0% (vol/vol) glutaraldehyde solution for 30 minutes at room temperature. Plates were rinsed extensively in distilled H2O and photographed.
Murine orthotopic mesothelioma model
Female Balb/c athymic nude (nu/nu) mice (4 weeks old) were obtained from Harlan Laboratories. All procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee, University of Colorado Denver Anschutz Medical Campus. One passage prior to use, puromycin was removed from the growth medium for luciferase expressing H226 (H226-luc) cells. H226-luc cells were collected by trypsinization, washed three times with sterile phosphate-buffered saline (PBS) and resuspended in sterile PBS at a concentration of 1.0 × 106 cells/100 µL. Female mice were anesthetized with isoflourane by inhalation, the left chest wall was cleaned with betadine followed by rinsing with 70% ethanol. A small skin incision in the left chest wall (approximately 5 mm in length) was made along the left lateral axillary line at the level of the xyphoid process. Subskin fat was dissected away until the left lung was visualized through the intercostal muscles and pleura. A one mL insulin syringe with a 30-gauge needle was used to inject 100 µl of the cell suspension (1×106 cells) into the pleural space and the skin incision was closed using NEXABAND Liquid topical tissue adhesive (Abbott Laboratories, North Chicago, IL). Following 6 weeks to allow the tumors to establish, the animals were injected with luciferin (300 mg/kg in 150 µl PBS). Approximately 15 minutes later, the mice were anesthetized and submitted to bioluminescence imaging (BLI) using a Xenogen IVIS Imaging System 50 Series (PerkinElmer, Waltham, MA). Tumor-bearing mice were randomized into groups (5 per group) and treated daily by oral gavage with ponatinib (20 mg/kg) or diluent alone as a control. Tumor burden was monitored every two weeks for the duration of the study.
Surgical mesothelioma cohort and preparation of a tissue microarray
We screened the archives of the Institute for Pathology, Heidelberg University, Germany, for available paraffin-embedded tissue of surgically obtained mesotheliomas treated between 2002 and 2010 in the Thoraxklinik at Heidelberg University, Germany. Usage of the human tissue was approved by the local ethics committee (No. 206/2005). All diagnoses were made by at least two experienced pulmonary pathologists according to the current (2004) WHO classification. Finally, a collection of 168 pleural mesotheliomas was used for tissue microarray (TMA) construction. Prior to TMA construction, a HE-stained slide of each block was analyzed in order to select tumor-containing regions. A TMA machine (AlphaMetrix Biotech, Rödermark, Germany) was used to extract tandem 1.0 mm cylindrical core sample from each tissue donor block.
Chromogenic-Silver in-situ Hybridization assay of formalin-fixed, paraffin-embedded tissues for FGFR1 gene copy number
A rapid fully automated Chromogenic-Silver in-situ Hybridization (CS-ISH) dual-color FGFR1 target assay was performed using the Ventana automated platform (Ventana, Medical Systems, Tucson, AZ). 4–5 µm thick sections were cut and mounted on superfrost Slides, which were then deparaffinized, pretreated with sodium citrate (90°C for 36 minutes, sodium citrate buffer pH 6, Ventana Medical System, Inc.) and digested (8 minutes, ISH-Protease2, Ventana Medical Systems, Inc.). The mixture of the FGFR1 DNP probe (Ventana Medical Systems, Inc.) and the chromosome 8 DIG probe (Ventana Medical Systems, Inc.) was denatured at 80°C for 12 minutes and subsequently hybridized for 6 hours at 44°C. Probes were detected using the respective antibodies and visualized with a chromogenic reagent conjugated antibody, resulting in a black signal for FGFR1 (ultraView SISH DNP Detection Kit, Ventana Medical Systems, Inc.) as well as a chromogenic reagent conjugated antibody detection kit, resulting in a red signal for chromosome 8 (ultraView Red ISH Detection Kit, Ventana Medical Systems, Inc.). After counterstaining with haematoxylin II (Ventana Medical Systems, Inc.) and a bluing reagent (Ventana Medical Systems, Inc.), coverslips were applied with mounting media (Cytoseal 60, Thermo Scientific). Samples were analysed under a 40× dry objective using standard bright-field microscopy. Two experienced observers (AvM, SP) evaluated the TMAs independently in a blinded manner and at least 100 tumour cells from each core TMA were evaluated. Low level amplification (LLA) was defined as more than two but less than nine black target signals compared to red reference signals whereas more than nine target signals were assigned as high level amplification (HLA).
In situ hybridization of formalin-fixed, paraffin-embedded tissues for FGFR1 mRNA
The mRNA in situ hybridization (ISH) assay was performed using the RNA scope® 2.0 assay system and the FGFR1 probe provided by Advanced Cell Diagnostics, Inc (Hayward, CA) according to the protocol provided by the company. ISH scores are generated and recorded using the following algorithm with a 400 × magnification setting on the microscope: 0, no staining; 1+, 1–3 dots/cell in >1% but <50% of the tumor cells; 2+, 1–3 dots per cell in >50% of the cells; 3+, clusters in <10% or 3–5 dots in >50% or >5 dots in <10% of tumor cells; 4+, clusters or >5 dots in >10% of the tumor cells.
RESULTS
Expression of FGF2-FGFR1 pathway components in mesothelioma cell lines
We previously reported that subsets of NSCLC and HNSCC cell lines co-express specific FGFs and FGFRs, thereby establishing an autocrine growth pathway (35, 36). To explore if components of FGFR autocrine signaling pathways are expressed in mesothelioma cell lines, gene expression data from the Cancer Cell Line Encyclopedia (CCLE; http://www.broadinstitute.org/ccle/home) was interrogated. Our previous study demonstrated that lung cancer cell lines H1703 and H520 cells serve as examples of high FGFR1 and either FGF2 or FGF9 expression while HCC4006 and H1648 cells are negative for these components (35). Expression levels of FGFR2–4 are generally low in the mesothelioma cell lines, but expression levels of FGFR1 and FGF2 are high in many, similar to expression levels observed in H1703 cells (Table S1). By contrast, no mesothelioma cell line expressed FGF9 mRNA at levels equal to that observed in H520 cells. The data indicate that components of an FGF2-FGFR1 pathway are expressed in pleural mesothelioma cell lines.
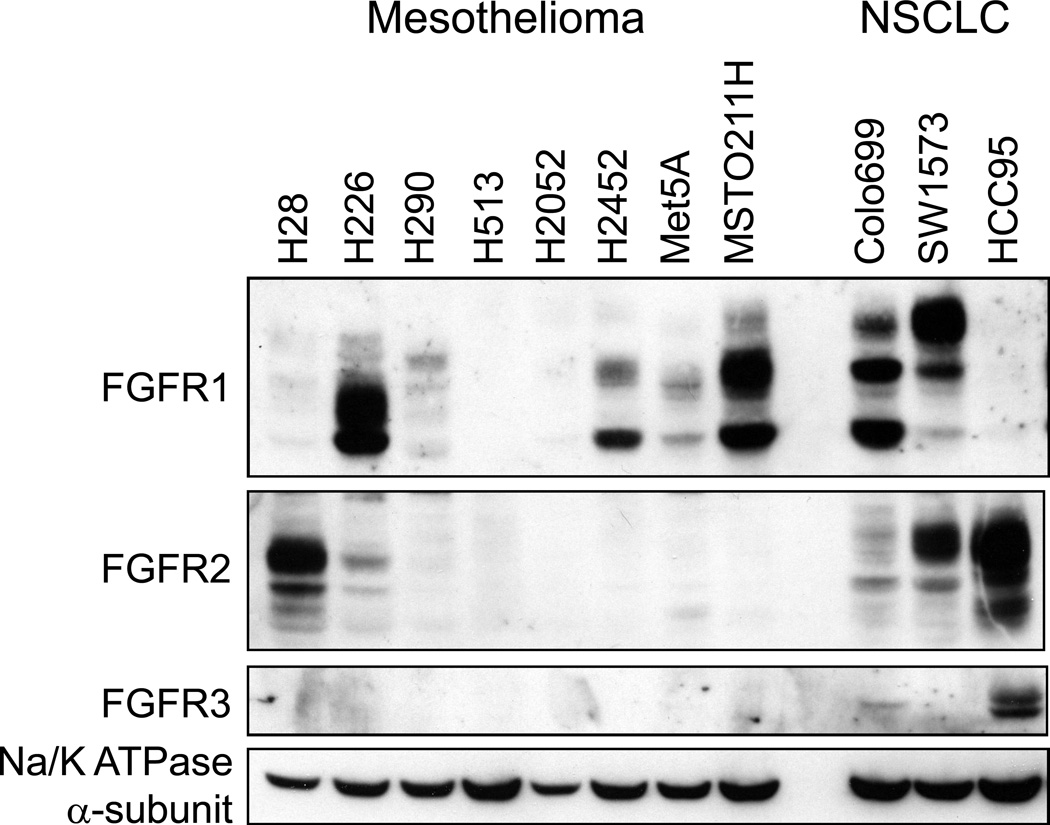
To validate these gene expression findings, a panel of 7 mesothelioma cell lines was employed for cell and molecular studies where five (H28, H226, H290, H513, and H2052) represent the epithelioid histology of MPM and two (H2452, MSTO211H) represent the biphasic variant. In addition, immortalized Met5A mesothelial cells were included. Immunoblot analysis of FGFR1–3 was performed using three lung cancer cell lines as positive controls for each of the FGFR proteins (Figure 1). While FGFR2 expression was limited to H28 cells and no mesothelioma cell line expressed detectable FGFR3, FGFR1 protein was expressed at levels equal to Colo699 and SW1573 lung cancer cells in H226 and MSTO211H cells with lower expression in H2452 and Met5A cells. Because FGFR1 gene copy number gain has been identified as a mechanism for increased FGFR1 expression in lung squamous cell carcinomas and HNSCC (31, 32, 34), the FGFR1 gene amplification status was measured in the mesothelioma cell lines using a fluorescence in situ hybridization (FISH) assay (see Materials and Methods). The FGFR1 FISH assay result for H226 cells is shown in Figure S1 and demonstrates a normal diploid FGFR1 gene status in these cells (FGFR1:CEP8 ratio = 1). H1703 lung cancer cells are shown as a positive control for focal FGFR1 gene amplification (FGFR1:CEP8 ratio = 3.5. The FGFR1 gene copy number status was similarly assessed for 5 additional mesothelioma cell lines and none exhibited FGFR1:CEP8 ratios > 2 (Table 1). To validate the FISH results, the relative FGFR1 gene copy number was assessed in CCLE SNP array datasets and presented in Table 1. Again, no mesothelioma cell line exhibited evidence for increased FGFR1 gene copy number. Thus, the data indicate that increased expression of FGFR1 in H226, MSTO211H and H2452 cells is not associated with FGFR1 gene amplification.
Figure 1. FGFR protein expression in mesothelioma cell lines.
Cell extracts were prepared from the indicated cell lines and aliquots were submitted to SDS-PAGE. Following electrophoretic transfer, the filters were probed for antibodies to FGFR1–3 and the α-subunit of NaK-ATPase as a loading control. Colo699 and SW1734 cells were employed as positive controls for FGFR1 and HCC95 cells are a positive control for expression of FGFR2 and FGFR3.
TABLE 1.
Ponatinib sensitivity associates with FGF2 and FGFR1 expression in mesothelioma cell lines
| Cell Line | Cell Line Morphology/ Histology |
Ponatinib IC50, nM |
FGFR1 protein |
FGFR1:C EP8 ratio (FISH) |
FGFR1 GCN |
FGF2 mRNA (QPCR) |
FGF2 protein (pg/µg cell protein) |
|---|---|---|---|---|---|---|---|
| H226 | Epithelial mesothelioma | 6 | 5.4 | 1.0 | 1.1 | 0.24±.07 | 438±15 |
| MSTO211H | Biphasic mesothelioma | 7 | 4.2 | 1.7 | 0.88 | 0.76±.22 | 888±139 |
| H2452 | Biphasic mesothelioma | 50 | 2.0 | 1.2 | 1.91 | 0.12±.02 | 249±54 |
| Met5A | Immortalized non-malignant mesothelial cells | 100 | 1.4 | ND | ND | 0.27±.05 | 191±6 |
| H290 | 200 | 1.3 | 0.9 | ND | 0.17±.06 | 96±5 | |
| H28 | mesothelioma | 500 | 1.1 | ND | 0.8 | 0.26±.07 | 133±15 |
| H513 | Epithelial mesothelioma | 500 | 0.1 | 1.2 | ND | 0.00±.00 | 0±4 |
| H2052 | mesothelioma | 500 | 0.4 | 0.8 | 0.7 | 0.97±.26 | 690±54 |
| H1703 | NSCLC | - | - | 3.5 | 3.0 | - | - |
The ponatinib IC50 values were calculated from the data in Figure 2 and FGFR1 protein levels were determined by immunoblotting and densitometry. The indicated cell lines were submitted to 2 color FISH assays for FGFR1 and CEP8 and the ratios are tabulated. The Cancer Cell Line Encyclopedia SNP data was interrogated and the relative FGFR1 gene copy number (GCN) is presented. FGF2 mRNA and protein were measured by quantitative RT-PCR (Materials and Methods) and ELISA (R&D Systems, Minneapolis, MN), respectively, with mean and SEM presented.
Measurement of FGF2 mRNA and protein by quantitative RT-PCR and ELISA, respectively, revealed generally high expression levels in the mesothelioma cell lines except H513 cells (Table 1), a finding consistent with the CCLE gene expression data in mesothelioma cell lines (Table S1). Measurement of FGF9 mRNA by quantitative RT-PCR revealed levels in the mesothelioma cell lines that were less than 1% of FGF9 mRNA levels in H520 cells (data not shown), again consistent with the findings in Table S1. Thus, these results indicate co-expression of FGFR1 or FGFR2 with FGF2 in 5 of 8 mesothelioma cell lines tested.
Expression of FGF2-FGFR1 pathway components in primary malignant mesothelioma specimens
We determined the expression of FGFR1 mRNA in a collection of 168 primary mesothelioma specimens with a commercially available in situ hybridization (ISH) assay suitable for formalin-fixed, paraffin-embedded tissues as described in the Materials and Methods. The findings in Table S2 revealed that 21 (12.5%) of the 168 evaluable tumors exhibited FGFR1 expression of ≥ 2+. FGFR1 mRNA positivity was distributed among epithelioid (n=9), biphasic (n=5), sarcomatoid (n=3) and mixed/not otherwise specified (n=4) histologies. While recent evidence indicates that the FGFR1 gene undergoes amplification or copy number gain in lung squamous cell carcinomas and HNSCC (31, 32, 34), none of the 168 tumors, regardless of FGFR1 mRNA expression levels, exhibited significant gains or amplification of the FGFR1 gene (Table S2).
To validate the findings with the MPM TMA, we interrogated published MPM mRNA expression and gene copy number data (37) deposited in GEO DataSets (GSE29354 and GSE29902). As shown in Table S3, 22/53 (42%) primary MPM tumors were positive for FGFR1 mRNA as defined by mRNA expression ≥ mean expression of the 53 tumors. These tumors included both epithelioid and biphasic histologies, but no sarcomatoid tumors. When the overlap of positivity for FGFR1 and FGF2 was considered, 8 tumors (15% of total) expressed both FGFR1 and FGF2. As with the malignant mesothelioma TMA (Table S2), none of the tumors exhibited evidence of increased FGFR1 gene copy number (Table S3). Combined, the primary malignant mesothelioma data indicate frequent expression of FGFR1 mRNA that is not associated with FGFR1 gene amplification or copy number gain.
FGFR1 and FGF2 form an autocrine signaling pathway in mesothelioma cell lines
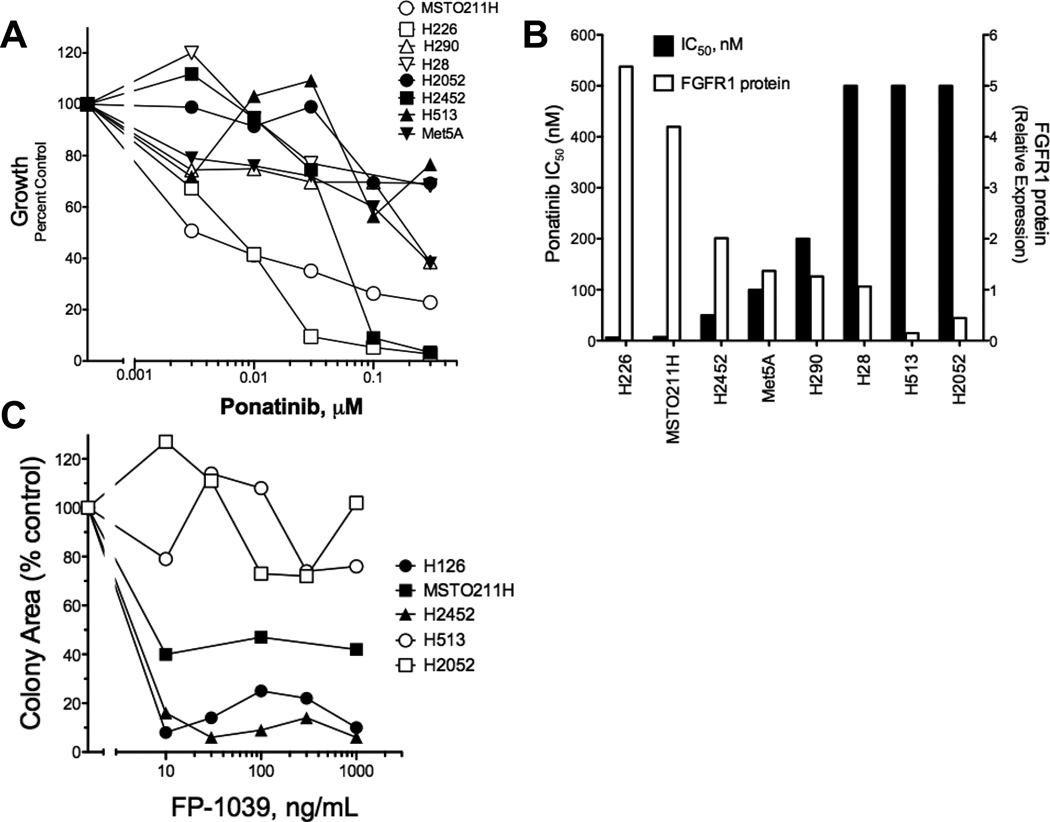
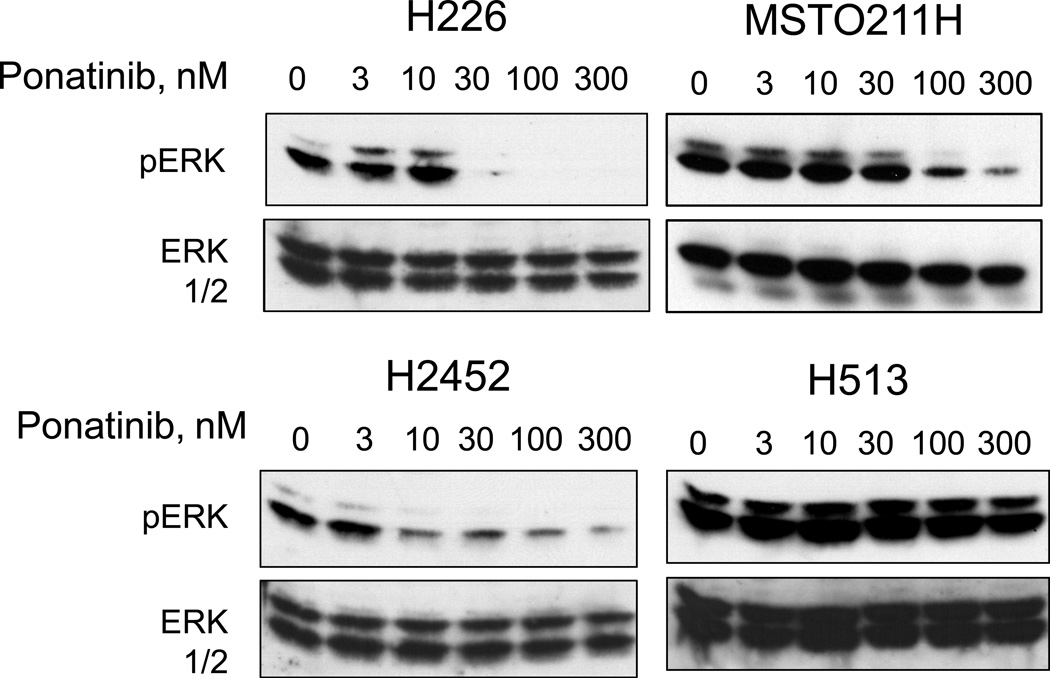
We tested the panel of mesothelioma cell lines for growth sensitivity to the multi-kinase TKI, ponatinib (38) which exhibits potent activity on FGFR1, 2 and 4 (IC50s = 2, 2 and 8 nM, respectively) and weaker inhibition of FGFR3 (18 nM) (39). The findings in Figure 2A demonstrate dose-dependent growth inhibition of H226, MSTO211H and H2452 cells with IC50 values ≤ 50 nM (Table 1). While H28 cells express high levels of FGFR2 and FGF2, growth was insensitive to ponatinib treatment. Pearson correlation analysis of FGFR1 protein levels and ponatinib IC50 values (Figure 2B) revealed a statistically significant association (r=−0.780, P=0.023). In addition to growth inhibition, ponatinib induced a dose-dependent inhibition of basal phospho-ERK in H226, MSTO211H and H2452 cells, but not H513 cells (Figure 3). We also tested the effect of ponatinib on the phosphorylation status of FGFR1 by immunoblot analyses using commercially-available antibodies to FGFR1-Y653/654 as well as by anti-phosphotyrosine immunoblotting following FGFR1 immune precipitation. In our hands, the sensitivity of these assays was insufficient to provide clear evidence for decreased FGFR1 tyrosine phosphorylation following ponatinib treatment in these cell lines.
Figure 2. Inhibition of in vitro growth of mesothelioma cell lines by the FGFR inhibitor, ponatinib.
A, Anchorage-independent growth (H2052, MSTO211H, H513, H226, Met5A, H2452) or cell proliferation assays over 7 days (H28, H290) were performed in the presence of the indicated concentrations of ponatinib as described in the Materials and Methods. B, The IC50 values were calculated with Prism software (GraphPad, San Diego, CA) and plotted versus FGFR1 protein expression measured by densitometry of immunoblots. C, Anchorage-independent growth of the indicated cell lines was measured in the presence of increasing concentrations of the FGF trap, FP-1039 and the data are presented as growth as a percent of control growth measured in the absence of FP-1039.
Figure 3. Inhibition of basal ERK activity in H226, MSTO211H and H2452 cells by ponatinib.
The indicated cell lines were incubated in serum-free medium for 2 hours in the presence of increasing concentrations of ponatinib. Cell extracts were prepared and immunoblotted for phospho-ERK and as described in the Materials and Methods. The filters were stripped and reprobed for total ERK to verify equal protein loading.
To define the requirement for autocrine FGF in the mesothelioma cell lines, we determined the effect of the ligand trap, FP-1039 (36, 40), on growth of a subset of the cell lines. As shown in Figure 2C, FP-1039 potently inhibited anchorage-independent growth of H226 and H2452 cells and more modestly the growth of MSTO211H cells. By contrast, growth of ponatinib-insensitive and FGFR1-negative H513 and H2052 cells was not influenced by the ligand trap. Thus, the data support an autocrine activity of FGF2 in mesothelioma cell lines similar to that previously defined in lung cancer and HNSCC cell lines (35, 36, 40, 41).
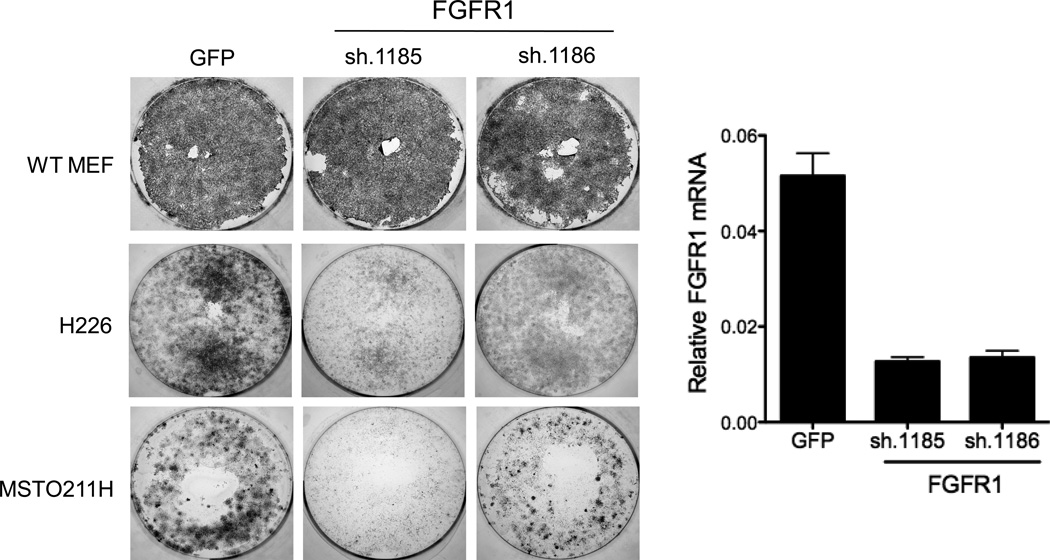
Besides FGFR1, ponatinib potently inhibits VEGFR1–3, PDGFRA, PDGFRB and RET (39). However, none of these RTKs are expressed at the mRNA level in H226, MSTO211H and H2452 cells (Table S1), suggesting that FGFR1 is a likely RTK driving growth of these cell lines and sensitivity to ponatinib. To directly test the requirement for FGFR1 in the ponatinib-sensitive cell lines, two independent FGFR1-targeting shRNAs in the pLKO.1 lentivirus vector were packaged and transduced into H226 and MSTO211H cells. Lentiviruses encoding an shRNA targeting GFP were used as a control for shRNA expression. The findings in Figure 4 show that the FGFR1-specific shRNAs reduced FGFR1 mRNA levels and inhibited clonogenic growth of H226 and MSTO211H cells relative to the GFP control shRNA. Thus, growth of H226 and MSTO211H cells is inhibited by direct RNAi-mediated silencing as well as by an FGFR inhibitor ponatinib.
Figure 4. Inhibition of clonogenic growth following RNAi-mediated silencing of FGFR1.
Lentiviral-encoded shRNAs targeting GFP as a non-silencing control or two independent shRNAs against FGFR1 were packaged and transduced into H226 and MSTO211H cells and then selected for puromycin resistance. Mouse embryo fibroblasts (MEFs) were similarly transduced as a test for the relative titer of the different lentiviral preparations. Following 8 days, the cultures were fixed, stained with crystal violet and photographed. Total RNA was prepared from replicate cultures and following reverse-transcription to cDNA, was submitted to a quantitative PCR assay for FGFR1 and GAPDH mRNAs as described in the Materials and Methods.
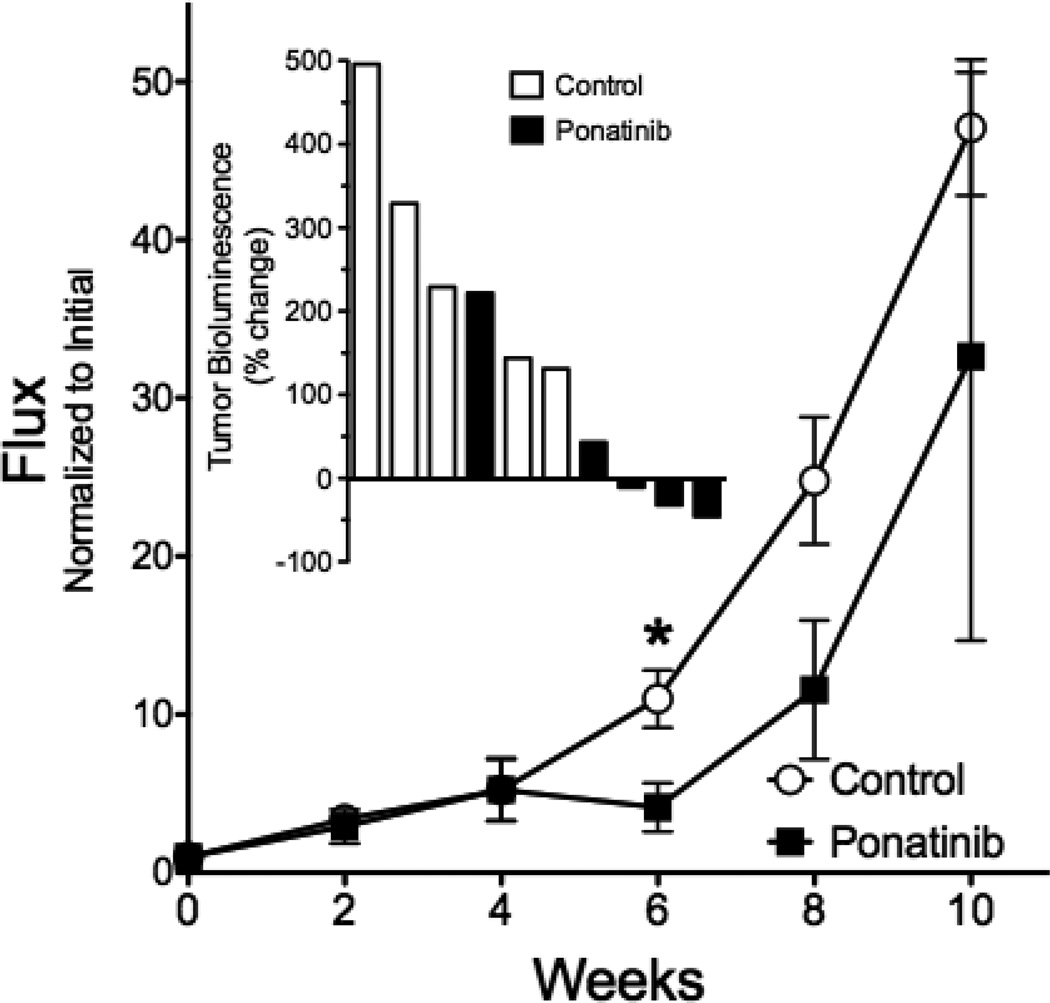
As an in vivo test of the ability of ponatinib to inhibit growth of FGFR1 dependent mesothelioma cells, we developed an orthotopic model of mesothelioma where H226 cells transduced with firefly luciferase were implanted into the pleural space of immune deficient mice. The tumor cells grow aggressively in the orthotopic site with local invasion into the lungs and heart (Figure S2). Following intrapleural injection and 6 weeks for tumor establishment, the mice were treated daily by oral gavage with diluent control or ponatinib at 20 mg/kg. Tumor size was monitored by bioluminescence imaging every two weeks. As shown in Figure 5 (inset), ponatinib induced modest tumor shrinkage in 3 of the 5 mice after 6 weeks of treatment, yielding a statistically significant inhibition of tumor size compared to the control treated mice. Importantly, growth of the ponatinib-treated tumors increased after 8 weeks of treatment and eventually grew at a rate equal to the control tumors. Thus, ponatinib inhibited the growth of H226 tumors in mice, although the response was transient and likely related to acquired resistance.
Figure 5. Effect of ponatinib on orthotopic growth of H226 cells in immune-deficient mice.
Luciferase-expressing H226 cells (106) were implanted into the pleural space of immune-deficient mice as described in the Materials and Methods. Six weeks later (t = 0 on graph), the tumor-bearing mice were randomized into groups of 5 mice and treated by oral gavage with ponatinib (20 mg/kg) or diluent daily. The mice were submitted to bioluminescence imaging (BLI) every two weeks to determine tumor burden and the data are presented as the photon flux normalized to the initial tumor measurement with the means and SEM shown; * indicates p<0.05 by student’s two-tailed t-test. The inset graph shows the percent change in tumor bioluminescence measured at six weeks of treatment relative to the bioluminescence observed at the initiation of treatment for the individual control and ponatinib-treated mice where 3 of 5 ponatinib-treated mice exhibited tumor shrinkage.
DISCUSSION
In light of the poor control of malignant mesothelioma by approved cytotoxic chemotherapeutic regimens, RTKs have been actively explored in MPM as alternative therapeutic targets (15, 19, 42, 43). While EGFR, VEGFRs, MET and PDGFRs have been a focus in these studies, our findings support the novel role of FGFR1 as a growth driver in a subset of MPM cell lines and suggest that FGFR-active TKIs may serve as therapeutics in this cancer. RNAi-mediated silencing of FGFR1 inhibited clonogenic growth of two FGFR1-expressing MPM cell lines, H226 and MSTO211H (Fig. 4) and the TKI, ponatinib, selectively inhibited growth of MPM cell lines expressing FGFR1 (Fig. 2). We previously demonstrated with RNAi-mediated silencing the requirement of FGF2 for H226 cell growth (35) and we validate this finding by showing sensitivity of FGFR1-dependent mesothelioma cell lines to the ligand trap, FP-1039. Combined, the data support the activity of an FGF-FGFR1 autocrine pathway in a subset of mesothelioma cell lines similar to that described in NSCLC and HNSCC (35, 36, 41).
FGFR1 is under extensive evaluation as a therapeutic target in NSCLC, especially of the squamous cell histology, based on the frequent gene amplification or copy number gain that has been established in these cancers (31, 32, 34). In lung cancer cell lines, FGFR1 gene copy number gain is associated with increased FGFR1 expression and sensitivity to TKIs that target this RTK (31, 34). While we observe significant FGFR1 protein expression in H226, MSTO211H and H2452 cells and association with ponatinib sensitivity (Figs. 1 and 2), FGFR1 gene amplification/copy number gain is not observed (Table 1 and Fig. S1). Likewise, elevated expression of FGFR1 mRNA is detected in primary MPM at a frequency ranging between 12.5 to 42%, but again, is not associated with altered FGFR1 gene copy number (Tables S2 and S3). Thus, distinct from squamous cell lung cancers and HNSCC, increased FGFR1 is not necessarily associated with gene amplification. This observation has obvious implications for future testing of the efficacy of FGFR TKIs in MPM patients since the biomarker being deployed for enrolling cancer patients in clinical trials of the FGFR-specific TKIs, BGJ-398 (NCT01004224) and AZD4547 (NCT00979134), is increased FGFR1 gene copy number. Based on our findings in the present study, we propose that expression of FGFR1 mRNA levels in primary tumor specimens should represent the biomarker of FGFR1 dependency in MPM. As a clinical test of this hypothesis, a trial of ponatinib in lung cancer is now opened at our institution entitled “A Phase II Study of Ponatinib in Cohorts of Patients With Lung Cancer Preselected Using Different Candidate Predictive Biomarkers” (NCT01935336). In this trial, lung cancer patients of all histologies are enrolled for ponatinib treatment based on tumor positivity for FGFR1 mRNA assessed by in situ hybridization, gene copy number gain by SISH or positivity for both FGFR1 mRNA and gene copy number gain. Enrolling malignant mesothelioma patients whose tumors are FGFR1 mRNA positive may represent a path forward for testing FGFR TKIs in this cancer as well.
The data in Figure 5 suggests rapid acquisition of ponatinib resistance in H226 cells propagated as orthotopic tumors. Chronic exposure of H226 cells to ponatinib cultured in vitro also results in outgrowth of resistant tumor cells, although no evidence for classical gatekeeper mutations or other secondary somatic mutations in FGFR1 was observed (data not shown). Thus, it is likely that FGFR TKIs will not provide cures when delivered as monotherapies to FGFR1-dependent MPM. The clinical experience with gefitinib and erlotinib in EGFR mutant lung cancer patients and crizotinib in ALK-rearranged lung cancer patients provides precedent for the limitations associated with TKI monotherapy due to acquired resistance (44–46). In fact, as reviewed by Glickman and Sawyers (45), durable control of solid tumors will likely require combinations of targeted agents similar to the present strategy where combinations of antiviral agents provide long-term control of HIV infections by preventing the outgrowth of drug-resistant virions. In addition to prevention of acquired resistance, combinations of TKIs may disrupt the function of RTK co-activation networks that are generally active in cancer cells (47) and MPM specifically (15). We are presently deploying functional genomics approaches to identify in an unbiased fashion additional signal pathways whose inhibition in combination with FGFR1 will achieve synergistic MPM growth inhibition. Thus, the goal is to build upon transient clinical efficacy with FGFR TKI monotherapy with eventual development of inhibitor cocktails to control malignant pleural mesothelioma.
Supplementary Material
Acknowledgements
The authors acknowledge publically available Cancer Cell Line Encyclopedia and GEO Dataset data obtained from malignant mesothelioma cell lines and primary tumors. The studies were supported by the Department of Defense (PRCRP Discovery Award CA110772) and the NIH (CA127105, Lung SPORE P50 CA58187, UC Cancer Center Support Grant P30 CA046934).
Footnotes
Disclosure of Potential Conflicts of Interest: L. Heasley is the recipient of a research contract with ARIAD Pharmaceuticals, Inc to complete pre-clinical studies with ponatinib.
Author’s Contributions
Conception and design: L. Heasley, S. Perner,
Development of methodology: K. Olszewski, L. Marek, D. Böhm,
Acquisition of data: T. Hinz, L. Marek, K. Olszewski, A. von Mässenhausen, E. Kleczko,
Analysis and interpretation of data: L. Heasley, S. Perner, T. Hinz, L. Marek, K. Olszewski, A. von Mässenhausen,
Writing, review and/or revision of the manuscript: L. Heasley, S. Perner, T. Hinz, L. Marek, K. Olszewski,
Technical or material support: D. Boehm, H. Hoffmann, A. Warth
REFERENCES
- 1.Raja S, Murthy SC, Mason DP. Malignant pleural mesothelioma. Curr Oncol Rep. 2011;13:259–264. doi: 10.1007/s11912-011-0177-9. [DOI] [PubMed] [Google Scholar]
- 2.van der Bij S, Koffijberg H, Burgers JA, Baas P, van de Vijver MJ, de Mol BA, et al. Prognosis and prognostic factors of patients with mesothelioma: a population-based study. Br J Cancer. 2012;107:161–164. doi: 10.1038/bjc.2012.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becklake MR, Bagatin E, Neder JA. Asbestos-related diseases of the lungs and pleura: uses, trends and management over the last century. Int J Tuberc Lung Dis. 2007;11:356–369. [PubMed] [Google Scholar]
- 4.Ceresoli GL, Locati LD, Ferreri AJ, Cozzarini C, Passoni P, Melloni G, et al. Therapeutic outcome according to histologic subtype in 121 patients with malignant pleural mesothelioma. Lung Cancer. 2001;34:279–287. doi: 10.1016/s0169-5002(01)00257-4. [DOI] [PubMed] [Google Scholar]
- 5.Yates DH, Corrin B, Stidolph PN, Browne K. Malignant mesothelioma in south east England: clinicopathological experience of 272 cases. Thorax. 1997;52:507–512. doi: 10.1136/thx.52.6.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogelzang NJ, Rusthoven JJ, Symanowski J, Denham C, Kaukel E, Ruffie P, et al. Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. J Clin Oncol. 2003;21:2636–2644. doi: 10.1200/JCO.2003.11.136. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 9.Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 10.Camidge DR, Doebele RC. Treating ALK-positive lung cancer--early successes and future challenges. Nat Rev Clin Oncol. 2012;9:268–277. doi: 10.1038/nrclinonc.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enomoto Y, Kasai T, Takeda M, Takano M, Morita K, Kadota E, et al. Epidermal growth factor receptor mutations in malignant pleural and peritoneal mesothelioma. J Clin Pathol. 2012;65:522–527. doi: 10.1136/jclinpath-2011-200631. [DOI] [PubMed] [Google Scholar]
- 12.Mezzapelle R, Miglio U, Rena O, Paganotti A, Allegrini S, Antona J, et al. Mutation analysis of the EGFR gene and downstream signalling pathway in histologic samples of malignant pleural mesothelioma. Br J Cancer. 2013;108:1743–1749. doi: 10.1038/bjc.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garland LL, Rankin C, Gandara DR, Rivkin SE, Scott KM, Nagle RB, et al. Phase II study of erlotinib in patients with malignant pleural mesothelioma: a Southwest Oncology Group Study. J Clin Oncol. 2007;25:2406–2413. doi: 10.1200/JCO.2006.09.7634. [DOI] [PubMed] [Google Scholar]
- 14.Govindan R, Kratzke RA, Herndon JE, 2nd, Niehans GA, Vollmer R, Watson D, et al. Gefitinib in patients with malignant mesothelioma: a phase II study by the Cancer and Leukemia Group B. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:2300–2304. doi: 10.1158/1078-0432.CCR-04-1940. [DOI] [PubMed] [Google Scholar]
- 15.Brevet M, Shimizu S, Bott MJ, Shukla N, Zhou Q, Olshen AB, et al. Coactivation of receptor tyrosine kinases in malignant mesothelioma as a rationale for combination targeted therapy. J Thorac Oncol. 2011;6:864–874. doi: 10.1097/jto.0b013e318215a07d. [DOI] [PubMed] [Google Scholar]
- 16.Jagadeeswaran R, Ma PC, Seiwert TY, Jagadeeswaran S, Zumba O, Nallasura V, et al. Functional analysis of c-Met/hepatocyte growth factor pathway in malignant pleural mesothelioma. Cancer Res. 2006;66:352–361. doi: 10.1158/0008-5472.CAN-04-4567. [DOI] [PubMed] [Google Scholar]
- 17.Honda M, Kanno T, Fujita Y, Gotoh A, Nakano T, Nishizaki T. Mesothelioma cell proliferation through autocrine activation of PDGF-betabeta receptor. Cell Physiol Biochem. 2012;29:667–674. doi: 10.1159/000176386. [DOI] [PubMed] [Google Scholar]
- 18.Perrone F, Jocolle G, Pennati M, Deraco M, Baratti D, Brich S, et al. Receptor tyrosine kinase and downstream signalling analysis in diffuse malignant peritoneal mesothelioma. Eur J Cancer. 2010;46:2837–2848. doi: 10.1016/j.ejca.2010.06.130. [DOI] [PubMed] [Google Scholar]
- 19.Nutt JE, O'Toole K, Gonzalez D, Lunec J. Growth inhibition by tyrosine kinase inhibitors in mesothelioma cell lines. Eur J Cancer. 2009;45:1684–1691. doi: 10.1016/j.ejca.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 20.Ou WB, Corson JM, Flynn DL, Lu WP, Wise SC, Bueno R, et al. AXL regulates mesothelioma proliferation and invasiveness. Oncogene. 2011;30:1643–1652. doi: 10.1038/onc.2010.555. [DOI] [PubMed] [Google Scholar]
- 21.Pinato DJ, Mauri FA, Lloyd T, Vaira V, Casadio C, Boldorini RL, et al. The expression of Axl receptor tyrosine kinase influences the tumour phenotype and clinical outcome of patients with malignant pleural mesothelioma. Br J Cancer. 2013;108:621–628. doi: 10.1038/bjc.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobson BA, De A, Kratzke MG, Patel MR, Jay-Dixon J, Whitson BA, et al. Activated 4E-BP1 represses tumourigenesis and IGF-I-mediated activation of the eIF4F complex in mesothelioma. Br J Cancer. 2009;101:424–431. doi: 10.1038/sj.bjc.6605184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitson BA, Jacobson BA, Frizelle S, Patel MR, Yee D, Maddaus MA, et al. Effects of insulin-like growth factor-1 receptor inhibition in mesothelioma. Thoracic Surgery Directors Association Resident Research Award. Ann Thorac Surg. 2006;82:996–1001. doi: 10.1016/j.athoracsur.2006.04.013. discussion -2. [DOI] [PubMed] [Google Scholar]
- 24.Ou WB, Hubert C, Corson JM, Bueno R, Flynn DL, Sugarbaker DJ, et al. Targeted inhibition of multiple receptor tyrosine kinases in mesothelioma. Neoplasia. 2011;13:12–22. doi: 10.1593/neo.101156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 26.di Martino E, L'Hote CG, Kennedy W, Tomlinson DC, Knowles MA. Mutant fibroblast growth factor receptor 3 induces intracellular signaling and cellular transformation in a cell type- and mutation-specific manner. Oncogene. 2009;28:4306–4316. doi: 10.1038/onc.2009.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutt A, Salvesen HB, Chen TH, Ramos AH, Onofrio RC, Hatton C, et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl Acad Sci U S A. 2008;105:8713–8717. doi: 10.1073/pnas.0803379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao RG, Jung J, Tchaicha J, Wilkerson MD, Sivachenko A, Beauchamp EM, et al. Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res. 2013;73:5195–5205. doi: 10.1158/0008-5472.CAN-12-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avet-Loiseau H, Li JY, Facon T, Brigaudeau C, Morineau N, Maloisel F, et al. High incidence of translocations t(11;14)(q13;q32) and t(4;14)(p16;q32) in patients with plasma cell malignancies. Cancer Res. 1998;58:5640–5645. [PubMed] [Google Scholar]
- 30.Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dutt A, Ramos AH, Hammerman PS, Mermel C, Cho J, Sharifnia T, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PloS one. 2011;6:e20351. doi: 10.1371/journal.pone.0020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goke F, Bode M, Franzen A, Kirsten R, Goltz D, Goke A, et al. Fibroblast growth factor receptor 1 amplification is a common event in squamous cell carcinoma of the head and neck. Mod Pathol. 2013;26:1298–1306. doi: 10.1038/modpathol.2013.58. [DOI] [PubMed] [Google Scholar]
- 33.Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70:2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marek L, Ware KE, Fritzsche A, Hercule P, Helton WR, Smith JE, et al. Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol Pharmacol. 2009;75:196–207. doi: 10.1124/mol.108.049544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall ME, Hinz TK, Kono SA, Singleton KR, Bichon B, Ware KE, et al. Fibroblast growth factor receptors are components of autocrine signaling networks in head and neck squamous cell carcinoma cells. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:5016–5025. doi: 10.1158/1078-0432.CCR-11-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bott M, Brevet M, Taylor BS, Shimizu S, Ito T, Wang L, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43:668–672. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gozgit JM, Wong MJ, Moran L, Wardwell S, Mohemmad QK, Narasimhan NI, et al. Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Ther. 2012;11:690–699. doi: 10.1158/1535-7163.MCT-11-0450. [DOI] [PubMed] [Google Scholar]
- 39.O'Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16:401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harding TC, Long L, Palencia S, Zhang H, Sadra A, Hestir K, et al. Blockade of nonhormonal fibroblast growth factors by FP-1039 inhibits growth of multiple types of cancer. Sci Transl Med. 2013;5:178ra39. doi: 10.1126/scitranslmed.3005414. [DOI] [PubMed] [Google Scholar]
- 41.Wynes MW, Hinz TK, Gao D, Martini M, Marek L, Ware KE, et al. FGFR1 mRNA and Protein Expression, not Gene Copy Number, Predict FGFR TKI Sensitivity Across All Lung Cancer Histologies. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014 doi: 10.1158/1078-0432.CCR-13-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ikuta K, Yano S, Trung VT, Hanibuchi M, Goto H, Li Q, et al. E7080, a multi-tyrosine kinase inhibitor, suppresses the progression of malignant pleural mesothelioma with different proangiogenic cytokine production profiles. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:7229–7237. doi: 10.1158/1078-0432.CCR-09-1980. [DOI] [PubMed] [Google Scholar]
- 43.Li Q, Wang W, Yamada T, Matsumoto K, Sakai K, Bando Y, et al. Pleural mesothelioma instigates tumor-associated fibroblasts to promote progression via a malignant cytokine network. Am J Pathol. 2011;179:1483–1493. doi: 10.1016/j.ajpath.2011.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doebele RC, Pilling AB, Aisner DL, Kutateladze TG, Le AT, Weickhardt AJ, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:1472–1482. doi: 10.1158/1078-0432.CCR-11-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glickman MS, Sawyers CL. Converting cancer therapies into cures: lessons from infectious diseases. Cell. 2012;148:1089–1098. doi: 10.1016/j.cell.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janne PA, Gray N, Settleman J. Factors underlying sensitivity of cancers to small-molecule kinase inhibitors. Nat Rev Drug Discov. 2009;8:709–723. doi: 10.1038/nrd2871. [DOI] [PubMed] [Google Scholar]
- 47.Xu AM, Huang PH. Receptor tyrosine kinase coactivation networks in cancer. Cancer Res. 2010;70:3857–3860. doi: 10.1158/0008-5472.CAN-10-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.