Abstract
Dendritic cells (DCs), macrophages (MPs) and monocytes are permissive to HIV. Whether they similarly process and present HIV epitopes to HIV-specific CD8 T cells is unknown despite the critical role of peptide processing and presentation for recognition and clearance of infected cells. Cytosolic peptidases degrade endogenous proteins originating from self or pathogens, exogenous antigens preprocessed in endolysosomes, thus shaping the peptidome available for endoplasmic reticulum (ER) translocation, trimming and MHC-I presentation. Here we compared the capacity of DCs, MPs and monocyte cytosolic extracts to produce epitope precursors and epitopes. We showed differences in the proteolytic activities and expression levels of cytosolic proteases between monocyte-derived DCs and MPs and upon maturation with LPS, R848 and CL097, with mature MPs having the highest activities. Using cytosol as a source of proteases to degrade epitope-containing HIV peptides, we showed by mass spectrometry that the degradation patterns of long peptides and the kinetics and amount of antigenic peptides produced differed among DCs, MPs and monocytes. Additionally, variable intracellular stability of HIV peptides prior to loading onto MHC may accentuate the differences in epitope availability for presentation by MHC-I between these subsets. Differences in peptide degradation led to 2- to 25-fold differences in the CTL responses elicited by the degradation peptides generated in DCs, MPs and monocytes. Differences in antigen processing activities between these subsets might lead to variations in the timing and efficiency of recognition of HIV-infected cells by CTLs and contribute to the unequal capacity of HIV-specific CTLs to control viral load.
Introduction
HIV infects CD4-expressing cell subsets, CD4 T lymphocytes, monocytes, DCs and MPs. Monocytes, MPs and CD4 T cells can be productively infected, spread virus and become viral reservoirs whereas DCs do not sustain productive infection but transmit HIV to CD4 T cells and present HIV-derived antigens to prime HIV-specific CD4 and CD8 T cells (1, 2). All four cell types have the capacity to present MHC-I HIV epitopes to CD8 T cells (3-7). However, whether HIV epitope-specific CD8 T cells equally recognize all infected cell subsets is not known, despite their critical role in the clearance of HIV-infected cells. MHC-I epitopes result from the intracellular degradation of proteins by the antigen processing machinery (8). Cytosolic self and pathogen-derived proteins and pathogens are degraded into peptides of variable lengths by proteasomes (9), and to various extents by one or multiple cytosolic peptidases such as leucine aminopeptidase (LAP) (10, 11), thimet oligopeptidase (TOP) (12, 13) or tripeptidyl peptidase II (TPPII) (14, 15). After transfer into the ER, ER-resident aminopeptidases ERAP1 (16-18) and ERAP2 (19, 20) can further trim peptides before or after loading onto MHC-I complexes. Exogenous antigens, such as free or antibody-coated pathogens can also be phagocytosed by target cells or by professional antigen presenting cells and degraded into peptides by cathepsins in endo-lysosomes before transfer to the cytosol for further trimming and cross-presentation by MHC-I (8). Thus the cytosol plays an important role in producing or destroying MHC-I epitopes in direct and cross-presentation pathways.
Differences in antigen processing activities among HIV-infectable cell subsets may affect epitope production and presentation to CTL. We previously showed that CD4 T cells and monocytes present different levels of cytosolic peptidase activities which alter the kinetics and amount of antigenic peptides produced (21). Moreover, different virus-specific CTL responses were stimulated by mouse DC lines and fibroblasts infected with lymphocytic choriomeningitis virus (LCMV) (22), or by mouse DCs and lung MPs infected with Influenza (23), supporting the hypothesis that different cell types may present different epitopes. Higher lysosomal activities and subsequent higher degradation of antigens by MPs compared to DCs have been proposed to contribute to the inability of MPs to prime CTL responses due to insufficient peptide presentation (24). However, cytosolic antigen processing activities involved in degrading incoming HIV have not been systematically compared in these cell subsets.
Beside intrinsic differences in antigen processing activities among cell subsets, external stimuli can alter the antigen processing machinery. During pathogen infection, multiple components of the antigen processing machinery, such as the immunoproteasome subunits (25), the proteasome activator PA28αβ complex and aminopeptidases (26, 27), are induced by interferon gamma which modifies the processing of various CTL epitopes (28-30). Bacteria, viruses, proinflammatory cytokines, CD40 ligand or TLR ligands such as LPS trigger maturation of DCs, alter proteasome composition (31-34) and in a few cases have been shown to alter the processing of a MHC-I peptide (28) and the cross-presentation of ovalbumin (35). TLR7/8 agonists are encoded by HIV, and other synthetic TLR agonists used as adjuvants in HIV vaccination in animal models (36, 37) can affect T cell responses (38). Assessing the effect of TLR agonists on the production of HIV-derived epitopes is important to anticipate their impact on epitope production during vaccination.
In this study, we compared activities and expression levels of the cytosolic antigen processing machinery in monocyte-derived DCs and MPs upon maturation with TLR4 and TLR7/8 ligands. Subset-specific differences in protease and peptidase activities led to variations in the degradation patterns of HIV peptides, the kinetics and amount of HIV epitopes produced, and the antigenicity of the degradation products. These results suggest that variable display of HIV peptides by infected cell subsets may lead to differences in the capacity of various CTLs to reduce viral load independently of their intrinsic immune functions.
Methods
Study participants
Buffy coats from anonymous blood donors were purchased from the Massachusetts General Hospital blood bank and approved for use by the Partners Human Research Committee under protocol 2005P001218 (Boston, MA).
Cell culture and HIV infection
Human peripheral blood mononuclear cells (PBMCs) were freshly isolated from buffy coats by Ficoll-Hypaque (Sigma-Aldrich, USA) density centrifugation. Monocytes were enriched from PBMCs using CD14+ immunomagnetic isolation kits according to the manufacturer's instructions (StemCell, Canada). DCs were differentiated from monocytes during a 6-day culture at 106 cells/mL in AIM-V media (Invitrogen, USA), supplemented with 1% HEPES (Sigma, USA), 1% human serum AB (Gemini Bio-Products, USA), 20ng/mL IL-4 (CellGenix, Germany) and 10ng/mL GM-CSF (CellGenix, Germany). On days 2 and 4, fresh IL-4 and GM-CSF were added. MPs were differentiated from monocytes during a 6-day culture at 106 cells/mL in low attachment flasks (Fisher, USA) in AIM-V media supplemented with 1% HEPES and 10% human serum AB as in (39). On day 6, maturation of DCs and MPs was induced by TLR ligand stimulation with 2μg/mL LPS, 1μg/mL CL097, or 1μg/mL R848 (Invivogen, USA) for 2 days. Where indicated, maturation with LPS or CL097 was induced for 5h, 24h and 48h. Immature and mature cells were stained with CD11c, CD83, CD80, CD86, HLA-ABC, HLA-DR, CD14 (BD Biosciences, USA) and BDCA-1 (Miltenyi Biotec, USA) to confirm differentiation, purity and maturation of cell populations (Supplemental figure 1).
EBV-immortalized HLA-A03/A11 B cells were maintained in RPMI medium, 10% FBS, 1% HEPES, 2mM L-glutamine, 50U/mL penicillin, and 50 μg/mL streptomycin (CellGro, USA). ATK9-, RK9-, KK9-, and QY9-specific CTL clones were maintained in the presence of 50U/mL IL-2, using 0.1μg/mL CD3-specific mAb 12F6, and irradiated PBMCs as stimulus for T cell proliferation.
Immature DCs and MPs were infected at day 6 with vesicular stomatitis virus glycoprotein (VSVg)-pseudotyped NL4.3 HIV-GFP-expressing virus in the presence or absence of Vpx (40). On day 1, day 3, and day 6 post-infection GFP expression in cells was analyzed by flow cytometry to measure the percentage of infection.
Preparation of cytosolic extracts
Cytosolic extracts from monocytes, DCs, and MPs were prepared by 0.125% digitonin permeabilization in ice-cold lysis buffer (50mM HEPES, 50mM potassium acetate, 5mM MgCl2, 1mM DTT, 1mM ATP, 0.5mM EDTA, 10% Glycerol, pH 7.4), followed by 17,762 rcf centrifugation at 4°C for 15 minutes to remove cell debris as previously done (21, 41). Protein concentration was measured by Bio-Rad protein assay according to the manufacturer's instructions.
Expression of the antigen processing machinery by Western blot
Proteins in cytosolic extracts were separated on a 4-12% Bis-Tris gradient gel (Invitrogen, USA), transferred to PVDF membranes (Millipore, USA), blocked with Odyssey Blocking Buffer (Li-cor Biosciences, USA), and probed with β-actin, s1, LAP (Abcam, USA), PA28α, α7, β1, β1i, β2, β5, β5i (Enzo Life Sciences, USA), Hsp90, TOP, ERAP1 (Santa Cruz, USA), ERAP2 (R&D Systems, USA), and TPPII (ProteinTech, USA) antibodies. Protein bands were visualized by dual infrared fluorophore Odyssey Infrared Imaging System, and densitometric quantification was performed (Li-cor Biosciences, USA). To exclude the effect of different cell sizes between DCs and MPs, we used actin as a loading control to compare protein expression levels and found Hsp90 to be another cytosolic protein not affected by cell type or maturation. In this study we used equivalent amounts of cytosolic extracts, as determined by actin normalization, to measure peptidase activities in extracts and to degrade HIV peptides.
Fluorescent measurement of proteolytic activities
The proteolytic activities of proteasomal caspase-like (cell: 50μM, extracts: 75μM Z-LLEAMC, EMD Millipore, Germany), tryptic (cell: 50μM, extracts: 2.5μM Boc-LRR-AMC, Bachem, USA), chymotryptic (cell: 50μM, extracts: 100μM Suc-LLVY-AMC, Bachem), as well as post-proteasomal aminopeptidases (cell: 5μM, extracts: 12.5μM H-Leu-AMC, Bachem), TOP (cell: 20μM, extracts: 5μM Mca-PLGPK-DNP, Bachem) and TPPII (cell and extracts: 100μM H-AAF-AMC, Bachem) were measured by cleavage of peptide-specific fluorogenic substrates. The specificity of reactions was checked by pre-incubating extracts or cells for 30 minutes with the relevant inhibitor of proteasome (10μM MG132, Enzo Life Sciences), aminopeptidases (12μM Bestatin, Sigma-Aldrich), TPPII (10μM Butabindide, Tocris), and TOP (10μM N-[(RS)-1-Carboxy-3-phenyl-propyl]-Ala-Ala-Phe-4-Abz-OH, Bachem) before the addition of substrate. For peptidase activities in live cells, 2×104 DCs or MPs in PBS/0.0025% digitonin were plated per well of a black 96-well plate. For cytosolic extracts, equivalent amounts as determined by actin normalization were used in reaction buffer (50mM HEPES, 50mM potassium acetate, 5mM MgCl2, 1mM DTT, 1mM ATP, 0.5mM EDTA, pH 7.4). The rate of fluorescence emission, which is proportional to the proteolytic activity, was measured every 5 minutes at 37°C in a Victor-3 Plate Reader (Perkin Elmer, USA) as in (21, 42).
In vitro epitope degradation and antigenicity assays
Highly purified HIV peptides (>98% pure) were purchased from the Massachusetts General Hospital peptide core facility or from BioSynthesis, USA. Peptides (2-4nmol) were digested with 30-90μg of cytosolic extracts at 37°C in 50μL of degradation buffer (50mM Tris-HCl, 137mM potassium acetate, 1mM MgCl2, and 1mM ATP, pH7.4) as in (41). Aliquots were taken at various time points, and the reaction was stopped with 2.5μL of 100% trifluoroacetic acid (TFA). Peptide fragments present in the digestion mix were purified by 20% trichloroacetic acid (TCA) precipitation and diluted in RPMI without serum. The pH was readjusted to 7.4 with NaOH. Chromium-51 (51Cr)-labeled HLA-matched B cells were pulsed with the purified digestion products (diluted to 0.4μg/mL for 5-ATK9-2 and 5-RK9-3, and 0.8μg/mL for 13-QY9-6) for 30 minutes at 37°C in the absence of serum and used as targets in a 4-hour 51Cr release assay with HLA-matched, epitope-specific CTL clones at a 4:1 effector-to-target ratio. The lysis percentages were compared with the lysis of B cells pulsed with undigested long peptides and optimal epitope titrations.
Mass spectrometry analysis of the degradation peptides
The identity of the peptides in the digestion mix was determined by in-house mass spectrometry analyses. Equal amounts of peptide degradation samples at different time points were injected into a Nano-HPLC (Eksigent) in line with an Orbitrap mass spectrometer (LTQ Orbitrap Discovery, Thermo) with a flow rate of 400nL/min. A Nano cHiPLC trap column (200μm × 0.5mm ChromXP c18-CL 5μm 120Å, Eksigent, USA) was used to remove salts from samples and peptides were separated on a Nano cHiPLC column (75μm × 15cm ChromXP c18-CL 5μm 300Å, Eksigent, USA) over a gradient of 2% to 40% buffer B (buffer A: 0.1% formic acid in water; buffer B: 0.1% formic acid in acetonitrile) and electrosprayed in the mass spectrometer. Mass spectra were recorded in the range of 370 to 2000Da. In MS/MS mode, the eight most intense peaks were selected with a window of 1Da and fragmented. The collision gas was helium, and the collision voltage was 35V. MS/MS spectra were searched against custom-made source peptide databases with Sequest and Proteome Discoverer (Version 1.3, Thermo Scientific, USA). The integrated area under a peak generated by a given peptide is proportional to the abundance of that peptide. Each degradation time point was run on the mass spectrometer at least twice.
Cytosolic stability of optimal epitopes
One nmol of highly purified peptide was degraded in 15μg of cytosolic extracts at 37°C in degradation buffer as in (43). Aliquots were taken at 0, 10, 30, and 60 minutes, and the reaction was stopped with 2.5μL of 100% TFA. The remaining peptide in the digestion mix at each time point was quantified by RP-HPLC. 100% represents the amount of peptide detected at time 0 calculated as the area under the peptide peak. A stability rate of each peptide was calculated by a nonlinear regression (one-phase exponential decay) of the degradation profile obtained over a 60-minute incubation.
Statistical analysis
Spearman's rank correlation coefficient was used to examine bivariate associations. The paired t-test and Wilcoxon signed rank test were used to compare measurements between groups. All p values are two-sided and p values of <0.05 were considered significant. In figures, p-value criteria are assigned as * p<0.05, ** p<0.01 and *** p<0.001. Statistical analyses were conducted using GraphPad Prism (GraphPad Prism Software, La Jolla, CA) and Microsoft Excel.
Results
TLR-mediated maturation differentially alters cytosolic antigen processing activities in DCs and MPs
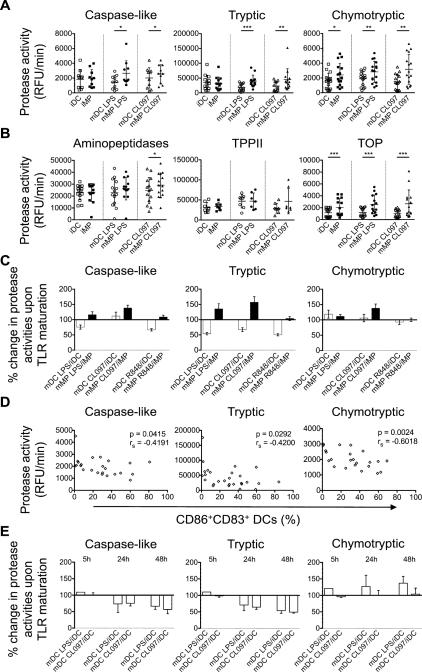
We first analyzed the cytosolic antigen processing activities in live, intact immature and mature monocyte-derived dendritic cells (iDCs, mDCs) and macrophages (iMPs, mMPs). Maturation was achieved by addition of LPS (TLR4 agonist), CL097 or R848 (TLR7/8 agonists) for two days. Proteasomal catalytic subunits (caspase-like, tryptic, and chymotryptic) and post-proteasomal activities (aminopeptidases, TPPII, and TOP) in live DCs and MPs were measured using protease-specific fluorogenic peptidic substrates, and fluorescence emission upon peptide hydrolysis was monitored over time as previously done (21, 42, 43).
iMPs displayed 1.8-fold higher proteasome chymotryptic activity than iDCs (p=0.0318), whereas proteasomal caspase-like and tryptic activities were comparable in both cell types. After LPS- or CL097-induced maturation, proteasome caspase-like, tryptic, and chymotryptic hydrolytic activities of MPs increased, whereas those in DCs decreased or remained unchanged, demonstrating significant differences for all three proteasomal activities between MPs and DCs (1.8-, 2.1-, and 1.5-fold higher in LPS-matured MPs than DCs, and 1.2-, 2.0-, and 2.1-fold higher in CL097-matured MPs, respectively) (Figure 1A). Similarly, significant higher proteasomal hydrolytic activities were observed in R848-matured MPs compared to R848-matured DCs (Supplemental figure 2A). Aminopeptidases and TPPII hydrolytic activities were comparable in both cell subsets regardless of the maturation stage (Figure 1B and Supplemental figure 2B). In contrast, iMPs and LPS-, CL097- and R848-matured MPs displayed approximately two-fold higher TOP activities than their DCs counterparts (Figure 1B, Supplemental figure 2B-2C).
Figure 1. Antigen processing activities in live intact human monocyte-derived DCs and MPs change differentially upon TLR-induced maturation.
A. Proteasomal caspase-like, tryptic, and chymotryptic activities were measured with peptidase-specific fluorogenic substrates in live iDC (□), iMP (■), mDC LPS (○), mMP LPS (●), mDC CL097 (△), and mMP CL097 (▲). Results are from n>10 healthy donors.
B. Post-proteasomal aminopeptidases, TPPII, and TOP activities were measured in the same cells. Results are from n>7 healthy donors. For (A) and (B), paired t-tests (chymotryptic, aminopeptidases) and Wilcoxon signed rank test (caspase-like, tryptic, TPPII, TOP) were performed (*p<0.05, **p<0.01, ***p<0.001), and error bars show SD.
C. Ratios of protease activities in TLR-matured over immature cells of the same donor were calculated and represented as mean ± SD for DCs (open bars) and MPs (solid bars). Results are from n>10 healthy donors.
D. Proteasomal activities in iDC and mDCs were plotted versus the percentage of CD86+ CD83+ DCs for each experiment. Surface expression was analyzed by flow cytometry. Comparison by Spearman test is indicated. n>23 measurements.
E. Ratios of protease activities in TLR-maturing DCs over immature cells of the same donor were calculated and represented as mean ± SD at the indicated time points post stimulation. Results are from n=2 healthy donors.
To compare how each TLR agonist alters cytosolic antigen processing activities in DCs or MPs, we calculated a ratio of activities in mature cells over their immature counterpart (Figure 1C). Both TLR4 and TLR7/8 stimulations increased proteasomal caspase-like, tryptic, and chymotryptic hydrolytic activities in MPs by 20% to 30%. In contrast, matured DCs showed decreased proteasomal activities upon maturation with a 30% lower caspase-like activity, and a 40% to 50% lower tryptic activity in LPS- and R848-matured DCs. In DCs caspase-like, tryptic, and chymotryptic hydrolytic activities inversely correlated with the expression of maturation markers CD83+ and CD86+ after LPS- or R848-induced maturation (rs=-0.4191, p=0.042; rs=-0.4200, p=0.029; and rs=-0.6018, p=0.002, respectively), which supports decreased proteasomal activities upon maturation of DCs (Figure 1D), as demonstrated by others in IFN-treated human primary DCs (44). To assess how TLR stimulation changes protease activities in maturing DCs, we measured proteasomal and aminopeptidases activities at 5h, 24h, and 48h post stimulation with LPS or CL097 and calculated a ratio of activities in maturing cells over their immature counterpart at each time point (Figure 1E and Supplemental figure 2D). In line with previous results, LPS- and CL097-maturing DCs showed 30% decreased proteasomal caspase-like and tryptic activities as early as 24h post stimulation. Further maturation with both TLR ligands decreased caspase-like activities by 40% and tryptic activities up to 50% compared to immature cells. Chymotryptic activities were increased as early as 5 hours upon maturation with LPS but not affected by CL097 at any time point. Aminopeptidases hydrolytic activities were unchanged upon maturation with LPS or CL097 (Supplemental figure 2D). We next analyzed whether HIV infection of DCs and MPs affected antigen processing activities. Single-round infection with VSVg-pseudotyped lentivirus expressing HIV-1 NL4.3 without Env resulted in infection rates of 23.7% and 14.4% 6 days post infection in MPs and DCs, respectively (data not shown). In line with results from TLR-matured cells, we detected 10% to 20% increased proteasomal caspase-like, tryptic, and chymotryptic hydrolytic activities in infected MPs. In contrast, infected DCs showed decreased proteasomal activities with 20% lower caspase-like activity, and 30% lower tryptic activities. Aminopeptidases activities did not change upon infection of both cell subsets (Supplemental figure 2E-2F). These results indicate that TLR-induced maturation triggered significant changes in proteasome and TOP activities of DCs and MPs in divergent ways, which corresponded to changes seen upon HIV infection.
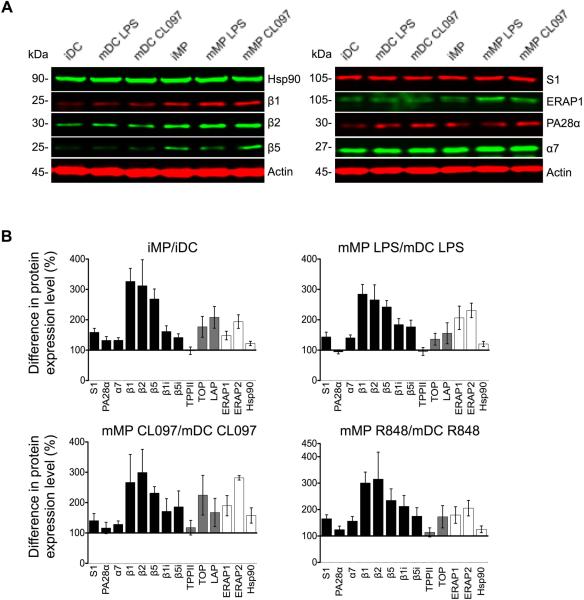
Expression of cytosolic proteases involved in antigen processing is higher in MPs than in DCs
To examine whether the elevated hydrolytic activities observed in mMPs compared to mDCs were due to different levels of peptidase expression, we analyzed cytosolic extracts by dual infrared fluorophore Western blotting, which allows for multiplex detection of signals over a wider quantifiable linear range than chemiluminescence (45) (Figure 2A). The constitutive proteasomal subunits responsible for caspase-like (β1), tryptic (β2), and chymotryptic activities (β5) in MPs were 326%, 315% and 269% that of DCs, and showed similar differences upon LPS maturation (285%, 266%, and 242%, respectively) (Figure 2B and Supplemental figure 3A). The expression of immunoproteasome catalytic subunits β1i and β5i in MPs was 162% and 142% that of DCs, respectively (Figure 2B and Supplemental figure 3B). The lack of a sensitive anti-β2i antibody prevented us from analyzing the tryptic immunoproteasome subunit. The levels of proteasomal core α7, lid S1, and regulator PA28α tended to be higher in MPs compared to DCs, but these differences were not always significant (Figure 2B and Supplemental figure 3B-3C). Post-proteasomal TOP and LAP expression was higher in MPs relative to DCs (177% and 208% in iMPs; 136% and 156% in LPS-matured mMPs, respectively), without reaching statistical significance (Figure 2B and Supplemental figure 3D). TPPII and chaperone heat shock protein 90 (Hsp90) levels were similar between DCs and MPs. The expression of ERAP1 and ERAP2 was higher in MPs compared to DCs, specifically for immature and LPS-matured cells (148% and 194% in iMPs; 206% and 231% in LPS-matured mMPs for ERAP1 and ERAP2 respectively, Figure 2B and Supplemental figure 3E). This analysis demonstrates that, regardless of cell maturation states and variability among donors, the higher expression of catalytic subunits of proteasomes, TOP and aminopeptidases likely contributes to elevated hydrolytic activities observed in MPs compared to DCs.
Figure 2. The expression of the antigen processing machinery is higher in MPs than in DCs.
A. Cytosolic extracts from immature, LPS-matured, or CL097-matured DCs and MPs were probed in Western blots for the expression of chaperone Hsp90, constitutive proteasome catalytic subunits β1, β2, and β5, proteasomal core α7, lid S1, regulator PA28α, and ER-resident aminopeptidase ERAP1, using actin as a loading control.
B. Signal intensities were normalized to actin and quantified for each cell type. The difference in protein expression levels in MPs over DCs from the same donor were calculated and represented as mean ± SEM. Proteasomal subunits (black bars), post-proteasomal peptidases (grey bars), ERAP1/2 and cytosolic Hsp90 (white bars) are shown.
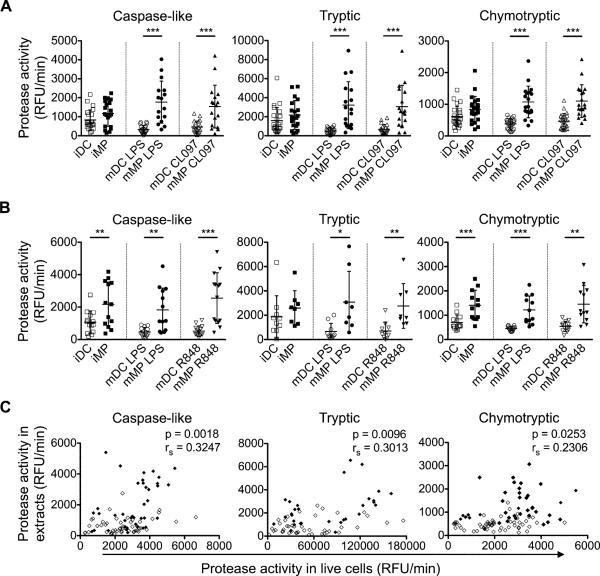
Cytosolic extracts of MPs produce more ATK9-containing antigenic peptides with faster kinetics than DCs extracts
We aimed to determine whether higher expression and activities of the antigen processing machinery in MPs altered the processing of HIV-derived epitopes. We previously developed an in vitro degradation assay to analyze epitope processing in cytosolic extracts (21, 41). A prerequisite to using this assay was to confirm that the differences in peptidase activities in live DCs and MPs were replicable in cytosolic extracts used for the epitope processing assay. Using equal amounts of extracts normalized to actin levels, we observed that LPS-matured MPs had 5.4-fold, 6.5-fold, and 2.8-fold higher caspase-like, tryptic, and chymotryptic activities, respectively, compared to LPS-matured DCs (p<0.0001). Similar trends were observed in extracts prepared from cells matured by CL097 and R848 (Figure 3A-3B). All three proteasomal activities measured in cells correlated significantly with their activities measured in extracts (rs=0.32, p=0.002, caspase-like; rs=0.30, p=0.010, tryptic; rs=0.23, p=0.025, chymotryptic; Figure 3C), thus validating the use of cytosolic extracts to compare epitope processing in DCs and MPs.
Figure 3. Changes in proteasomal activities in cell extracts reflect those observed in live intact DCs and MPs.
A. Proteasomal caspase-like, tryptic, and chymotryptic activities were measured in cytosolic extracts from iDC (□), iMP (■), LPS-matured DC (○) and MP (●), CL097-matured DC (△) and MP (▲) using protease-specific fluorogenic substrates. Results are from n>16 healthy donors.
B. Proteasomal caspase-like, tryptic, and chymotryptic activities were measured as described in (A) including cytosolic extracts from R848-matured mDC (▽) and mMP (▼). Results are from n>13 healthy donors and show mean ± SD. Paired t-tests (caspase-like, tryptic (B), chymotryptic) and Wilcoxon signed rank test (tryptic (A)) were performed (*p<0.05, **p<0.01, ***p<0.001).
C. The proteasomal hydrolytic activities measured in live intact immature or mature DCs (◇) and MPs (◆) were plotted against their activities in corresponding extracts. A partial correlation on Spearman ranked data was performed to control for cell type-dependent effects. n>73 measurements.
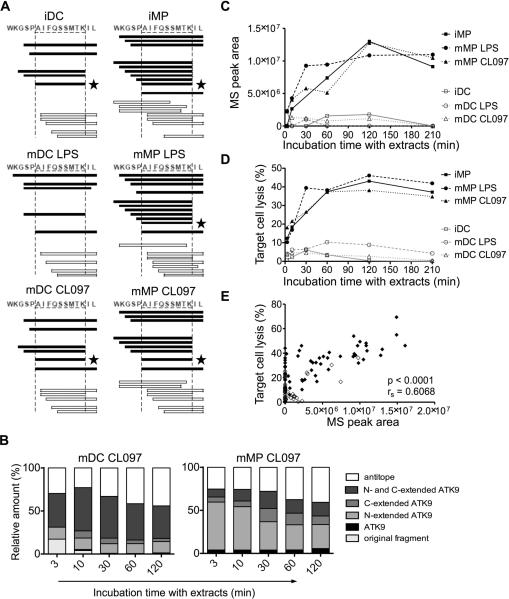
We used cytosolic extracts from DCs and MPs to degrade a synthetic 16-mer peptide representing HIV-1 reverse transcriptase (RT), 5-ATK9-2 (WKGSPAIFQSSMTKIL, aa 153-168), which contains the HLA-A03/A11-restricted ATK9 epitope (AIFQSSMTK, aa 158-166) (46). Degradation products were identified by mass spectrometry (21, 41, 42). The degradation of 5-ATK9-2 led to production of optimal ATK9 and N-extended ATK9 that could be further trimmed into ATK9, N- and C-extended ATK9, and also fragments lacking intact ATK9, called antitopes (Figure 4A and Supplemental figure 4). MP extracts produced a large variety of N-extended ATK9 at each degradation time point and optimal epitope ATK9 was detectable within 3 minutes in MP extracts, whereas it appeared after 10 minutes in DC extracts (Supplemental figure 4A-4B). To assess and compare the production of all peptides over time in each cell subset, we calculated for each time point the contribution of each category of peptides (epitope, antitopes, extended epitopes, original fragment 5-ATK9-2) to the total degradation products (Figure 4B). Each peptide is defined by a peak and area under peak, which we previously showed to be proportional to the amount of the corresponding peptide (42). In MP extracts matured with CL097, optimal ATK9 and N-extended ATK9 represented the major products detected as early as 3 minutes and throughout the degradation, whereas in DC extracts similarly matured, longer peptides were produced in accordance with lower hydrolytic activities. Regardless of the maturation state of the cells, MP extracts always produced more N-extended and optimal ATK9 than DC extracts. The area of the peak corresponding to ATK9 detected by mass spectrometry increased over the course of 210 minutes of degradation in MPs extracts to up to 11.4-fold higher than during the degradation in DC extracts (Figure 4C). To independently confirm the difference in production of ATK9 epitope between DCs and MPs, we measured the antigenicity of the degradation peptides produced over 210 minutes as previously done (21, 41, 42). Degradation peptides were purified and pulsed onto an HLA-A11 B cell line used as target cells in a killing assay with ATK9-specific CTLs. Degradation peptides generated in MPs became increasingly antigenic over the course of the experiment, with lysis % plateauing around 120 minutes at 38% lysis for CL097-matured MP digests, whereas it reached only 3% lysis for CL097-matured DC digests – a 12.6-fold difference in antigenicity of the degradation products (Figure 4D). The antigenicity of the degradation products measured by CTL killing assay correlated strongly with the production of optimal ATK9 epitope detected by mass spectrometry (rs=0.6068, p<0.0001), providing additional validation of the quantification of epitope production by mass spectrometry (Figure 4E). These results show that the processing of ATK9 epitope from HIV RT-derived precursor peptide, 5-ATK9-2, was faster and yielded more antigenic peptides in MP than in DC cytosolic extracts.
Figure 4. MP extracts produce HLA-11-restricted ATK9 epitope more efficiently than DCs.
A. Four nmol of 5-ATK9-2 (aa 153-168 in HIV-1 RT) were degraded with 30μg of cytosolic extracts from immature, LPS-matured, and CL097-matured DCs and MPs for 3, 10, 30, 60, 120, and 210 minutes. Peptides containing epitope HLA-A11-restricted ATK9 (black bars), or no epitope (white bars) were identified by mass spectrometry. Optimal epitope ATK9 (★) is indicated. The degradation profiles in DCs and MPs extracts are shown at 120 minutes.
B. All degradation products of 5-ATK9-2 generated in CL097-matured DC and MP were identified by mass spectrometry at each time point. The peak area of each identified peptide was calculated with Proteome Discoverer. Peptides were grouped into optimal ATK9 epitope (black), fragments containing N-terminal (light grey), C-terminal (medium grey) or N- and C-terminal extensions (dark grey) of ATK9, and antitopes that are lacking intact ATK9 (white). The contribution of each category of peptides to the total intensity of all degradation products is shown at each time point.
C. Kinetics of optimal ATK9 production by iDC (□), iMP (■), LPS-matured DC (○) and MP (●), CL097-matured DC (△) and MP (▲) detected by mass spectrometry.
D. 5-ATK9-2 degradation products were purified and pulsed onto HLA-A03/A11 B cell targets. ATK9-specific CTL responses were assessed by 51Cr release assay. For (A-D), data are representative of three independent experiments with extracts from different healthy donors.
E. ATK9 peak areas in DCs (◇) or MPs (◆) detected by mass spectrometry in three independent experiments were plotted against the target cell lysis values generated by the corresponding degradation peptides. Spearman correlation test was performed; n=106 data points. For (C-E) the 30 minutes degradation time points for iMP were lost.
The kinetics and amount of HIV optimal epitopes produced in cytosolic extracts from monocytes, DCs and MPs vary among epitopes
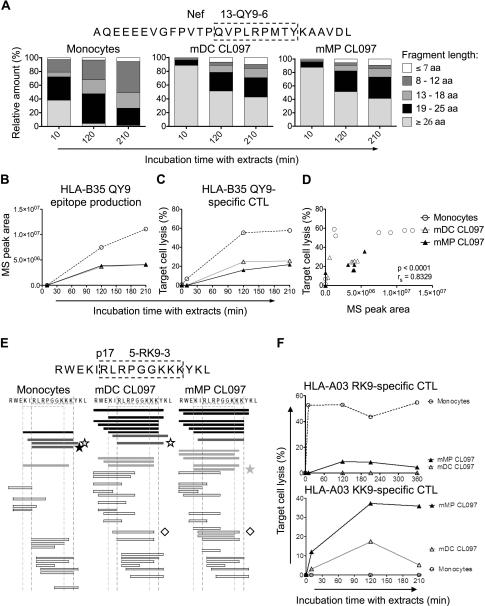
To investigate whether differential peptidase activities in DCs and MPs impacted the processing of other HIV-1 epitopes, we next analyzed the processing of i) a 28-mer Nef-derived precursor peptide 13-QY9-6 (AQEEEEVGFPVTPQVPLRPMTYKAAVDL, aa 60-87 in Nef) which contains HLA-B35-restricted QY9 and 16 other epitopes, and also ii) a 17-mer p17 fragment 5-RK9-3 (RWEKIRLRPGGKKKYKL, aa 15-31) containing HLA-A03-restricted RK9 and 6 other epitopes (Figure 5) (47).
Figure 5. Extracts from monocytes, DCs and MPs generate variable amounts of Nef and p17-derived antigenic peptides at different rates.
A. Two nmol of 13-QY9-6 (aa 60-87 in Nef) were degraded with 45μg of cytosolic extracts from monocytes, mDC CL097, and mMP CL097 for 10, 120, and 210 minutes. Degradation products identified by mass spectrometry were grouped according to their lengths of fragments: longer than 26 aa (light grey), 19-25 aa (black), 13-18 aa (medium grey), 8-12 aa (dark grey) and fragments equal or shorter than 7 aa (white). The contribution of each category of peptides to the total intensity of all degradation products is shown at each time point.
B. QY9 production in extracts from monocytes (○), mDC CL097 (△), and mMP CL097 (▲) detected by mass spectrometry.
C. The 13-QY9-6 degradation products were purified and pulsed onto HLA-B35 B cell targets. QY9-specific CTL responses were assessed by a 51Cr release assay. For (A-C), data are representative of three independent experiments with different healthy donors.
D. Mass spectrometry peak areas of peptide QY9 produced in extracts from monocytes (○), mDC CL097 (△) and mMP CL097 (▲) from two independent experiments were plotted against their respective target cell lysis values from the 51Cr release assay. Spearman correlation test was performed; n=27 data points.
E. Two nmol of 5-RK9-3 (aa 15-31 in Gag p17) were degraded in 90μg of cytosolic extracts from monocytes, CL097-matured DC and MP for 10, 120, 210, and 360 minutes. Peptides encompassing both RK9 and KK9 epitopes (black bars), RK9 only (dark grey bars), KK9 only (light grey bars), or no epitopes (white bars) were identified by mass spectrometry. Optimal epitopes A03-RK9 (★), A03-KK9 ( ), A03-RY10 (☆), and B27-IK9 (◇) are indicated. The degradation profiles of 5-RK9-3 in extracts from monocytes, mDC CL097, and mMP CL097 are shown at 120 minutes.
), A03-RY10 (☆), and B27-IK9 (◇) are indicated. The degradation profiles of 5-RK9-3 in extracts from monocytes, mDC CL097, and mMP CL097 are shown at 120 minutes.
F. The 5-RK9-3 degradation products in extracts from monocytes (○), CL097-matured DC (△) and MP (▲) were purified and pulsed onto HLA-A03 B cells, and specific CTL responses against A03-RK9 (top) and A03-KK9 (bottom) were measured by 51Cr release assay. For (E) and (F) data are representative of three independent experiments with extracts from different healthy donors.
MP and DC extracts similarly degraded 13-QY9-6 at a slow rate with 85% of the total peak intensity contributed by peptides longer than 26aa after 10 minutes, while in 10 minutes monocyte extracts yielded 60% of 8-25aa that were later trimmed to 8-12aa long peptides (Figure 5A). HLA-B35-restricted QY9 epitope (QVPLRPMTY, aa 73-81) was efficiently generated by all three of the cell types, with monocyte extracts producing significantly higher amounts (2.7-fold after 210 minutes) than DC and MP extracts (Figure 5B). These differences in QY9 production were independently verified in a killing assay with QY9-specific CTL, where the antigenicity of the degradation fragments from monocyte extracts was 2.2- and 2.7-fold greater than that of DC and MP extracts, respectively (Figure 5C). There was little difference in antigenicity of peptides generated with DC and MP extracts (26% and 22% lysis, respectively), and no significant differences were observed among the different maturation states induced by TLR4, 7 and 8 stimulation (data not shown). The antigenicity of the degradation products correlated strongly with the detection of optimal QY9 epitope by mass spectrometry (rs=0.8329, p<0.0001; results from two independent experiments) (Figure 5D). In contrast to A11-ATK9, A03-QY9 was similarly produced in DCs and MPs. Differences in QY9 epitope production in monocytes, as compared to DCs or MPs, may be due to differences in activities of peptidases involved in QY9 production or destruction, such as aminopeptidases which are significantly higher in monocytes than in DC and MP (data not shown).
We had previously shown that peptide degradation patterns and kinetics of epitope production contributes to immunodominance patterns in HIV infection (41). In PBMC or monocyte extracts immunodominant HLA-A03 RK9 (RLRPGGKKK, aa 20-28) and RY10 (RLRPGGKKKY, aa 20-29 in p17) (48) are processed faster and accumulate while subdominant overlapping HLA-A03 KK9 (KIRLRPGGK, aa 18-26) tends to be destroyed (41). Whether this hierarchy of epitope production exists in all cell subsets is not known. We used cytosolic extracts from monocytes, DCs and MPs to degrade Gag p17 peptide fragment 5-RK9-3 containing the three overlapping CTL epitopes (KK9, KIRLRPGGK, aa 18-26; RK9, RLRPGGKKK, aa 20-28; and RY10, RLRPGGKKKY, aa 20-29) (Figure 5E). Mass spectrometry detection of the fragments produced in monocyte extracts after 120 minutes of degradation were in line with our previous study (21), as more RK9-containing fragments were generated, compared to fragments containing KK9. In contrast, extracts from CL097-matured DCs and MPs produced more KK9-containing fragments than RK9-containing fragments (Figure 5E). The higher aminopeptidase activities of monocytes may lead to frequent cleavages between KI or IR residues, destroying KK9 and preserving RK9, as we previously showed in cytosolic extracts from PBMCs, monocytes and CD4 T cells (21). The differences in RK9 and KK9 production were independently verified in a killing assay using RK9-specific and KK9-specific CTLs. 5-RK9-3 degradation peptides in monocyte extracts resulted in significant higher target cell lysis by RK9-specific CTL than KK9-specific CTL (53% and 0% lysis by RK9- and KK9-specific CTL, respectively, at 120 minutes), whereas the opposite trend was observed for the degradation products of CL097-matured DC and MP extracts (17% and 37% lysis by KK9 CTL for DC and MP extracts; 0% and 9% lysis by RK9 CTL, respectively, at 120 minutes) (Figure 5F). No significant differences were detected within DC or MP subsets upon LPS, CL097 or R848 maturation (data not shown). This suggests that even for overlapping epitopes restricted by the same HLA, different infectable myeloid subsets may present variable ratios of epitopes, and therefore epitope-specific CTLs will be better suited to clear some subsets than others.
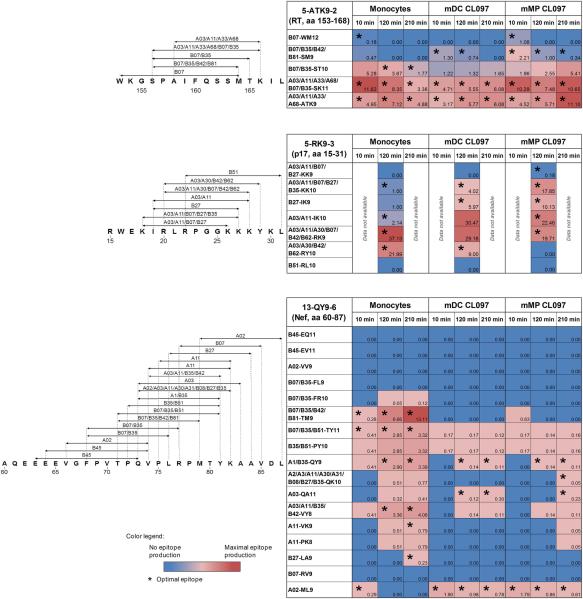
Figure 6 summarizes peptide production for the 29 epitopes studied. Five epitopes (17%) were produced as optimal epitopes and eleven (38%) as N-extended epitopes in all three subsets, but only five epitopes were detected at the same time point in all subsets. Thus due to heterogeneity in peptidase activities among subsets, peptides presented by HIV-infectable subsets may differ in their timing of presentation, amount and length (optimal or extended epitopes), all of which could affect recognition by T cells.
Figure 6. Variable production of 29 HIV-1 epitopes in cytosolic extracts of monocytes, DCs and MPs.
The maps show the location of epitopes (arrows) within sequences 5-ATK9-2 (aa 153-168 in RT), 5-RK9-3 (aa 15-31 in Gag p17), and 13-QY9-6 (aa 60-87 in Nef). The tables show a summary of the relative amount of optimal epitopes and corresponding N-terminal extensions detected by mass spectrometry in the degradation products of 5-ATK9-2, 5-RK9-3, and 13-QY9-6 in monocytes, mDC CL097, and mMP CL097 extracts after 10, 120, 210 minutes of digestion. Numbers represent the contribution of optimal epitopes and N-extended epitopes to the total intensity of all degradation products at each time point. The presence of optimal epitopes is indicated (*). For each epitope the data are representative of three mass spectrometry analyses from independent experiments.
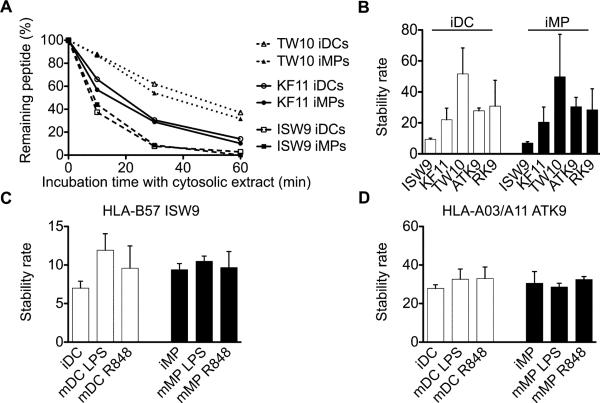
Intracellular stabilities of different HIV epitopes are highly variable but displays similar hierarchies in DCs and MPs
We previously showed that epitope stability in PBMC cytosol is highly variable among epitopes and contributes to defining the amount of epitopes present at the cell surface (43). In order to analyze the cytosolic epitope stability in extracts of DCs and MPs, we followed the degradation of three HLA-B57-restricted optimal epitopes located in p24 Gag (KF11, KAFSPEVIPMF, aa 30-40; ISW9, ISPRTLNAW, aa 15-23; TW10, TSTLQEQIGW, aa 108-117) (47, 49) in extracts of iDCs and iMPs. Using RP-HPLC analysis to follow the disappearance of each peptide (43), we showed that peptide ISW9 was rapidly degraded in extracts of both cell subsets with less than 50% epitope remaining after 10 minutes (half-life 7 minutes in iDCs, 9.4 minutes in iMPs). In contrast, TW10 and KF11 were more stable with 87% and 60% epitope remaining after 10 minutes incubation in cytosolic extracts, which corresponds to cytosolic half-lives of 51.7 and 22.1 minutes in iDCs, and 49.8 and 20.4 minutes in iMPs respectively (Figure 7A). To rank epitopes we calculated a stability rate as a nonlinear regression (one-phase exponential decay) of the degradation profile obtained over a 60-minute reaction as described in our previous study (43) (Figure 7B). TW10, RK9 and ATK9 were more stable than KF11 and ISW9 in accordance to our previously published results in PBMC cytosolic extracts (43). Maturation of DCs and MPs with LPS or R848 did not affect the cytosolic half-lives of the epitopes tested (Figure 7C-7D). These results demonstrate that the respective intracellular stability of different optimal HIV epitopes is highly variable in DCs and MPs, but follows a specific hierarchy that is comparable in both subsets. The combined effects of differential epitope production and stability among various subsets is likely to be a major factor in the relative presentation of different epitopes by different cell types and to modulate epitope-specific CTL recognition of infected cell subsets.
Figure 7. Respective intracellular stability of different HIV epitopes is highly variable but displays similar hierarchy in DCs and MPs.
A. One nmol of HLA-B57-restricted KF11 (○), HLA-B57-restricted ISW9 (□), and HLAB57-restricted TW10 epitope (△) were degraded in 15μg of DC or MP extracts (open or solid symbols, respectively). Degradation products were analyzed by RP-HPLC after 10, 30, and 60 minutes. 100% represents the amount of peptide detected by RP-HPLC at time 0, calculated as the surface area under the peptide peak.
B. The cytosolic stability rate of optimal epitopes KF11, ISW9, TW10, ATK9, and RK9 was calculated by a nonlinear regression (one-phase exponential decay) of the degradation profile obtained over a 60-minute incubation in iDC and iMP extracts. Bars represent the mean ± SD of three independent experiments with extracts from different healthy donors.
Optimal epitopes ISW9 (C) and ATK9 (D) were degraded in extracts of immature or mature DCs and MPs. The cytosolic stability rate was calculated as described in (B) and results represent the mean ± SD of three independent experiments.
Discussion
The capacity of cell subsets to degrade proteins into epitopes is likely to have an impact on the capacity of CD8 T cells to clear infected cells, but is less well defined than intrinsic differences in T cell functions. Here we uncovered differences in the capacity of DCs, MPs and monocytes to degrade HIV proteins into epitopes.
We find that HIV-infectable cell subsets have heterogeneous epitope processing activities, with MPs harboring the highest proteasome activities (specifically after maturation with TLR4 or 7/8 ligands), monocytes the highest aminopeptidase activities, and CD4 T cells the lowest cytosolic peptidase activities ((21, 42), Figure 1 and data not shown). Higher peptidase activities were often linked to higher expression of peptidases but could also be due to the expression of additional subset-specific peptidases such as the monocyte-specific serine protease or other cell-specific peptidases yet to be identified (50).
Differences in values of peptidase activities between intact live cells and cell extracts may be due to variations in uptake of substrates between DCs and MPs, differences in ratio between peptidase and substrate between extracts and intracellular volumes of live cells, and the fact that proteasome activities measured in live cells include both cytosolic and nuclear proteasomes. However despite differences in values the alterations of peptidase activities induced by TLR ligands and infection occured similarly in live cells and extracts used for in vitro degradation.
The degradation of a protein into epitopes is a multistep process involving one or several proteases and peptidases. Some HIV epitopes are produced solely by the proteasome whereas others require proteasome and ERAP, or proteasome, aminopeptidase and TPPII, TOP or Nardilysin processing (12, 41, 42, 51, 52). We showed that the cleavability of epitope-flanking and intraepitopic motifs by various peptidases defines how efficiently an epitope is produced and destroyed, and is variable amongst epitopes located in the same protein (41-43). The substrate preference will define the requirement for specific peptidases to make or degrade peptides, and eventually determine whether epitopes may be equally or differently produced and presented among HIV-infectable subsets. The faster and higher production of the proteasome- and aminopeptidase-dependent epitope A11-ATK9 in MPs corresponds to higher proteasome and aminopeptidase activities measured in MPs compared to DCs. Since proteasomes and aminopeptidases are involved in the processing of most epitopes, the degradation patterns of peptides between DCs, MPs and monocytes are likely to differ as evidenced by the variable kinetics or amount of peptide produced for the 29 epitopes studied so far.
If all infected subsets display sufficient amount of peptides, they may be recognized and cleared by CD8 T cells and this broader recognition of infected targets may contribute to the superior antiviral function of these T cells -independently of their polyfunctionality, avidity and TCR (53-59). However, if peptide presentation by one subset is slower or below threshold of detection for a specific TCR, these cells may produce and propagate virus before immune recognition occurs. Due to low infectivity of dendritic cells and insufficient amount of cells to sort infected cells we were not able to directly compare the endogenous processing and presentation of HIV epitopes by DCs and MPs to CTL clones. However, the similar modulations of antigen processing activities observed in HIV-infected and in TLR-stimulated DCs and MPs suggest that our observations are relevant to epitope presentation in the context of HIV infection.
The impact of differences in epitope production among infectable subsets will be in part determined by TCR avidity for a specific peptide, and also by the amount of peptides displayed by each infected cell. The expression of specific HLA alleles or allomorphs in HIV-infected persons has been correlated with spontaneous control or progression of HIV (60-63). Pereyra et al. have recently found that the course of infection is better predicted by a combination of the specificities of the epitopes recognized and the HLA type compared to HLA type alone (64). One possible explanation for this association –in addition to variability in T cell functions or peptide avidity– may be a better presentation of epitopes linked to control by all infectable subsets.
The cytosol is the place where HIV enters or traffics before assembly and budding, and is critical for the degradation of incoming virions or newly produced proteins (4, 65). However, HIV may also be endocytosed and subjected to degradation by cathepsins (6, 65-69). MPs have higher cathepsin activities than DCs (24) and CD4 T cells (Dinter et al., manuscript in preparation). These differences in peptidase activities of the cross-presentation pathway may further contribute to differences in epitopes available for presentation and cross-presentation. Because many experimental vaccines deliver protein/peptide immunogens to endocytic compartments via viral vectors or nanoparticles, assessing how these vaccine peptides are degraded in the context of vaccination is necessary to ensure that the peptides produced and presented by HIV-infected cells are also presented by DCs after vaccination. DC subsets such as Langherans cells, BDCA3+ DCs or even monocyte-derived DCs receiving a vaccine against a given pathogen prime T cell responses that may differ in specificity, magnitude, and phenotype (70-72). Further variations come from a combination of factors including DC subsets, adjuvants (73), immunomodulating cytokines or TLR ligands included in the vaccine (36, 37, 74), variations in endolysosomal peptidase activities among DC subsets (75, 76), and receptor-mediated targeting (72, 77-79). Approaches to HIV peptide immunogens include focus on conserved areas to induce immune pressure crippling HIV replication or covering the diversity of HIV through chimera of variants (80, 81). In addition to sequence coverage of HIV and ways to induce sustainable polyfunctional responses, identifying HIV protein areas efficiently presented by all infected cells may help to improve vaccine design by expanding the recognition of target cells.
Supplementary Material
Acknowledgements
The authors would like to thank Drs F. Pereyra, D. Heckerman, B. Walker and D. Kaufmann for stimulating discussions about this manuscript.
The project was funded by grants AI084753 and AI084106 from NIAID to SLG. Jens Dinter is funded in part by a Ph.D. training fellowship from the Ernst Schering Foundation.
Footnotes
Competing interests
All authors have no conflicting financial interests.
References
- 1.Stebbing J, Gazzard B, Douek DC. Where does HIV live? N Engl J Med. 2004;350:1872–1880. doi: 10.1056/NEJMra032395. [DOI] [PubMed] [Google Scholar]
- 2.Eisele E, Siliciano RF. Redefining the Viral Reservoirs that Prevent HIV-1 Eradication. Immunity. 2012;37:377–388. doi: 10.1016/j.immuni.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujiwara M, Takiguchi M. HIV-1-specific CTLs effectively suppress replication of HIV-1 in HIV-1-infected macrophages. Blood. 2007 doi: 10.1182/blood-2006-07-037481. [DOI] [PubMed] [Google Scholar]
- 4.Buseyne F, Le Gall S, Boccaccio C, Abastado JP, Lifson JD, Arthur LO, Riviere Y, Heard JM, Schwartz O. MHC-I-restricted presentation of HIV-1 virion antigens without viral replication. Nat Med. 2001;7:344–349. doi: 10.1038/85493. [DOI] [PubMed] [Google Scholar]
- 5.Lubong Sabado R, Kavanagh DG, Kaufmann DE, Fru K, Babcock E, Rosenberg E, Walker B, Lifson J, Bhardwaj N, Larsson M. In vitro priming recapitulates in vivo HIV-1 specific T cell responses, revealing rapid loss of virus reactive CD4 T cells in acute HIV-1 infection. PLoS One. 2009;4:e4256. doi: 10.1371/journal.pone.0004256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larsson M, Fonteneau JF, Lirvall M, Haslett P, Lifson JD, Bhardwaj N. Activation of HIV-1 specific CD4 and CD8 T cells by human dendritic cells: roles for cross-presentation and non-infectious HIV-1 virus. Aids. 2002;16:1319–1329. doi: 10.1097/00002030-200207050-00003. [DOI] [PubMed] [Google Scholar]
- 7.Severino ME, Sipsas NV, Nguyen PT, Kalams SA, Walker BD, Johnson RP, Yang OO. Inhibition of human immunodeficiency virus type 1 replication in primary CD4(+) T lymphocytes, monocytes, and dendritic cells by cytotoxic T lymphocytes. J Virol. 2000;74:6695–6699. doi: 10.1128/jvi.74.14.6695-6699.2000. {Wagner, 1998 #13} [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 9.Grant EP, Michalek MT, Goldberg AL, Rock KL. Rate of antigen degradation by the ubiquitin-proteasome pathway influences MHC class I presentation. J Immunol. 1995;155:3750–3758. [PubMed] [Google Scholar]
- 10.Georgiadou D, Hearn A, Evnouchidou I, Chroni A, Leondiadis L, York IA, Rock KL, Stratikos E. Placental leucine aminopeptidase efficiently generates mature antigenic peptides in vitro but in patterns distinct from endoplasmic reticulum aminopeptidase 1. J Immunol. 2010;185:1584–1592. doi: 10.4049/jimmunol.0902502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Towne CF, York IA, Neijssen J, Karow ML, Murphy AJ, Valenzuela DM, Yancopoulos GD, Neefjes JJ, Rock KL. Leucine aminopeptidase is not essential for trimming peptides in the cytosol or generating epitopes for MHC class I antigen presentation. J Immunol. 2005;175:6605–6614. doi: 10.4049/jimmunol.175.10.6605. [DOI] [PubMed] [Google Scholar]
- 12.Kessler JH, Khan S, Seifert U, Le Gall S, Chow KM, Paschen A, Bres-Vloemans SA, de Ru A, van Montfoort N, Franken KL, Benckhuijsen WE, Brooks JM, van Hall T, Ray K, Mulder A, Doxiadis, van Swieten PF, Overkleeft HS, Prat A, Tomkinson B, Neefjes J, Kloetzel PM, Rodgers DW, Hersh LB, Drijfhout JW, van Veelen PA, Ossendorp F, Melief CJ. Antigen processing by nardilysin and thimet oligopeptidase generates cytotoxic T cell epitopes. Nat Immunol. 2011;12:45–53. doi: 10.1038/ni.1974. [DOI] [PubMed] [Google Scholar]
- 13.York IA, Mo AXY, Lemerise K, Zeng W, Shen Y, Abraham CR, Saric T, Goldberg A, Rock KL. The cytosolic endopeptidase, thimet oligopeptidase, destroys antigenic peptides and limits the extent of MHC class I antigen presentation. Immunity. 2003;18:429–440. doi: 10.1016/s1074-7613(03)00058-x. [DOI] [PubMed] [Google Scholar]
- 14.Firat E, Huai J, Saveanu L, Gaedicke S, Aichele P, Eichmann K, van Endert P, Niedermann G. Analysis of direct and cross-presentation of antigens in TPPII knockout mice. J Immunol. 2007;179:8137–8145. doi: 10.4049/jimmunol.179.12.8137. [DOI] [PubMed] [Google Scholar]
- 15.Reits E, Neijssen J, Herberts C, Benckhuijsen W, Janssen L, Drijfhout JW, Neefjes J. A major role for TPPII in trimming proteasomal degradation products for MHC class I antigen presentation. Immunity. 2004;20:495–506. doi: 10.1016/s1074-7613(04)00074-3. [DOI] [PubMed] [Google Scholar]
- 16.Serwold T, Gaw S, Shastri N. ER aminopeptidases generate a unique pool of peptides for MHC class I molecules. Nat Immunol. 2001;2:644–651. doi: 10.1038/89800. [DOI] [PubMed] [Google Scholar]
- 17.York IA, Chang SC, Saric T, Keys JA, Favreau JM, Goldberg A, Rock KL. The ER aminopeptidase ERAPI enhances or limits antigen presentation by trimming epitopes to 8-9 residues. Nat Immunol. 2002;3:1177–1184. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- 18.Hammer GE, Gonzalez F, Champsaur M, Cado D, Shastri N. The aminopeptidase ERAAP shapes the peptide repertoire displayed by major histocompatibility complex class I molecules. Nat Immunol. 2006;7:103–112. doi: 10.1038/ni1286. [DOI] [PubMed] [Google Scholar]
- 19.Saveanu L, Carroll O, Lindo V, Del Val M, Lopez D, Lepelletier Y, Greer F, Schomburg L, Fruci D, Niedermann G, van Endert PM. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol. 2005;6:689–697. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 20.Tanioka T, Hattori A, Masuda S, Nomura Y, Nakayama H, Mizutani S, Tsujimoto M. Human leukocyte-derived arginine aminopeptidase. The third member of the oxytocinase subfamily of aminopeptidases. J Biol Chem. 2003;278:32275–32283. doi: 10.1074/jbc.M305076200. [DOI] [PubMed] [Google Scholar]
- 21.Lazaro E, Godfrey SB, Stamegna P, Ogbechie T, Kerrigan C, Zhang M, Walker BD, Le Gall S. Differential HIV epitope processing in monocytes and CD4 T cells affects cytotoxic T lymphocyte recognition. J Infect Dis. 2009;200:236–243. doi: 10.1086/599837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butz EA, Bevan MJ. Differential presentation of the same MHC class I epitopes by fibroblasts and dendritic cells. J Immunol. 1998;160:2139–2144. [PubMed] [Google Scholar]
- 23.Crowe SR, Turner SJ, Miller SC, Roberts AD, Rappolo RA, Doherty PC, Ely KH, Woodland DL. Differential antigen presentation regulates the changing patterns of CD8+ T cell immunodominance in primary and secondary influenza virus infections. J Exp Med. 2003;198:399–410. doi: 10.1084/jem.20022151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delamarre L, Pack M, Chang H, Mellman I, Trombetta ES. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science. 2005;307:1630–1634. doi: 10.1126/science.1108003. [DOI] [PubMed] [Google Scholar]
- 25.Griffin TA, Nandi D, Cruz M, Fehling HJ, Kaer LV, Monaco JJ, Colbert RA. Immunoproteasome assembly: cooperative incorporation of interferon gamma (IFN-gamma)-inducible subunits. J Exp Med. 1998;187:97–104. doi: 10.1084/jem.187.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beninga J, Rock KL, Goldberg A. Interferon-gamma can stimulate post-proteasomal trimming of the N terminus of an antigenic peptide by inducing leucine aminopeptidase. J Biol Chem. 1998;273:18734–18742. doi: 10.1074/jbc.273.30.18734. [DOI] [PubMed] [Google Scholar]
- 27.Saric T, Chang SC, Hattori A, York IA, Markant S, Rock KL, Tsujimoto M, Goldberg A. An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat Immunol. 2002;3:1169–1176. doi: 10.1038/ni859. [DOI] [PubMed] [Google Scholar]
- 28.Morel S, Levy F, Burlet-Schiltz O, Brasseur F, Probst-Kepper M, Peitrequin AL, Monsarrat B, van Velthoven R, Cerottini JC, Boon T, Gairin JE, Van den Eynde BJ. Processing of some antigens by the standard proteasome but not by the immunoproteasome results in poor presentation by dendritic cells. Immunity. 2000;12:107–117. doi: 10.1016/s1074-7613(00)80163-6. [DOI] [PubMed] [Google Scholar]
- 29.Sijts AJ, Standera S, Toes RE, Ruppert T, Beekman NJ, van Veelen PA, Ossendorp FA, Melief CJ, Kloetzel PM. MHC class I antigen processing of an adenovirus CTL epitope is linked to the levels of immunoproteasomes in infected cells. J Immunol. 2000;164:4500–4506. doi: 10.4049/jimmunol.164.9.4500. [DOI] [PubMed] [Google Scholar]
- 30.van Hall T, Sijts A, Camps M, Offringa R, Melief C, Kloetzel PM, Ossendorp F. Differential influence on cytotoxic T lymphocyte epitope presentation by controlled expression of either proteasome immunosubunits or PA28. J Exp Med. 2000;192:483–494. doi: 10.1084/jem.192.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Schuler-Thurner B, Schuler G, Huber C, Seliger B. Bipartite regulation of different components of the MHC class I antigen-processing machinery during dendritic cell maturation. Int. Immunol. 2001;13:1515–1523. doi: 10.1093/intimm/13.12.1515. [DOI] [PubMed] [Google Scholar]
- 32.Macagno A, Gilliet M, Sallusto F, Lanzavecchia A, Nestle FO, Groettrup M. Dendritic cells up-regulate immunoproteasomes and the proteasome regulator PA28 during maturation. Eur J Immunol. 1999;29:4037–4042. doi: 10.1002/(SICI)1521-4141(199912)29:12<4037::AID-IMMU4037>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 33.Macagno A, Kuehn L, de Giuli R, Groettrup M. Pronounced up-regulation of the PA28a/b proteasome regulator subunit but little increase in the steady-state content of immunoproteasome during dendritic cell maturation. Eur J Immunol. 2001;31:3271–3280. doi: 10.1002/1521-4141(200111)31:11<3271::aid-immu3271>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 34.Ossendorp F, Fu N, Camps M, Granucci F, Gobin SJ, van den Elsen PJ, Schuurhuis D, Adema GJ, Lipford GB, Chiba T, Sijts A, Kloetzel PM, Ricciardi-Castagnoli P, Melief CJ. Differential expression regulation of the alpha and beta subunits of the PA28 proteasome activator in mature dendritic cells. J Immunol. 2005;174:7815–7822. doi: 10.4049/jimmunol.174.12.7815. [DOI] [PubMed] [Google Scholar]
- 35.Crespo MI, Zacca ER, Nunez NG, Ranocchia RP, Maccioni M, Maletto BA, Pistoresi-Palencia MC, Moron G. TLR7 triggering with polyuridylic acid promotes cross-presentation in CD8alpha+ conventional dendritic cells by enhancing antigen preservation and MHC class I antigen permanence on the dendritic cell surface. J Immunol. 2013;190:948–960. doi: 10.4049/jimmunol.1102725. [DOI] [PubMed] [Google Scholar]
- 36.Wille-Reece U, Flynn BJ, Lore K, Koup RA, Kedl RM, Mattapallil JJ, Weiss WR, Roederer M, Seder RA. HIV Gag protein conjugated to a Toll-like receptor 7/8 agonist improves the magnitude and quality of Th1 and CD8+ T cell responses in nonhuman primates. Proc Natl Acad Sci U S A. 2005;102:15190–15194. doi: 10.1073/pnas.0507484102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wille-Reece U, Wu CY, Flynn BJ, Kedl RM, Seder RA. Immunization with HIV-1 Gag protein conjugated to a TLR7/8 agonist results in the generation of HIV-1 Gag-specific Th1 and CD8+ T cell responses. J Immunol. 2005;174:7676–7683. doi: 10.4049/jimmunol.174.12.7676. [DOI] [PubMed] [Google Scholar]
- 38.Lore K, Betts MR, Brenchley JM, Kuruppu J, Khojasteh S, Perfetto S, Roederer M, Seder RA, Koup RA. Toll-like receptor ligands modulate dendritic cells to augment cytomegalovirus- and HIV-1-specific T cell responses. J Immunol. 2003;171:4320–4328. doi: 10.4049/jimmunol.171.8.4320. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez-Garcia M, Porichis F, de Jong OG, Levi K, Diefenbach TJ, Lifson JD, Freeman GJ, Walker BD, Kaufmann DE, Kavanagh DG. Expression of PD-L1 and PD-L2 on human macrophages is up-regulated by HIV-1 and differentially modulated by IL-10. J Leukoc Biol. 2011;89:507–515. doi: 10.1189/jlb.0610327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pertel T, Reinhard C, Luban J. Vpx rescues HIV-1 transduction of dendritic cells from the antiviral state established by type 1 interferon. Retrovirology. 2011;8:49. doi: 10.1186/1742-4690-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Gall S, Stamegna P, Walker BD. Portable flanking sequences modulate CTL epitope processing. J Clin Invest. 2007;117:3563–3575. doi: 10.1172/JCI32047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang SC, Martin E, Shimada M, Godfrey SB, Fricke J, Locastro S, Lai NY, Liebesny P, Carlson JM, Brumme CJ, Ogbechie OA, Chen H, Walker BD, Brumme ZL, Kavanagh DG, Le Gall S. Aminopeptidase Substrate Preference Affects HIV Epitope Presentation and Predicts Immune Escape Patterns in HIV-Infected Individuals. J Immunol. 2012;188:5924–5934. doi: 10.4049/jimmunol.1200219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lazaro E, Kadie C, Stamegna P, Zhang SC, Gourdain P, Lai NY, Zhang M, Martinez SM, Heckerman D, Le Gall S. Variable HIV peptide stability in human cytosol is critical to epitope presentation and immune escape. J. Clin. Invest. 2011;121:2480–2492. doi: 10.1172/JCI44932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lattanzi L, Rozera C, Marescotti D, D'Agostino G, Santodonato L, Cellini S, Belardelli F, Gavioli R, Ferrantini M. IFN-alpha boosts epitope cross-presentation by dendritic cells via modulation of proteasome activity. Immunobiology. 2011;216:537–547. doi: 10.1016/j.imbio.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 45.Mathews ST, Plaisance EP, Kim T. Imaging systems for westerns: chemiluminescence vs. infrared detection. Methods Mol Biol. 2009;536:499–513. doi: 10.1007/978-1-59745-542-8_51. [DOI] [PubMed] [Google Scholar]
- 46.Walker BD, Flexner C, Birch-Limberger K, Fisher L, Paradis TJ, Aldovini A, Young R, Moos B, Scholey RT. Long-term culture and fine specificity of human cytotoxic T-lymphocyte clones reactive with human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1989;86:9514–9518. doi: 10.1073/pnas.86.23.9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Llano A, Williams A, Olvera A, Silva-Arrieta S, Brander C. Best-Characterized HIV-1 CTL Epitopes: The 2013 Update. In: Yusim K, Korber B, Brander C, Barouch DH, de Boer R, Haynes BF, Koup RA, Moore JP, Walker BD, Watkins DI, editors. HIV Molecular Immunology 2013. Los Alamos National Laboratory, Theoretical Biology and Biophysics; Los Alamos, New Mexico: 2013. pp. 3–25. [Google Scholar]
- 48.Yu XG, Addo MM, Rosenberg ES, Rodriguez WR, Lee PK, Fitzpatrick CA, Johnston MN, Strick D, Goulder PJ, Walker BD, Altfeld M. Consistent patterns in the development and immunodominance of human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T-cell responses following acute HIV-1 infection. J Virol. 2002;76:8690–8701. doi: 10.1128/JVI.76.17.8690-8701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brumme ZL, John M, Carlson JM, Brumme CJ, Chan D, Brockman MA, Swenson LC, Tao I, Szeto S, Rosato P, Sela J, Kadie CM, Frahm N, Brander C, Haas DW, Riddler SA, Haubrich R, Walker BD, Harrigan PR, Heckerman D, Mallal S. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PLoS One. 2009;4:e6687. doi: 10.1371/journal.pone.0006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chateau MT, Robert-Hebmann V, Devaux C, Lazaro JB, Canard B, Coux O. Human monocytes possess a serine protease activity capable of degrading HIV-1 reverse transcriptase in vitro. Biochem Biophys Res Commun. 2001;285:863–872. doi: 10.1006/bbrc.2001.5252. [DOI] [PubMed] [Google Scholar]
- 51.Lopez D, Gil-Torregrosa BC, Bergman C, Del Val M. Sequential cleavage by metallopeptidases and proteasomes is involved in processing HIV-1 ENV epitope for endogenous MHC class I antigen presentation. J Immunol. 2000;164:5070–5077. doi: 10.4049/jimmunol.164.10.5070. [DOI] [PubMed] [Google Scholar]
- 52.Tenzer S, Wee E, Burgevin A, Stewart-Jones G, Friis L, Lamberth K, Chang CH, Harndahl M, Weimershaus M, Gerstoft J, Akkad N, Klenerman P, Fugger L, Jones EY, McMichael AJ, Buus S, Schild H, van Endert P, Iversen AK. Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance. Nat Immunol. 2009;10:636–646. doi: 10.1038/ni.1728. [DOI] [PubMed] [Google Scholar]
- 53.Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, Asher TE, Samri A, Schnuriger A, Theodorou I, Costagliola D, Rouzioux C, Agut H, Marcelin AG, Douek D, Autran B, Appay V. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Almeida JR, Sauce D, Price DA, Papagno L, Shin SY, Moris A, Larsen M, Pancino G, Douek DC, Autran B, Saez-Cirion A, Appay V. Antigen sensitivity is a major determinant of CD8+ T-cell polyfunctionality and HIV-suppressive activity. Blood. 2009;113:6351–6360. doi: 10.1182/blood-2009-02-206557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong T, Stewart-Jones G, Chen N, Easterbrook P, Xu X, Papagno L, Appay V, Weekes M, Conlon C, Spina C, Little S, Screaton G, van der Merwe A, Richman DD, McMichael AJ, Jones EY, Rowland-Jones SL. HIV-specific cytotoxic T cells from long-term survivors select a unique T cell receptor. J Exp Med. 2004;200:1547–1557. doi: 10.1084/jem.20032044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen H, Ndhlovu ZM, Liu D, Porter LC, Fang JW, Darko S, Brockman MA, Miura T, Brumme ZL, Schneidewind A, Piechocka-Trocha A, Cesa KT, Sela J, Cung TD, Toth I, Pereyra F, Yu XG, Douek DC, Kaufmann DE, Allen TM, Walker BD. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nat Immunol. 2012;13:691–700. doi: 10.1038/ni.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, Van Baarle D, Kostense S, Miedema F, McLaughlin M, Ehler L, Metcalf J, Liu S, Connors M. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol. 2002;3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 59.Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, Weeks KA, Rood JE, Berkley AM, Sacha JB, Cogliano-Shutta NA, Lloyd M, Roby G, Kwan R, McLaughlin M, Stallings S, Rehm C, O'Shea MA, Mican J, Packard BZ, Komoriya A, Palmer S, Wiegand AP, Maldarelli F, Coffin JM, Mellors JW, Hallahan CW, Follman DA, Connors M. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity. 2008;29:1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Altfeld M, Kalife ET, Qi Y, Streeck H, Lichterfeld M, Johnston MN, Burgett N, Swart ME, Yang A, Alter G, Yu X, Meier A, Rockstroh JK, Allen TM, Jessen H, Rosenberg E, Carrington M, Walker B. HLA class I alleles that contribute strongly to the initial CD8+ T cell responses against HIV-1 are associated with delayed progression to AIDS. Plos Med. 2006;3:1851–1863. doi: 10.1371/journal.pmed.0030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, International HIV Controllers Study The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trachtenberg E. Advantage of rare HLA supertype in HIV disease progression. Nat Medicine. 2003;9:930–937. doi: 10.1038/nm893. [DOI] [PubMed] [Google Scholar]
- 63.Kaslow RA, Carrington M, Apple R, Park L, Munoz A, Saah AJ, Goedert JJ, Winkler C, O'Brien SJ, Rinaldo CRJ, Detels R, Blattner W, Phair J, Erlich H, Mann DL. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Medicine. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 64.Pereyra F, Heckerman D, Carlson JM, Kadie C, Soghoian DZ, Karel D, Goldenthal A, Davis OB, DeZiel CE, Lin T, Peng J, Piechocka-Trocha A, Carrington M, Walker BD. HIV control is mediated in part by CD8+ T-cell targeting of specific epitopes. Journal of Virology. 2014 doi: 10.1128/JVI.01004-14. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sabado RL, Babcock E, Kavanagh DG, Tjomsland V, Walker BD, Lifson JD, Bhardwaj N, Larsson M. Pathways utilized by dendritic cells for binding, uptake, processing and presentation of antigens derived from HIV-1. Eur J Immunol. 2007;37:1752–1763. doi: 10.1002/eji.200636981. [DOI] [PubMed] [Google Scholar]
- 66.Tjomsland V, Ellegard R, Burgener A, Mogk K, Che KF, Westmacott G, Hinkula J, Lifson JD, Larsson M. Complement opsonization of HIV-1 results in a different intracellular processing pattern and enhanced MHC class I presentation by dendritic cells. Eur J Immunol. 2013;43:1470–1483. doi: 10.1002/eji.201242935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beignon AS, McKenna K, Skoberne M, Manches O, Dasilva I, Kavanagh DG, Larsson M, Gorelick RJ, Lifson JD, Bhardwaj N. Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptor-viral RNA interactions. J Clin Invest. 2005;115:3265–3275. doi: 10.1172/JCI26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jouve M, Sol-Foulon N, Watson S, Schwartz O, Benaroch P. HIV-1 buds and accumulates in “nonacidic” endosomes of macrophages. Cell host & microbe. 2007;2:85–95. doi: 10.1016/j.chom.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 69.Raposo G, Moore M, Innes D, Leijendekker R, Leigh-Brown A, Benaroch P, Geuze H. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic. 2002;3:718–729. doi: 10.1034/j.1600-0854.2002.31004.x. [DOI] [PubMed] [Google Scholar]
- 70.Palucka K, Banchereau J, Mellman I. Designing vaccines based on biology of human dendritic cell subsets. Immunity. 2010;33:464–478. doi: 10.1016/j.immuni.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bozzacco L, Trumpfheller C, Siegal FP, Mehandru S, Markowitz M, Carrington M, Nussenzweig MC, Piperno AG, Steinman RM. DEC-205 receptor on dendritic cells mediates presentation of HIV gag protein to CD8+ T cells in a spectrum of human MHC I haplotypes. Proc Natl Acad Sci U S A. 2007;104:1289–1294. doi: 10.1073/pnas.0610383104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chatterjee B, Smed-Sorensen A, Cohn L, Chalouni C, Vandlen R, Lee BC, Widger J, Keler T, Delamarre L, Mellman I. Internalization and endosomal degradation of receptor-bound antigens regulate the efficiency of cross presentation by human dendritic cells. Blood. 2012;120:2011–2020. doi: 10.1182/blood-2012-01-402370. [DOI] [PubMed] [Google Scholar]
- 73.Quinn KM, Yamamoto A, Costa A, Darrah PA, Lindsay RW, Hegde ST, Johnson TR, Flynn BJ, Lore K, Seder RA. Coadministration of polyinosinic:polycytidylic acid and immunostimulatory complexes modifies antigen processing in dendritic cell subsets and enhances HIV gag-specific T cell immunity. J Immunol. 2013;191:5085–5096. doi: 10.4049/jimmunol.1301730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wille-Reece U, Flynn BJ, Lore K, Koup RA, Miles AP, Saul A, Kedl RM, Mattapallil JJ, Weiss WR, Roederer M, Seder RA. Toll-like receptor agonists influence the magnitude and quality of memory T cell responses after prime-boost immunization in nonhuman primates. J Exp Med. 2006;203:1249–1258. doi: 10.1084/jem.20052433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McCurley N, Mellman I. Monocyte-derived dendritic cells exhibit increased levels of lysosomal proteolysis as compared to other human dendritic cell populations. PLoS One. 2010;5:e11949. doi: 10.1371/journal.pone.0011949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dudziak D, Kamphorst AO, Heidkamp GF, Buchholz VR, Trumpfheller C, Yamazaki S, Cheong C, Liu K, Lee HW, Park CG, Steinman RM, Nussenzweig MC. Differential antigen processing by dendritic cell subsets in vivo. Science. 2007;315:107–111. doi: 10.1126/science.1136080. [DOI] [PubMed] [Google Scholar]
- 77.Cohn L, Chatterjee B, Esselborn F, Smed-Sorensen A, Nakamura N, Chalouni C, Lee BC, Vandlen R, Keler T, Lauer P, Brockstedt D, Mellman I, Delamarre L. Antigen delivery to early endosomes eliminates the superiority of human blood BDCA3+ dendritic cells at cross presentation. J Exp Med. 2013 doi: 10.1084/jem.20121251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Idoyaga J, Lubkin A, Fiorese C, Lahoud MH, Caminschi I, Huang Y, Rodriguez A, Clausen BE, Park CG, Trumpfheller C, Steinman RM. Comparable T helper 1 (Th1) and CD8 T-cell immunity by targeting HIV gag p24 to CD8 dendritic cells within antibodies to Langerin, DEC205, and Clec9A. Proc Natl Acad Sci U S A. 2011;108:2384–2389. doi: 10.1073/pnas.1019547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Flamar AL, Xue Y, Zurawski SM, Montes M, King B, Sloan L, Oh S, Banchereau J, Levy Y, Zurawski G. Targeting concatenated HIV antigens to human CD40 expands a broad repertoire of multifunctional CD4+ and CD8+ T cells. Aids. 2013;27:2041–2051. doi: 10.1097/QAD.0b013e3283624305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ferguson AL, Mann JK, Omarjee S, Ndung'u T, Walker BD, Chakraborty AK. Translating HIV sequences into quantitative fitness landscapes predicts viral vulnerabilities for rational immunogen design. Immunity. 2013;38:606–617. doi: 10.1016/j.immuni.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stephenson KE, Barouch DH. A global approach to HIV-1 vaccine development. Immunol Rev. 2013;254:295–304. doi: 10.1111/imr.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.