Abstract
Immature dendritic cells (iDCs) in genital and rectal mucosa may be one of the first cells to come into contact with HIV-1 during sexual transmission of virus. HIV-1 activates the host complement system, which results in opsonization of virus by inactivated complement fragments, for example, iC3b. We investigated antiviral and inflammatory responses induced in human iDCs after exposure to free HIV-1 (F-HIV), complement-opsonized HIV-1 (C-HIV), and complement and Ab–opsonized HIV-1 (CI-HIV). F-HIV gave rise to a significantly higher expression of antiviral factors such as IFN-β, myxovirus resistance protein A, and IFN-stimulated genes, compared with C-HIV and CI-HIV. Additionally, F-HIV induced inflammatory factors such as IL-1β, IL-6, and TNF-α, whereas these responses were weakened or absent after C-HIV or CI-HIV exposure. The responses induced by F-HIV were TLR8-dependent with subsequent activation of IFN regulatory factor 1, p38, ERK, PI3K, and NF-κB pathways, whereas these responses were not induced by C-HIV, which instead induced activation of IFN regulatory factor 3 and Lyn. This modulation of TLR8 signaling was mediated by complement receptor 3 and led to enhanced infection. The impact that viral hijacking of the complement system has on iDC function could be an important immune evasion mechanism used by HIV-1 to establish infection in the host.
Introduction
Dendritic cells (DCs) bridge the innate and adaptive immune response and play an important role in maintaining tolerance (1). DCs may also represent early target cells during sexual transmission of HIV-1 in the genital and rectal mucosa (2). Although they are crucial for the induction of HIV-specific immune responses (3), they can also facilitate the transmission of HIV-1 to CD4+ T cells in the submucosa and lymph nodes (4). The initial interactions between HIV-1 and DCs will program the activation of these cells via different pattern recognition receptors (PRRs), such as TLR8 and DC-SIGN, and influence the DC functionality and the viral infection (5). Other elements present at the site of infection, for example, innate factors such as complement proteins and other immune cells, will also shape the DC response to the virus.
The complement system can be activated through different pathways and is vital for both innate and adaptive immune responses (6). Generally, complement activation by pathogens leads to recruitment of inflammatory cells, opsonization and destruction of the pathogen, augmentation of B-cell responses (6), and Ag presentation by DCs (7). All of these mechanisms are important in both protecting the body from autoimmune diseases and the clearance of many pathogens, including influenza (8). Consequently, some pathogens have developed immune evasion strategies where they escape the complement attack by taking advantage of the properties of different complement components (9). HIV-1 can stop the complement cascade through host-derived complement inhibitors, incorporated into the viral envelope, and becomes coated in complement fragments iC3b and C3d (10–12). Most interactions between HIV-1 and the host will be with opsonized virus as complement proteins, and after seroconversion HIV-specific Abs are present in almost all body fluids (10).
Complement components have been studied for their ability to influence the level of HIV-1 infection in immune cells such as DCs and T cells (10, 13, 14). In DCs, complement opsonization of HIV-1 leads to enhanced infection via complement receptor (CR) 3 (13–15), and our previous studies suggest that this may in part be due to enhanced viral uptake and altered Ag presentation machinery, which guides more virions into the cell cytosol (16, 17).
In this study, we have examined events and signaling cascades, with a focus on early inflammatory and antiviral responses activated in immature DCs (iDCs) by free HIV-1 (F-HIV) and complement-opsonized HIV-1 (C-HIV) and the underlying cellular mechanisms responsible for the enhanced infection in iDCs induced by C-HIV. Our study shows that whereas F-HIV induced antiviral and inflammatory responses in iDCs, complement opsonization resulted in a different response pattern via a CR3 dependent process. The activation of antiviral and inflammatory responses by F-HIV was dependent on TLR8 signaling with subsequent activation of IFN regulatory factor (IRF) 1, ERK, p38, and NF-κB signaling and IRF7 and PI3K protein expression. C-HIV induced a different signaling pattern, with elevated activation of IRF3 and the tyrosine protein kinase Lyn, as well as enhanced infection of the cells.
Our study clearly shows the impact that viral hijacking of the complement system can have on the functionality of DCs, which could be an important part of HIV-1 pathogenesis, and is to our knowledge the first study to show that TLR-induced antiviral responses can be suppressed by CR3 engagement.
Materials and Methods
DC exposure assay
Monocyte-derived DCs were propagated from PBMCs derived from buffy coats, healthy volunteers, or three individuals diagnosed with systemic lupus erythematosus (SLE) according to classification criteria (18) with rs1143679 (R77H) mutation in CD11b (19) (Ethical Permits M173-07 and M75-08/2008) as described previously (19, 20).
HIV-1BaL/SUPT1-CCR5 CL.30 (lots 4143, 4235, 4238, and 4243) was produced using chronically infected cultures of the ACVP/BCP cell line (no. 204), originally derived by infecting SUPT1-CCR5 CL.30 cells (Dr. J. Hoxie, University of Pennsylvania) with an infectious stock of HIV-1BaL (National Institutes of Health AIDS Research and Reference Reagent Program, catalog no. 416, lot no. 59155). Virus was purified and concentrated as previously described (21) and aliquots were frozen in liquid nitrogen vapor. All virus preparations were assayed for infectivity.
Virus was incubated with RPMI 1640 (Sigma-Aldrich, St. Louis, MO) to generate free virus (F-HIV), single-donor human serum supplemented with 25% veronal buffer to generate complement-opsonized virus (C-HIV), or single-donor human serum supplemented with 25% veronal buffer, 0.02 μg/ml HIV-specific IgG, and 20 μg/ml γ globulins to generate virus opsonized with complement and Abs (CI-HIV). DCs (1 × 106) were incubated with multiplicities of infection (MOIs) of 0.08–8 of F-HIV, C-HIV, CI-HIV, or mock (RPMI 1640).
Ligands and inhibitors
The following ligands and inhibitors were added to the DCs 0.5 h prior to virus exposure: 10–30 mM NH4Cl, 5 μg/ml TLR8 agonist CL097, 1–10 μM PI3K inhibitor LY294002 (InvivoGen, San Diego, CA), 2.5–25 μM p38 inhibitor SB203580 (Cayman Chemicals, Ann Arbor, MI), 0.5–5 μM ERK inhibitor OU126 (Sigma-Aldrich, Stockholm, Sweden), 1–5 μM TLR8 inhibitory peptide IRS957 or control peptide (CyberGene, Stockholm, Sweden), 1 μg/ml neutrophil inhibitory factor (NIF; R&D Systems, Minneapolis, MN), 20 μg/ml CD11b mAb (KIM75, gift from M. Robinson, UCB Celltech), 20 μg/ml CD18 mAb (KIM185, gift from Prof. Thomas Vorup-Jensen, Aarhus University, Aarhus, Denmark), or 20 μg/ml matching isotype control mAbs.
Total RNA extraction, reverse transcription, and real-time quantitative PCR
Total RNA was extracted using an RNeasy Mini kit (Qiagen, Manchester, U.K.) and cDNA produced by SuperScript III reverse transcriptase first-strand cDNA synthesis kit (Invitrogen, Carlsbad, CA). Real-time quantitative PCR (qPCR) was performed with Fast SYBR Green Master mix (Applied Biosystems, Foster City, CA) using a 7900HT Fast Real-Time PCR System with 7900 System SDS 2.3 software (Applied Biosystems). Primers targeting β-actin and GAPDH were used as housekeeping genes for reference as described by Vandesompele et al. (22). Primers were purchased from CyberGene (Solna, Sweden). To compensate for variation between plates, values were normalized as described by Rieu and Powers (23).
ELISA and Bead array
Levels of IL-1β (Mabtech, Nacka Strand, Sweden), CXCL8 (BioSite, Täby, Sweden), and IFN-β (R&D Systems, Abingdon, U.K.) were determined by ELISA according to manufacturers’ protocols. p24 levels were determined by an in-house p24 ELISA assay as described previously (17). Protein levels of IL-1β, IL-6, IL-10, IL-12p70, TNF-α, and CXCL8 in cell supernatants were assessed using a Cytometric Bead Array (BD Biosciences, Stockholm, Sweden) according to the manufacturer’s protocols.
Tandem mass spectroscopy proteomics analysis
iDCs (1 × 106 cells/ml) were challenged with mock, F-HIV, C-HIV, or CI-HIV (MOI of 8) and cultured for 24 h. The cells were lysed in SDS lysis buffer, incubated for 5 min at 95°C, sonicated, and 100 μg cell lysate was digested with trypsin into peptides and labeled with iTRAQ (AB Sciex, Foster City, CA) according to the manufacturer’s protocol as described previously (24). The peptides were fractionated by reverse phase liquid chromatography and analyzed by a liquid chromatography–tandem mass spectroscopy using an Easy nLC coupled to a LTQ Orbitrap Velos MS (Thermo Scientific, West Palm Beach, FL). Two independent technical replicates were performed for each sample.
Nuclear translocation and Western blot
iDCs (1 × 106 cells/ml) were exposed to F-HIV, C-HIV, or CI-HIV or mock for 0.5, 1, 3, or 6 h. For nuclear translocation the cells were fixed, permeabilized, stained with rabbit anti–NF-κB p65 (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-IRF1 (Cell Signaling Technology, Danvers, MA), or mouse anti-IRF3 (Invitrogen, Stockholm, Sweden) Abs followed by Alexa Fluor 488–conjugated anti-rabbit (Abcam, Cambridge, U.K.) or Rhodamine Red-X–conjugated anti-mouse (Molecular Probes, Eugene, OR) secondary Abs and fluorescence mounting medium containing DAPI (Vector Laboratories, Peterborough, U.K.). The cells were photographed using a Zeiss LSM 700 microscope (Carl Zeiss, Stockholm, Sweden) and colocalization was assessed using Volocity software (PerkinElmer, Waltham, MA). Western blots were performed using rabbit mAbs against myxovirus protein A (MxA; Novus Biologicals, Littleton, CO), p-Lyn (Abcam), Lyn (Millipore), p-ERK, ERK, p-p38, p38, PI3K p85, IRF1, IRF3, IRF7, NF-κB p65, and actin followed by HRP-conjugated anti-rabbit Ab (Cell Signaling Technology).
Statistical analysis
Results were tested for statistical significance using a repeated measures ANOVA followed by a Bonferroni posttest, and p < 0.05 was considered statistically significant.
Results
Complement opsonization of HIV-1 decreased inflammatory responses of iDCs
Monocyte-derived DCs (0.5 × 106), a model for submucosal iDCs (25), were exposed to F-HIV, C-HIV, CI-HIV, or mock inoculum at MOIs ranging from 0.08 to 8. Expression of inflammatory factors was evaluated at gene and protein levels. mRNA levels of IL-1β and IL-6, determined by qPCR, were significantly increased for F-HIV but not for C-HIV or CI-HIV compared with mock-treated samples (Fig. 1A, 1B). F-HIV elicited the strongest response with an ∼470-fold increase for IL-1β and an ∼220-fold increase for IL-6 compared with mock. In addition to IL-1β and IL-6, an array of cytokines including TNF-α, CXCL8, and IL-10 also exhibited mRNA profiles where F-HIV gave rise to a higher response than C-HIV and CI-HIV (Table I). Additionally, all cytokines except CXCL8 also exhibited this profile on protein level (Fig. 1C). Quantitative proteomics demonstrated significant upregulation of several inflammatory factors, for example, CD40, TRAF1, and TNFAIP2 for iDCs exposed to F-HIV but not C-HIV and CI-HIV at 24 h after exposure (Table II). The inflammatory response profile was similar even when virus amounts were titrated down to an MOI of 0.08 (Supplemental Fig. 1A) and confirmed for different strains of HIV-1 (Supplemental Fig. 1B). Additionally, we investigated whether the purity of the viral preparation affected the results by adding preparations containing only microvesicles and other virus production “byproducts” to the virus preparations prior to opsonization and found that the responses were not affected by virus preparation purity (Supplemental Fig. 1C).
FIGURE 1.
Complement opsonization of HIV-1 results in reduced inflammatory responses in DCs compared with F-HIV. DCs (1 × 106/ml) were exposed to an MOI of 8 of F-HIV, C-HIV, or CI-HIV or were mock treated and evaluated for expression of IL-1β (A) and IL-6 (B) by lysing the cells and performing qPCR. Concentrations of IL-1β, IL-6, CXCL8, IL-12, and TNF-α were also measured in supernatants collected from DC cultures 24 h after exposure using a flow Cytometric Bead Array (C). qPCR values have been normalized with the mock value set to 1. Data are shown as means ± SEM of 4–12 independent experiments. *p < 0.05 compared with F-HIV.
Table I. mRNA expression of inflammatory, antiviral, and innate signaling genes in DCs exposed to F-HIV, C-HIV, and CI-HIV.
| Gene |
Peak (h) |
F-HIV |
C-HIV |
CI-HIV |
|---|---|---|---|---|
| Inflammatory genes | ||||
| IL-6 | 6 | 220* | 3.0† | 2.0† |
| TNF-α | 6 | 37* | 9.0† | 9.0† |
| CCL3 | 12 | 38* | 10† | 7.0† |
| IL-10 | 24 | 3.0 | 2.0 | 2.0 |
| Antiviral genes | ||||
| IFIT3 | 6 | 8.6* | 1.3† | 1.4† |
| APOBEC3A | 6 | 7.3* | 0.9† | 0.7† |
| APOBEC3B | 24 | 1.4 | 0.9 | 1.2 |
| APOBEC3G | 24 | 1.3 | 0.8 | 0.9 |
| Tetherin | 24 | 2.4* | 2.9* | 2.0* |
| TRIM5α | 48 | 0.7 | 1.2 | 1.4 |
| TRIM22 | 48 | 0.7 | 1.5 | 1.5 |
| Innate signaling genes | ||||
| IRF5 | — | 0.8 | 1.1 | 1.3 |
| IRF9 | — | 0.8 | 0.6 | 0.7 |
| TLR8 | 48 | 1.3 | 1.6 | 1.3 |
| MyD88 | 48 | 2.2* | 3.3* | 3.3* |
| NOD2 | 48 | 1.2 | 1.4 | 1.7 |
| MAVS | 48 | 1.3 | 2.1 | 2.3 |
DCs (1 × 106/ml) were exposed to an MOI of 8 of F-HIV, C-HIV, or CI-HIV or were mock treated for 3, 6, 12, 24, or 48 h and expression of genes associated with inflammatory and antiviral responses and innate signaling was determined by qPCR. Values have been normalized with mock set at 1 and are shown as the mean values of three to six independent experiments.
*p < 0.05, upregulation compared with mock, †p < 0.05 compared with F-HIV.
–, no significant difference in expression between time points.
Table II. Protein expression of inflammatory and antiviral factors in DCs 24 h after exposure to F-HIV, C-HIV, or CI-HIV.
| F-HIV |
C-HIV |
CI-HIV |
||
|---|---|---|---|---|
| Inflammatory factors | ||||
| CCL5 | C-C motif chemokine 5 | 2.40* | 1.40 | 1.50 |
| CCL17 | C-C motif chemokine 17 | 2.60* | 1.63 | 2.10 |
| CCL22 | C-C motif chemokine 22 | 1.73 | 1.33 | 1.83 |
| EBI3 | IL-27 subunit β | 4.33* | 3.93 | 4.17 |
| CD40 | TNFR superfamily member 5 | 2.27 | 1.63 | 1.63 |
| TRAF1 | TNFR-associated factor 1 | 5.40* | 3.87 | 4.07 |
| TNFAIP2 | TNF-α–induced protein 2 | 3.83* | 2.50 | 2.13 |
| NFKB1 | NF-κB p105 subunit | 2.27* | 2.07* | 2.10* |
| NFKB2 | NF-κB p100 subunit | 2.33* | 2.07* | 2.09* |
| Antiviral factors | ||||
| MX1 | IFN-induced GTP-binding protein Mx1 | 7.83* | 4.50 | 3.40 |
| MX2 | IFN-induced GTP-binding protein Mx2 | 4.83* | 2.90 | 2.20 |
| GBP1 | IFN-induced guanylate-binding protein 1 | 2.83* | 1.90 | 1.43 |
| GBP2 | IFN-induced guanylate-binding protein 2 | 1.8* | 1.43 | 1.23 |
| IFIT1 | IFN-induced protein with tetratricopeptide repeats 1 | 5.77* | 2.97 | 2.50 |
| IFIT2 | IFN-induced protein with tetratricopeptide repeats 2 | 6.07 | 3.90 | 2.97 |
| IFIT3 | IFN-induced protein with tetratricopeptide repeats 3 | 7.33* | 4.47 | 3.40 |
| IFIT4 | IFN-induced protein with tetratricopeptide repeats 4 | 2.65 | 1.80 | 1.55 |
| PKR | IFN-induced, dsRNA-activated protein kinase | 3.17 | 1.70 | 1.57 |
| IFIH1 | Isoform 1 of IFN-induced helicase C domain–containing protein 1 | 3.97* | 2.43 | 1.90 |
| ISG15 | Ubiquitin-like protein ISG15 | 15.73* | 8.9 | 6.00 |
DCs (1 × 106/ml) were exposed to an MOI of 8 of F-HIV, C-HIV, or CI-HIV or were mock treated for 24 h and expression of proteins associated with inflammatory and antiviral responses was determined by mass spectroscopy proteomics. Values have been normalized with mock set at 1 and are shown as the mean fold increase compared with mock of three independent experiments.
*p < 0.05 upregulation compared with mock.
Complement opsonization of HIV-1 decreased antiviral responses and enhanced infection of iDCs
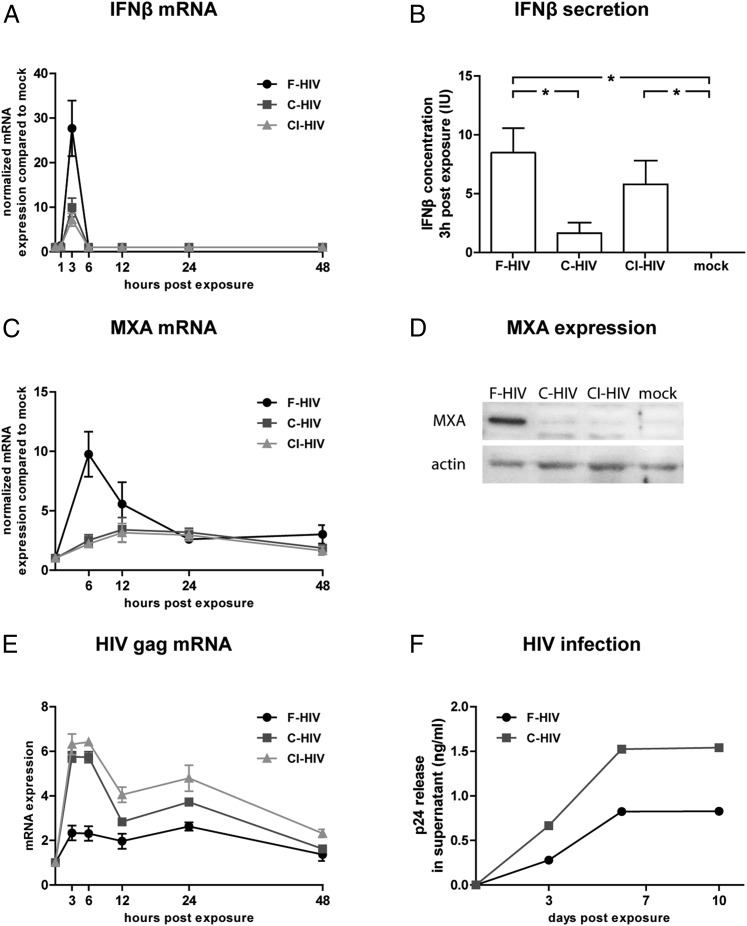
IFN-β mRNA and protein expression levels were significantly upregulated at 3 h in all virus-exposed groups, with F-HIV giving rise to a significantly greater response than C-HIV and CI-HIV (Fig. 2A, 2B). IFN-β mRNA and protein was only detected at 3 h after exposure. Additionally, we could not detect any expression of IFN-α, IFN-γ, or IFN-λ at any time point (data not shown). mRNA levels of the IFN-stimulated gene (ISG) MxA peaked 6 h after viral exposure, and F-HIV gave rise to significantly higher expression than C-HIV (Fig. 2C). These differences were also seen for the MxA protein levels in iDCs 12 h after exposure, which when assessed with Western blot gave a clear MxA protein band for F-HIV, whereas C-HIV and CI-HIV elicited no or weak bands (Fig. 2D). mRNA levels of antiviral factors IFIT3 and APOBEC3A also exhibited similar profiles to IFN-β and MxA, with the highest levels induced by F-HIV and suppressed levels in C-HIV or CI-HIV, whereas no considerable effect on APOBEC3B, APOBEC3G, tetherin (BSTA-2), TRIM5, or TRIM22 mRNA levels were observed in iDCs for any HIV-exposed groups (Table I). Quantitative proteomics of iDCs exposed to F-HIV for 24 h demonstrated significant upregulation of several antiviral factors, for example, MxA, MX2, IFIT1, IFIT2, IFIT3, IFIT4, GBP1, GBP2, IFIH1, and ISG15, whereas iDCs exposed to C-HIV and CI-HIV did not (Table II). Previous studies and our own (13–15, 26, 27) have shown that complement opsonization elevates HIV infection of iDCs and mucosa DCs. We found that C-HIV gave rise to higher levels of HIV gag transcript (5) compared with F-HIV (Fig. 2E), indicative of a higher initial infection. We have previously shown that complement opsonization enhanced viral uptake by iDCs but not viral binding at 4°C (17). iDCs were incubated with virus for 3 h at 4°C to obtain equal amounts of bound F-HIV and C-HIV (confirmed by p24 ELISA), and the productive infection was assessed at days 3, 7, and 10 after exposure. C-HIV gave rise to a higher infection (measured as p24 release in supernatant) compared with F-HIV (Fig. 2F). The antiviral response profile was similar even when virus amounts were titrated down to an MOI of 0.08 (Supplemental Fig. 1A) and was confirmed for different strains of HIV-1 (Supplemental Fig. 1B).
FIGURE 2.
Complement opsonization of HIV-1 inhibited antiviral responses and increased infection in DCs. DCs (1 × 106/ml) were exposed to F-HIV, C-HIV, and CI-HIV at an MOI of 8 or mock treated and evaluated for expression of IFN-β (A and B) and MxA (C and D) by lysing the cells and performing qPCR (A and C), measuring cytokine concentration in the supernatants (B), or by Western blot (D). (E) Levels of the early HIV transcript gag were determined by qPCR. (F) DCs were exposed to F-HIV or C-HIV for 3 h at 4°C, washed, and infection was assessed by measuring levels of HIV p24 release in supernatants 3, 7, and 10 d after exposure using ELISA. qPCR values have been normalized with mock values set to 1. Data are shown as means ± SEM (A–C and E) or representative results (D and F) of 3–12 independent experiments. *p < 0.05.
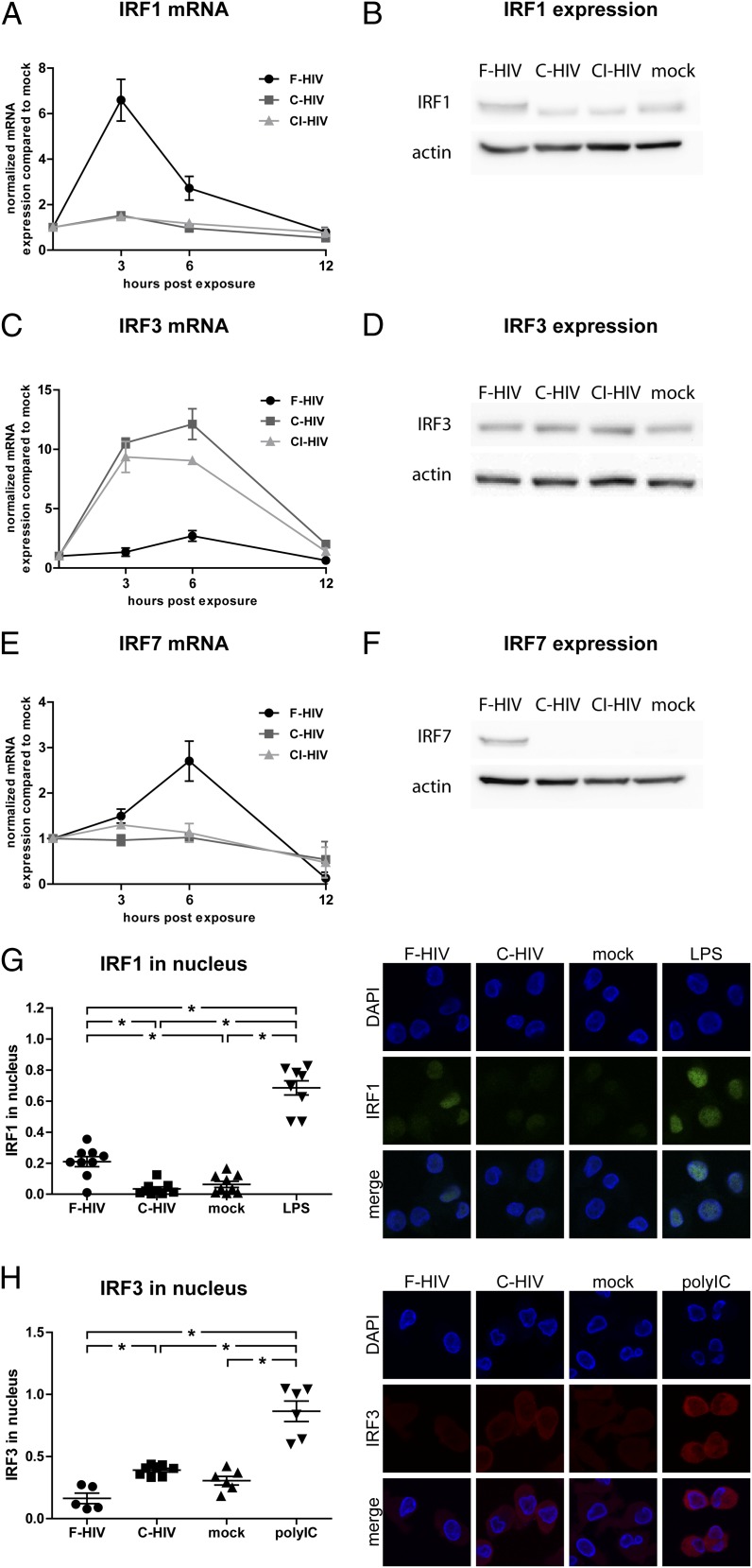
Complement opsonization of HIV-1 suppressed IRF1 and enhanced IRF3 activation
Harman et al. (28) have previously shown that HIV-1 induces IRF1 and IRF7 and that several ISGs in iDCs are activated via IRF1, and therefore we examined the IRFs triggered by F-HIV and C-HIV. Our data confirmed that IRF1 and IRF7 mRNA and protein expression were induced by F-HIV. However, C-HIV and CI-HIV did not activate these transcription factors (Fig. 3A, 3B, 3E, 3F). In contrast, C-HIV and CI-HIV induced mRNA and protein expression of IRF3, whereas F-HIV had less protein expression of this transcription factor than did mock-treated iDCs (Fig. 3C, 3D). Additionally, F-HIV induced nuclear translocation of IRF1, but not IRF3, whereas C-HIV induced nuclear translocation of IRF3 but not IRF1 (Fig. 3G, 3H). We examined additional IRFs and factors involved in signaling and found no major differences in mRNA levels of IRF5, IRF9, TLR8, MyD88, NOD2, or MAVS in the HIV-exposed groups (Table I).
FIGURE 3.
Complement opsonization of HIV-1 suppressed IRF1 and IRF7 but activated IRF3 signaling. DCs (1 × 106/ml) were exposed to F-HIV, C-HIV, and CI-HIV at an MOI of 8 or were mock treated. mRNA and protein expression of IRF1 (A and B), IRF3 (C and D), and IRF7 (E and F) was assessed using qPCR (A, C, and E) and Western blot (B, D, and F). DCs were exposed to F-HIV, C-HIV, or mock treated and the fraction of IRF1 (G) and IRF7 (H) in the cell nuclei was quantified using a confocal microscope with LPS and polyinosinic-polycytidylic acid as positive controls. Images show representative cells stained with DAPI (nuclei, blue) and IRF1 (green) or IRF3 (red) (original magnification ×40). qPCR values have been normalized with the mock value set to 1. Data are shown as means ± SEM for graphs and as representative results for confocal and Western blot images of three to six independent experiments. *p < 0.05.
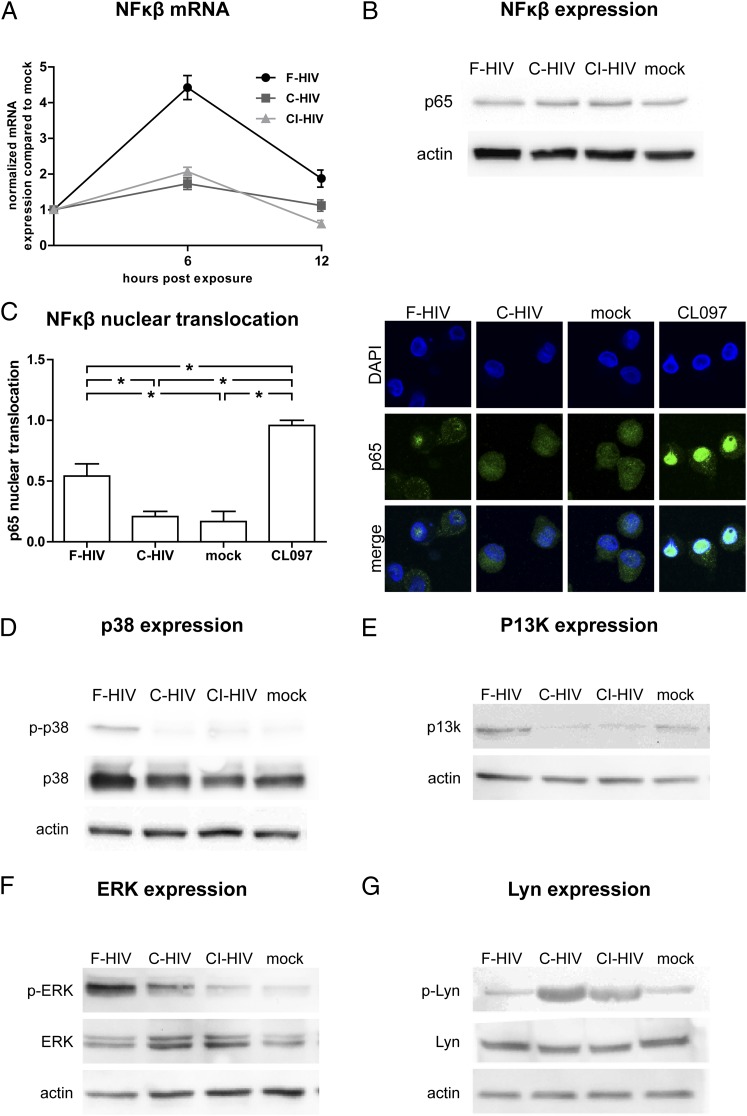
Complement opsonization of HIV-1 suppressed phosphorylation of ERK and p38 and nuclear translocation of NF-κB and induced phosphorylation of Lyn
Besides the IRFs, multiple signaling events and factors are involved in the induction of antiviral and inflammatory responses in DCs and in their regulation (29, 30), and we therefore examined the effects HIV had on some of these molecules. We found that F-HIV increased the expression of NF-κB mRNA and activated NF-κB p65 nuclear translocation whereas C-HIV and CI-HIV did not (Fig. 4A–C). F-HIV induced phosphorylation of the MAPKs ERK and p38 and increased the protein expression of PI3K, whereas C-HIV and CI-HIV had no or low effects (Fig. 4D–F). In contrast, C-HIV and CI-HIV, but not F-HIV, induced the phosphorylation of the tyrosine protein kinase Lyn (Fig. 4G).
FIGURE 4.
Complement opsonization of HIV-1 suppressed NF-κβ, p38, and ERK signaling but activated Lyn. DCs (1 × 106/ml) were exposed to F-HIV, C-HIV, and CI-HIV at an MOI of 8 or were mock treated. NF-κβ mRNA expression was measured by qPCR (A), NF-κβ p65 subunit protein expression was measured using Western blot (B), and translocation of p65 (green) into the nucleus (blue) was evaluated using a confocal microscope with TLR8 agonist CL097 as a positive control (original magnification ×40) (C). Protein expression and phosphorylation of p38 (D), PI3K (E), ERK (F), and Lyn (G) were examined using Western blot. qPCR values have been normalized with the mock value set to 1. Data are shown as means ± SEM for graphs and as representative results for confocal and Western blot images of three to six independent experiments. *p < 0.05.
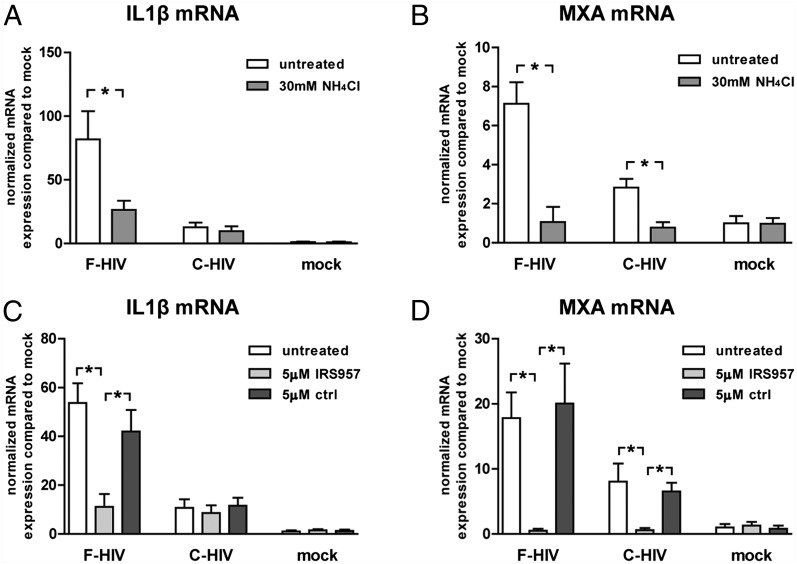
TLR8 signaling was required for inflammatory and antiviral responses in iDCs
In the light of previous studies that demonstrated that TLR8 recognizes HIV-derived pathogen-associated molecular patterns in human DCs (5) and that NOD2 can recognize ssRNA (31), we examined the role of these PRRs in the HIV-1–induced inflammatory and antiviral responses. Endosomal acidification is a necessity for activation of endosomal TLRs, and neutralization of acidification blocks proteolysis of the endosomal cargo, such as viruses, and subsequently the TLR8 activation (32, 33). To examine the role of TLR8 in our system we used the weak base NH4Cl to inhibit acidification of the endosomal compartment. Pretreatment of iDCs with NH4Cl led to failure of HIV to activate inflammatory and antiviral responses (Fig. 5A, 5B). The TLR8 inhibitory oligonucleotide IRS957 gave the same outcome as NH4Cl with decreased inflammatory and antiviral responses (Fig. 5C, 5D), and the same profile was seen when the TLR8 agonist CL097 was used instead of free HIV (Supplemental Fig. 2). The neutralizing mAb b12 or fusion inhibitor C34 had no effect on responses induced by either F-HIV or C-HIV (Supplemental Fig. 3), excluding the involvement of cytosolic PRRs such as NOD2 in our system.
FIGURE 5.
Complement opsonization of HIV-1 suppressed TLR8 activation in DCs. DCs (1 × 106/ml) were pretreated with the weak base NH4Cl, the TLR8 specific inhibitory oligonucleotide IRS957, or control before exposure to an MOI of 8 for F-HIV or C-HIV or were mock treated for 6 h. mRNA levels of IL-1β and MxA for DCs pretreated with NH4Cl (A and B) or IRS957 (C and D) were assessed by qPCR. Values have been normalized with mock values set to 1, and data are shown as means ± SEM of three to eight independent experiments. *p < 0.05.
Inhibition of inflammatory and antiviral responses in iDCs by C-HIV was mediated by CR3 activation
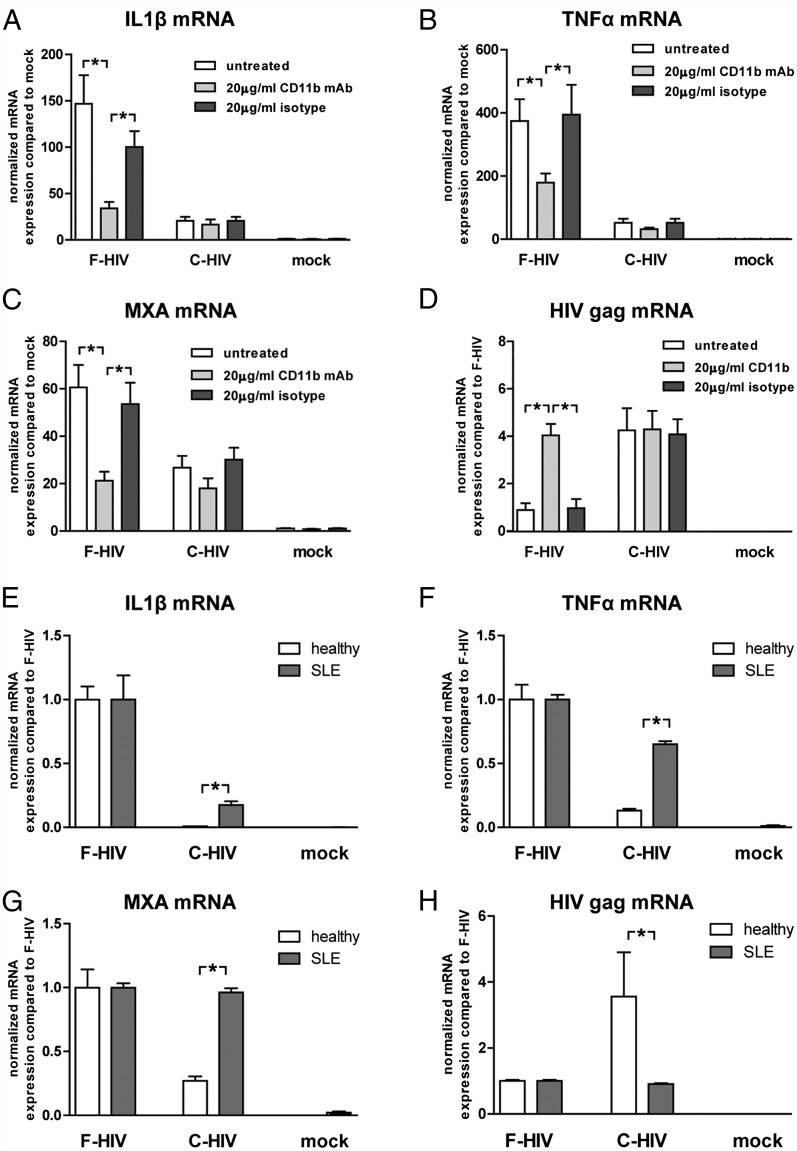
We examined the role of CR3 in the immune suppression induced by C-HIV, as this receptor has been proven to impair responses against other pathogens (34, 35). Targeting CD11b, the α-integrin chain of CR3, with an activating Ab before adding F-HIV gave the same decreased immune response as seen for C-HIV, decreasing activation of both the inflammasome, as measured by IL-1β and TNF-α, and the antiviral responses, as measured by MxA (Fig. 6A–C). Additionally, this also enhanced the level of HIV gag transcripts for F-HIV to the level of C-HIV (Fig. 6D). To further examine the role of CD11b in the CR3-mediated suppression of responses, we used DCs propagated from blood derived from patients diagnosed with SLE, with the rs1143697 (R77H) mutation in CD11b. This mutation can result in decreased expression of CD11b on APCs and has been shown to impair the repression of inflammatory responses when CR3 is targeted by iC3b-coated RBCs (36, 37). We found that complement opsonization of HIV suppressed IL-1β and TNF-α mRNA expression less efficiently in DCs derived from SLE patients (Fig. 6E, 6F). Additionally, MxA and HIV gag transcript levels in these cells were not affected by complement opsonization of the virus (Fig. 6G, 6H). To verify the role of CR3 using other factors, we used the CR3-activating mAb KIM185 (38), which binds to and activates CR3 via the β-chain CD18, and NIF, which binds to and activates CR3 via the CD11b I domain (39). The KIM185 mAb and NIF gave a similar suppressive effect on IL-1β induction by F-HIV as the CD11b-activating mAb (KIM75) (Supplemental Fig. 4A, 4B), whereas they had no effect or very little effect on the antiviral MxA responses (Supplemental Fig. 4A, 4B). We assessed the effects that blocking complement activation by heat inactivation of the fresh serum had on HIV-1–induced activation of iDCs. HIV opsonized with heat-inactivated serum gave similar inflammatory and antiviral responses and HIV gag transcript levels as the free virions (Supplemental Fig. 4C, 4D). To further verify the role of iC3b in the CR3 activation, HIV was opsonized using C3-depleted serum or C9-depleted serum as control. The opsonization in control C9-depleted serum inhibited inflammatory and antiviral responses, whereas HIV opsonized using C3 depleted serum did not (Supplemental Fig. 4F).
FIGURE 6.
Complement opsonization of HIV-1 suppressed inflammation and antiviral responses in DCs through CR3 activation. DCs (1 × 106/ml) were pretreated with an activating CD11b Ab or isotype control mAb before exposure to an MOI of 8 of F-HIV or C-HIV or were mock treated for 6 h and mRNA levels of IL-1β, TNF-α, MxA, and HIV gag transcript (A–D) were assessed by qPCR. DCs derived from healthy individuals or from SLE patients with impairing mutation in CD11b (rs1143679:RH77) were exposed to an MOI of 8 of F-HIV or C-HIV or were mock treated for 6 h and mRNA levels for IL-1β, TNF-α, MxA, and HIV gag transcript (E-H) were measured using qPCR. Values have been normalized with mock or F-HIV values set to 1 and data are shown as means ± SEM of three to eight independent experiments. *p < 0.05.
Activating CR3 with NIF or KIM185 prior to activating the TLR8 signaling pathway using the TLR8 agonist CL097 suppressed inflammatory responses in the DCs, whereas there were no effects on the antiviral responses (Supplemental Fig. 2).
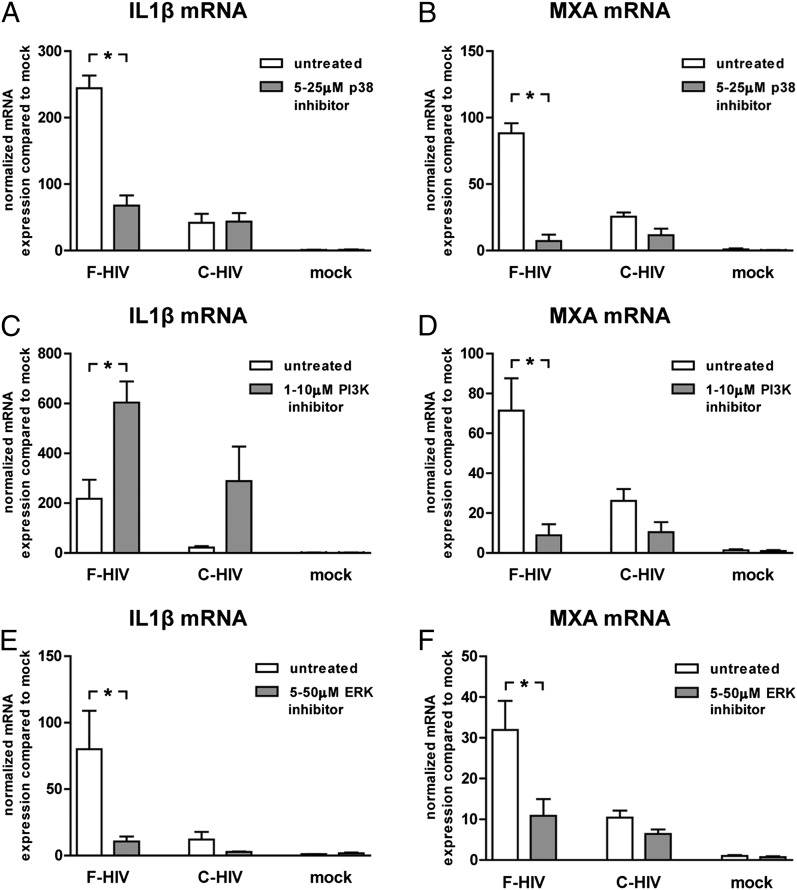
Inhibition of ERK and p38 decreased inflammation and antiviral responses, whereas PI3K inhibition increased inflammation but decreased antiviral responses in iDCs
We blocked different signaling pathways implicated in the HIV-1–induced activation of inflammatory and antiviral responses. Block of MAPK p38 decreased the inflammatory and antiviral responses induced by F-HIV in iDCs, but had no significant effect on C-HIV (Fig. 7A, 7B). PI3K inhibition elevated the inflammatory responses induced by both F-HIV and C-HIV (Fig. 7C), whereas it decreased the antiviral responses induced by F-HIV (Fig. 7D). Additionally, the block of PI3K decreased the level of early gag transcripts induced by C-HIV (Supplemental Fig. 4E). ERK block highly suppressed the inflammatory and antiviral responses induced by F-HIV (Fig. 7E, 7F).
FIGURE 7.
Inhibition of ERK and p38 decreased HIV-1 induced inflammation and antiviral responses, whereas PI3K inhibition increased inflammation but decreased antiviral responses in DCs. DCs (1 × 106/ml) were preincubated with inhibitors for p38 (SB203580) (A and B), PI3K (LY294002) (C and D), or ERK (OU126) (E and F) prior to exposure to F-HIV or C-HIV at an MOI of 8 or were mock treated for 6 h. mRNA expression of IL-1β and MxA was determined by qPCR. Values have been normalized with mock values set at 1 and data are shown as means ± SEM of four to six independent experiments. *p < 0.05.
Discussion
The fact that the engagement of CR3 on DCs by C-HIV promotes viral infection of the DCs and aids subsequent infection of CD4 T cells has been known for some time, but the mechanistic events behind the enhanced infection have remained unknown (13–15, 26, 27). Our study suggests that the difference in replication efficiency between free and complement-opsonized virus depends on the reduced antiviral response and proinflammatory cytokine production and altered signaling profile induced by complement opsonization of HIV-1, and this could play an important role in virus pathogenesis.
Serum components, especially C3 and its fragments, deposit on bacterial and viral surfaces and interact with different CRs, ensuring an efficient phagocytosis of the pathogens by macrophages and DCs (17, 40). CR3 has many different ligand binding sites, with the primary domain on CD11b, the α-chain of CR3, responsible for recognition of, for example, iC3b, ICAM1, and fibrinogen (40). In the the case for C-HIV, it has been shown that iC3b/CR3 interaction enhances HIV infection in iDCs (13–15, 35). We found that targeting CD11b, with C-HIV or CD11b-activating Ab KIM75 led to both suppressed inflammatory and antiviral responses as well as an increase in the production of early viral transcripts. Of note, the KIM75 Ab has a limited impact on the amount of virus that binds to the DCs (17), which indicates that the increase in early transcript production is a result of enhanced infection, rather than a reflection of the amount of virus taken up by the cells.
NIF, an activator of CR3, only affected the inflammatory responses, which could be due to the different ligand binding sites for NIF and iC3b on CR3 (39). When we used DCs derived from individuals with SLE with mutation in the CD11b that impairs this integrin’s function (36, 37), complement opsonization of HIV no longer suppressed the antiviral responses and the inflammatory responses were partly restored. Taken together, our results indicate that the suppression of responses achieved by C-HIV is due to binding and signaling via CR3 and not routing of virions to compartments void of PRRs.
DCs are equipped to distinguish between different kinds and densities of complement proteins. For instance, apoptotic cells in the body are opsonized with iC3b, facilitating their removal by internalization via CR3/CR4 (41, 42) while avoiding induction of inflammation and promoting self-tolerance (40, 43, 44). The similarity in opsonization pattern between HIV-1 and apoptotic cells may be a mechanism used by the virus to avoid immune activation and may therefore be one reason for the failure to initiate adequate inflammatory and antiviral responses as well as the poor humoral immune response against this pathogen seen in vivo.
The TLR8 signaling cascade activates several signaling pathways, that is, IRFs, NF-κB, and MAPKs, including ERK, JNK, and p38 (29), leading to inflammatory and antiviral responses. The secretion of the proinflammatory factor IL-1β requires instigation of the inflammasome by an activator. We found that F-HIV but not C-HIV induced the secretion of IL-1β in iDCs and PKR, recently identified as activator of the inflammasome, was upregulated by F-HIV, indicating that it could be the activator of the NALP3 inflammasome (45, 46).
All investigated cytokines showed a markedly higher mRNA expression in DCs exposed to free virus compared with when the virus was complement opsonized. Whereas IL-1β, IL-6, and TNF-α retained this profile at the protein level, no significant difference in CXCL8 protein levels could be detected between the virus-exposed groups, possibly due to a negative feedback loop such as microRNA-146 limiting its secretion (47).
Activation of MAPKs is an early event in the activation of TLR signaling and we found, in accordance with previous studies by others, that free HIV induced phosphorylation of the MAPKs p38 and ERK in iDCs (48, 49). However, these responses were more limited for the complement-opsonized virus, recalling previous observations with C3-opsonized Francisella tularensis (34). Harman et al. (28) have previously shown that both IRF1 and IRF7 are upregulated in HIV-infected p24+ DCs and that HIV induces activation of ISGs in an IFN type I–independent manner via IRF1. Additionally, ISGs and IRF7 have been shown to be activated by HIV Tat in APCs (50). Our study confirms these findings, with TLR8 signaling triggered by free HIV leading to upregulation of IRF1 and IRF7. We could also detect an early but transient IFN-β production in DCs, which was not seen by Harman et al. (28). Of note, the neutralization of this small amount of IFN-β produced by the HIV-exposed DCs did not affect the antiviral response and expression of ISGs (data not shown), which supports an alternative mechanism to the type I IFN induction of the antiviral responses involving activation via IRF1 (28). Free HIV did not affect IRF3, in accordance with previous studies (28). When HIV was complement opsonized, however, IRF1 and IRF7 were suppressed, and instead IRF3 was activated. IRF3 signaling normally has a fundamental role in type I IFN and ISG expression and can limit viral replication. Phosphorylation of constitutively expressed IRF3 is usually activated by upstream enzymes such as TBK1 and Ikkε (51). Degradation of IRF3 is required for a productive infection in T cells (52), but which role IRF3 plays in the enhanced infection of DCs induced by C-HIV remains to be determined.
More and more evidence points to a cross-talk between CR3 and TLR pathways with the TLR involved in the inside–out activation of CR3 and CR3 regulating TLR activation (53). So far, cross-talk between CR3 and TLR2, TLR4, and TLR9 has been described (34, 54, 55). In human cells such as monocytes, it has been shown that CR3 ligand binding capacity is activated by TLR2 inside–out signaling, and the cross-talk between these receptors might involve Rac1, PI3K, and cytohesin-1 (56). In our setting, with the involvement of TLR8 and CR3, we saw a similar regulatory role of PI3K as previously described for other TLRs, as blocking this pathway gave rise to elevated inflammation both for F-HIV and C-HIV (57). PI3K signaling is activated by multiple receptors, intersects different pathways, and is important for the induction and the regulation of both TLR- and CR3-mediated responses. In human macrophages, C3-opsonized F. tularensis induces signaling via TLR2 and CR3, with the CD11b signaling occurring via Lyn kinase, PI3K, and inhibitor of MAPK phosphorylation MKP-1 (34). This pathway seems to fit our system where activation of CR3 by C-HIV led to Lyn phosphorylation, and inhibition of PI3K led to enhanced inflammation and suppressed infection. The Lyn/PI3K pathway’s suppressive role in inflammatory responses induced by TLR stimuli has been documented in different systems (34, 54), and the role of Lyn in signaling events downstream of TLRs has been studied in DCs (58–60). Dai et al. (34) established that PI3K can be activated by both the TLR2 and CR3 pathways in human macrophages (34), which indicates that this kinase may be one link between the TLR8 and CR3 signaling pathways in DCs. The importance of Lyn as a regulator of immune activation is clear from the hyperactive DCs found in Lyn−/− mice, which secrete high levels of cytokines associated with autoimmunity (61, 62).
Myeloid DCs are normally not easily infected by HIV-1 and productive infection has been shown to exploit the TLR8 signaling cascade activated by the virus (5). The activation of the transcription factors IRF1, and IRF7 by free HIV via the TLR8 pathway, might lead to binding to their ISRE site in the HIV LTR regions, which exist in most HIV-1 strains and are known to promote an efficient infection (63). Our findings clearly show that simultaneously targeting TLR8 and CR3 enhanced the ability of HIV to replicate, and that this occurred when IRF1 and IRF7 activation was not induced but IRF3 was active, which suggests that IRF3 binds to an ISRE site in the HIV LTR and gives rise to enhanced viral replication while avoiding activation of antiviral responses in the iDCs.
The complement system is vital for the regulation of tolerance, and complement factors such as C3 have been shown to be pivotal in transplant settings (64). In this study we show that viral hijacking of the complement system leads to modulation of TLR8-mediated responses via CR3 (Fig. 8) and the impact this has on the function of DCs could be one important immune evasion mechanism used by HIV-1 in vivo, allowing it to act almost unnoticed by the immune system and providing a window of time to establish infection.
FIGURE 8.
Proposed signaling pathways in DCs exposed to F-HIV and C-HIV. F-HIV triggers TLR8 signaling and the activation of p38, ERK, and NF-κB pathways, leading to the production of inflammatory cytokines. TLR8 activation by F-HIV also leads to the activation of IRF1 and the production of IRF7 and antiviral factors/ISGs. C-HIV engages CR3, leading to CR3/TLR8 cross-talk involving Lyn and PI3K. This suppresses the inflammatory and antiviral responses and induces IRF3 activation, leading to enhanced viral transcription.
Supplementary Material
Acknowledgments
We thank the Biological Products Core of the AIDS and Cancer Virus Program, Leidos Biomecial Research, Inc., Frederick National Laboratory (Frederick, MD) for providing infectious and inactivated HIV-1, and Prof. T. Geijtenbeek for the early gag transcript protocol. We also thank Max Abou and Stuart McCorrister of the National Microbiology Laborabory for the technical support.
This work was supported by grants from the Swedish Research Council, the Swedish Physicians against AIDS Research Foundation, the Swedish International Development Cooperation Agency, the Swedish International Development Cooperation Agency Special Assistant to the Resident Coordinator, VINNMER for Vinnova, and Linköping University Hospital Research Fund Grant C-ALF (all to M.L.). This work was also supported in part by the Swedish Society of Medicine (to M.L.) with federal funds from the National Cancer Institute, National Institutes of Health Contract HHSN261200800001E (to J.D.L.) and the Swedish Society for Medical Research (to C.S.).
The online version of this article contains supplemental material.
- C-HIV
- complement-opsonized HIV-1
- CI-HIV
- complement and Ab–opsonized HIV-1
- CR
- complement receptor
- DC
- dendritic cell
- F-HIV
- free HIV-1
- iDC
- immature dendritic cell
- IRF
- IFN regulatory factor
- ISG
- IFN-stimulated gene
- MOI
- multiplicity of infection
- MxA
- myxovirus resistance protein A
- NIF
- neutrophil inhibitory factor
- PRR
- pattern recognition receptor
- qPCR
- real-time quantitative PCR
- SLE
- systemic lupus erythematosus.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Steinman R. M. 2007. Lasker Basic Medical Research Award. Dendritic cells: versatile controllers of the immune system. Nat. Med. 13: 1155–1159 [DOI] [PubMed] [Google Scholar]
- 2.Hladik F., McElrath M. J.. 2008. Setting the stage: host invasion by HIV. Nat. Rev. Immunol. 8: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lubong Sabado R., Kavanagh D. G., Kaufmann D. E., Fru K., Babcock E., Rosenberg E., Walker B., Lifson J., Bhardwaj N., Larsson M.. 2009. In vitro priming recapitulates in vivo HIV-1 specific T cell responses, revealing rapid loss of virus reactive CD4 T cells in acute HIV-1 infection. PLoS ONE 4: e4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sewell A. K., Price D. A.. 2001. Dendritic cells and transmission of HIV-1. Trends Immunol. 22: 173–175 [DOI] [PubMed] [Google Scholar]
- 5.Gringhuis S. I., van der Vlist M., van den Berg L. M., den Dunnen J., Litjens M., Geijtenbeek T. B.. 2010. HIV-1 exploits innate signaling by TLR8 and DC-SIGN for productive infection of dendritic cells. Nat. Immunol. 11: 419–426 [DOI] [PubMed] [Google Scholar]
- 6.Carroll M. C. 2004. The complement system in regulation of adaptive immunity. Nat. Immunol. 5: 981–986 [DOI] [PubMed] [Google Scholar]
- 7.van Montfoort N., de Jong J. M., Schuurhuis D. H., van der Voort E. I., Camps M. G., Huizinga T. W., van Kooten C., Daha M. R., Verbeek J. S., Ossendorp F., Toes R. E.. 2007. A novel role of complement factor C1q in augmenting the presentation of antigen captured in immune complexes to CD8+ T lymphocytes. J. Immunol. 178: 7581–7586 [DOI] [PubMed] [Google Scholar]
- 8.Ochsenbein A. F., Pinschewer D. D., Odermatt B., Carroll M. C., Hengartner H., Zinkernagel R. M.. 1999. Protective T cell-independent antiviral antibody responses are dependent on complement. J. Exp. Med. 190: 1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Favoreel H. W., Van de Walle G. R., Nauwynck H. J., Pensaert M. B.. 2003. Virus complement evasion strategies. J. Gen. Virol. 84: 1–15 [DOI] [PubMed] [Google Scholar]
- 10.Stoiber H., Banki Z., Wilflingseder D., Dierich M. P.. 2008. Complement-HIV interactions during all steps of viral pathogenesis. Vaccine 26: 3046–3054 [DOI] [PubMed] [Google Scholar]
- 11.Stoiber H., Pruenster M., Ammann C. G., Dierich M. P.. 2005. Complement-opsonized HIV: the free rider on its way to infection. Mol. Immunol. 42: 153–160 [DOI] [PubMed] [Google Scholar]
- 12.Saifuddin M., Parker C. J., Peeples M. E., Gorny M. K., Zolla-Pazner S., Ghassemi M., Rooney I. A., Atkinson J. P., Spear G. T.. 1995. Role of virion-associated glycosylphosphatidylinositol-linked proteins CD55 and CD59 in complement resistance of cell line-derived and primary isolates of HIV-1. J. Exp. Med. 182: 501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bajtay Z., Speth C., Erdei A., Dierich M. P.. 2004. Cutting edge: productive HIV-1 infection of dendritic cells via complement receptor type 3 (CR3, CD11b/CD18). J. Immunol. 173: 4775–4778 [DOI] [PubMed] [Google Scholar]
- 14.Bouhlal H., Chomont N., Réquena M., Nasreddine N., Saidi H., Legoff J., Kazatchkine M. D., Bélec L., Hocini H.. 2007. Opsonization of HIV with complement enhances infection of dendritic cells and viral transfer to CD4 T cells in a CR3 and DC-SIGN-dependent manner. J. Immunol. 178: 1086–1095 [DOI] [PubMed] [Google Scholar]
- 15.Pruenster M., Wilflingseder D., Bánki Z., Ammann C. G., Muellauer B., Meyer M., Speth C., Dierich M. P., Stoiber H.. 2005. C-type lectin-independent interaction of complement opsonized HIV with monocyte-derived dendritic cells. Eur. J. Immunol. 35: 2691–2698 [DOI] [PubMed] [Google Scholar]
- 16.Tjomsland V., Ellegård R., Burgener A., Mogk K., Che K. F., Westmacott G., Hinkula J., Lifson J. D., Larsson M.. 2013. Complement opsonization of HIV-1 results in a different intracellular processing pattern and enhanced MHC class I presentation by dendritic cells. Eur. J. Immunol. 43: 1470–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tjomsland V., Ellegård R., Che K., Hinkula J., Lifson J. D., Larsson M.. 2011. Complement opsonization of HIV-1 enhances the uptake by dendritic cells and involves the endocytic lectin and integrin receptor families. PLoS ONE 6: e23542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan E. M., Cohen A. S., Fries J. F., Masi A. T., McShane D. J., Rothfield N. F., Schaller J. G., Talal N., Winchester R. J.. 1982. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 25: 1271–1277 [DOI] [PubMed] [Google Scholar]
- 19.Enocsson H., Wetterö J., Skogh T., Sjöwall C.. 2013. Soluble urokinase plasminogen activator receptor levels reflect organ damage in systemic lupus erythematosus. Transl. Res. 162: 287–296 [DOI] [PubMed] [Google Scholar]
- 20.Sabado R. L., Babcock E., Kavanagh D. G., Tjomsland V., Walker B. D., Lifson J. D., Bhardwaj N., Larsson M.. 2007. Pathways utilized by dendritic cells for binding, uptake, processing and presentation of antigens derived from HIV-1. Eur. J. Immunol. 37: 1752–1763 [DOI] [PubMed] [Google Scholar]
- 21.Rossio J. L., Esser M. T., Suryanarayana K., Schneider D. K., Bess J. W., Jr., Vasquez G. M., Wiltrout T. A., Chertova E., Grimes M. K., Sattentau Q., et al. 1998. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 72: 7992–8001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F.. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rieu I., Powers S. J.. 2009. Real-time quantitative RT-PCR: design, calculations, and statistics. Plant Cell 21: 1031–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burgener A., Mogk K., Westmacott G., Plummer F., Ball B., Broliden K., Hasselrot K.. 2012. Salivary basic proline-rich proteins are elevated in HIV-exposed seronegative men who have sex with men. AIDS 26: 1857–1867 [DOI] [PubMed] [Google Scholar]
- 25.Elkord E., Williams P. E., Kynaston H., Rowbottom A. W.. 2005. Human monocyte isolation methods influence cytokine production from in vitro generated dendritic cells. Immunology 114: 204–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilflingseder D., Banki Z., Garcia E., Pruenster M., Pfister G., Muellauer B., Nikolic D. S., Gassner C., Ammann C. G., Dierich M. P., et al. 2007. IgG opsonization of HIV impedes provirus formation in and infection of dendritic cells and subsequent long-term transfer to T cells. J. Immunol. 178: 7840–7848 [DOI] [PubMed] [Google Scholar]
- 27.Tjomsland V., Ellegård R., Kjölhede P., Wodlin N. B., Hinkula J., Lifson J. D., Larsson M.. 2013. Blocking of integrins inhibits HIV-1 infection of human cervical mucosa immune cells with free and complement-opsonized virions. Eur. J. Immunol. 43: 2361–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harman A. N., Lai J., Turville S., Samarajiwa S., Gray L., Marsden V., Mercier S. K., Jones K., Nasr N., Rustagi A., et al. 2011. HIV infection of dendritic cells subverts the IFN induction pathway via IRF-1 and inhibits type 1 IFN production. Blood 118: 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown J., Wang H., Hajishengallis G. N., Martin M.. 2011. TLR-signaling networks: an integration of adaptor molecules, kinases, and cross-talk. J. Dent. Res. 90: 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weichhart T., Säemann M. D.. 2008. The PI3K/Akt/mTOR pathway in innate immune cells: emerging therapeutic applications. Ann. Rheum. Dis. 67(Suppl. 3): iii70–iii74 [DOI] [PubMed] [Google Scholar]
- 31.Sabbah A., Chang T. H., Harnack R., Frohlich V., Tominaga K., Dube P. H., Xiang Y., Bose S.. 2009. Activation of innate immune antiviral responses by Nod2. Nat. Immunol. 10: 1073–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uematsu S., Akira S.. 2007. Toll-like receptors and Type I interferons. J. Biol. Chem. 282: 15319–15323 [DOI] [PubMed] [Google Scholar]
- 33.Kuznik A., Bencina M., Svajger U., Jeras M., Rozman B., Jerala R.. 2011. Mechanism of endosomal TLR inhibition by antimalarial drugs and imidazoquinolines. J. Immunol. 186: 4794–4804 [DOI] [PubMed] [Google Scholar]
- 34.Dai S., Rajaram M. V., Curry H. M., Leander R., Schlesinger L. S.. 2013. Fine tuning inflammation at the front door: macrophage complement receptor 3-mediates phagocytosis and immune suppression for Francisella tularensis. PLoS Pathog. 9: e1003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hajishengallis G., Shakhatreh M. A., Wang M., Liang S.. 2007. Complement receptor 3 blockade promotes IL-12-mediated clearance of Porphyromonas gingivalis and negates its virulence in vivo. J. Immunol. 179: 2359–2367 [DOI] [PubMed] [Google Scholar]
- 36.Zhou Y., Wu J., Kucik D. F., White N. B., Redden D. T., Szalai A. J., Bullard D. C., Edberg J. C.. 2013. Multiple lupus-associated ITGAM variants alter Mac-1 functions on neutrophils. Arthritis Rheum. 65: 2907–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rhodes B., Fürnrohr B. G., Roberts A. L., Tzircotis G., Schett G., Spector T. D., Vyse T. J.. 2012. The rs1143679 (R77H) lupus associated variant of ITGAM (CD11b) impairs complement receptor 3 mediated functions in human monocytes. Ann. Rheum. Dis. 71: 2028–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gjelstrup L. C., Andersen S. H., Petersen S. V., Enghild J. J., Blom A. M., Vorup-Jensen T., Thiel S.. 2011. The role of higher-order protein structure in supporting binding by heteroclitic monoclonal antibodies: the monoclonal antibody KIM185 to CD18 also binds C4-binding protein. Mol. Immunol. 49: 38–47 [DOI] [PubMed] [Google Scholar]
- 39.Thornton B. P., Vĕtvicka V., Pitman M., Goldman R. C., Ross G. D.. 1996. Analysis of the sugar specificity and molecular location of the beta-glucan-binding lectin site of complement receptor type 3 (CD11b/CD18). J. Immunol. 156: 1235–1246 [PubMed] [Google Scholar]
- 40.Cummings K. L., Waggoner S. N., Tacke R., Hahn Y. S.. 2007. Role of complement in immune regulation and its exploitation by virus. Viral Immunol. 20: 505–524 [DOI] [PubMed] [Google Scholar]
- 41.Sohn J. H., Bora P. S., Suk H. J., Molina H., Kaplan H. J., Bora N. S.. 2003. Tolerance is dependent on complement C3 fragment iC3b binding to antigen-presenting cells. Nat. Med. 9: 206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skoberne M., Somersan S., Almodovar W., Truong T., Petrova K., Henson P. M., Bhardwaj N.. 2006. The apoptotic-cell receptor CR3, but not αvβ5, is a regulator of human dendritic-cell immunostimulatory function. Blood 108: 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amarilyo G., Verbovetski I., Atallah M., Grau A., Wiser G., Gil O., Ben-Neriah Y., Mevorach D.. 2010. iC3b-opsonized apoptotic cells mediate a distinct anti-inflammatory response and transcriptional NF-κB-dependent blockade. Eur. J. Immunol. 40: 699–709 [DOI] [PubMed] [Google Scholar]
- 44.Verbovetski I., Bychkov H., Trahtemberg U., Shapira I., Hareuveni M., Ben-Tal O., Kutikov I., Gill O., Mevorach D.. 2002. Opsonization of apoptotic cells by autologous iC3b facilitates clearance by immature dendritic cells, down-regulates DR and CD86, and up-regulates CC chemokine receptor 7. J. Exp. Med. 196: 1553–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pontillo A., Silva L. T., Oshiro T. M., Finazzo C., Crovella S., Duarte A. J.. 2012. HIV-1 induces NALP3-inflammasome expression and interleukin-1β secretion in dendritic cells from healthy individuals but not from HIV-positive patients. AIDS 26: 11–18 [DOI] [PubMed] [Google Scholar]
- 46.Lu B., Nakamura T., Inouye K., Li J., Tang Y., Lundbäck P., Valdes-Ferrer S. I., Olofsson P. S., Kalb T., Roth J., et al. 2012. Novel role of PKR in inflammasome activation and HMGB1 release. Nature 488: 670–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taganov K. D., Boldin M. P., Chang K. J., Baltimore D.. 2006. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 103: 12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anand A. R., Prasad A., Bradley R. R., Deol Y. S., Nagaraja T., Ren X., Terwilliger E. F., Ganju R. K.. 2009. HIV-1 gp120-induced migration of dendritic cells is regulated by a novel kinase cascade involving Pyk2, p38 MAP kinase, and LSP1. Blood 114: 3588–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilflingseder D., Müllauer B., Schramek H., Banki Z., Pruenster M., Dierich M. P., Stoiber H.. 2004. HIV-1-induced migration of monocyte-derived dendritic cells is associated with differential activation of MAPK pathways. J. Immunol. 173: 7497–7505 [DOI] [PubMed] [Google Scholar]
- 50.Kim N., Kukkonen S., Martinez-Viedma Mdel. P., Gupta S., Aldovini A.. 2013. Tat engagement of p38 MAP kinase and IRF7 pathways leads to activation of interferon-stimulated genes in antigen-presenting cells. Blood 121: 4090–4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hiscott J. 2007. Triggering the innate antiviral response through IRF-3 activation. J. Biol. Chem. 282: 15325–15329 [DOI] [PubMed] [Google Scholar]
- 52.Okumura A., Alce T., Lubyova B., Ezelle H., Strebel K., Pitha P. M.. 2008. HIV-1 accessory proteins VPR and Vif modulate antiviral response by targeting IRF-3 for degradation. Virology 373: 85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hajishengallis G., Lambris J. D.. 2010. Crosstalk pathways between Toll-like receptors and the complement system. Trends Immunol. 31: 154–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keck S., Freudenberg M., Huber M.. 2010. Activation of murine macrophages via TLR2 and TLR4 is negatively regulated by a Lyn/PI3K module and promoted by SHIP1. J. Immunol. 184: 5809–5818 [DOI] [PubMed] [Google Scholar]
- 55.Han C., Jin J., Xu S., Liu H., Li N., Cao X.. 2010. Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nat. Immunol. 11: 734–742 [DOI] [PubMed] [Google Scholar]
- 56.Harokopakis E., Albzreh M. H., Martin M. H., Hajishengallis G.. 2006. TLR2 transmodulates monocyte adhesion and transmigration via Rac1- and PI3K-mediated inside-out signaling in response to Porphyromonas gingivalis fimbriae. J. Immunol. 176: 7645–7656 [DOI] [PubMed] [Google Scholar]
- 57.Fukao T., Koyasu S.. 2003. PI3K and negative regulation of TLR signaling. Trends Immunol. 24: 358–363 [DOI] [PubMed] [Google Scholar]
- 58.Napolitani G., Bortoletto N., Racioppi L., Lanzavecchia A., D’Oro U.. 2003. Activation of src-family tyrosine kinases by LPS regulates cytokine production in dendritic cells by controlling AP-1 formation. Eur. J. Immunol. 33: 2832–2841 [DOI] [PubMed] [Google Scholar]
- 59.Johnsen I. B., Nguyen T. T., Ringdal M., Tryggestad A. M., Bakke O., Lien E., Espevik T., Anthonsen M. W.. 2006. Toll-like receptor 3 associates with c-Src tyrosine kinase on endosomes to initiate antiviral signaling. EMBO J. 25: 3335–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanjuan M. A., Rao N., Lai K. T., Gu Y., Sun S., Fuchs A., Fung-Leung W. P., Colonna M., Karlsson L.. 2006. CpG-induced tyrosine phosphorylation occurs via a TLR9-independent mechanism and is required for cytokine secretion. J. Cell Biol. 172: 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scapini P., Hu Y., Chu C. L., Migone T. S., Defranco A. L., Cassatella M. A., Lowell C. A.. 2010. Myeloid cells, BAFF, and IFN-γ establish an inflammatory loop that exacerbates autoimmunity in Lyn-deficient mice. J. Exp. Med. 207: 1757–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lamagna C., Scapini P., van Ziffle J. A., DeFranco A. L., Lowell C. A.. 2013. Hyperactivated MyD88 signaling in dendritic cells, through specific deletion of Lyn kinase, causes severe autoimmunity and inflammation. Proc. Natl. Acad. Sci. USA 110: E3311–E3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Lint C., Amella C. A., Emiliani S., John M., Jie T., Verdin E.. 1997. Transcription factor binding sites downstream of the human immunodeficiency virus type 1 transcription start site are important for virus infectivity. J. Virol. 71: 6113–6127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baruah P., Simpson E., Dumitriu I. E., Derbyshire K., Coe D., Addey C., Dyson J., Chai J. G., Cook T., Scott D., Botto M.. 2010. Mice lacking C1q or C3 show accelerated rejection of minor H disparate skin grafts and resistance to induction of tolerance. Eur. J. Immunol. 40: 1758–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.