Abstract
Our recent study of microRNA (miRNA) expression signatures in prostate cancer (PCa) has revealed that all members of the miR-23b/27b/24-1 cluster are significantly downregulated in PCa tissues. The aim of this study was to investigate the effectiveness of these clustered miRNAs as a disease progression marker and to determine the functional significance of these clustered miRNAs in PCa. Expression of the miR-23b/27b/24-1 cluster was significantly reduced in PCa tissues. Kaplan-Meier survival curves showed that low expression of miR-27b predicted a short duration of progression to castration-resistant PCa. Gain-of-function studies using mature miR-23b, miR-27b, and miR-24-1 significantly inhibited cell proliferation, migration and invasion in PCa cells (PC3 and DU145). To identify the molecular targets of these miRNAs, we carried out gene expression and in silico database analyses. GOLM1 was directly regulated by miR-27b in PCa cells. Elucidation of the molecular targets and pathways regulated by the tumor-suppressive microRNAs should shed light on the oncogenic and metastatic processes in PCa.
Keywords: microRNA, miR-23b, miR-27b, miR-24-1, castration-resistant prostate cancer
INTRODUCTION
Prostate cancer (PCa) is the most frequently diagnosed cancer and the second leading cause of cancer-related deaths among men in developed countries [1]. Androgen signaling through the androgen receptor (AR) is an important oncogenic pathway for PCa progression. Most patients are initially responsive to androgen deprivation therapy (ADT), but their cancers eventually become resistant to ADT and progress to castration-resistant prostate cancer (CRPC). Although prostate-specific antigen (PSA) has been used for monitoring CRPC, outcomes in PCa patients are diverse, even within the same risk group, because of the heterogeneity of PCa cells [2-4]. Thus, identification of effective biomarkers for detection of CRPC is needed. With currently available therapies, CRPC is difficult to treat, and most clinical trials for advanced PCa have shown limited benefits, with disease progression and metastasis to bone or other sites [5, 6]. Therefore, understanding the molecular mechanisms of androgen-independent signaling and metastatic signaling pathways underlying PCa using current genomic approaches would help to improve therapies for and prevention of the disease.
A growing body of evidence indicates that microRNAs (miRNAs) also contribute to the initiation, development, and metastasis of various types of cancers [7]. Many human cancers show aberrant expression of miRNAs that can function as either tumor suppressors or oncogenes. Therefore, identification of aberrantly expressed miRNAs is the first step toward elucidating miRNA-mediated oncogenic pathways in human cancers. Based on this point, our group previously established miRNA expression signatures and identified novel oncogenic pathways regulated by tumor-suppressive microRNAs in several types of cancers, including PCa [8-11].
In a recent study from our laboratory, we found that the miR-23b/27b/24-1 cluster was significantly downregulated in PCa [11]. Some miRNAs are located in close proximity on the human genome; these are termed clustered miRNAs. In the human genome, 247 human miRNAs have been found to be clustered at 64 sites at inter-miRNA distances less than 5000 bp [12, 13]. We previously reported that miR-1-1/133a-2 and miR-1-2/133a-1 formed clusters in different chromosomal loci in the human genome (20q13.33 and 18q11.2, respectively) and that these clusters function as tumor suppressors, targeting several oncogenic genes in human cancers, including PCa [13-15]. More recently, we showed that the miR-143/145 cluster, located at the 5q32 locus, acts as a tumor-suppressive miRNA cluster in renal cell carcinoma and PCa [16, 17].
In this study, we hypothesized that the miR-23b/27b/24-1 cluster functioned as a tumor suppressor by targeting several oncogenic genes involved in specific cancer-related pathways in PCa. Elucidation of the molecular targets regulated by the tumor-suppressive miR-23b/27b/24-1 cluster will provide new insights into the potential molecular mechanisms of PCa oncogenesis and metastasis and will facilitate the development of novel diagnostic and therapeutic strategies for the treatment of PCa.
RESULTS
Expression of the miR-23b/27b/24-1 cluster in PCa specimens and cell lines
The public miRNA database (miRbase: release 21) shows that miR-23b/27b/24-1 cluster is located in close proximity on the human chromosome 9q22.32 region within 900 base pairs. The mature sequence of miR-23b is 5'-AUCACAUUGCCAGGGAUUACC-3', miR-27b is 5'-UUCACAGUGGCUAAGUUCUGC-3' and miR-24-1 is 5'-UGGCUCAGUUCAGCAGGAACAG-3'. This clustered miRNAs are involved in C9orf3 in their host gene.
We evaluated the expression levels of the clustered miRNAs (miR-23b, miR-27b and miR-24-1) in noncancerous tissues (n = 41) and PCa tissues (n = 49). In patients from whom normal prostate tissues were collected, the median PSA level was 7.3 ng/mL (range: 4.3–35.5 ng/mL). In contrast, in patients from whom PCa tissues were collected, PSA levels were quite high, with a median of 244 ng/mL (range: 3.45–3750 ng/mL). Thirty-nine PCa patients had progressive disease classified as N1 or M1 according to TNM classification (Table 1).
Table 1. Patients' characteristics.
| Characteristic | PCa (n = 49) | non-PCa (n = 41) |
|---|---|---|
| Age (years) | ||
| Median (range) | 73 (58-88) | 63 (52-85) |
| PSA (ng/ml) | ||
| Median (range) | 244 (3.45-3750) | 7.3 (4.3-35.5) |
| cT stage | ||
| T2c | 1 | |
| T3a | 12 | |
| T3b | 18 | |
| T4 | 18 | |
| cN stage | ||
| N0 | 15 | |
| N1 | 34 | |
| cM stage | ||
| M0 | 19 | |
| M1 | 30 | |
| Gleason Score | ||
| 3+4 | 1 | |
| 4+3 | 3 | |
| 4+4 | 30 | |
| 4+5 | 14 | |
| 5+4 | 0 | |
| 5+5 | 1 |
The expression levels of miR-23b, miR-27b, and miR-24-1 were significantly downregulated (P < 0.0001) in tumor tissues as compared to corresponding noncancerous tissues (Figure 1A). Spearman's rank test showed positive correlations between the expression of miR-23b and miR-27b (R = 0.905 and P < 0.0001) and the expression of miR-23b and miR-24-1 (R = 0.888 and P < 0.0001). Similarly, the expression of miR-24-1 was positively correlated with that of miR-27b (R = 0.965 and P < 0.0001; Figure 1B).
Figure 1. Expression levels of clustered miR-23b/27b/24-1 in PCa specimens.
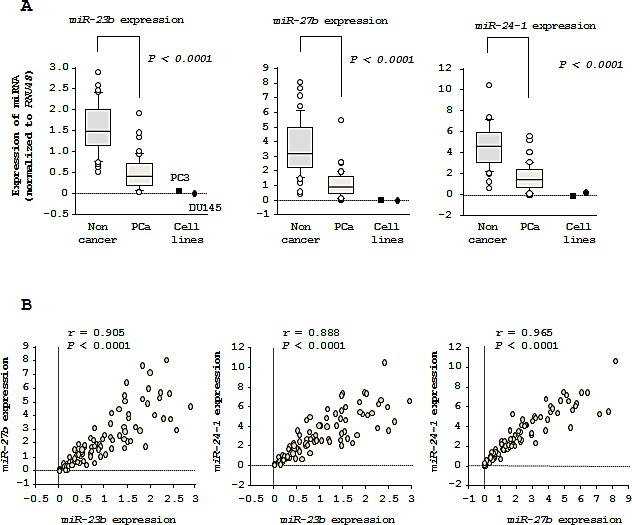
(A) Expression levels of miR-23b, miR-27b, and miR-24-1 in PCa clinical specimens. RNU48 was used for normalization. (B) Correlations among the relative expression levels of miR-23b/miR-27b, miR-23b/miR-24-1, and miR-27b/miR-24-1.
Correlations between miR-23b/27b/24-1 expression and clinicopathological features in PCa specimens
Among 49 PCa patients, 47 patients underwent ADT with luteinizing hormone-releasing hormone (LHRH) agonists and anti-androgens (Supplemental Table 1). A total of 16 ADT-treated patients progressed to CRPC over a median follow-up of 15.6 months. For patients with high versus low miR-23b/27b/24-1 expression, the risk of progression to CRPC was evaluated using the Kaplan-Meier method and log rank test for significant separation of survival curves. Low expression of miR-27b was found to be associated with shorter progression-free interval (P = 0.0346; Figure 2B). However, neither miR-23b nor miR-24-1 predicted the time to CRPC in these PCa patients (P = 0.321 and P = 0.231, respectively; Figure 2A and 2C). The multivariate Cox proportional hazards model was used to assess independent predictors of time to progression to CRPC, including Gleason score, cT stage, cN stage, cM stage, PSA, age, and miR-27b expression. miR-27b expression was the only independent prognostic factor of patient outcomes for PCa patients treated with ADT (hazard ratio [HR]: 0.255, 95% confidence interval [CI]: 0.069-0.944, P = 0.0407; Table 2).
Figure 2. Correlations of miR-23b/27b/24-1 expression and clinicopathological features in PCa specimens.
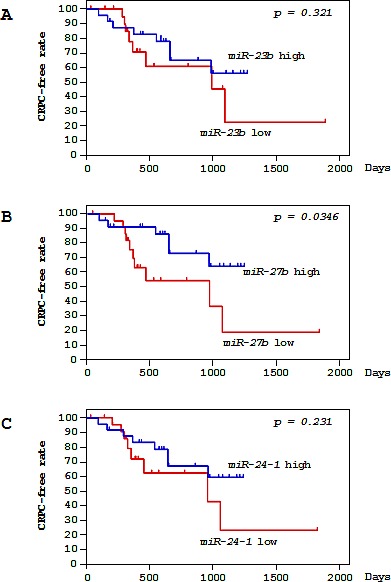
Kaplan-Meier survival curves for CRPC progression-free survival based on (A) miR-23b, (B) miR-27b, and (C) miR-24-1 expression in PCa patients. P-values were calculated using the log-rank test.
Table 2. Cox proportional analysis for the prediction of CRPC progression-free survival.
| Covariant | HR | 95% CI | P-value |
|---|---|---|---|
| miR-27b | 0.255 | 0.069-0.944 | 0.0407 |
| Gleason score | 2.003 | 0.747-5.374 | 0.1674 |
| cT stage | 2.111 | 0.490-9.098 | 0.3162 |
| cN stage | 2.28 | 0.372-13.975 | 0.3729 |
| cM stage | 1.021 | 0.203-5.124 | 0.9799 |
| PSA | 1.001 | 1.000-1.001 | 0.0719 |
| Age | 1.045 | 0.949-1.150 | 0.3674 |
Effects of restoring miR-23b/27b/24-1 expression on cell proliferation, migration, and invasion in PC3 and DU145 PCa cells
To investigate the functional effects of the miR-23b/27b/24-1 cluster, we performed gain-of-function studies using miRNA transfection in 2 PCa cell lines (PC3 and DU145).
As observed using XTT assays, cell proliferation was significantly inhibited in miR-27b and miR-24-1 transfectants as compared with mock- or miR-control-transfected PC3 cells. However, the miR-23b transfectant did not exhibit reduced cell proliferation in PC3 cells. In contrast, inhibition of cell proliferation was only observed in the miR-24-1 transfectant in DU145 cells (Figure 3A).
Figure 3. Effects of miR-23b/27b/24-1 transfection on cell proliferation, migration, and invasion in PC3 and DU145 cells.
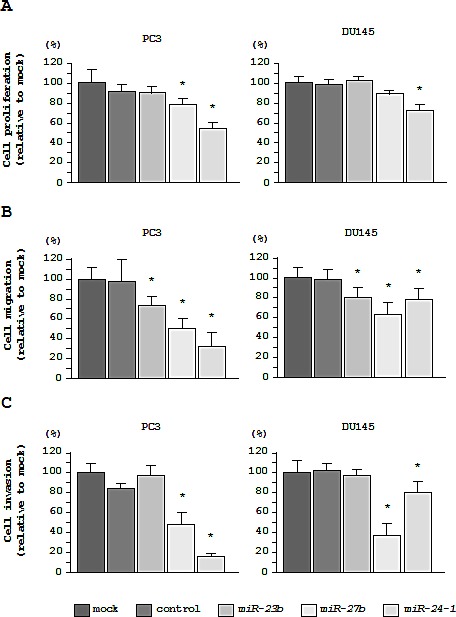
(A) Cell proliferation 72 h after transfection with miR-23b/27b/24-1 was determined using XTT assays. (B) Cell migration activity 48 h after transfection with miR-23b/27b/24-1 was determined using migration assays. (C) Cell invasion activity 48 h after transfection with miR-23b/27b/24-1 was determined using Matrigel invasion assays. *P < 0.005.
In cell migration assays, transfection with each of the 3 miRNAs significantly inhibited cancer cell migration in all cell lines (Figure 3B).
In cell invasion assays, transfection with miR-27b and miR-24-1 inhibited cell invasion in PC3 and DU145 cells. However, transfection with miR-23b did not inhibit cell invasion in either PC3 or DU145 cells (Figure 3C).
In this study, we investigated the synergistic effects of miR-23b/27b/24-1 cluster in PCa cells. We performed XTT and migration assays using transfection of all possible combinations of these clustered miRNAs. However, we did not find synergistic effects in these assays (Supplemental Figure 1).
Identification of target genes regulated by the miR-23b/27b/24-1 cluster in PCa
To gain further insights into the molecular pathways and targets regulated by the tumor-suppressive miR-23b/27b/24-1 cluster in PCa, we first identified putative target genes of miR-23b, miR-27b, and miR-24-1 by searching the TargetScan database. This analysis identified 4206, 4075, and 4321 putative target genes for miR-23b, miR-27b, and miR-24-1, respectively.
Next, we investigated the expression statuses of these putative targets in PCa clinical specimens and examined gene expression profiles in the GEO database (accession numbers GSE29079) to evaluate upregulated genes in PCa specimens. Among the 4206 putative target genes of miR-23b, 147 genes were significantly upregulated in PCa specimens compared to noncancerous prostate tissues. In a similar analysis, among 4075 and 4321 putative targets of miR-27b and miR-24-1, 157 and 139 genes were upregulated in PCa tissues, respectively.
Furthermore, we performed genome-wide gene expression analysis using PC3 cells and selected genes that were downregulated following transfection with miR-23b/27b/24-1 as compared with the miR-control. Entries from the gene expression data were approved by GEO, and were assigned GEO accession number GSE47657. When we integrated all of these analysis results, a total of 34, 52, and 56 genes were identified as targets of miR-23b, miR-27b, and miR-24-1, respectively (Supplemental Tables 2-4). Our strategy for selection of miR-23b/27b/24-1 cluster-targeted genes is shown in Figure 4.
Figure 4. Flow chart of the strategy for analysis of miR-23b/27b/24-1 cluster target genes.
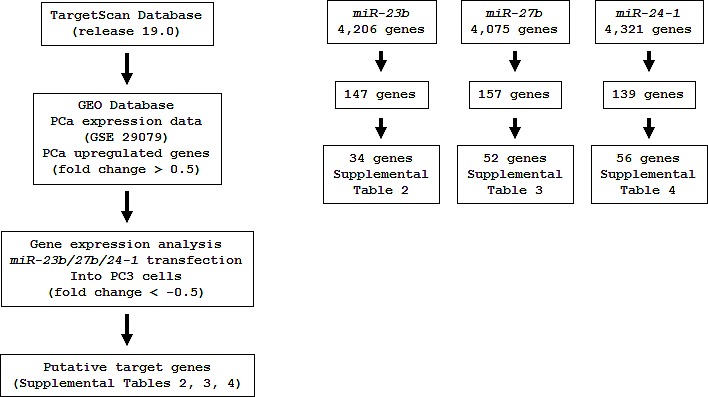
A total of 4206, 4075 and 4321 genes were selected as putative miR-23b, 27b and 24-1 target genes, respectively by TargetScan database analysis. We then analyzed the expression levels of these candidate genes by using available data sets of GEO (GSE 29079). The analyses showed that 147, 157 and 139 genes were significantly upregulated in PCa specimens compared with normal specimens. Furthermore, expression analysis data of miR-23b, miR-27b and miR-24-1 transfectants of PC3 cells were merged. Finally, 34, 52 and 56 genes were putative candidate genes regulated by miR-23b, miR-27b and miR-24-1, respectively.
GOLM1 was a direct target of miR-27b in PCa cells
We performed real-time RT-qPCR and western blotting in PC3 and DU145 cells to investigate whether restoration of miR-27b altered the expression of the GOLM1 gene and GOLM1 protein. The mRNA and protein expression levels of GOLM1/GOLM1 were significantly repressed in miR-27b transfectants as compared with mock- or miR-control-transfected cells (Figures 5A and 5B).
Figure 5. Downregulation of GOLM1 expression by miR-27b in PC3 and DU145 cells.
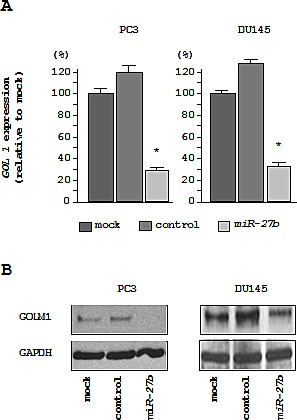
(A) GOLM1 mRNA expression 72 h after transfection with miR-27b. GUSB was used as an internal control. *P < 0.005. (B) GOLM1 protein expression 72 h after transfection with miR-27b. GAPDH was used as a loading control.
Therefore, we next performed luciferase reporter assays in PC3 cells to determine whether GOLM1 mRNA had target sites for miR-27b. The TargetScan database predicted that 2 putative miR-27b binding sites existed in the 3'UTR of GOLM1 (positions 79-86 and 364-370). We used vectors encoding a partial wild-type sequence of the 3'UTR of GOLM1 mRNA, including the predicted miR-27b target site or a vector lacking the miR-27b target site. We found that the luminescence intensity was significantly reduced by cotransfection with miR-27b and the vector carrying the wild-type 3'UTR of GOLM1. On the other hand, the luminescence intensity was not decreased when the seed sequence of the target site was deleted from the vectors (Figure 6).
Figure 6. Regulation of GOLM1 expression by miR-27b.

miR-27b binding sites in GOLM1 mRNA. Luciferase reporter assay using the two vectors encoding putative miR-27b target sites of the GOLM1 3'-UTR (positions 79-86 and 364-370) for both wild-type and mutant constructs. *P < 0.005.
GOLM1 expression in PCa specimens
Among 90 PCa and non-PCa samples, we selected RNA samples that could be used for reverse transcription analysis of GOLM1 mRNA expression. Finally, 11 PCa samples and 10 non-PCa samples were subjected to GOLM1 mRNA expression analysis in this study. RT-qPCR showed that miR-27b expression was significantly lower in PCa samples compared with non-PCa samples (P = 0.0012, Figure 7A). Moreover, the expression of GOLM1 was significantly higher in PCa tissues compared with normal tissues (P = 0.0006, Figure 7A). Spearman's rank test showed that the lower expression of GOLM1 was correlated with higher miR-27b expression (r = -0.695 and P = 0.0019, Figure 7B).
Figure 7. Expression of GOLM1 and miR-27b in PCa specimens.
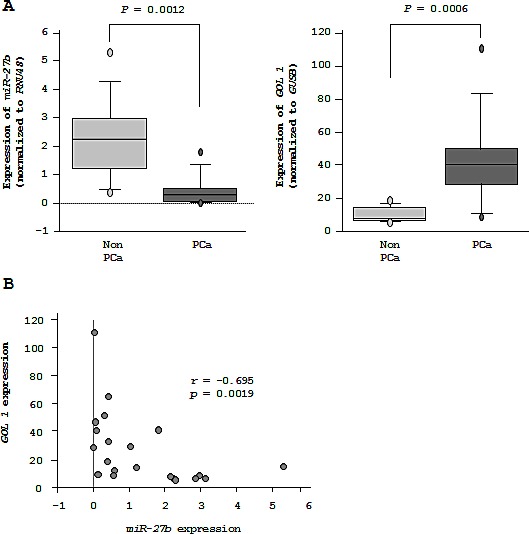
(A) miR-27b and GOLM1 mRNA expression in 10 non-PCa specimens and 11 clinical PCa specimens. (B) Inverse correlation between GOLM1 mRNA and miR-27b expression.
DISCUSSION
Aberrantly expressed miRNAs disrupt the tightly regulated RNA networks in cancer cells, triggering cancer development and metastasis. Therefore, identification of the differentially expressed miRNAs in cancer cells is the first step to elucidating novel miRNA-mediated pathways in cancer. Expression signatures of various types of cancer tissues are important sources for the study of miRNAs in cancer. Based on the observed miRNA signatures, our group has identified tumor-suppressive miRNAs in PCa [11, 15, 17]. In this study, we focused on the miR-23b/27b/24-1 cluster because all of these miRNAs have been reported as downregulated miRNAs in our PCa signatures [11]. This is the first report that aimed to investigate the functional significance of all members of the miR-23b/27b/24-1 cluster in PCa. We confirmed the downregulation of the miR-23b/27b/24-1 cluster in PCa tissues with sets of independent clinical specimens. Elucidation of several miRNA signatures in PCa has demonstrated that some members of this miRNA cluster are expressed at low levels in cancer tissues [18-20]. From our analysis, we found that the miRNAs within the miR-23b/27b/24-1 cluster were regulated by the same transcriptional control mechanism in the human genome. A recent report showed that the transcription factor AP-1 directly binds to the promoter region of miR-23b and reduces its expression in MDA-MB-231 cells [21]. Interestingly, the expression of AP-1 (c-Jun and c-Fos) is associated with aggressive and androgen-independent prostate cancer [22, 23]. While the mechanism silencing the expression of the miR-23b/27b/24-1 cluster in PCa cells is still unknown, analysis of the detailed molecular mechanisms will be necessary in future studies.
Current PCa screening methods are based on the measurement of serum PSA, and a definite diagnosis is established by ultrasound-guided prostate needle biopsies [24, 25]. PSA is the most common marker for detection of PCa and for following the course of CRPC or metastatic PCa. However, the course of PCa progression and clinical outcomes of PCa patients can differ even in patients with the same PSA value, Gleason score, and pathological stage. Thus, it is crucial to identify more sensitive biomarkers for improvement of PCa prognosis. Several groups have described the independent biochemical recurrence of prediction markers based on the expression levels of miRNAs, such as miR-21, miR-145, miR-200a, and miR-30d [26-29], using radical prostatectomy specimens. If the expression status of miRNAs derived from needle biopsies at the initial visit can predict the possibility of progression to CRPC or metastasis, physicians can make more accurate treatment decisions for PCa patients. To investigate this, we analyzed the expression levels of miRNAs within the miR-23b/27b/24-1 cluster and their associations with the clinicopathological features of PCa patients. Surprisingly, the expression status of miR-27b indicated that this miRNA was a good prognostic marker for time to progression to CRPC. A large-scale cohort study will be necessary to determine whether miR-27b is an effective marker for CRPC-free interval.
Our present data demonstrated that restoration of the miR-23b/27b/24-1 cluster significantly inhibited cancer cell proliferation, migration, and invasion in both androgen-dependent and –independent PCa cells, suggesting that the miR-23b/27b/24-1 cluster functioned as a tumor suppressor in PCa. Several studies have reported that these miRNAs have tumor-suppressive roles in PCa cells, similar to the results of our present study. For example, miR-23b directly controls the proto-oncogenes Src kinase and Akt, and overexpression of miR-23b inhibits proliferation, migration, invasion, cell cycle arrest, and apoptosis [30]. Another report has shown that miR-23b and miR-27b are downregulated in metastatic and CRPC tumors and that ectopic expression of these miRNAs suppresses cell invasion and migration in CRPC cell lines [31]. In contrast, the expression of miR-23b and miR-27b is highly upregulated in human breast cancer, and knockdown of miR-23b and miR-27b substantially represses breast cancer growth [32]. Interestingly, the expression status of the miR-23b/27b/24-1 cluster is not consistent among different types of cancers. Elucidation of the mechanisms controlling the expression of clustered miRNAs in each cancer is an important theme in this developing field. To date, few reports have described the functional significance of miR-27b and miR-24-1 in PCa cells.
Identification of the targets regulated by the tumor-suppressive miR-23b/27b/24-1 cluster is important for clarifying our understanding of PCa oncogenesis and metastasis. Based on this view, we identified target genes of the miR-23b/27b/24-1 cluster using a combination of in silico analysis and gene expression analysis with miR-23b, miR-27b, and miR-24-1 transfectants. Using this strategy, we succeeded in proving that the tumor-suppressive miR-1/133a or miR-143/145 cluster regulates oncogenic genes in various cancers [15-17]. In the present study, we identified putative target genes regulated by miRNAs in the miR-23b/27b/24-1 cluster. Identification of these target genes will contribute to elucidation of novel cancer networks in PCa.
Finally, we focused on the GOLM1 gene because this gene has been identified as a putative target of tumor-suppressive miR-27b and because miR-27b is a predictor of time to CRPC. Furthermore, our recent study demonstrated that the tumor-suppressive miR-143/145 cluster commonly targets GOLM1 and that silencing of GOLM1 significantly inhibits the migration and invasion of PCa cells [17]. Our present data clearly showed that GOLM1 was directly regulated by tumor-suppressive miR-27b in PCa cells. The GOLM1/GP73/GOLPH2 protein is encoded by the GOLM1 gene, located on human chromosome 9q21.33. GOLM1 has been shown to be overexpressed in human PCa tissue [33, 34], lung adenocarcinoma [35], and hepatocellular carcinoma [36]. However, while a number of studies have demonstrated that GOLM1 is expressed in cancer cells, the exact molecular mechanisms mediating GOLM1 function remain unclear. Additionally, we investigated whether GOLM1 contributed to the oncogenesis and metastasis of PCa. Therefore, elucidation of the regulatory networks of GOLM1-mediated signaling pathways will provide important information for the development of new therapeutic strategies against cancer cell metastasis. Further research is needed to reveal the oncogenic functions of GOLM1 that are regulated by the tumor-suppressive miR-143/145 cluster or by miR-27b in PCa.
In conclusions, downregulation of the miR-23b/27b/24-1 cluster was a frequent event in PCa, and these clustered miRNAs functioned as tumor suppressors. Furthermore, miR-27b expression was a good disease progression marker in PCa. Elucidation of the molecular targets and pathways regulated by the tumor-suppressive miR-23b/27b/24-1 cluster should shed light on the oncogenic and metastatic processes in PCa and lead to the development of more effective strategies for future therapeutic interventions in patients with PCa.
METHODS
Patients and clinical prostate specimens
Clinical prostate specimens were obtained from patients admitted to Teikyo University Chiba Medical Centre Hospital from 2008 to 2013. All patients had elevated PSA levels and had undergone transrectal prostate needle biopsy. Prostatic cancerous tissues (PCa, n = 49) and noncancerous tissues (non-PCa, n = 41) were used in this study. The patients' backgrounds and clinicopathological characteristics are summarized in Supplemental Table 1. Written consent for tissue donation for research purposes was obtained from each patient before tissue collection. Investigation has been conducted in accordance with the ethical standards and according to the Declaration of Helsinki and according to national and international guidelines. The protocol was approved by the Institutional Review Board of Teikyo University.
For pathological justification of tissue composition, a pair of needle biopsy specimens was collected from the same region as from patients in this study, and one was subjected to pathological justification; no cancerous tissue was found in non-PCa specimens. CRPC was defined according to European Association of Urology guidelines [37].
Cell culture
Human prostate cancer cells (PC3 and DU145 cells) were obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum in a humidified atmosphere of 5% CO2 and 95% air at 37°C.
RNA isolation
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. The quality of RNA was confirmed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) as described previously [15, 38, 39].
Quantitative real-time reverse transcription polymerase chain reaction (RT-qPCR)
The procedure for PCR quantification was carried out as previously described [15, 38, 39]. The expression levels of miR-23b (Assay ID: 000400), miR-27b (Assay ID: 000409), and miR-24-1 (Assay ID: 000402) were analyzed by TaqMan quantitative real-time PCR (TaqMan MicroRNA Assay; Applied Biosystems) and normalized to RNU48 (Assay ID: 001006). TaqMan probes and primers for GOLM1 (P/N: Hs00213061_m1), GAPDH (P/N: Hs02758991_g1), and GUSB (P/N: Hs00939627_m1) as an internal control were obtained from Applied Biosystems (Assay-On-Demand Gene Expression Products). The ΔΔCt method was applied for calculation of the relative quantities of target genes. All reactions were performed in triplicate and included negative control reactions that lacked cDNA.
Transfections with mature miRNA and small-interfering RNA (siRNA)
The following mature miRNA species were used in this study: Ambion Pre-miR miRNA precursor for hsa-miR-23b (product ID: PM10711), hsa-miR-27b (product ID: PM10750), and hsa-miR-24-1 (product ID: PM12902; Applied Biosystems). RNAs were incubated with OPTI-MEM (Invitrogen) and Lipofectamine RNAiMAX reagent (Invitrogen). The transfection procedures and transfection efficiencies of miRNA in PC3 and DU145 cells were reported previously [15, 38, 39].
Cell proliferation, migration, and invasion assays
To investigate the functional significance of the miR-23b/27b/24-1 cluster, we performed cell proliferation, migration, and invasion assays using PC3 and DU145 cells. The experimental procedures were performed as described in our previous studies [15, 38, 39].
Genome-wide gene expression and in silico analyses for the identification of genes regulated by the miR-23b/27b/24-1 cluster
To gain further insights into the specific genes affected by the miR-23b/27b/24-1 cluster, we performed a combination of in silico and genome-wide gene expression analyses. First, genes regulated by the miR-23b/27b/24-1 cluster were listed using the TargetScan database as described previously [15, 38, 39]. Next, to identify upregulated genes in PCa, we analyzed a publicly available gene expression data set in the Gene Expression Omnibus (GEO, accession number: GSE29079). Finally, we performed genome-wide gene expression analysis using PC3 cells transfected with miR-23b, miR-27b, or miR-24-1. A SurePrint G3 Human GE 60K Microarray (Agilent Technologies) was used for expression profiling of each miRNA transfectant in comparison with negative control miRNA transfectants. Finally, downregulated mRNAs that contained target sites for each miRNA (miR-23b/27b/24-1) were listed as putative target genes for these miRNAs.
Western blotting
Cells were harvested 72 h after transfection, and lysates were prepared. Twenty micrograms of protein lysate from each sample was separated on Mini-PROTEAN TGX gels (Bio-Rad, Hercules, CA, USA) and transferred to polyvinylidene difluoride membranes. Immunoblotting was performed with rabbit anti-GOLM1 antibodies (1:250, HPA010638, Atlas Antibodies, Stockholm, Sweden), and anti-GAPDH antibodies (1:1000, ab8245, Abcam) were used as an internal loading control. Membranes were washed and incubated with anti-rabbit IgG horseradish peroxidase (HRP)-linked antibodies (7074, Cell Signaling Technology, Danvers, MA, USA). Complexes were visualized with Clarity Western ECL Substrate (Bio-Rad, Hercules, CA, USA). The experimental procedures were performed as described in our previous studies [15, 38, 39].
Plasmid construction and dual-luciferase reporter assay
Partial wild-type sequences of the GOLM1 3' untranslated region (UTR) or those with 2 deleted miR-27b target sites (positions 79-86 and 364-370 of the GOLM1 3'UTR) were inserted between the XhoI–PmeI restriction sites in the 3'UTR of the hRluc gene in the psiCHECK-2 vector (C8021; Promega, Madison, WI, USA). The protocol for vector construction was described previously [15, 38, 39].
Statistical analysis
The relationships between two groups and the numerical values obtained by real-time RT–qPCR were analyzed using Mann–Whitney U-tests. Spearman's rank test was used to evaluate the correlation between the expressions levels of miR-23b, miR-27b, and miR-24-1. Relationships among more than 3 variables and numerical values were analyzed using Mann–Whitney U-tests after Bonferroni adjustment. Survival analysis was evaluated by the Kaplan-Meier method and log-rank tests. A multivariate Cox proportional hazards model was used to establish independent factors for survival. The chi-square test was used to analyze the relationship between miR-23b/27b/24-1 expression and clinicopathological characteristics. The Kaplan-Meier method and the log-rank test were performed using Stat Mate software (version 4.01, ATMS Co., Tokyo, Japan), and other analyses were performed using Expert StatView (version 5, SAS Institute Inc., Cary, NC, USA).
SUPPLEMENTARY MATERIAL FIGURE AND TABLES
Acknowledgments
This study was supported by the KAKENHI 24592590 (C), 23592351 (C) and 25292333 (B).
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Kelly WK, Scher HI, Mazumdar M, Vlamis V, Schwartz M, Fossa SD. Prostate-specific antigen as a measure of disease outcome in metastatic hormone-refractory prostate cancer. Journal of clinical oncology. 1993;11(4):607–615. doi: 10.1200/JCO.1993.11.4.607. [DOI] [PubMed] [Google Scholar]
- 3.Scher HI, Kelly WM, Zhang ZF, Ouyang P, Sun M, Schwartz M, Ding C, Wang W, Horak ID, Kremer AB. Post-therapy serum prostate-specific antigen level and survival in patients with androgen-independent prostate cancer. Journal of the National Cancer Institute. 1999;91(3):244–251. doi: 10.1093/jnci/91.3.244. [DOI] [PubMed] [Google Scholar]
- 4.Colloca G. Prostate-specific antigen kinetics as a surrogate endpoint in clinical trials of metastatic castration-resistant prostate cancer: a review. Cancer treatment reviews. 2012;38(8):1020–1026. doi: 10.1016/j.ctrv.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Chi KN, Bjartell A, Dearnaley D, Saad F, Schroder FH, Sternberg C, Tombal B, Visakorpi T. Castration-resistant prostate cancer: from new pathophysiology to new treatment targets. European urology. 2009;56(4):594–605. doi: 10.1016/j.eururo.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 6.Sturge J, Caley MP, Waxman J. Bone metastasis in prostate cancer: emerging therapeutic strategies. Nature reviews Clinical oncology. 2011;8(6):357–368. doi: 10.1038/nrclinonc.2011.67. [DOI] [PubMed] [Google Scholar]
- 7.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nature reviews Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 8.Kikkawa N, Hanazawa T, Fujimura L, Nohata N, Suzuki H, Chazono H, Sakurai D, Horiguchi S, Okamoto Y, Seki N. miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC) British journal of cancer. 2010;103(6):877–884. doi: 10.1038/sj.bjc.6605811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nohata N, Hanazawa T, Kikkawa N, Sakurai D, Fujimura L, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nakagawa M, Katayama A, Harabuchi Y, Okamoto Y, Seki N. Tumour suppressive microRNA-874 regulates novel cancer networks in maxillary sinus squamous cell carcinoma. British journal of cancer. 2011;105(6):833–841. doi: 10.1038/bjc.2011.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hidaka H, Seki N, Yoshino H, Yamasaki T, Yamada Y, Nohata N, Fuse M, Nakagawa M, Enokida H. Tumor suppressive microRNA-1285 regulates novel molecular targets: aberrant expression and functional significance in renal cell carcinoma. Oncotarget. 2012;3(1):44–57. doi: 10.18632/oncotarget.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuse M, Kojima S, Enokida H, Chiyomaru T, Yoshino H, Nohata N, Kinoshita T, Sakamoto S, Naya Y, Nakagawa M, Ichikawa T, Seki N. Tumor suppressive microRNAs (miR-222 and miR-31) regulate molecular pathways based on microRNA expression signature in prostate cancer. Journal of human genetics. 2012;57(11):691–699. doi: 10.1038/jhg.2012.95. [DOI] [PubMed] [Google Scholar]
- 12.Chhabra R, Dubey R, Saini N. Cooperative and individualistic functions of the microRNAs in the miR-23a~27a~24-2 cluster and its implication in human diseases. Molecular cancer. 2010;9:232. doi: 10.1186/1476-4598-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nohata N, Hanazawa T, Enokida H, Seki N. microRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget. 2012;3(1):9–21. doi: 10.18632/oncotarget.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshino H, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Nishiyama K, Nohata N, Seki N, Nakagawa M. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. British journal of cancer. 2011;104(5):808–818. doi: 10.1038/bjc.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kojima S, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nohata N, Fuse M, Ichikawa T, Naya Y, Nakagawa M, Seki N. Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. British journal of cancer. 2012;106(2):405–413. doi: 10.1038/bjc.2011.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshino H, Enokida H, Itesako T, Kojima S, Kinoshita T, Tatarano S, Chiyomaru T, Nakagawa M, Seki N. The tumor-suppressive microRNA-143/145 cluster targets hexokinase-2 in renal cell carcinoma. Cancer science. 2013 doi: 10.1111/cas.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kojima S, Enokida H, Yoshino H, Itesako T, Chiyomaru T, Kinoshita T, Fuse M, Nishikawa R, Goto Y, Naya Y, Nakagawa M, Seki N. The tumor-suppressive microRNA-143/145 cluster inhibits cell migration and invasion by targeting GOLM1 in prostate cancer. Journal of human genetics. 2014;59(2):78–87. doi: 10.1038/jhg.2013.121. [DOI] [PubMed] [Google Scholar]
- 18.Carlsson J, Davidsson S, Helenius G, Karlsson M, Lubovac Z, Andren O, Olsson B, Klinga-Levan K. A miRNA expression signature that separates between normal and malignant prostate tissues. Cancer cell international. 2011;11(1):14. doi: 10.1186/1475-2867-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer research. 2007;67(13):6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 20.Szczyrba J, Loprich E, Wach S, Jung V, Unteregger G, Barth S, Grobholz R, Wieland W, Stohr R, Hartmann A, Wullich B, Grasser F. The microRNA profile of prostate carcinoma obtained by deep sequencing. Molecular cancer research. 2010;8(4):529–538. doi: 10.1158/1541-7786.MCR-09-0443. [DOI] [PubMed] [Google Scholar]
- 21.Pellegrino L, Stebbing J, Braga VM, Frampton AE, Jacob J, Buluwela L, Jiao LR, Periyasamy M, Madsen CD, Caley MP, Ottaviani S, Roca-Alonso L, El-Bahrawy M, Coombes RC, Krell J, Castellano L. miR-23b regulates cytoskeletal remodeling, motility and metastasis by directly targeting multiple transcripts. Nucleic Acids Res. 2013;41(10):5400–5412. doi: 10.1093/nar/gkt245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards J, Krishna NS, Mukherjee R, Bartlett JM. The role of c-Jun and c-Fos expression in androgen-independent prostate cancer. The Journal of pathology. 2004;204(2):153–158. doi: 10.1002/path.1605. [DOI] [PubMed] [Google Scholar]
- 23.Kajanne R, Miettinen P, Tenhunen M, Leppa S. Transcription factor AP-1 promotes growth and radioresistance in prostate cancer cells. International journal of oncology. 2009;35(5):1175–1182. doi: 10.3892/ijo_00000434. [DOI] [PubMed] [Google Scholar]
- 24.Mohler JL, Kantoff PW, Armstrong AJ, Bahnson RR, Cohen M, D'Amico AV, Eastham JA, Enke CA, Farrington TA, Higano CS, Horwitz EM, Kawachi MH, Kuettel M, Lee RJ, Macvicar GR, Malcolm AW, et al. Prostate cancer, version 1. 2014. Journal of the National Comprehensive Cancer Network. 2013;11(12):1471–1479. doi: 10.6004/jnccn.2013.0174. [DOI] [PubMed] [Google Scholar]
- 25.Heidenreich A, Bastian PJ, Bellmunt J, Bolla M, Joniau S, van der Kwast T, Mason M, Matveev V, Wiegel T, Zattoni F, Mottet N. EAU guidelines on prostate cancer. part 1: screening, diagnosis, and local treatment with curative intent-update 2013. European urology. 2014;65(1):124–137. doi: 10.1016/j.eururo.2013.09.046. [DOI] [PubMed] [Google Scholar]
- 26.Li T, Li RS, Li YH, Zhong S, Chen YY, Zhang CM, Hu MM, Shen ZJ. miR-21 as an independent biochemical recurrence predictor and potential therapeutic target for prostate cancer. The journal of urology. 2012;187(4):1466–1472. doi: 10.1016/j.juro.2011.11.082. [DOI] [PubMed] [Google Scholar]
- 27.Avgeris M, Stravodimos K, Fragoulis EG, Scorilas A. The loss of the tumour-suppressor miR-145 results in the shorter disease-free survival of prostate cancer patients. British journal of cancer. 2013;108(12):2573–2581. doi: 10.1038/bjc.2013.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barron N, Keenan J, Gammell P, Martinez VG, Freeman A, Masters JR, Clynes M. Biochemical relapse following radical prostatectomy and miR-200a levels in prostate cancer. Prostate. 2012;72(11):1193–1199. doi: 10.1002/pros.22469. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi N, Uemura H, Nagahama K, Okudela K, Furuya M, Ino Y, Ito Y, Hirano H, Inayama Y, Aoki I, Nagashima Y, Kubota Y, Ishiguro H. Identification of miR-30d as a novel prognostic maker of prostate cancer. Oncotarget. 2012;3(11):1455–1471. doi: 10.18632/oncotarget.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Majid S, Dar AA, Saini S, Arora S, Shahryari V, Zaman MS, Chang I, Yamamura S, Tanaka Y, Deng G, Dahiya R. miR-23b represses proto-oncogene Src kinase and functions as methylation-silenced tumor suppressor with diagnostic and prognostic significance in prostate cancer. Cancer research. 2012;72(24):6435–6446. doi: 10.1158/0008-5472.CAN-12-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishteiwy RA, Ward TM, Dykxhoorn DM, Burnstein KL. The microRNA -23b/-27b cluster suppresses the metastatic phenotype of castration-resistant prostate cancer cells. PLoS one. 2012;7(12):e52106. doi: 10.1371/journal.pone.0052106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin L, Wessely O, Marcusson EG, Ivan C, Calin GA, Alahari SK. Prooncogenic factors miR-23b and miR-27b are regulated by Her2/Neu, EGF, and TNF-alpha in breast cancer. Cancer research. 2013;73(9):2884–2896. doi: 10.1158/0008-5472.CAN-12-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kristiansen G, Fritzsche FR, Wassermann K, Jager C, Tolls A, Lein M, Stephan C, Jung K, Pilarsky C, Dietel M, Moch H. GOLPH2 protein expression as a novel tissue biomarker for prostate cancer: implications for tissue-based diagnostics. British journal of cancer. 2008;99(6):939–948. doi: 10.1038/sj.bjc.6604614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W, Wang X, Li B, Lu J, Chen G. Diagnostic significance of overexpression of Golgi membrane protein 1 in prostate cancer. Urology. 2012;80(4):952.e951–957. doi: 10.1016/j.urology.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 35.Zhang F, Gu Y, Li X, Wang W, He J, Peng T. Up-regulated Golgi phosphoprotein 2 (GOLPH2) expression in lung adenocarcinoma tissue. Clinical biochemistry. 2010;43(12):983–991. doi: 10.1016/j.clinbiochem.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 36.Bachert C, Fimmel C, Linstedt AD. Endosomal trafficking and proprotein convertase cleavage of cis Golgi protein GP73 produces marker for hepatocellular carcinoma. Traffic (Copenhagen, Denmark) 2007;8(10):1415–1423. doi: 10.1111/j.1600-0854.2007.00621.x. [DOI] [PubMed] [Google Scholar]
- 37.Heidenreich A, Bastian PJ, Bellmunt J, Bolla M, Joniau S, van der Kwast T, Mason M, Matveev V, Wiegel T, Zattoni F, Mottet N. EAU Guidelines on Prostate Cancer. Part II: Treatment of Advanced, Relapsing, and Castration-Resistant Prostate Cancer. European urology. 2014;65(2):467–479. doi: 10.1016/j.eururo.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Kinoshita T, Nohata N, Hanazawa T, Kikkawa N, Yamamoto N, Yoshino H, Itesako T, Enokida H, Nakagawa M, Okamoto Y, Seki N. Tumour-suppressive microRNA-29s inhibit cancer cell migration and invasion by targeting laminin-integrin signalling in head and neck squamous cell carcinoma. British journal of cancer. 2013;109(10):2636–2645. doi: 10.1038/bjc.2013.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nohata N, Hanazawa T, Kinoshita T, Inamine A, Kikkawa N, Itesako T, Yoshino H, Enokida H, Nakagawa M, Okamoto Y, Seki N. Tumour-suppressive microRNA-874 contributes to cell proliferation through targeting of histone deacetylase 1 in head and neck squamous cell carcinoma. British journal of cancer. 2013;108(8):1648–1658. doi: 10.1038/bjc.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


