Abstract
Objective
The purpose of this study was to determine the prevalence of vancomycin-resistant Staphylococcus aureus isolated from clinical samples in Shiraz hospitals.
Methods
From March to December 2012, 100 S. aureus isolates (mainly from wound and blood) were collected from three hospitals in Shiraz, south of Iran. After identification of Staphylococcus aureus by biochemical, microbiological and molecular methods, antibiotic susceptibility testing was performed by Kirby-Bauer disc diffusion test for 13 different antibiotics. Vancomycin-resistant Staphylococcus aureus isolates were determined by vancomycin agar screening test and PCR for vancomycin resistant genes (vanA and vanB).
Results
The lowest and highest resistance was seen for quinupristin-dalfopristin (n=1) and ampicillin (n=95), respectively. Vancomycin agar screening test showed that 37 isolates can grow on these media. Further study by PCR also detected vanA and/or vanB genes in all of these strains. Also, 19 isolates showed either vanA or vanB but were susceptible according to vancomycin agar screening test. In total, vanA and vanB resistant genes were detected in 34% and 37% of clinical isolates, respectively.
Conclusion
The results showed that the frequency of vancomycin resistance genes (vanA, vanB) is very high in Staphylococcus aureus strains isolated from patients in south of Iran. Thus, urgent interventions are needed to keep the emergence and transmission of these isolates to a minimum.
Keywords: Staphylococcus aureus, Vancomycin Resistance, vanA, vanB, Iran
Introduction
Staphylococcus aureus is a major human pathogen which is known for more than a century. Primarily as a colonizer in one-third of general population, S. aureus can also cause life-threatening infections both in the community and healthcare settings. Staphylococcal infections range in severity from uncomplicated skin and soft tissue infections (such as folliculitis) to the more severe infections like necrotizing pneumonia and endocarditis.1,2
Resistance of staphylococci to antimicrobial agents is an issue of worldwide concern with a history of almost 70 years. Penicillin was successfully used to treat S. aureus until 1942, when penicillin-resistant S. aureus appeared.3 In 1961, methicillin-resistant S. aureus (MRSA) was reported from England and is now a common cause of hospital-acquired infections.4
Vancomycin is the main antimicrobial agent available to treat serious infections caused by MRSA. In May 1996, the first documented clinical infection due to S. aureus with the intermediate resistance to vancomycin (minimum inhibitory concentration [MIC] equal to 8 µg/ml) was reported from Japan.5 Later, vancomycin-intermediate S. aureus (VISA) strains were isolated in USA, Australia, Europe and other Asian countries.2
The first clinical MRSA isolate exhibiting high-level resistance to glycopeptides (vancomycin MIC>256 µg/ml; teicoplanin MIC=128 µg/ml) due to acquisition of the vanA operon was detected in 2002 from Michigan.6 Although there are few reports of vancomycin-resistant Staphylcoccus aureus (VRSA) worldwide, it seems that Iran is a hot spot region for the emergence of these isolates.7-10
The present study aimed to determine the antimicrobial susceptibility patterns of S. aureus strains isolates from patients in Shiraz hospitals in south of Iran with emphasis to the possible presence of vancomycin resistance.
Methods
A total number of 220 staphylococcal samples were collected from laboratories of Shiraz hospitals (Shahid-Faghihi, Namazi and MRI) from March to December 2012. The strains were collected from various clinical specimens including pus, pharynx, sputum, wound swabs, skin, urine and blood. One hundred S. aureus isolates were selected after identification by standard biochemical and microbiological tests, including Gram staining, oxidase, catalase, coagulase, latex agglutination, motility, DNase, haemolysis and mannitol fermentation tests.11
To confirm the strains, PCR amplification of the nuc gene was performed for all isolates with these primers; nucF 5’GCGATTGATGGTGATACGGTT3’ and nucR 5’AGCCAAGCCTTGACGAACTAAAGC 3’, according to Brakstad et al. (1992).12
Staphylcoccus aureus ATCC 29213 and Enterococcus faecalis ATCC 29212 strains were used as vancomycin-susceptible controls. Vancomycin-resistant Enterococcus faecalis ATCC 51299 was used as a positive control.
Disc agar diffusion (DAD) test was carried out using Kirby-Bauer method according to CLSI procedure with the following discs, which are all from MAST company, Liverpool, UK: penicillin G (10µg), ampicillin (10 µg), amoxicillin (20 µg), vancomycin (30 µg), clindamycin (2 µg), rifampin (5 µg), ciprofloxacin (5 µg), oxacillin (1 µg), tetracycline (30 µg), erythromycin (15 µg), linezolid (30 µg), quinupristin-dalfopristin (15 µg), and teicoplanin (30 µg). Mueller–Hinton agar plates were overlaid with the inoculum (turbidity equivalent to that of a 0.5 McFarland standard) of the S. aureus clinical strains. The zone diameter of bacterial growth inhibition surrounding the disc was measured and compared with a standard for each drug. S. aureus ATCC 25923 was used as quality control strain for the DAD test.13
Vancomycin agar screen plates were prepared In-house by addition of 6 mg/L vancomycin to brain heart infusion (BHI) agar (Merck, Germany). Inoculum suspension was prepared by transferring colonies from overnight growth on nutrient agar plate to sterile saline to produce a suspension that matches the turbidity of a 0.5 McFarland standard. Then, 0.1 ml of this suspension was spread on BHI agar containing 6 mg/L of vancomycin (BHI6V), the vancomycin agar screen plate, and was incubated for 24h at 35ºC in ambient air. E.fecalis ATCC 29212 was used as a vancomycin-susceptible and E.fecalis ATCC 51299 was used as a vancomycin-resistant control, according to the CLSI guideline.14
Genes encoding the vancomycin resistance determinants, vanA and vanB, were investigated by PCR using specific primers (Table 1).15 PCR amplification was carried out in a 20µl reaction mixture with each primer as the following steps: an initial denaturation step at 98°C for 2 min; followed by 35 cycles of 98°C for 10 sec, 50°C for 1 min and 72°C for 90 sec for vanA gene, and an initial denaturation step at 94°C for 10 min; followed by 30 cycles of 94°C for 30 sec, 50°C for 45 sec and 72°C for 30 sec for vanB gene, then finally elongation step at 72°C for 10 min. The PCR products were electrophoresed in a 1.5% agarose gel which was stained with ethidium bromide and visualized by using UV transillumination.15
Table 1. Primers used in this study.
| Target | Primer | Sequence (5’to3’) | Product size (bp) | Reference |
|---|---|---|---|---|
| nuc | Forward | GCG ATT GAT GGT GAT ACG GTT | 279 | 12 |
| Reverse | AGC CAA GCC TTG ACG AAC TAA AGC | |||
| vanA | Forward | ATG AAT AGA ATA AAA GTT GC | 1032 | 15 |
| Reverse | TCA CCC CTT TAA CGC TAA TA | |||
| vanB | Forward | GTG ACA AAC CGG AGG CGA GGA | 430 | 15 |
| Reverse | CCG CCA TCC TCC TGC AAA AAA | |||
| (ddl E. faecium) | Forward | CAGTGCATGTGCCATGGATA | 429 | 16 |
| Reverse | ACTTCGGCTGGAATCTGCAT | |||
| (ddl E. faecalis) | Forward | CAGAAGTGAAGAGCACGATG | 688 | 16 |
| Reverse | AGGTAAAGTCGTACGGACAT |
This PCR was performed to rule out the contamination by Enterococcus spp. Oligonucleotide primers were directed to the ddlE. faecalis, and ddlE. faecium genes are shown in table 1. The ddl primers yielded a product of 429bp for E. faecalis and 688 bp for E. faecium. PCR amplification was programmed as follows: 10 min at 95°C; 30 cycles of 30 s at 94°C, 30 s at 58°C, and 30 s at 72°C; and 10 min at 72°C. Samples were held at 4°C until the products could be analyzed. Ten-microliter samples of the PCR products were electrophoresed through a 1.5% agarose gel for 45 min at 150 V. The gels were stained with ethidium bromide and photographed under UV light.16
DNA Sequencing was carried out on PCR products by Takapoozist Company in Iran. The sequences were aligned and compared with reference sequences achieved using GenBank with the BLAST system.
Results
In this study, 100 strains were isolated from patients and all were confirmed as S. aureus by amplification of nuc gene. The age of patients ranged from 5 to 60 years with an average age of 30.4 ± 42.4 and 56% out of them were male. The number of isolated staphylococci from each ward and their antimicrobial resistance rates are detailed in table 2. Most of the isolates were from internal medicine (n=29), surgery (n=28) and ICU (n=16) wards. Resistance rates were highest for ampicillin (n=95) and lowest for quinupristin-dalfopristin (n=1) (Table 2).
Table 2. Antimicrobial resistance rates of S. aureus isolates in different wards.
| Antimicrobial Agent | Resistant rates (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Internal Medicine (n=29) |
Surgery (n=28) |
ICU (n=16) |
NICU (n=7) |
Dermatology (n=7) |
Outpatient (n=7) |
Emergency (n=4) |
Dialysis (n=2) |
Total (n=100) |
|
| Ampicillin | 93.1 | 96.4 | 100 | 100 | 100 | 71.4 | 100 | 100 | 95 |
| Penicillin G | 100 | 92.9 | 100 | 100 | 85.7 | 71.4 | 100 | 50 | 94 |
| Amoxicillin | 96.1 | 89.3 | 100 | 100 | 71.4 | 85.7 | 75 | 100 | 92 |
| Tetracycline | 51.8 | 50 | 50 | 57.1 | 28.6 | 14.3 | 75 | 0 | 47 |
| Ciprofloxacin | 48.3 | 42.9 | 50 | 71.4 | 28.6 | 28.6 | 75 | 0 | 46 |
| Erythromycin | 37.9 | 46.4 | 43.7 | 71.4 | 28.6 | 28.6 | 75 | 50 | 44 |
| Oxacillin | 44.8 | 46.4 | 43.7 | 57.1 | 28.6 | 28.6 | 75 | 0 | 44 |
| Clindamycin | 34.5 | 35.7 | 43.7 | 57.1 | 28.6 | 28.6 | 75 | 0 | 38 |
| Rifampin | 37.9 | 17.9 | 31.2 | 14.3 | 28.6 | 28.6 | 75 | 0 | 29 |
| Vancomycin | 0 | 3.7 | 0 | 14.3 | 14.3 | 0 | 0 | 0 | 3 |
| Linezolid | 0 | 3.7 | 0 | 0 | 14.3 | 0 | 0 | 0 | 2 |
| Teicoplanin | 0 | 0 | 0 | 0 | 14.3 | 0 | 0 | 0 | 1 |
| Synercid* | 0 | 0 | 0 | 0 | 14.3 | 0 | 0 | 0 | 1 |
* Synercid = Quinupristin-dalfopristin
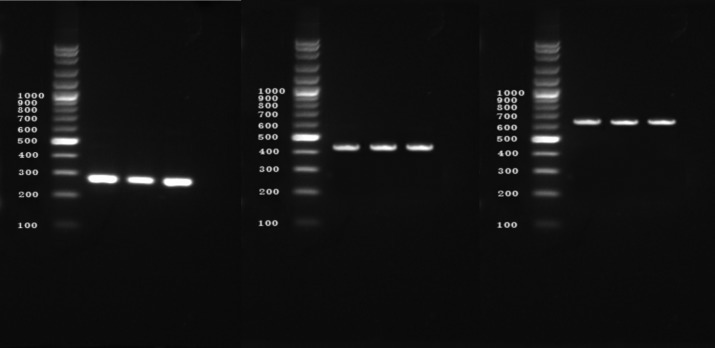
Amplification of nuc, ddlE. faecalis and ddlE. faecium targets produced distinct bands corresponding to their respective molecular sizes that were easily recognizable (Fig. 1)
Figure 1.
Agarose gel electrophoresis of PCR-amplified nuc and ddl genes (E.faecalis and E. faecium). Lanes: M, 100-bp ladder; 1-3: S. aureus isolates showing 279 bp nuc amplicon; 4-6: S. aureus isolates showing 429 bp ddl genes (E.faecalis) amplicon,7-9 S. aureus isolates showing 688 bp ddl genes (E. faecium) amplicon.
There were 3 van gene containing S. aureus (VRSA) strains according to disc diffusion test; one strain was isolated from a 35-year old female patient in dermatology ward. It was resistant to all antibiotics tested except ciprofloxacin. Two other VRSA isolates were also found; one isolated from blood of a patient in NICU and another from a wound of a patient in surgical ward (Data not shown).
Screening for vancomycin resistance showed that 37% isolates can grow on BHI6V. Further studies by PCR also detected vanA and/or vanB genes in all of these strains (Fig. 2). Also, 19 isolates showed either vanA or vanB but were susceptible according to vancomycin agar screening test. Totally, vanA and vanB resistant genes were detected in 34% and 37% of clinical isolates, respectively (Table 3).
Figure 2.
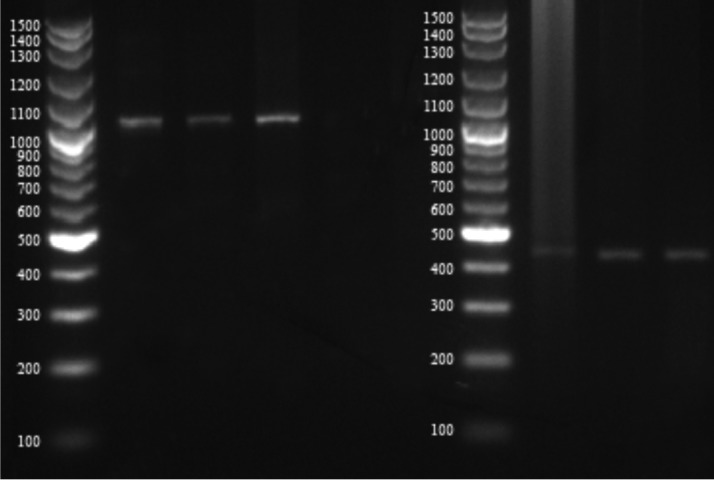
Agarose gel electrophoresis of PCR-amplified vancomycin resistance genes (vanA and vanB). Lanes: M, 100-bp ladder; 1-3: S. aureus isolates showing 1032 bp vanA amplicon; 4-6: S. aureus isolates showing 430 bp vanB amplicon.
Table 3. Results of vanA and vanB PCR compared to vancomycin screening agar test.
| vanA | vanB | (vanA + vanB) | |||||
|---|---|---|---|---|---|---|---|
| + | – | + | – | + | – | ||
| Screening agar | + | 28 | 24 | 28 | 24 | 19 | 33 |
| – | 6 | 39 | 9 | 37 | 0 | 48 | |
| Total | 34 | 63 | 37 | 61 | 19 | 81 | |
Discussion
During the past decade VRSA did not spread rapidly and there were only a few reports of this superbug. Until the end of 2012, 33 cases of vanA-type VRSA have been reported worldwide: 13 from the United States, 16 from India, 3 from Iran (2 from Tehran, 1 from Mashhad) and 1 from Pakistan.7 Limited spread of VRSA is attributed to the highly-costly vanA operon for S. aureus, which can be acquired from enterococcal conjugation.17
Recent articles show that VRSA is now reported in at least four continents (i.e. Asia, Europe, North and South America).7,18,19 The European VRSA was isolated in May 2013 from pus of the toe amputation wound of a 74-year-old female in a Portuguese hospital. The patient had multiple co-morbidities and her culture grew Pseudomonas aeruginosa, vancomycin-resistant Enterococcus faecalis, and methicillin-resistant VRSA.18 There is also another report from Brazil which describes a 35-year-old male with a history of diabetes mellitus and Sezary syndrome who had blood culture positive for methicillin-resistant VRSA. Vancomycin resistant E. faecalis was also isolated from the patient and he died despite vancomycin therapy.19 Unfortunately, we do not have enough clinical information regarding our VRSA strains.
In contrast to previous studies, we found a high number of vanA and/or vanB-VRSA confirmed by PCR which were phenotypically susceptible to vancomycin. Recently, Banerjee et al also found two vanA-VISA strains which none of these two expressed vanA operon. This finding can at least partly explain our results.20
There is only one report of vanB-VRSA in the literature. Also, VRSA isolates simultaneously with vanA and vanB genes have been found only in the mentioned study.21 In a study from India, Chakraborty and colleagues found eight VRSA strains from 30 S. aureus isolated from pus samples. All eight isolates had simultaneously vanA and vanB genes and were also resistant to vancomycin according to vancomycin macro-broth dilution. Compared to this study, we found all types of vancomycin resistance including vanA, vanB and vanA+vanB and some of our isolates were phenotypically susceptible to vancomycin.
In our study, about 40% of the isolates harbored at least one of the van genes. There is a possibility that these infections were caused by dissemination of a few clones of VRSA circulating in south of Iran but, we can neither confirm nor exclude this possibility.
Previously, all VRSA isolates were considered to be susceptible to newer antimicrobial agents such as linezolid and quinupristin-dalfopristin. Saravolatz et al reported that all VRSA reported in the United States were susceptible to these agents.22 In our study, one of the vanA-VRSA isolates from dermatology ward was resistant to linezolid and quinupristin-dalfopristin. Further studies of this isolate with E-test also confirmed our findings (Data not shown). Interestingly, this isolate was susceptible to ciprofloxacin which is unusual and has been also reported in a study which has reported VRSA from Kolkata.23,24 Recently, Alzolibani et al have reported VRSA among children with atopic dermatitis in Saudi Arabia but the authors did not confirm their isolates by PCR of van genes.25 Although we found several VRSA in our study, these isolates remain very rare and are not usually detected.26,27
Conclusion
Our results showed that the frequency of vancomycin resistance genes (vanA, vanB) is very high in Staphylococcus aureus strains isolated from patients in Shiraz hospitals and multidrug-resistant VRSA is also emerging. Thus urgent interventions are needed to keep the emergence and spread of these isolates to a minimum.
Acknowledgements
The authors wish to thank laboratory personals of Shahid-Faghihi, Namazi and MRI hospitals in Shiraz for cooperation in sample collection. The study was conducted with the financial help of Jahrom University of Medical Sciences.
References
- 1.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 2010. May;375(9725):1557-1568. 10.1016/S0140-6736(09)61999-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howden BP, Davies JK, Johnson PD, Stinear TP, Grayson ML. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev 2010. Jan;23(1):99-139. 10.1128/CMR.00042-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jevons MP. ‘‘Celbenin’’-resistant Staphylococci. BMJ 1961;1:124-125 . 10.1136/bmj.1.5219.124-a [DOI] [Google Scholar]
- 4.Deresinski S. Methicillin-resistant Staphylococcus aureus: an evolutionary, epidemiologic, and therapeutic odyssey. Clin Infect Dis 2005. Feb;40(4):562-573. 10.1086/427701 [DOI] [PubMed] [Google Scholar]
- 5.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother 1997. Jul;40(1):135-136. 10.1093/jac/40.1.135 [DOI] [PubMed] [Google Scholar]
- 6.Chang S, Sievert DM, Hageman JC, Boulton ML, Tenover FC, Downes FP, et al. Vancomycin-Resistant Staphylococcus aureus Investigative Team . Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N Engl J Med 2003. Apr;348(14):1342-1347. 10.1056/NEJMoa025025 [DOI] [PubMed] [Google Scholar]
- 7.Askari E, Tabatabai SM, Arianpoor A, Naderi Nasab M. VanA-positive vancomycin–resistant Staphylococcus aureus: systematic search and review of reported cases. Infect Dis Clin Pract 2013;21:91-93 . 10.1097/IPC.0b013e31826e8199 [DOI] [Google Scholar]
- 8.Aligholi M, Emaneini M, Jabalameli F, Shahsavan S, Dabiri H, Sedaght H. Emergence of high-level vancomycin-resistant Staphylococcus aureus in the Imam Khomeini Hospital in Tehran. Med Princ Pract 2008;17(5):432-434. 10.1159/000141513 [DOI] [PubMed] [Google Scholar]
- 9.Dezfulian A, Aslani MM, Oskoui M, Farrokh P, Azimirad M, Dabiri H, et al. Identification and characterization of a high vancomycin-resistant Staphylococcus aureus harboring vanA gene cluster isolated from diabetic foot ulcer. Iran J Basic Med Sci 2012. Mar;15(2):803-806. [PMC free article] [PubMed] [Google Scholar]
- 10.Azimian A, Havaei SA, Fazeli H, Naderi M, Ghazvini K, Samiee SM, et al. Genetic characterization of a vancomycin-resistant Staphylococcus aureus isolate from the respiratory tract of a patient in a university hospital in northeastern Iran. J Clin Microbiol 2012. Nov;50(11):3581-3585. 10.1128/JCM.01727-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bannerman TL. Staphylococcus, Micrococcus, and other catalase positive cocci that grow aerobically. In: Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH, ed. Manual of clinical microbiology. ASM Press: Washington DC, 2003; 8: 384–404. [Google Scholar]
- 12.Brakstad OG, Aasbakk K, Maeland JA. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J Clin Microbiol 1992. Jul;30(7):1654-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. 2011. Performance standards for antimicrobial susceptibility testing, 21st informational supplement. CLSI document M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial disk susceptibility tests. Approved standard, 11th ed. CLSI document M02-A11. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 15.Clark NC, Cooksey RC, Hill BC, Swenson JM, Tenover FC. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob Agents Chemother 1993. Nov;37(11):2311-2317. 10.1128/AAC.37.11.2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willems RJ, Top J, van Schaik W, Leavis H, Bonten M, Sirén J, et al. Restricted gene flow among hospital subpopulations of Enterococcus faecium. MBio 2012;3(4):e00151-e12. 10.1128/mBio.00151-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Périchon B, Courvalin P. Glycopeptide resistance, In: Dougherty TJ, Pucci MJ, editors. Antibiotic discovery and development. New York, US: Springer; 2012. p. 515-542. [Google Scholar]
- 18.Melo-Cristino J, Resina C, Manuel V, Lito L, Ramirez M. First case of infection with vancomycin-resistant Staphylococcus aureus in Europe. Lancet 2013. Jul;382(9888):205. 10.1016/S0140-6736(13)61219-2 [DOI] [PubMed] [Google Scholar]
- 19.ProMED-mail. Staphylococcus aureus, MRSA - Brazil: (SP) vancomycin resistant (VRSA). ProMED-mail 2013; 30 Jun: 20130630.1800166. Available at: http://www.promedmail.org/direct.php?id=20130630.1800166 Accessed July 26 2013.
- 20.Banerjee T, Anupurba S. Colonization with vancomycin-intermediate Staphylococcus aureus strains containing the vanA resistance gene in a tertiary-care center in north India. J Clin Microbiol 2012. May;50(5):1730-1732. 10.1128/JCM.06208-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakraborty SP. KarMahapatra S, Bal M, Roy S. Isolation and identification of vancomycin resistant Staphylococcus aureus from post operative pus sample. Am J Med Sci 2011;4:152-168 [Google Scholar]
- 22.Saravolatz LD, Pawlak J, Johnson L, Bonilla H, Saravolatz LD, II, Fakih MG, et al. In vitro activities of LTX-109, a synthetic antimicrobial peptide, against methicillin-resistant, vancomycin-intermediate, vancomycin-resistant, daptomycin-nonsusceptible, and linezolid-nonsusceptible Staphylococcus aureus. Antimicrob Agents Chemother 2012. Aug;56(8):4478-4482. 10.1128/AAC.00194-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gould IM. Clinical activity of anti-Gram-positive agents against methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother 2011. May;66(Suppl 4):iv17-iv21. 10.1093/jac/dkr073 [DOI] [PubMed] [Google Scholar]
- 24.Saha B, Singh AK, Ghosh A, Bal M. Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia). J Med Microbiol 2008. Jan;57(Pt 1):72-79. 10.1099/jmm.0.47144-0 [DOI] [PubMed] [Google Scholar]
- 25.Alzolibani AA, Al Robaee AA, Al Shobaili HA, Bilal JA, Issa Ahmad M, Bin Saif G. Documentation of vancomycin-resistant Staphylococcus aureus (VRSA) among children with atopic dermatitis in the Qassim region, Saudi Arabia. Acta Dermatovenerol Alp Pannonica Adriat 2012. Sep;21(3):51-53. [DOI] [PubMed] [Google Scholar]
- 26.Al-Yaqoubi M, Elhag K. Susceptibilities of common bacterial isolates from oman to old and new antibiotics. Oman Med J 2008. Jul;23(3):173-178. [PMC free article] [PubMed] [Google Scholar]
- 27.Hamid ME, Mustafa FY, Alwaily A, Abdelrahman S, Al Azragi T. Prevalence of bacterial pathogens in Aseer region, Kingdom of Saudi Arabia: emphasis on antimicrobial susceptibility of Staphylococcus aureus. Oman Med J 2011. Sep;26(5):368-370. 10.5001/omj.2011.91 [DOI] [PMC free article] [PubMed] [Google Scholar]