Abstract
MicroRNAs manifest significant functions in brain neural stem cell (NSC) self-renewal and differentiation through the post-transcriptional regulation of neurogenesis genes. Let-7b is expressed in the mammalian brain and regulates NSC proliferation and differentiation by targeting the nuclear receptor TLX, which is an essential regulator of NSC self-renewal. Whether let-7b and TLX act as important regulators in retinal progenitor cell (RPC) proliferation and differentiation remains unknown. Here, our data show that let-7b and TLX play important roles in controlling RPC fate determination in vitro. Let-7b suppresses TLX expression to negatively regulate RPC proliferation and accelerate the neuronal and glial differentiation of RPCs. The overexpression of let-7b downregulates TLX levels in RPCs, leading to reduced RPC proliferation and increased neuronal and glial differentiation, whereas antisense knockdown of let-7b produces robust TLX expression,enhanced RPC proliferation and decreased differentiation. Moreover, the inhibition of endogenous TLX by small interfering RNA suppresses RPC proliferation and promotes RPC differentiation. Furthermore, overexpression of TLX rescues let-7b-induced proliferation deficiency and weakens the RPC differentiation enhancement caused by let-7b alone. These results suggest that let-7b, by forming a negative feedback loop with TLX, provides a novel model to regulate the proliferation and differentiation of retinal progenitors in vitro.
Retinal progenitor cells (RPCs) are a subtype of undifferentiated cells that retain the ability to self-renew and can generate both retinal neuronal and glial lineages. Although much effort has been devoted to understanding the properties of retinal progenitor cells, the mechanisms of the regulation underlying RPC proliferation and differentiation are not well characterized. Numerous papers have described MicroRNAs (miRNAs) as 22-nucleotide (nt) single-stranded non-coding RNAs that participate in various biological functions, including processes involved in development, pathophysiology, cancer biology, immunology, and cell fate decision1,2,3,4. As genetic regulators of stem cell fate determination, miRNAs regulate gene expression at the post-transcriptional level by mediating complementary sequences for mRNA degradation or translational repression5,6. Analyses of miRNA expression profiles have demonstrated tissue- and stage-specific miRNAs, including the lethal-7 (let-7) family of miRNAs, miR-124, miR-9, miR-137, miR-184, and miR-128, which regulate neurogenesis3. Among them, the let-7 gene was first discovered in Caenorhabditis elegans and is highly conserved across species in both its sequence and function as a key developmental regulator7,8. As a member of the let-7 family, let-7b was demonstrated to be expressed in the brain and retina and upregulated upon neuronal differentiation of mouse and human embryonic carcinoma cells and retinal progenitors7,9,10,11. In addition, the overexpression of let-7b in brain neural stem cells (NSCs) was found to lead to decreased proliferation but to promote both neuronal and glial differentiation by targeting the stem cell regulator TLX12,13. However, whether let-7b plays an important role in the proliferation and differentiation of neural retinal progenitors remains unclear.
TLX (NR2E1), an orphan nuclear receptor expressed in vertebrate forebrains and highly expressed in the adult brain, is an essential intrinsic regulator that is involved in both maintaining the proliferation and inhibiting the differentiation of embryonic and adult neural stem cells14,15. A previous study demonstrated that TLX contributed to normal brain-eye development and appeared to play a role in the control of human behavioral disorders15. In addition, TLX was identified to work in parallel with other pathways to control the decision of whether to continue proliferating or to differentiate into neurons; early disruption in progenitor cell proliferation/differentiation has long-lasting consequences on the formation of later generated structures16. According to recent studies, TLX, which is highly expressed in brain neural stem cells, plays a critical role in neural development by regulating cell cycle progression and the exit of neural stem cells in the developing brain14,16. Furthermore, TLX is expressed in retinal progenitor cells and thus underlies the fundamental developmental program of retinal organization and controls the generation of the appropriate number of retinal progenies17. Nevertheless, the function of TLX in RPC proliferation and differentiation remains to be characterized.
In the present study, we investigated the interaction between let-7b and TLX during RPC proliferation and differentiation in vitro and found that let-7b was expressed at low levels in undifferentiated RPC cultures; however, its expression was strengthened during differentiation. In addition, the expression levels of TLX were decreased gradually upon differentiation. Increasing the level of let-7b downregulated the expression of TLX, leading to decreased RPC proliferation and accelerated neuronal and glial differentiation, whereas knockdown of let-7b resulted in increased TLX expression and enhanced RPC proliferation. Furthermore, the overexpression of TLX led to downregulated let-7b expression and reduced RPC differentiation. The present study suggests that let-7b and TLX act as important regulators in regulating the proliferation and differentiation of RPCs.
Results
The expression levels of let-7b and its target gene TLX during RPC differentiation
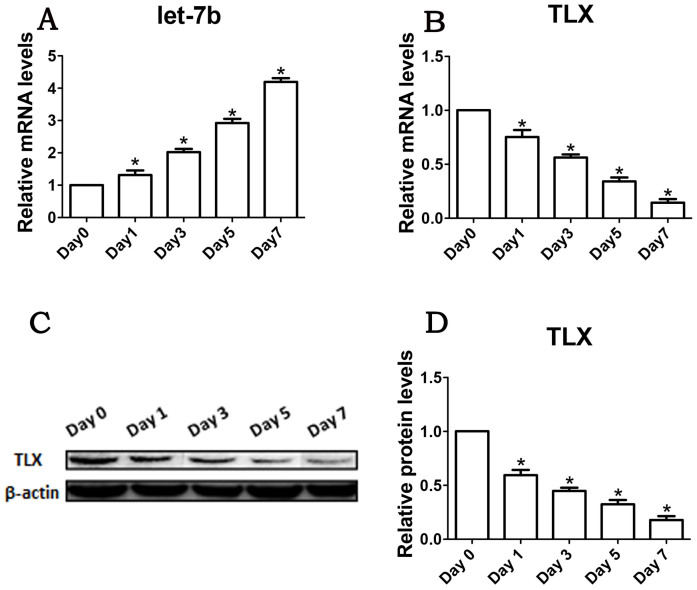
Recently, let-7b was identified to function as a regulator in brain neural stem cell biology. TLX was demonstrated to be the target gene of let-7b and to interact with let-7b in cell fate determination. To investigate the expression tendency of let-7b and TLX during RPC differentiation, the expression levels were evaluated using a qPCR analysis. As shown in Fig. 1, let-7b is expressed in RPCs isolated from the fresh retinal tissue of postnatal day one GFP transgenic C57BL/6 mice (d0). The expression of let-7b is progressively increased during RPC differentiation (Fig. 1A), however, and is accompanied by the noticeably decreased expression of TLX (Fig. 1B). Western blot analysis was also performed to evaluate the protein levels of TLX that decreased during RPC differentiation (Fig. 1C and D). These results suggested that let-7b and TLX might function as important regulators in RPC differentiation.
Figure 1. The expression levels of let-7b and TLX in RPCs during differentiation.
Seven days after RPCs were cultured under differentiation conditions, the expression levels of let-7b and TLX were analyzed by qPCR and Western blotting. (A, B): The qPCR results showed that the expression level of let-7b gradually increased, whereas TLX exhibited the reciprocal trend during 7-day differentiation of RPCs. (C, D): Western blot results showed that the protein level of TLX was downregulated during the same differentiation time course. Day 0 represents the undifferentiated RPC state and was normalized as 1. Western bands were scanned and normalized to the internal control β-actin. Error bars indicate the standard deviation of the mean; *p < 0.05 by Student's t-test. Full-length blots/gels are presented in Supplementary Figure 5.
Let-7b inhibits TLX expression in RPC culture
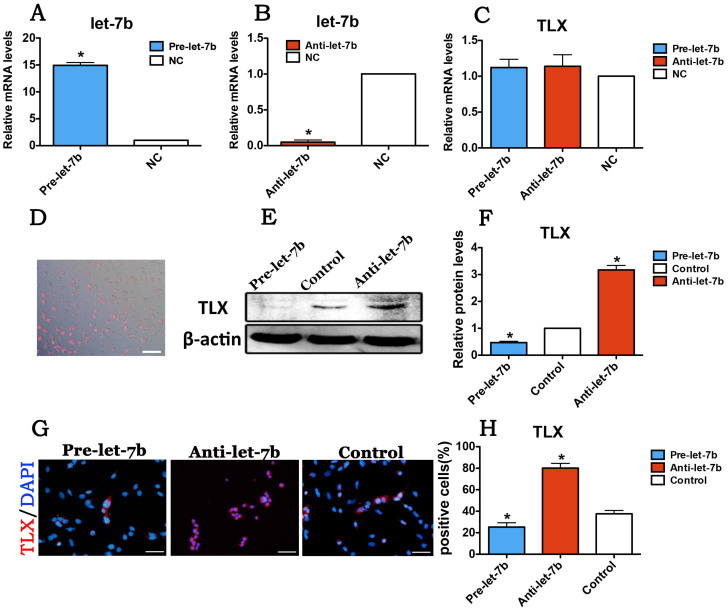
qPCR was used to detect the expression levels of let-7b and TLX in RPCs treated with let-7b mimics or let-7b inhibitor. And the transfection of let-7b mimics into RPCs led to an approximately 15-fold increased expression level of let-7b (Fig. 2A), whereas obvious cytotoxicity was not detected in transfected cells (Supplementary Fig. 4).
Figure 2. Let-7b regulated the expression of TLX in RPCs during proliferation.
The expression levels of let-7b and TLX in RPCs, which were transfected with let-7b mimics or let-7b inhibitor, were analyzed. (A, B): The qPCR results displayed that the expression of let-7b was sharply upregulated with let-7b mimics transfection and significantly downregulated with let-7b inhibitor treatment in RPCs. (C): The mRNA level of TLX in RPCs showed no obvious change when treated with let-7b mimics or let-7b inhibitor. (D): A high level of cy3 expression in the transfected RPCs was revealed using fluorescence microscopy. (E–H): the results of Western blotting and immunostaining are consistent with those of qPCR. Error bars indicate the standard deviation of the mean. *p < 0.05 by Student's t-test. Scale bars: 200 μm (D), 50 μm (G). Full-length blots/gels are presented in Supplementary Figure 6.
The expression of let-7b was repressed more than 90% when treated with let-7b inhibitor in RPCs (Fig. 2B). However, no obvious change was noted in the mRNA expression level of TLX in the let-7b mimics or the let-7b inhibitor treatment group (Fig. 2C), suggesting that let-7b regulated the expression of TLX through inhibiting translation. In the present study, a transfection efficiency of more than 90% was detected by fluorescent microscopy (Fig. 2D). In addition, the Western blot data further demonstrated the above results according to that level of TLX, which was significantly repressed in let-7b mimic-treated RPC cultures but increased by approximately 3-fold in the cells treated with let-7b inhibitor (Fig. 2E and F). Additionally, the immunostaining results exhibited an increased rate of TLX-positive cells with let-7b inhibitor treatment compared with controls in RPC cultures under proliferation conditions (Fig. 2G and H). Collectively, these data indicate that let-7b negatively regulates TLX expression in RPC cultures.
Let-7b inhibits RPC proliferation
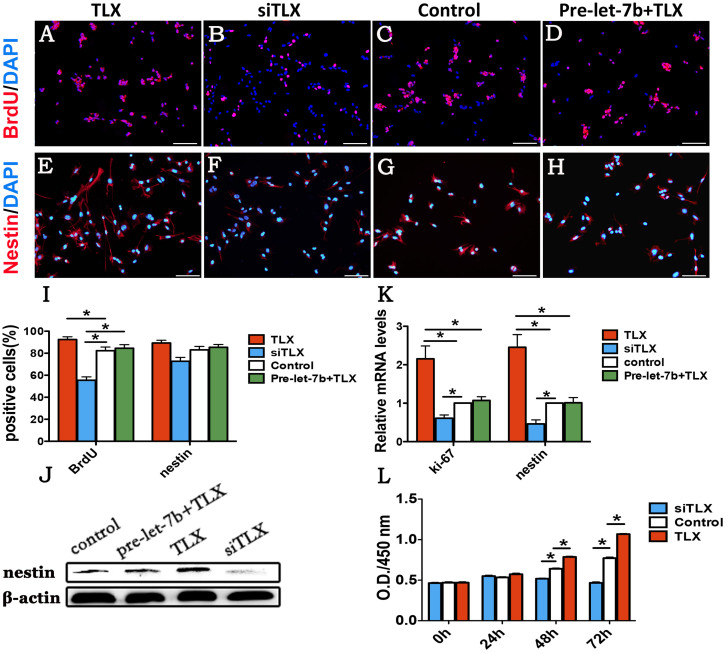
To test whether let-7b regulates RPC proliferation, RPCs transfected with let-7b mimics or let-7b inhibitor were cultured under proliferation conditions. The effect of let-7b on RPC proliferation was first investigated by immunostaining analysis. As revealed in the immunostaining results, at baseline, over 80% of the RPCs expressed the cell proliferation marker 5-bromodeoxyuridine (BrdU) (Fig. 3C, and I), and over 80% of the RPCs expressed the retinal progenitor marker nestin (Fig. 3G, and I), which is indicative of active proliferation. Following 4 days under different treatments, the proportion of BrdU-positive cells was found to be higher than that in the control group when treated with let-7b inhibitor (Fig. 3A, and I). In contrast, a decrease in the BrdU-positive cell percentage was quite apparent in the cell group treated with let-7b mimics (Fig. 3B, and I). Additionally, the percentage of nestin-immunoreactive cells was decreased or increased in RPCs treated with let-7b mimics or let-7b inhibitor, respectively, compared with the control cells (Fig. 3E, F, and I). The results of Western blotting showed a notable decrease in the nestin protein level that was attributable to the overexpression of let-7b in the RPC cultures (Fig. 3J), and an increased nestin level was detected by inhibiting let-7b in RPC cultures. Additionally, qPCR data showed that the expression levels of nestin and ki-67 were notably downregulated (>2-fold) in the let-7b mimics-treated RPC cultures and upregulated in let-7b inhibitor-treated RPC cultures (Fig. 3K). Furthermore, from the results of co-transfecting both let-7b and TLX in RPCs (Fig. 3I, K and J), we found that the negative effect of let-7b overexpression on the RPC proliferation capacity was considerably rescued by TLX treatment. To evaluate the effects of let-7b on RPC proliferation, CCK8 analysis was also performed on RPC cultures, which were treated with let-7b mimics or inhibitor under proliferation conditions. The data indicated that no obvious difference in the proliferation capacity was observed between the first two days of culture; thereafter, an inhibited expansion ability was recorded for RPC cultures treated with let-7b mimics, while the let-7b inhibitor enhanced the proliferation of RPCs (Fig. 3L). These results suggest that let-7b can negatively regulate RPC proliferation and that TLX can negatively affect the function of let-7b acting on RPC expansion.
Figure 3. Let-7b inhibits RPC proliferation.
(A–H): Four days after RPCs were cultured under proliferation conditions, the cell proliferation marker BrdU and retinal progenitor marker nestin were evaluated by immunostaining analysis. The cells treated with let-7b mimics or let-7b inhibitor were immunostained with antibodies against BrdU and nestin, respectively. Cell nuclei were counterstained with DAPI. (I): Quantification of cell proliferation and retinal progenitors using the percentage of BrdU- and nestin-positive cells. The expression of BrdU was decreased in the let-7b mimics-treated RPCs and increased when RPCs were treated with let-7b inhibitor; the overexpression of TLX rescued the effects of let-7b on RPC proliferation in the RPCs. (J): The Western blot results displayed that the expression level of nestin was obviously reduced in the let-7b mimics-treated RPC cultures and increased in the RPC cultures when treated with let-7b inhibitor; the downregulation of nestin by let-7b was blocked by overexpression of TLX. (K): qPCR analysis of the expression level of ki-67 and nestin was consistent with the results shown above. (L): The proliferation ability of RPCs treated with let-7b mimics and let-7b inhibitor were assessed using CCK-8 analysis. The proliferation ability of RPCs was obviously increased when treated with let-7b inhibitor and decreased when treated with let-7b mimics in 48- and 72-h cultures under proliferation conditions. Error bars indicate the standard deviation of the mean; *p < 0.05 by Student's t-test. Scale bars: 100 μm. Full-length blots/gels are presented in Supplementary Figure 7.
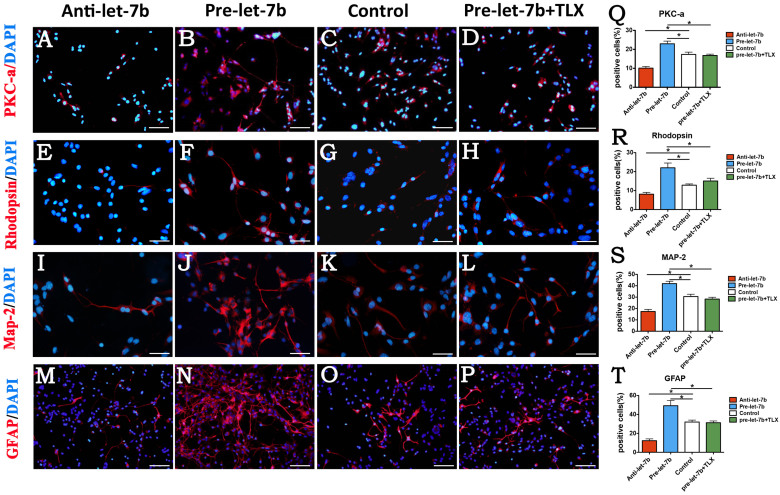
Let-7b accelerates RPC differentiation
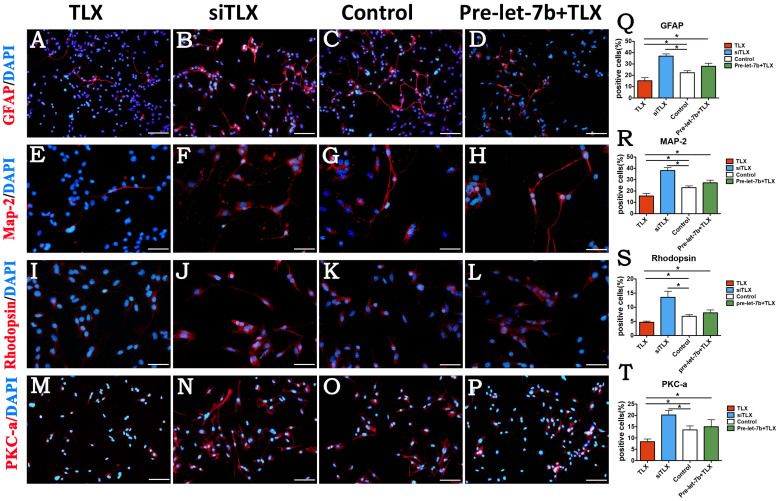
After 7 days of culture under differentiation conditions, the cells treated with let-7b mimics, let-7b inhibitor, let-7b mimics plus TLX clone, and negative control were fixed and immunostained with antibodies against PKC-α, rhodopsin, MAP-2, and GFAP. Immunostaining analyses showed that in let-7b mimics-treated RPC cultures compared with control cells, both retinal neuronal and glial cell markers, including PKC-α (22.94% vs 17.33%, respectively,) (Fig. 4B, C, and Q), rhodopsin (22.04% vs 12.87%, respectively) (Fig. 4F, G, and R), MAP-2 (41.88% vs 30.52%, respectively) (Fig. 4J, K, and S), and GFAP (49.23% vs 31.96%, respectively) (Fig. 4N, O, and T), were significantly increased. In contrast, in let-7b inhibitor-treated RPC cultures compared with control cells, the inhibition of let-7b resulted in a substantial decrease in PKC-α (10.11% vs 17.33%, respectively) (Fig. 4A, C, and Q), rhodopsin (8.12% vs 12.87%, respectively) (Fig. 4E, G, and R), MAP-2 (17.40% vs 30.52%, respectively) (Fig. 4I, K, and S), and GFAP (12.26% vs 31.96%, respectively) (Fig. 4 M, O and T). Furthermore, the overexpression of TLX inhibited the effect of let-7b on RPC differentiation (Fig. 4D, H, L, P, Q, R, S, and T). The effects of let-7b on RPC differentiation-related marker expression were also evaluated by qPCR analysis: let-7b mimics-treated RPC cultures exhibited higher levels of PKC-α, rhodopsin, MAP-2, and GFAP than did the control cells (Supplementary Fig. 1A, B, C, and D). However, when the cells were treated with let-7b inhibitor, the levels of PKC-α, rhodopsin, MAP-2, and GFAP were strongly downregulated. The combined effects of treatment with let-7b mimics plus TLX on RPC differentiation were also detected in this study. The qPCR results showed that the addition of TLX obviously diminished the promotion of RPC differentiation caused by let-7b treatment (Supplementary Fig. 1A, B, C, and D). Moreover, it has been indicated in our study that let-7b may prefer to promote the differentiation of rhodopsin (Supplementary Fig. 3), which is the specific marker of rod cells. These results indicate that let-7b remarkably enhances the differentiation of RPCs toward both neuronal and glial cell lineages, whereas TLX weakens the enhancement of differentiation caused by let-7b treatment in RPC cultures.
Figure 4. Let-7b accelerates RPCs differentiation.
(A–P): RPCs were treated with let-7b mimics, let-7b inhibitor, or let-7b mimics plus TLX clone under differentiation conditions for 7 days, fixed, and immunolabeled with antibodies against PKC-α, rhodopsin, MAP-2, and GFAP. (Q–T): The percentages of PKC-α-, rhodopsin-, MAP-2-, and GFAP-positive cells were evaluated to investigate RPC differentiation. The ratio of PKC-α, rhodopsin, MAP-2, and GFAP-positive cells displayed a significant increase in the let-7b mimics-treated RPC cultures and obvious decrease when treated with let-7b inhibitor; however, TLX blocked the enhancement that was caused by overexpression of let-7b in RPC cultures. Error bars indicate the standard deviation of the mean; *p < 0.05 by Student's t-test. Abbreviations: PKC-α, protein kinase C alpha; MAP-2, microtubule-associated protein-2; GFAP, glial fibrillary acidic protein. Scale bars: 100 μm (A–D), 50 μm (E–L), 200 μm (M–P).
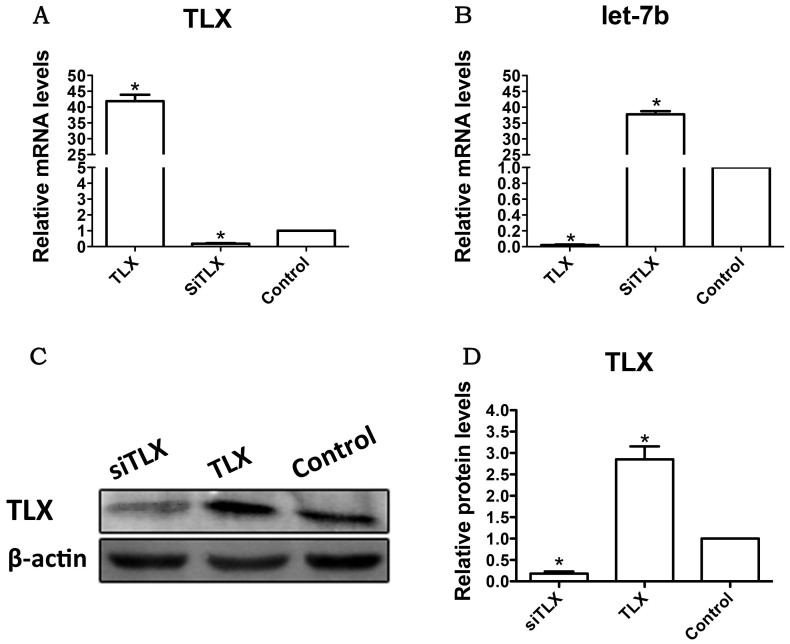
TLX regulates RPC proliferation and differentiation
As one target gene of let-7b, TLX has been identified to play an important role during the proliferation and differentiation of brain neural stem cells14. Whether TLX is also an important regulator in the proliferation and differentiation of RPCs was investigated in this study. The qPCR data showed that the level of TLX was remarkably downregulated when treated with siTLX, but was raised to approximately 43-fold in TLX-treated RPCs (Fig. 5A). In contrast, the expression level of let-7b was significantly upregulated in the range of 35- to 40-fold in the siTLX-treated RPC cultures but showed a very large decrease in cells treated with TLX (Fig. 5B); these findings were consistent with the results from western blot analyses (Fig. 5C and D).
Figure 5. Effects of the overexpression or inhibition of TLX on the expression of let-7b in the RPC cultures.
(A): The qPCR results revealed that the expression level of TLX decreased sharply with siTLX treatment but increased approximately 40-fold in the TLX clone-treated RPC cultures compared with the control. (B–D): The expression level of let-7b was markedly increased in siTLX-treated RPC cultures and decreased in the TLX clone-treated cells, a result that was consistent with that from Western blot analysis. Error bars indicate the standard deviation of the mean; *p < 0.05 by Student's t-test. Full-length blots/gels are presented in Supplementary Figure 8.
Under proliferation conditions, the immunostaining data revealed that siTLX-treated RPC cultures displayed fewer BrdU-positive and nestin-positive cells compared with the control cells (Fig. 6B, C, F, G and I). Additionally, compared with the control cells, the overexpression of TLX obviously increased the percentage of BrdU-positive cells (92.48% vs 82,32%) and slightly increased the percentage of nestin-immunoreactive cells (89.25% vs 83.08%) (Fig. 6A, C, E, G and I). Furthermore, the qPCR results showed that the expression levels of nestin and ki-67 were downregulated by approximately 3- and 2-fold, respectively, in siTLX-treated RPC cultures and were increased by approximately 2.5- and 2-fold, respectively, in cells treated with TLX compared with control cells (Fig. 6K). The protein expression level of nestin (Fig. 6J) exhibited the same trend with the qPCR results. Our CCK8 analysis also demonstrated a positive role for TLX in promoting RPC proliferation (Fig. 6L). To define the role of TLX in RPC differentiation, RPCs treated with the TLX clone or siTLX were cultured under differentiation conditions for 7 days; the cells were then fixed and immunostained with antibodies against PKC-α (Fig. 7A, B, and C), rhodopsin (Fig. 7E, F, and G), MAP-2 (Fig. 7I, J, and K), and GFAP (Fig. 7M, N, and O). The data showed that the percentages of PKC-α-immunoreactive (20.17% vs 13.59%), rhodopsin-immunoreactive (13.51% vs 6.81%), MAP-2-immunoreactive (38.10% vs 22.99%), and GFAP-immunoreactive (36.95% vs 22.27%) cells displayed remarkable increases when treated with siTLX but that these levels decreased sharply under TLX clone treatment (Fig. 7Q, R, S, and T). In addition, with siTLX treatment, the expression levels of GFAP, rhodopsin, MAP-2, and PKC-α in the RPC cultures manifested large increases by about 2-, 1.7-, 2.5- and 1.7-fold, respectively, in qPCR analyses, but their expression levels were apparently decreased in the TLX-treated groups (Supplementary Fig. 2A, B, C, and D). These results suggest that TLX could enhance RPC proliferation and inhibit RPC differentiation.
Figure 6. TLX promotes the proliferation of RPCs.
(A–H): After 4 days of culture under proliferation conditions, the RPCs treated with siTLX or TLX clone were immunostained with antibodies against BrdU and nestin, respectively. (I): The percentage of BrdU -positive cells were decreased in the siTLX-treated RPCs and increased when treated with TLX clone. Co-transfection of TLX clone together with let-7b mimics inhibited the improvement of the proliferation of RPCs, which was caused by TLX treatment alone, however, the percentage of nestin-positive cells showed no obvious change. (J): The protein level of nestin detected by Western blotting appeared to increase with TLX clone treatment alone but was weakened when co-transfected with TLX and let-7b mimics together. Additionally, treatment with siTLX markedly decreased the protein level of nestin in the RPC cultures. (K): The mRNA expression levels of ki-67 and nestin were analyzed by qPCR and showed that the transfection of TLX clone raised the expression of ki-67 and nestin by approximately 2-fold; however, siTLX treatment decreased their expression levels compared with the control. The qPCR results showed that the overexpression of let-7b inhibited the promotion of TLX acting on the expression of ki-67 and nestin. (L): The proliferation ability of the RPCs transfected with siTLX and TLX clone were assessed using CCK-8 analysis. The expansion capacity of the cells exhibited an obvious improvement in the TLX clone-treated cultures, whereas a weakened proliferation capacity was detected with the siTLX-treated cells under proliferation conditions. Error bars indicate the standard deviation of the mean; *p < 0.05 by Student's t-test. Scale bars: 100 μm. Full-length blots/gels are presented in Supplementary Figure 9.
Figure 7. TLX inhibits the differentiation of RPCs.
(A–T): RPCs transfected with TLX and immunostaining with antibodies against GFAP, MAP-2, rhodopsin, and PKC-α, the proportions of GFAP-, MAP-2-, rhodopsin-and PKC-α- immunoreactive cells were detected to be higher in the siTLX-treated cultures than in the control cells, but their positive percentage decreased by TLX clone treatment. Treatment with let-7b rescued the reduction, which was caused by the overexpression of TLX. Error bars indicate the standard deviation of the mean; *p < 0.05 by Student's t-test. Scale bars: 200 μm (A–B), 50 μm (E–L), 100 μm (M–P).
Under proliferation conditions, the results from immunostaining (Fig. 6D, H, and I), western blot (Fig. 6J), and qPCR analyses (Fig. 6K) showed that treatment with let-7b mimics inhibited the proliferative promotion induced by TLX overexpression. Additionally, when RPCs were cultured under differentiation conditions, treatment with let-7b mimics rescued the inhibition of RPC differentiation caused by TLX overexpression according to the results of immunostaining (Fig. 7. Q, R, S and T) and qPCR (Supplementary Fig. 2 A, B, C, and D), indicating that TLX may be an important target gene for let-7b in RPC proliferation and differentiation.
Taken together, our data demonstrate that the overexpression of let-7b inhibits the proliferation of RPCs but accelerates the neuronal and glial differentiation of RPCs. Additionally, TLX could enhance RPC proliferation and negatively regulate the effects of let-7b on RPC cultures, suggesting that let-7b and TLX play important roles in regulating RPC fate determination.
Discussion
Let-7b, a member of the let-7 gene family, is a heterochronic switch gene that participates in important biological functions in cell fate decision7,8,9. The expression of let-7b is upregulated significantly during the differentiation of brain neural stem cells (NSCs)13. Increased let-7b expression in neural stem/progenitor cells results in the suppression of their proliferation and the promotion of their neuronal differentiation12,13. In addition, a recent report indicated that let-7b might be an important regulator of the early-to-late developmental transition in retinal progenitors in vivo11. These results suggest that let-7b plays an important role in coordinating the proliferation and differentiation of stem/progenitor cells. However, whether let-7b serves as a potential regulator in RPC proliferation and differentiation remains to be unraveled.
In the present study, we showed that let-7b was expressed at a low level during the proliferation of RPCs but that its expression was gradually increased during the differentiation of RPCs. The latter result is consistent with the expression of let-7b in brain NSCs13. Under proliferative conditions, the overexpression of let-7b in retinal progenitors largely downregulated the expression level of the cell proliferative marker ki-67, indicating that the overexpression of let-7b resulted in the proliferative deficiency of RPCs. However, although the immunoreactivity intensity of nestin, a widely employed marker of retinal progenitor cells18, became faded, the proportion of nestin-positive cells in RPC cultures exhibited no great change after treatment with let-7b mimics. Additionally, the expression of RPC differentiation-related markers, including Map2 and GFAP, were not detected in the RPC cultures treated with let-7b (data not shown). These results demonstrate that the proliferation capacity of RPCs was obviously diminished by let-7b treatment but that the RPCs were maintained in an undifferentiated progenitor state, indicating that let-7b serves as a negative regulator in the proliferation of RPCs.
Previous studies have demonstrated that let-7b plays an important role in NSC differentiation11,13. The extensive involvement of let-7b in the differentiation of RPCs has been explored in the present study. Our data demonstrated that let-7b overexpression could promote RPC differentiation under differentiation conditions, although we failed to induce neural differentiation by overexpressing let-7b alone in cultured RPCs. Because each miRNA potentially targets various mRNAs19, it may often fine-tune gene expression to coordinate cell behaviors but does not primarily regulate genes20. In the present study, the expression levels of RPC differentiation-related markers, including MAP-2 (a marker for pan-neurons), PKC-α (a marker for bipolar neurons), and rhodopsin (a marker for photoreceptors), were increased in let-7b-treated RPC cultures, indicating that the overexpression of let-7b could enhance the neuronal differentiation of RPCs. Regarding the glial differentiation of RPCs, the expression of GFAP, which is positively correlated with the glia population21, was also detected in the present study and showed that the expression of GFAP was upregulated in the RPC cultures treated with let-7b mimics, indicating that let-7b also promotes the glial differentiation of RPCs. Conversely, with let-7b inhibitor treatment, the potential of RPCs to differentiate toward both neuronal and glial cells was diminished. These findings suggest that let-7b plays an important role in promoting the neuronal and glial differentiation of RPCs.
Let-7b has been shown to target multiple key molecules such as TLX13, which is essential in maintaining the undifferentiated and proliferative state of both embryonic and adult NSCs. The abundant expression of TLX was detected in the neuroepithelium, retina and nasal epithelium22,23. TLX may act as an important regulator in early neural progenitor cell self-renewal and prevent neurogenesis24. Additionally, TLX underlies a fundamental developmental program of retinal organization and controls the generation of retinal progenies and development of glial cells during the protracted period of retinogenesis17. In the present study, the expression level of TLX was high under proliferation conditions and dramatically declined during the differentiation of RPCs, a finding that was consistent with a previous report demonstrating that TLX is rich in NSCscultures during proliferation but sharply downregulated upon differentiation13, indicating that TLX may serve as an important regulator in the proliferation and differentiation of RPCs.
In the current study, TLX was abundantly expressed under proliferation conditions and obviously decreased upon the differentiation of RPCs, with a concomitant increased expression of let-7b. The inhibition of TLX suppressed RPC proliferation and promoted RPC differentiation into retinal neuronal and glial cells. Conversely, the overexpression of TLX enhanced RPC proliferation and inhibited RPC differentiation, suggesting that TLX plays an important role in RPC proliferation and differentiation. In addition, the relationship between let-7b and TLX expression resembles that between miR-31 and its target gene Satb224. In both cases, the miRNA targets are preferentially expressed at high levels, while the targeting miRNA expression is low. In turn, the levels of these targets are decreased as their targeting miRNAs increase. In the present study, the expression level of TLX is high when its targeting let-7b expression is low in RPC cultures under proliferation conditions, and TLX expression is decreased as let-7b is upregulated significantly upon differentiation. These data support the previous report showing that increased miRNA expression suppresses its target genes to regulate cell fate transition25.
A single miRNA may control multiple target genes26. However, one or more key targets may be associated with specific cellular contexts among these targets. Let-7b has been indicated to act as an important upstream regulator to negatively modulate TLX expression by directly targeting its 3′untranslated region (UTR) in brain NSC fate determination13. In the current study, we showed that let-7b positively regulated RPC differentiation and negatively regulated both RPC proliferation and TLX expression, whereas TLX promoted RPC proliferation, inhibited RPC differentiation, and downregulated let-7b expression. In addition, the co-transfection of let-7b mimics and TLX into RPCs rescued the let-7b-induced RPC proliferation deficiency and reversed the increased neuronal differentiation caused by let-7b treatment alone. Moreover, the overexpression of let-7b in RPC cultures downregulated the expression of TLX. In contrast, antisense knockdown of let-7b increased TLX expression at the protein level but not the mRNA level, suggesting that let-7b regulates the expression of TLX in RPC cultures by inhibiting the translation of mRNA. These data demonstrate that TLX may be an excellent candidate as a let-7b molecular target by forming a negative regulatory loop with let-7b in regulating RPC cell fate determination.
In summary, our data suggest that let-7b and its target TLX play important roles in RPC proliferation and differentiation: the expression levels of TLX are downregulated by the overexpression of let-7b, leading to proliferative deficiency and accelerated neuronal and glial differentiation; however, the decreased expression of let-7b contributes to the accelerated expression of TLX and enhanced RPC proliferation. Future studies are required to address how miRNAs orchestrate the control of RPC proliferation and differentiation through precise transitions between the progenitors and their differentiated cells. Nevertheless, the data in the present study clearly demonstrate that let-7b, by forming a negative feedback loop with TLX, provides a novel model for regulating the proliferation and differentiation of retinal progenitors in vitro.
Methods
Retinal progenitor cell (RPC) isolation, culture and differentiation in vitro
RPCs were isolated from fresh retinal tissue of postnatal-day-one GFP transgenic C57BL/6 mice (a gift from Dr. Masaru Okabe, University of Osaka, Japan)27,28. The cells were seeded into flasks at a density of 2 × 105 cells/ml with proliferation medium containing advanced DMEM/F12 (Invitrogen, Carlsbad, CA, USA), 1% N2 neural supplement (Invitrogen), 2 mM L-glutamine (Invitrogen), 100 U/ml penicillin-streptomycin (Invitrogen) and 20 ng/ml epidermal growth factor (EGF, Invitrogen)29,30. Every two or three days, the proliferation medium was changed, and the cells were passaged at regular intervals of three or four days. For differentiation, the cells were seeded at a density of 1 × 105 cells/ml with differentiation medium, to which 10% fetal bovine serum (FBS) (Invitrogen) was added while EGF was removed; the cultures were then allowed to grow for 7 days. The culture medium was renewed every two days. All of the animals were handled according to the ARVO animal usage standards and following the approval by the animal care and use committee of the Schepens Eye Research Institute, where the original derivation of the cells was performed.
Transfection
Let-7b oligonucleotides (let-7b mimics (let-7b precursor molecules, whose structures are similar to the intracellular let-7b precursor but with chemical modification and optimization, could be processed into mature miRNA by mimicking the let-7b natural shearing process), let-7b inhibitor, and negative control) and small interfering RNAs (siTLX and negative control) were synthesized by Biomics Biotech Co., Ltd (Nantong, China). The Myc-DDK-tagged ORF cDNA clone of TLX was synthesized by OriGene Technologies, Inc (Beijing, China). For the transfection of RNA duplexes, a 20 nM final concentration in Opti-MEM of let-7b miRNA mimics, let-7b inhibitor, and negative control, siRNA TLX, or TLX clone was mixed with Lipofectamine 2000 (Invitrogen) in serum-free media and incubated at room temperature for 20 min. The mixture was then added to the cells in 6-well plates at 60%–70% degrees of fusion. The medium was replaced by proliferation or differentiation medium 6 hours later. For long-term detection under differentiation culture conditions, let-7b oligonucleotides, small interfering RNAs or the cDNA clones were repeatedly transfected every 3 days. The cells were harvested at the indicated time points after transfection for total protein or RNA extraction. The oligonucleotide sequence of siTLX was as follows: 5′-GAGGAGCAUUCGAAGGAAUTT-3′.
Immunocytochemistry
RPCs were seeded on 18-mm glass coverslips (VWR, West Chester, PA, USA) coated with laminin (Sigma-Aldrich, Saint Louis, MO, USA) in 12-well plates. After 2 days of culture, when the cells were 50–70% confluent, cells were transfected with let-7b mimics, let-7b inhibitor, siTLX, or TLX clone for the indicated time points; the cells were then fixed with 4% paraformaldehyde (Sigma-Aldrich), permeabilized with 0.3% Triton X-100 (Sigma-Aldrich) in PBS, and blocked with 10% normal goat serum (Invitrogen)31. Next, the cells were incubated with rabbit polyclonal anti-TLX (1:50; Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse monoclonal anti-ki-67 (1:200; BD, San Jose, CA, USA), mouse monoclonal anti-nestin (1:200; BD, San Jose, CA, USA), rabbit monoclonal anti-MAP-2 (1:200; Epitomics, Burlingame, CA, USA), mouse monoclonal anti-PKC-α (1:200; BD, San Jose, CA, USA), mouse monoclonal anti-rhodopsin (1:100; Chemicon, Billerica, MA, USA), or mouse monoclonal anti-GFAP (1:200; Chemicon) at 4°C overnight. Thereafter, the cells were incubated with fluorescently labeled secondary antibodies (1:800; Alexa Fluor546- goat anti-mouse/rabbit, BD, San Jose, CA, USA). The cells were then rinsed 3 times in PBS, and the cell nucleus was counterstained with 4′, 6-diamidino-2-phenylindole (DAPI; Invitrogen). Negative controls were performed in parallel using the same protocol but without the primary antibody. Immunoreactive cells were visualized and imaged using a fluorescence microscope (Olympus BX51, Japan). Additionally, the percentage of positive cells was collected using Image-Pro Plus 6.0 (Media Cybernetics, Bethesda, MD, USA), which is automated software used for counting after merging images of immunopositive cells with nuclei stained with DAPI and immunopositive cells treated with primary antibodies.
5-Bromo-2-deoxyuridine (BrdU) incorporation Cells were cultured for 12 h in the presence of 10 μM BrdU (Sigma-Aldrich, Poole, UK), fixed in 4% paraform- aldehyde for 15 min at room temperature and incubated with blocking buffer (PBS containing 10% NGS, 0.3% Triton X- 100, and 100 μg/ml RNaseA) for 60 min at room temperature. Cells were then washed in PBS, incubated with 2M HCl for 30 min at room temperature, and washed with Hanks' Balanced Salt Solution (HBSS) followed by PBS at room temperature. After overnight incubation at 4°C with anti-BrdU antibody (1:200, Santa Cruz Biotechnology, Santa Cruz, CA, USA) in blocking buffer, cells were washed in PBS, and incubated with fluorescent-conjugated secondary antibody (Alexa Fluor488-goat anti-mouse, 1:800) for 1 h at room temperature. Cell nuclei were counterstained with with 4′, 6-diamidino-2-phenylindole (DAPI; Invitrogen). Immunoreactive cells were visualized and imaged using a fluorescence microscope (Olympus BX51, Japan).
Western blot analysis
After the transfection of miRNAs into RPCs, the total proteins of the cells were harvested at specified time points. A BCA kit (Pierce, Rockford, IL, USA) and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were used to analyze the protein concentrations and separate the proteins, respectively24. Following SDS-PAGE, the proteins were transferred to 0.22 mm polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). Next, the membranes were blocked with 5% nonfat milk, and they were then incubated with rabbit polyclonal anti-TLX (Santa Cruz Biotechnology, 1:200), mouse monoclonal anti-nestin (BD, 1:500), and mouse anti-β-actin (Sigma, 1:0000) at 37°C for 2 hours. The membranes were next incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Sigma, 1:5000). Protein expression images were visualized using Odyssey V 3.0 image scanning (LI-COR, Lincoln, NE, USA). Quantification of the densitometric intensities of the protein bands was performed using the Bandscan 5.0 software, and the values were normalized against β-actin for each sample.
RNA isolation and quality controls
Total RNA was extracted from each sample using Trizol reagent (Invitrogen). DNase I was used to digest and eliminate any contaminating genomic DNA. The extracted total RNA concentration and purity were assessed spectrophotometrically at the ODs of 260 nm and 280 nm. The samples with OD260/280 nm ratios between 1.9 and 2.1 were used for cDNA synthesis.
Reverse transcription and quantitative polymerase chain reaction (qPCR)
One microgram of total RNA extracted from RPCs was reverse transcribed using the PrimeScript™ RT reagent kit (Perfect Real Time; TaKaRa, Dalian, China)28. After reverse transcription, 1 μl of cDNA diluted 10-fold in nuclease-free water (Invitrogen) was used as a template for qPCR, which was performed on an Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, Foster, CA, USA) in a 20-μl total volume containing 10 μl of 2 × Power SYBR Green PCR Master Mix (Applied Biosystems), 10 μl of diluted cDNA, and 300 nM of gene-specific primers. The primer sequences are shown in Table 1. The PCR efficiency of the reaction was measured with primers using serial dilutions of the cDNA (1:1, 1:5, 1:25, 1:125, 1:625, and 1:3,125). Each sample was tested in triplicate. The relative mRNA or miRNA expression was analyzed using the Pfaffl method32. The relative mRNA or miRNA levels were expressed as the fold change relative to the untreated controls after being normalized to the expression of β-actin or 5S rRNA, respectively.
Table 1. Primers used for qPCR.
| Genes | Accession no. | Forward (5′-3′) | Reverse (5′-3′) | Annealing temperature (°C) | Product size (base pairs) |
|---|---|---|---|---|---|
| Nestin | NM_016701 | aactggcacctcaagatgt | tcaagggtattaggcaagggg | 60 | 235 |
| Ki-67 | X82786 | cagtactcggaatgcagcaa | cagtcttcaggggctctgtc | 60 | 170 |
| TLX | NM_152229.2 | cgattagacgccactgaa | ggtatctggtatgaatgtagc | 60 | 166 |
| Map2 | NM_001039934 | agaaaatggaagaaggaatgactg | acatggatcatctggtaccttttt | 60 | 112 |
| PKC-α | NM_011101 | cccattccagaaggagatga | ttcctgtcagcaagcatcac | 60 | 212 |
| GFAP | NM_010277 | agaaaaccgcatcaccattc | tcacatcaccacgtccttgt | 60 | 184 |
| Rhodopsin | NM_145383 | tcaccaccaccctctacaca | tgatccaggtgaagaccaca | 60 | 216 |
| β-actin | NM_007393 | agccatgtacgtagccatcc | ctctcagctgtggtggtgaa | 60 | 152 |
Cell viability
The effect of let-7b and TLX on RPC proliferation was assessed using the cell counting kit (CCK-8; Dojindo, Kumamoto, Japan)33. Generally, RPCs were suspended at a final concentration of 1 × 104 cells/well and cultured in 96-well plates. After treatment with let-7b mimics, let-7b inhibitor, negative controls, TLX clone, or siTLX, the CCK-8 solution was added to each well at days 0, 1, 2 and 3 of the culture period. After the cells were incubated for another 4 h at 37°C according to the reagent instructions, the absorbance at 450 nm was measured using an ELISA microplate reader (ELX800, BioTeK, USA). The cell viability was directly proportional to the absorbance at 450 nm; therefore, the viability was expressed as the A490 value.
Cytotoxicity analysis
Observations to determine cytotoxicity were determined by measuring release of the cytosolic enzyme lactate dehydrogenase (LDH) into culture medium upon cell lysis using CytoTox 96 Non-Radioactive Cytotoxicity Assay kit (Promega, USA). Cytotoxicity (%) was expressed as the percent of LDH released into the medium out of the total LDH activity.
Statistical analyses
The experimental statistics presented in the present study are expressed as the means ± standard derivation (SD). Each experiment was repeated at least three times unless otherwise specified. Statistical significance of the differences in RPC expression between the experimental and control groups was analyzed using Student's t-test (P < 0.05 was deemed to indicate statistical significance).
Author Contributions
N.N.: conception and design, collection and assembly of the data, data interpretation and manuscript writing; D.D.Z., Q.X., J.Z.C., Z.W., X.Y.W., M.Y.Z. and J.J.: collection and assembly of data; X.Q.F., Y.D. and M.L.: conception and design, data analysis and interpretation; P.G.: conceptual and design, data analysis and interpretation, financial support, and manuscript writing.
Supplementary Material
Supporting information
Acknowledgments
The authors are grateful to Dr. Henry Klassen and Dr. Michael J. Young for the original mouse RPCs. This research was supported by the National Natural Science Foundations of China (81070737, 31300810), Science and Technology Commission of Shanghai (13DZ0s00303), the Education Commission of Shanghai (11YZ47).
References
- Ambros V. The functions of animal microRNAs. Nature 431, 350–355, 10.1038/nature02871 (2004). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- Kawahara H., Imai T. & Okano H. MicroRNAs in Neural Stem Cells and Neurogenesis. Front in Neurosci 6, 30, 10.3389/fnins.2012.00030 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R. W. & Sontheimer E. J. Origins and Mechanisms of miRNAs and siRNAs. Cell 136, 642–655, 10.1016/j.cell.2009.01.035 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asuelime G. E. & Shi Y. The little molecules that could: a story about microRNAs in neural stem cells and neurogenesis. Front in Neurosci 6, 176, 10.3389/fnins.2012.00176 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M. F. & Shi Y. Dynamic Roles of microRNAs in Neurogenesis. Front in Neurosci 6, 71, 10.3389/fnins.2012.00071 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A. E. et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, 86–89, 10.1038/35040556 (2000). [DOI] [PubMed] [Google Scholar]
- Reinhart B. J. et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906, 10.1038/35002607 (2000). [DOI] [PubMed] [Google Scholar]
- Sempere L. F. et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5, R13, 10.1186/gb-2004-5-3-r13 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn F. G. et al. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J 21, 415–426, 10.1096/fj.06-6130com (2007). [DOI] [PubMed] [Google Scholar]
- La Torre A., Georgi S. & Reh T. A. Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc Natl Acad Sci U S A 110, E2362–2370, 10.1073/pnas.1301837110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J., Kim I., Chada K. & Morrison S. J. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell 135, 227–239, 10.1016/j.cell.2008.09.017 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C. et al. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci U S A 107, 1876–1881, 10.1073/pnas.0908750107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. et al. Nuclear receptor TLX regulates cell cycle progression in neural stem cells of the developing brain. Mol Endocrinol 22, 56–64, 10.1210/me.2007-0290 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams B. S. et al. Pathological aggression in “fierce” mice corrected by human nuclear receptor 2E1. J Neurosci 25, 6263–6270, 10.1523/jneurosci.4757-04.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K. et al. The Tlx gene regulates the timing of neurogenesis in the cortex. J Neurosci 24, 8333–8345, 10.1523/jneurosci.1148-04.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki T. et al. Tlx, an orphan nuclear receptor, regulates cell numbers and astrocyte development in the developing retina. J Neurosci 24, 8124–8134, 10.1523/jneurosci.2235-04.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D. et al. Nestin is required for the proper self-renewal of neural stem cells. Stem cells 28, 2162–2171, 10.1002/stem.541 (2010). [DOI] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B. & Bartel D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20, 10.1016/j.cell.2004.12.035 (2005). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233, 10.1016/j.cell.2009.01.002 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhams P. L., Basco E., Hajos F., Csillag A. & Balazs R. Radial glia in the developing mouse cerebral cortex and hippocampus. Anat Embryol (Berl) 163, 331–343 (1981). [DOI] [PubMed] [Google Scholar]
- Monaghan A. P., Grau E., Bock D. & Schutz G. The mouse homolog of the orphan nuclear receptor tailless is expressed in the developing forebrain. Development 121, 839–853 (1995). [DOI] [PubMed] [Google Scholar]
- Yu R. T., McKeown M., Evans R. M. & Umesono K. Relationship between Drosophila gap gene tailless and a vertebrate nuclear receptor Tlx. Nature 370, 375–379, 10.1038/370375a0 (1994). [DOI] [PubMed] [Google Scholar]
- Deng Y. et al. Effects of a miR-31, Runx2, and Satb2 regulatory loop on the osteogenic differentiation of bone mesenchymal stem cells. Stem cells Dev 22, 2278–2286, 10.1089/scd.2012.0686 (2013). [DOI] [PubMed] [Google Scholar]
- Farh K. K. et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 310, 1817–1821, 10.1126/science.1121158 (2005). [DOI] [PubMed] [Google Scholar]
- Zhao C., Sun G., Li S. & Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol 16, 365–371, 10.1038/nsmb.1576 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe M., Ikawa M., Kominami K., Nakanishi T. & Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett 407, 313–319 (1997). [DOI] [PubMed] [Google Scholar]
- Xia J. et al. An in vitro comparison of two different subpopulations of retinal progenitor cells for self-renewal and multipotentiality. Brain Res 1433, 38–46, 10.1016/j.brainres.2011.11.054 (2012). [DOI] [PubMed] [Google Scholar]
- Gu P. et al. Isolation of retinal progenitor and stem cells from the porcine eye. Mol Vis 13, 1045–1057 (2007). [PMC free article] [PubMed] [Google Scholar]
- Gu P., Yang J., Wang J., Young M. J. & Klassen H. Sequential changes in the gene expression profile of murine retinal progenitor cells during the induction of differentiation. Mol Vis 15, 2111–2122 (2009). [PMC free article] [PubMed] [Google Scholar]
- Hu Y. et al. An in vitro comparison study: the effects of fetal bovine serum concentration on retinal progenitor cell multipotentiality. Neurosci Lett 534, 90–95, 10.1016/j.neulet.2012.11.006 (2013). [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29, e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Yang J., Gu P. & Klassen H. Effects of glial cell line-derived neurotrophic factor on cultured murine retinal progenitor cells. Mol Vis 16, 2850–2866 (2010). [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information