Abstract
The alpha subunits of voltage-gated calcium channels (Cavs) are large transmembrane proteins responsible for crucial physiological processes in excitable cells. They are assisted by three auxiliary subunits that can modulate their electrical behavior. Little is known about the evolution and roles of the various subunits of Cavs in nonbilaterian animals and in nonanimal lineages. For this reason, we mapped the phyletic distribution of the four channel subunits and reconstructed their phylogeny. Although alpha subunits have deep evolutionary roots as ancient as the split between plants and opistokonths, beta subunits appeared in the last common ancestor of animals and their close-relatives choanoflagellates, gamma subunits are a bilaterian novelty and alpha2/delta subunits appeared in the lineage of Placozoa, Cnidaria, and Bilateria. We note that gene losses were extremely common in the evolution of Cavs, with noticeable losses in multiple clades of subfamilies and also of whole Cav families. As in vertebrates, but not protostomes, Cav channel genes duplicated in Cnidaria. We characterized by in situ hybridization the tissue distribution of alpha subunits in the sea anemone Nematostella vectensis, a nonbilaterian animal possessing all three Cav subfamilies common to Bilateria. We find that some of the alpha subunit subtypes exhibit distinct spatiotemporal expression patterns. Further, all six sea anemone alpha subunit subtypes are conserved in stony corals, which separated from anemones 500 MA. This unexpected conservation together with the expression patterns strongly supports the notion that these subtypes carry unique functional roles.
Keywords: voltage-gated calcium channel, ion channel, Cnidaria, Nematostella vectensis, evolution of nervous system
Introduction
Voltage-gated Ca2+ channels (Cav) play a fundamental role in synaptic transmission and muscle contraction in Bilateria, a group which comprise the vast majority of animal species (Catterall et al. 2005; Tyson and Snutch 2013; Simms and Zamponi 2014). Although much is known about the physiological roles of these channels in Bilateria, little is known about their function or their tissue distribution in nonbilaterian animals. Among these limited data, there are indications for Cav playing a role in neuronal and muscular function in cnidarians such as sea anemones and jellyfish (Anderson 1987; Holman and Anderson 1991; Jeziorski et al. 1998). Strong phylogenetic support places the Cnidaria as sister to the Bilateria (Erwin et al. 2011; Ryan et al. 2013), suggesting that they share a common ancestor, which possessed muscles and a nervous system. Nevertheless, recent surprising findings emphasize the independent evolution of striated muscle (Steinmetz et al. 2012) and Na+-permeable voltage-gated channels (Gur Barzilai et al. 2012) in Cnidaria and Bilateria. Better understanding of these features in Cnidaria may help us grasp the complexity of the nervous and muscular systems of ancient animals that lived more than half a billion years ago.
In bilaterians, the pore-forming α subunits of Cavs (α1), which are responsible for conducting the Ca2+ ions (fig. 1), are divided into two broad groups based on physiology: Low-voltage-activated (LVA) channels that open near resting potential and high-voltage-activated (HVA) channels that require a sizable depolarization to open (Catterall et al. 2005). The LVA channels are also termed T type or Cav3. The HVA group is further subdivided by their biophysical and pharmacological properties into L type (Cav1) and a third category of N, P/Q, and R types (Cav2). Two major protostome groups, the Ecdysozoa and Lophotrochozoa, possess three Cavs, one each from the Cav1–3 groups (Liebeskind et al. 2011; Cai and Clapham 2012), whereas vertebrates have ten Cav channels resulting from the two rounds of genome duplications that expanded the repertoire of many vertebrate genes (Jegla et al. 2009). In contrast to our understanding of Cav evolution in Bilateria, we still lack much knowledge regarding the evolution of these channels in nonbilaterian and nonanimal phyla. We know from recent studies that α subunits of Cavs can be found in the representatives of early-branching groups as choanoflagellates and algae and that they are distantly related to calcium channels of fungi (Verret et al. 2010; Liebeskind et al. 2012), yet we do not know much about the evolution of the Cav1, 2, and 3 subfamilies. Furthermore, bilaterian Cavs contain auxiliary subunits (α2δ, β, and γ; fig. 1), and the phyletic distribution and evolutionary history of the gene families encoding them are unknown. To address these gaps in knowledge, we analyzed in this work the phyletic distribution of the various Cav subunits and reconstructed their phylogenies. Additionally, we used the sea anemone Nematostella vectensis to study Cav spatiotemporal expression patterns to better understand the functionality of these channels in nonbilaterian organisms.
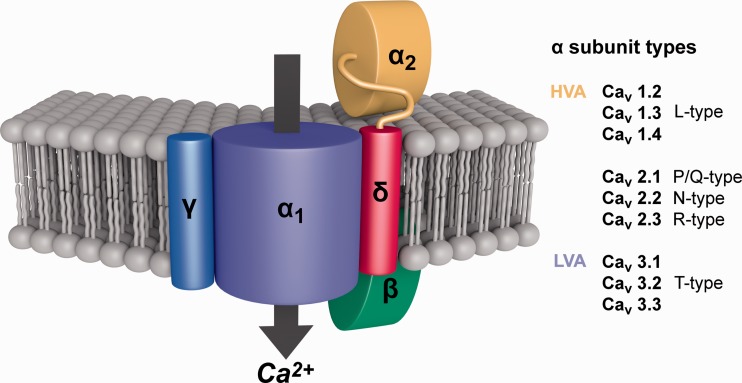
Fig. 1.—
Schematic representation of a Cav channel (α1) with its three auxiliary (β, α2/δ, γ) subunits. The α1 subunit forms a voltage-gated calcium-permeating channel that functions alone. The trafficking of the α1 subunit and its biophysical properties are influenced by the other subunits. The α2 and δ subunits are distinct proteins made from a common precursor protein and linked through a disulfide bond. The β subunit is intracellular whereas the others are transmembrane proteins. On the right is a table of the three α1 Cav channel types. Figure adapted from Khosravani and Zamponi (2006) Voltage-Gated Calcium Channels and Idiopathic Generalized Epilepsies. Physiological Reviews, volume 86, issue 3, p. 945 by permission of the American Physiological Society.
Materials and Methods
Identification of Cav Homologs and Phylogenetic Analysis
Putative Cav homologs were detected in GenBank (nr), Broad Institute and Joint Genome Institute databases through BLAST. Transcript clusters were translated to proteins. Accession numbers of proteins used in all phylogenetic analyses can be found in supplementary table S1, Supplementary Material online. Protein models were aligned using MUSCLE and low-quality alignment regions were removed by TrimAl (Edgar 2004; Capella-Gutierrez et al. 2009). ProtTest was used to find the most suitable model for phylogeny reconstruction (Abascal et al. 2005) and this model was used to reconstruct a maximum-likelihood tree with PhyML (Guindon et al. 2010). Support values were calculated using 100 bootstrap replicates. A Bayesian tree was constructed using MrBayes version 3.1.2 with the same model. The run lasted 5,000,000 generations and every 100th generation was sampled. We estimated that the Bayesian analysis reached convergence when the potential scale reduction factor reached 1.0.
RNA Isolation and Polymerase Chain Reaction Amplification of Cav Transcript Fragments from N. vectensis
Total RNA was isolated from planulae (4 days old) and adult polyps (>5 months old) of N. vectensis using Trizol (Life Technologies, USA) according to the manufacturer's instructions. The purified RNA was used as a template for reverse transcription reaction using the SuperScript III reverse transcriptase (Life Technologies) and random primers (New England Biolabs, USA) according to manufacturer instructions. Advantage2 DNA polymerase mix (Clontech) was used for polymerase chain reaction (PCR) under high-stringency conditions: 94 °C for 2:00 min, 35× (94 °C for 20 s, 65 °C for 20 s, 72 °C for 1 min) and 72 °C for 5 min. The amplified fragments were 1–1.5 kb long (for primer list, see supplementary table S2, Supplementary Material online) and they were purified using Illustra PCR purification kit (GE Healthcare, UK) and ligated into pGEM-T (Promega). The resulting plasmids were sequenced from both ends (performed at MicroSynth, Switzerland) and were used as templates for producing RNA probes for in situ hybridization (ISH).
In Situ Hybridization
For ISH experiments, N. vectensis larvae were fixed at 48–168 h postfertilization in ice-cold 3.7% formaldehyde in one-third of seawater with 0.2% glutaraldehyde for 90 s and then in 3.7% formaldehyde in one-third of seawater without glutaraldehyde for additional 60 min. Antisense RNA probes for ISH were generated and labeled by using the T7 or SP6 MEGAscript kits (Life Technologies) and an RNA labeling mix with digoxygenin (Roche, Germany). The ISH procedure was performed as described previously (Genikhovich and Technau 2009). The stained samples were photographed in a Nikon Eclipse 80i microscope with differential interference contrast optics connected to a Nikon Digital Sight DS-U2 camera.
Results
Distribution and Evolution of α1 Subunits of Cav Channels
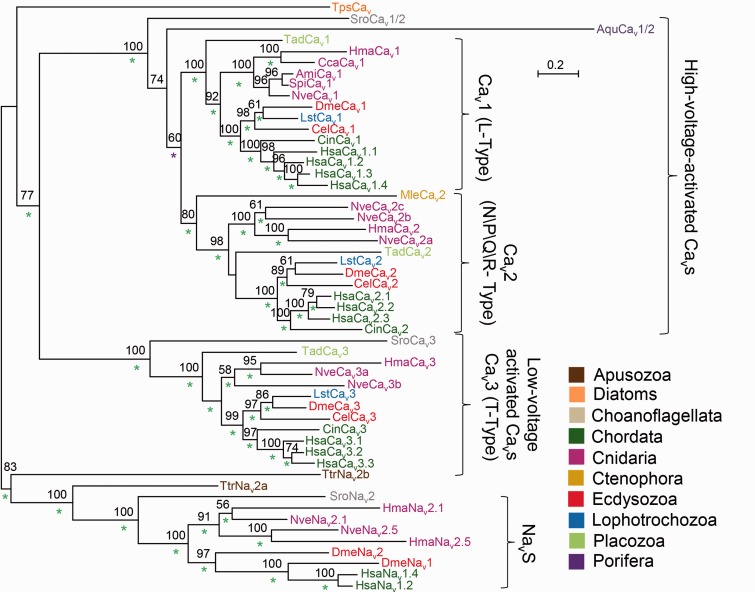
We collected from publicly available genomic and transcriptomic databases (see Methods and Materials) the putative protein sequences of bilaterian, cnidarians, placozoan (Trichoplax), poriferan (sponges), and ctenophore (comb jellies) Cav α subunits. In addition, we searched for such homologs in all available data from nonanimal groups. We reconstructed a phylogeny of these proteins, which included only complete models (all four channel domains present) to increase its accuracy (fig. 2). The earliest branching group where we could find homologs of Cav α1 subunits was green algae, such as Chlamydomonas and Micromonas; however, as their sequences are highly derived and as they also include some characteristics of Nav channels (Verret et al. 2010; Liebeskind et al. 2012) we did not include them in our phylogenetic analysis. Another organism where we found Cav α1 subunit homologs is Thecamonas trahens, a member of the Apusozoa, which according to recent phylogeny is a sister group to all opisthokonts (fungi, animals, and their close protozoan relatives) (Derelle and Lang 2012). However, our phylogeny indicates that both complete Thecamonas protein models are clustered with α subunits of voltage-gated sodium channels (Nav1) and their close homologs (Nav2) rather than with Cavs. The next group we could find Cav homologs in was Choanoflagellata, a protist sister group of animals: The genome of the choanoflagellate Salpingoeca rosetta (Fairclough et al. 2013) contains two α-subunit homologs, with one of them clustering with HVA channels and the other with LVA channels (fig. 2). In the genome of another choanoflagellate, Monosiga brevicollis (King et al. 2008), we could detect only a short gene fragment encoding a partial Cav of only 258 amino acids, suggesting either a partial gene deletion or a technical problem in genome assembly at this specific region (data not shown). We found an HVA homolog in the genome of the sponge Amphimedon queenslandica (Srivastava et al. 2010) and a homolog of Cav2 in the genome of the ctenophore Mnemiopsis leidyi (Ryan et al. 2013). In cnidarians and the placozoan Trichoplax adhaerens (Srivastava et al. 2008), we found representatives of all three Cav α1 subunit families.
Fig. 2.—
Phylogeny of Cav α1 subunits. A maximum-likelihood phylogenetic tree was constructed with the LG model (+G, +F). Bootstrap support values above 50% are indicated above branches. Posterior probability (PP) values of a Bayesian tree constructed with the WAG model are indicated by a green (PP = 1.0), or purple (0.95 ≤ PP < 1.0) asterisk. Abbreviations of species names are: Aqu, Amphimedon queenslandica (sponge); Ami, Acropora millepora (stony coral); Cel, Caenorhabditis elegans (nematode); Cin, Ciona intestinalis (tunicate); Cca, Cyanea capillata (jellyfish); Dme, Drosophila melanogaster (fruit fly); Hsa, Homo sapiens (human); Hma, Hydra magnipapillata (hydra); Lst, Lymnaea stagnalis (pond snail); Mle, Mnemiopsis leidyi (comb jelly); Nve, Nematostella vectensis (starlet anemone); Spi, Stylophora pistillata (stony coral); Sro, Salpingoeca rosetta (choanoflagellate); Tad, Trichoplax adhaerens (placozoan); Tps, Thalassiosira pseudonana (diatom); Ttr, Thecamonas trahens.
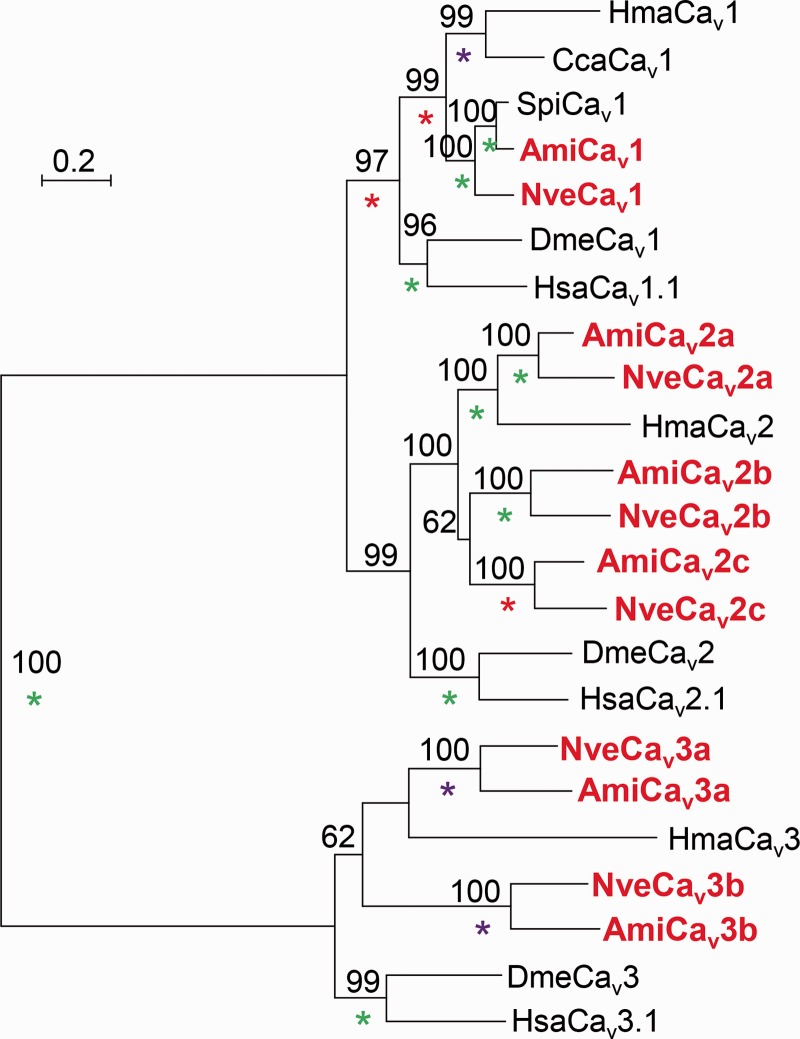
In the genome of N. vectensis (Putnam et al. 2007), we identified one Cav1 gene, three Cav2 genes, and two Cav3 genes. Interestingly, in the transcriptomic data available for the stony coral Acropora millepora (Meyer et al. 2009; Moya et al. 2012) we identified partial transcript models which demonstrate clear orthologous relationships with each of the Cav genes of N. vectensis (fig. 3). Taking into account the Acropora and Nematostella approximate separation time (Shinzato et al. 2011), it is likely that these orthologs are conserved in both lineages for about 500 Myr. Thus, similar to vertebrates, but not protostomes, Cav channel genes duplicated in Cnidaria.
Fig. 3.—
Phylogeny of Cav α1 subunit subtypes from Nematostella vectensis and Acropora millepora. Sequences from these two species appear in red bold. A maximum likelihood phylogenetic tree was constructed with the LG model (+I, +G, +F). Bootstrap support values above 50% are indicated above branches. Posterior probability (PP) values of a Bayesian tree constructed with the WAG model are indicated by a green (PP = 1.0), purple (0.95 ≤ PP < 1.0), or red (0.9 ≤ PP < 0.95) asterisk. Abbreviations of species names are: Ami, Acropora millepora (stony coral); Cca, Cyanea capillata (jellyfish); Dme, Drosophila melanogaster (fruit fly); Hsa, Homo sapiens (human); Hma, Hydra magnipapillata (hydra); Nve, Nematostella vectensis (starlet anemone); Spi, Stylophora pistillata (stony coral).
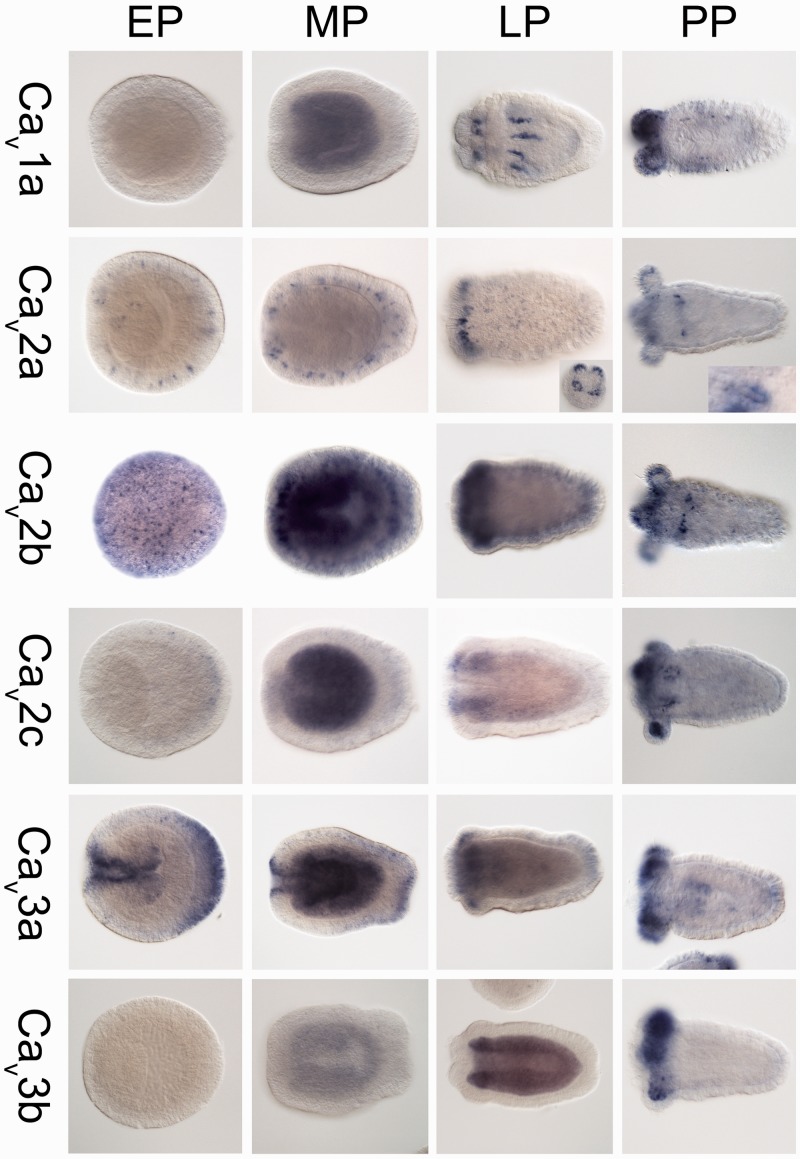
Localization of the transcripts of the six Cav α1 subunits of Nematostella by ISH demonstrated complex spatiotemporal expression patterns. Cav1a expression starts only from the mid-planula stage, where it is diffused in the endoderm. Later in the late-planula and primary polyp stages the expression becomes concentrated in regions along the mesenteries and in the endoderm and ectoderm of the tentacle buds, regions which also include muscles and neurons (Marlow et al. 2009; Renfer et al. 2010; fig. 4). In contrast, Cav2a is expressed already from the early planula stage and is specific to nematocytes, the stinging cells which typify Cnidaria (Kass-Simon and Scappaticci 2002; Zenkert et al. 2011). This is clearly evident by the stain-free nematocyst (stinging capsule) structure which is surrounded by stained cytoplasm (fig. 4 and supplementary fig. S1, Supplementary Material online). Cav2b, Cav2c, and the two Cav3 genes of Nematostella also exhibit distinct developmental expression patterns (fig. 4), but it is more difficult to postulate on their identity.
Fig. 4.—
The spatiotemporal expression of the Cav α subunits subtype from Nematostella vectensis as determined by ISH. Gene expression is indicated by blue staining. The insets show the expression of Cav2a in nematocytes (stinging cells). In all pictures (not including insets), the oral end of all larvae is to the left. Abbreviations of developmental stages are: EP, early planula; MP, mid-planula; LP, late planula (tentacle buds are noticeable); and PP, primary polyp.
Distribution and Evolution of Auxiliary Subunits of Cav Channels
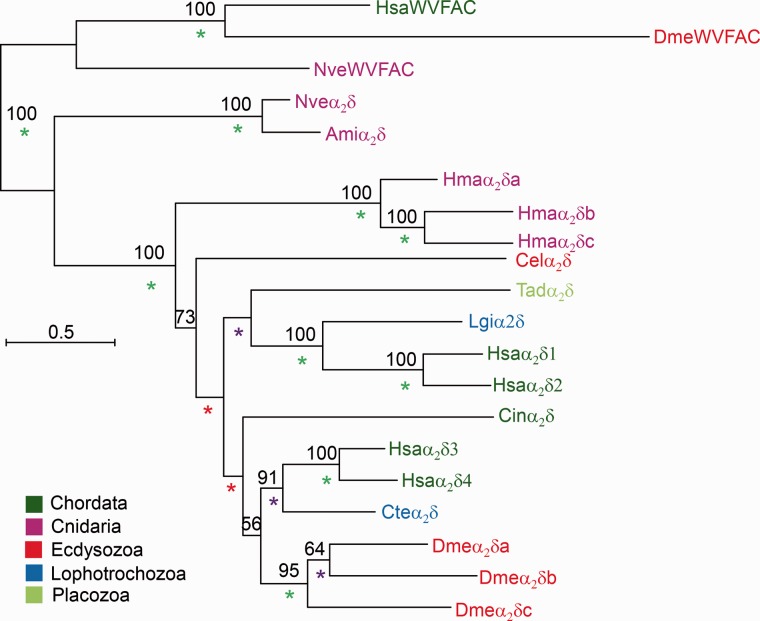
As with the study of α1 subunits, we collected putative protein homologs of the α2δ, β, and γ subunits of Cav channels from transcriptomic and genomic databases and reconstructed their phylogenies. We could not find α2δ homologs in sponges and ctenophores or in any nonanimal group. We detected single α2δ genes in Trichoplax, Acropora, and Nematostella. In the cnidarian Hydra magnipapillata there are three α2δ genes, suggesting a lineage-specific expansion (fig. 5). We rooted the phylogenetic tree of the α2δ subunits by using the clade of human cache domain containing 1 protein (VWFAC1) and its cnidarian homologs as an outgroup. The VWFAC1 proteins are similar in sequence (∼40% similarity) to α2δ subunits and seem to be highly conserved in most animals, including cnidarians, arthropods, and vertebrates. However, to the best of our knowledge, the function of these proteins in bilaterians is currently unknown.
Fig. 5.—
Phylogeny of Cav α2δ subunits. A maximum-likelihood phylogenetic tree was constructed with the LG model (+I, +G, +F). Bootstrap support values above 50% are indicated above branches. Posterior probability (PP) values of a Bayesian tree constructed with the WAG model are indicated by a green (PP = 1.0), purple (0.95 ≤ PP < 1.0), or red (0.9 ≤ PP < 0.95) asterisk. Abbreviations of species names are: Ami, Acropora millepora (stony coral); Cel, Caenorhabditis elegans (nematode); Cin, Ciona intestinalis (tunicate); Cte, Capitella teleta (annelid worm); Dme, Drosophila melanogaster (fruit fly); Hsa, Homo sapiens (human); Hma, Hydra magnipapillata (hydra); Lgi, Lottia gigantea (owl limpet); Nve, Nematostella vectensis (starlet anemone); Tad, Trichoplax adhaerens (placozoan).
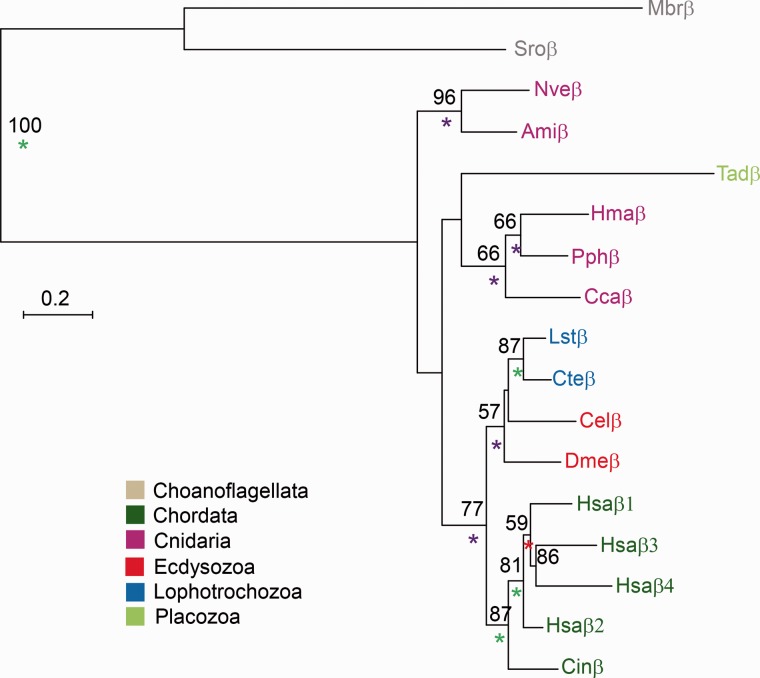
Unlike α2δ subunits which seem to be metazoan-specific, β subunits can be found in choanoflagellates (fig. 6). We also detected a single β subunit in each cnidarian species and in Trichoplax. This suggests that β subunits appeared in the common ancestor of choanoflagellates and animals but were lost independently in sponges and ctenophores.
Fig. 6.—
Phylogeny of Cav β subunits. A maximum-likelihood phylogenetic tree was constructed with the LG model (+G). Bootstrap support values above 50% are indicated above branches. Posterior probability (PP) values of a Bayesian tree constructed with the WAG model are indicated by a green (PP = 1.0), purple (0.95 ≤ PP < 1.0), or red (0.9 ≤ PP < 0.95) asterisk. Abbreviations of species names are: Ami, Acropora millepora (stony coral); Cel, Caenorhabditis elegans (nematode); Cin, Ciona intestinalis (tunicate); Cca, Cyanea capillata (jellyfish); Cte, Capitella teleta (annelid worm); Dme, Drosophila melanogaster (fruit fly); Hsa, Homo sapiens (human); Hma, Hydra magnipapillata (hydra); Lst, Lymnaea stagnalis (pond snail); Mbr, Monosiga brevicollis (choanoflagellate); Nve, Nematostella vectensis (starlet anemone); Pph, Physalia physalis (hydrozoan cnidarian); Sro, Salpingoeca rosetta (choanoflagellate); Tad, Trichoplax adhaerens (placozoan).
We could not detect any γ subunits in nonvertebrate species but a single homolog in the hemichordate Saccoglossus kowalevskii and a single homolog in each of the two annelids, Capitella teleta and Helobdella robusta (supplementary table S1, Supplementary Material online). These protein models show only modest similarity to γ subunits of vertebrates (41–46% similarity) but contain a typical claudin-2 domain and have a similar length to their vertebrate homologs (250–300 amino acids). Moreover, when we used them for a reciprocal BLAST query against the human proteome, the best scoring hits were γ subunits of Cav, suggesting that these might be true homologs. The extremely patchy phyletic distribution of γ subunits strongly suggests that they were lost independently in multiple lineages. However, there are some indications that other proteins with claudin domains can also influence the expression levels and function of Cav channels in invertebrates (Simske 2013) and it is possible that high sequence variability of such subunits is masking common ancestral origins.
Discussion
The current work of mapping the phyletic distribution of the four subunits of Cav channels and the analysis of their phylogeny together with results of previous research (Verret et al. 2010; Liebeskind et al. 2011; Gur Barzilai et al. 2012; Liebeskind et al. 2012) allows us to reconstruct their evolutionary history. It seems that the first α1 subunits of Cavs appeared very early in eukaryote evolution, already in the common ancestor of Viridiplantae (plants and green algae), Apusozoa, and Opisthokonta. However, it was lost in multiple lineages, such as all extant land plants, Amoebozoa and fungi. The finding of HVA and LVA Cav channels in S. rosetta indicates that these Cav subfamilies already separated in the ancestor of choanoflagellates and animals. Intriguingly, choanoflagellates possess a primordial neurosecretory apparatus (Burkhardt et al. 2011) and we hypothesize that Cav channels may play a role in its function. When HVA channels further diverged to Cav1 and Cav2 is harder to determine, as the order of divergence of sponges and ctenophores is still under debate (Philippe et al. 2011; Ryan et al. 2013). Our phylogeny indicates that the single Cav of the sponge A. queenslandica is a sister clade to all other animal α1 subunits of HVA channels, whereas the single α1 subunit of the ctenophore M. leidyi clusters with the Cav2 subfamily. This suggests that possibly Cav2-like characteristics already appeared in the ancient HVA Cavs prior to the divergence of ctenophores. Alternatively, the divergence of Cav1 and Cav2 might have happened before the divergence of ctenophores from the rest of the animals, and the lineage of M. leidyi lost Cav1. Gene loss is a general trend in the evolution of Cavs, as both sponges and ctenophores seem to have independently lost the Cav3 subfamily (fig. 2). Such losses of channel genes in animal lineages may be suspected as artifacts due to technical reasons, such as errors in gene annotation, genome assembly, and/or insufficient sequencing depth. However, it is noteworthy that the sequencing depth of the genomes of Amphimedon (9-fold by Sanger sequencing; Srivastava et al. 2010) and Mnemiopsis (12-fold coverage by 454 sequencing; Ryan et al. 2013) is more than adequate. Moreover, our searches in the recently sequenced genomes of the ctenophore Pleurobrachia bachei (Moroz et al. 2014) and the sponge Oscarella carmela (available through the Compagen website; Hemmrich and Bosch 2008) supported the above scenario in which Cav3 subfamily and β subunits were lost in sponges and ctenophores.
An intriguing question is what might be the functional value of Cav channels in light of the fact that Nav2 channels are also voltage-gated channels conducting mostly calcium ions (Zhou et al. 2004; Gur Barzilai et al. 2012). The answer to this question might lie in the vastly different voltage-sensitivities of Nav2 and HVA Cav channels, as the latter opens only in relatively high voltages and therefore are not well-suited for conducting neuronal action potentials (Tyson and Snutch 2013; Simms and Zamponi 2014).
Our analyses suggest that auxiliary subunits were gradually added during evolution to the Cav complex: β subunits appeared already in the ancestor of choanoflagellates and animals, α2δ appeared only later in the common ancestor of placozoans, cnidarians and bilaterians, whereas γ subunits might have appeared only in the bilaterian lineage. As auxiliary subunits can increase the complexity of electrical signaling (Lacerda et al. 1991; Obermair et al. 2005; Dolphin 2012), this trend at the genetic level could support increasing complexity at the neuronal level. However, we also notice that all subunits other than α2δ were lost in some lineages, demonstrating a highly plastic evolution of the Cav complex.
Our finding by using ISH techniques that each Cavs α1 subunit of Nematostella occupies a distinct spatiotemporal expression domain (fig. 4) raises the possibility that each of these subunits has acquired a specialized role. The expression of Cav1a in muscles and/or motor neurons is in accordance with previous works that recorded L-type calcium currents in the muscles of sea anemones and isolated transcripts encoding Cav1 from motor neuron-rich regions of a jellyfish bell (Holman and Anderson 1991; Jeziorski et al. 1998). The expression of Nematostella Cav2a in nematocytes fits a previous report on the isolation of transcripts encoding Cav channels from the stinging cells of the cnidarian Physalia physalis (Bouchard et al. 2006) and reports of Cav-dependence of nematocyst action (Gitter et al. 1994; Watson and Hessinger 1994). The notion of specialization of cnidarian Cavs α1 subunits is also strongly supported by our finding that the six Cav α subunit subtypes are highly conserved for about 500 Myr in the lineage of sea anemone and reef-building corals (fig. 3). The expansion and specialization of Cav α subunits is part of a wider trend that seems to be true also for other ion channel families in Cnidaria, such as voltage-gated potassium channels (Jegla et al. 2012; Martinson et al. 2014) and Nav2 channels (Gur Barzilai et al. 2012).
Supplementary Material
Supplementary figure S1 and tables S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
Y.M. is grateful to Ulrich Technau (University of Vienna) for providing the experimental infrastructure required to perform parts of this work. H.Z. is grateful to Mikhail Matz for access to databases.
Literature Cited
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Anderson P. Properties and pharmacology of a TTX insensitive Na+ current in neurones of the jellyfish Cyanea Capillata. J Exp Biol. 1987;133:231–248. [Google Scholar]
- Bouchard C, et al. Cloning and functional expression of voltage-gated ion channel subunits from cnidocytes of the Portuguese Man O’War Physalia physalis. J Exp Biol. 2006;209:2979–2989. doi: 10.1242/jeb.02314. [DOI] [PubMed] [Google Scholar]
- Burkhardt P, et al. Primordial neurosecretory apparatus identified in the choanoflagellate Monosiga brevicollis. Proc Natl Acad Sci U S A. 2011;108:15264–15269. doi: 10.1073/pnas.1106189108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Clapham DE. Ancestral Ca2+ signaling machinery in early animal and fungal evolution. Mol Biol Evol. 2012;29:91–100. doi: 10.1093/molbev/msr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA, Perez-Reyes E, Snutch TP, Striessnig J. International Union of Pharmacology. XLVIII. Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacol Rev. 2005;57:411–425. doi: 10.1124/pr.57.4.5. [DOI] [PubMed] [Google Scholar]
- Derelle R, Lang BF. Rooting the eukaryotic tree with mitochondrial and bacterial proteins. Mol Biol Evol. 2012;29:1277–1289. doi: 10.1093/molbev/msr295. [DOI] [PubMed] [Google Scholar]
- Dolphin AC. Calcium channel auxiliary alpha2delta and beta subunits: trafficking and one step beyond. Nat Rev Neurosci. 2012;13:542–555. doi: 10.1038/nrn3311. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin DH, et al. The Cambrian conundrum: early divergence and later ecological success in the early history of animals. Science. 2011;334:1091–1097. doi: 10.1126/science.1206375. [DOI] [PubMed] [Google Scholar]
- Fairclough SR, et al. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 2013;14:R15. doi: 10.1186/gb-2013-14-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genikhovich G, Technau U. In situ hybridization of starlet sea anemone (Nematostella vectensis) embryos, larvae, and polyps. 2009 doi: 10.1101/pdb.prot5282. Cold Spring Harb Protoc. 2009:pdb.prot5282. [DOI] [PubMed] [Google Scholar]
- Gitter A, Oliver D, Thurm U. Calcium-and voltage-dependence of nematocyst discharge in Hydra vulgaris. J Comp Physiol A. 1994;175:115–122. [Google Scholar]
- Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Gur Barzilai M, et al. Convergent evolution of sodium ion selectivity in metazoan neuronal signaling. Cell Rep. 2012;2:242–248. doi: 10.1016/j.celrep.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmrich G, Bosch TC. Compagen, a comparative genomics platform for early branching metazoan animals, reveals early origins of genes regulating stem-cell differentiation. Bioessays. 2008;30:1010–1018. doi: 10.1002/bies.20813. [DOI] [PubMed] [Google Scholar]
- Holman M, Anderson P. Voltage-activated ionic currents in myoepithelial cells isolated from the sea anemone Calliactis tricolor. J Exp Biol. 1991;161:333–346. [Google Scholar]
- Jegla T, et al. Expanded functional diversity of shaker K(+) channels in cnidarians is driven by gene expansion. PLoS One. 2012;7:e51366. doi: 10.1371/journal.pone.0051366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegla TJ, Zmasek CM, Batalov S, Nayak SK. Evolution of the human ion channel set. Comb Chem High Throughput Screen. 2009;12: 2–23. doi: 10.2174/138620709787047957. [DOI] [PubMed] [Google Scholar]
- Jeziorski MC, Greenberg RM, Clark KS, Anderson PA. Cloning and functional expression of a voltage-gated calcium channel alpha1 subunit from jellyfish. J Biol Chem. 1998;273:22792–22799. doi: 10.1074/jbc.273.35.22792. [DOI] [PubMed] [Google Scholar]
- Kass-Simon G, Scappaticci AA. The behavioral and developmental physiology of nematocysts. Can J Zool. 2002;80:1772–1794. [Google Scholar]
- King N, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerda AE, et al. Normalization of current kinetics by interaction between the alpha 1 and beta subunits of the skeletal muscle dihydropyridine-sensitive Ca2+ channel. Nature. 1991;352:527–530. doi: 10.1038/352527a0. [DOI] [PubMed] [Google Scholar]
- Liebeskind BJ, Hillis DM, Zakon HH. Evolution of sodium channels predates the origin of nervous systems in animals. Proc Natl Acad Sci U S A. 2011;108:9154–9159. doi: 10.1073/pnas.1106363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebeskind BJ, Hillis DM, Zakon HH. Phylogeny unites animal sodium leak channels with fungal calcium channels in an ancient, voltage-insensitive clade. Mol Biol Evol. 2012;29:3613–3616. doi: 10.1093/molbev/mss182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlow HQ, Srivastava M, Matus DQ, Rokhsar D, Martindale MQ. Anatomy and development of the nervous system of Nematostella vectensis, an anthozoan cnidarian. Dev Neurobiol. 2009;69:235–254. doi: 10.1002/dneu.20698. [DOI] [PubMed] [Google Scholar]
- Martinson AS, et al. Functional evolution of Erg potassium channel gating reveals an ancient origin for IKr. Proc Natl Acad Sci U S A. 2014;111:5712–5717. doi: 10.1073/pnas.1321716111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E, et al. Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx. BMC Genomics. 2009;10:219. doi: 10.1186/1471-2164-10-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz LL, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510:109–114. doi: 10.1038/nature13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A, et al. Whole transcriptome analysis of the coral Acropora millepora reveals complex responses to CO(2)-driven acidification during the initiation of calcification. Mol Ecol. 2012;21:2440–2454. doi: 10.1111/j.1365-294X.2012.05554.x. [DOI] [PubMed] [Google Scholar]
- Obermair GJ, et al. The Ca2+ channel alpha2delta-1 subunit determines Ca2+ current kinetics in skeletal muscle but not targeting of alpha1S or excitation-contraction coupling. J Biol Chem. 2005;280:2229–2237. doi: 10.1074/jbc.M411501200. [DOI] [PubMed] [Google Scholar]
- Philippe H, et al. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 2011;9:e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Renfer E, Amon-Hassenzahl A, Steinmetz PR, Technau U. A muscle-specific transgenic reporter line of the sea anemone, Nematostella vectensis. Proc Natl Acad Sci U S A. 2010;107:104–108. doi: 10.1073/pnas.0909148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JF, et al. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 2013;342:1242592. doi: 10.1126/science.1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, et al. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature. 2011;476:320–323. doi: 10.1038/nature10249. [DOI] [PubMed] [Google Scholar]
- Simms BA, Zamponi GW. Neuronal voltage-gated calcium channels: structure, function, and dysfunction. Neuron. 2014;82:24–45. doi: 10.1016/j.neuron.2014.03.016. [DOI] [PubMed] [Google Scholar]
- Simske JS. Claudins reign: the claudin/EMP/PMP22/gamma channel protein family in. Tissue Barriers. 2013;1(3):e25502. doi: 10.4161/tisb.25502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Srivastava M, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz PR, et al. Independent evolution of striated muscles in cnidarians and bilaterians. Nature. 2012;487:231–234. doi: 10.1038/nature11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson JR, Snutch TP. Molecular nature of voltage-gated calcium channels: structure and species comparison. Wiley Interdiscip Rev Membr Transp Signal. 2013;2:181–206. [Google Scholar]
- Verret F, Wheeler G, Taylor AR, Farnham G, Brownlee C. Calcium channels in photosynthetic eukaryotes: implications for evolution of calcium-based signalling. New Phytol. 2010;187:23–43. doi: 10.1111/j.1469-8137.2010.03271.x. [DOI] [PubMed] [Google Scholar]
- Watson GM, Hessinger DA. Evidence for calcium channels involved in regulating nematocyst discharge. Comp Biochem Physiol Comp Physiol. 1994;107:473–481. doi: 10.1016/0300-9629(94)90028-0. [DOI] [PubMed] [Google Scholar]
- Zenkert C, Takahashi T, Diesner MO, Ozbek S. Morphological and molecular analysis of the Nematostella vectensis cnidom. PLoS One. 2011;6:e22725. doi: 10.1371/journal.pone.0022725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Chung I, Liu Z, Goldin AL, Dong K. A voltage-gated calcium-selective channel encoded by a sodium channel-like gene. Neuron. 2004;42:101–112. doi: 10.1016/s0896-6273(04)00148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.