Abstract
Discoba (Excavata) is an ancient group of eukaryotes with great morphological and ecological diversity. Unlike the other major divisions of Discoba (Jakobida and Euglenozoa), little is known about the mitochondrial DNAs (mtDNAs) of Heterolobosea. We have assembled a complete mtDNA genome from the aggregating heterolobosean amoeba, Acrasis kona, which consists of a single circular highly AT-rich (83.3%) molecule of 51.5 kb. Unexpectedly, A. kona mtDNA is missing roughly 40% of the protein-coding genes and nearly half of the transfer RNAs found in the only other sequenced heterolobosean mtDNAs, those of Naegleria spp. Instead, over a quarter of A. kona mtDNA consists of novel open reading frames. Eleven of the 16 protein-coding genes missing from A. kona mtDNA were identified in its nuclear DNA and polyA RNA, and phylogenetic analyses indicate that at least 10 of these 11 putative nuclear-encoded mitochondrial (NcMt) proteins arose by direct transfer from the mitochondrion. Acrasis kona mtDNA also employs C-to-U type RNA editing, and 12 homologs of DYW-type pentatricopeptide repeat (PPR) proteins implicated in plant organellar RNA editing are found in A. kona nuclear DNA. A mapping of mitochondrial gene content onto a consensus phylogeny reveals a sporadic pattern of relative stasis and rampant gene loss in Discoba. Rampant loss occurred independently in the unique common lineage leading to Heterolobosea + Tsukubamonadida and later in the unique lineage leading to Acrasis. Meanwhile, mtDNA gene content appears to be remarkably stable in the Acrasis sister lineage leading to Naegleria and in their distant relatives Jakobida.
Keywords: Discoba, endosymbiotic gene transfer, horizontal gene transfer, codon usage bias, plant-type RNA editing, split ribosomal protein gene
Introduction
Among the three supergroups of eukaryotes, Excavata is by far the least well characterized (Adl et al. 2012; He et al. 2014). This includes the Discoba, the only excavates to possess respiratory competent mitochondria and mitochondrial DNA (mtDNA) (Simpson et al. 2006). Nonetheless, it is already apparent that the mtDNAs of the three to four major branches of Discoba—Jakobida, Euglenozoa, Heterolobosea, and probably Tsukubamonadida (Kamikawa et al. 2014)—exhibit a level of diversity unsurpassed by any other major eukaryotic lineage (Gray et al. 2004). For example, Jakobida have the most bacterial-like, gene-rich mtDNAs known, with 60–66 protein-coding genes with known functions (Burger et al. 2013). These include genes for a classical bacterial RNA polymerase, whereas all other known mtDNAs use a nuclear-encoded viral-type RNA polymerase (Lang et al. 1997). Meanwhile, euglenozoan mtDNAs are difficult to compare with those of other species due to extensive gene fragmentation (Flegontov et al. 2011), RNA editing (insertion/deletion) (Lukeš et al. 2005; Hajduk and Ochsenreiter 2010), and/or trans-splicing (Kiethega et al. 2011; Moreira et al. 2012).
Much less is known about the mtDNAs of Heterolobosea, which have only been characterized for two closely related species—Naegleria gruberi and Naegleria fowleri (Fritz-Laylin et al. 2011; Herman et al. 2013). These are among the most gene-rich mtDNAs outside of the jakobids, with 42 annotated and 4 hypothetical protein-coding genes (open reading frames [ORFs]). Naegleria gruberi is also the first organism outside of land plants found to encode DYW-type pentatricopeptide repeat (PPR) proteins (Knoop and Rüdinger 2010). These proteins are involved in organellar RNA editing (Lurin et al. 2004; Fujii and Small 2011; Yagi et al. 2013), and two sites of C-to-U RNA editing have been verified in the N. gruberi mitochondrion (Rüdinger et al. 2011). Lastly, the mtDNA of the only known tsukubamonad, Tsukubamonas globosa, has a gene content roughly similar to that of the two Naegleria species (Kamikawa et al. 2014).
Heterolobosea is a vast group consisting almost exclusively of unicellular amoebas or amoeboflagellates (Page and Blanton 1985; Harding et al. 2013). The only molecularly well-characterized taxon is Naegleria due to the medical importance of the opportunistic pathogen N. fowleri, which can cause fatal primary amoebic meningoencephalitis in humans (Visvesvara et al. 2007). The only completely sequenced heterolobosean is N. gruberi, a close relative of N. fowleri and a model organism for research on the microtubule cytoskeleton (Fritz-Laylin et al. 2010). Otherwise, there are no full genome sequences available for any other heterolobosean despite their abundance and ecological diversity (Cavalier-Smith and Nikolaev 2008; Yubuki and Leander 2008; Park and Simpson 2011; Pánek et al. 2012; Harding et al. 2013).
The Acrasidae are the only even quasi-multicellular heteroloboseans (Brown, Kolisko, et al. 2012). These are common soil microbes that spend most of their life cycle as free-living amebae. However, when food is depleted, the amebae can aggregate and cooperate to form small multicellular tree-like fruiting bodies (Brown, Silberman, et al. 2012). As this aggregative behavior resembles that of the dictyostelid slime molds (Amoebozoa), dictyostelids were originally classified together with acrasids as their sister taxon in the family Acrasidae (Olive 1970). However, Olive (1970) also noted striking differences in the morphology of their amoeboid stages. Molecular phylogenies now clearly assign the dictyostelids to the eukaryotic supergroup Amorphea, along with Metazoa and Fungi, and place acrasids on the opposite side of the tree in Heterolobosea (Adl et al. 2012; He et al. 2014).
We have assembled a complete Acrasis kona mtDNA genome using a combination of shot-gun and Sanger sequencing with long range polymerase chain reaction (PCR), along with a draft nuclear genome and transcriptome. The A. kona mtDNA has lost nearly 40% of the protein-coding genes identified in Naegleria mtDNA, most of which are found in A. kona nuclear DNA and polyA transcripts. In place of these missing genes, over one-fourth of the A. kona mtDNA consists of novel ORFs, while the remaining protein-coding sequences exhibit extensive reorganization, such as gene splitting and transposition and gene cluster reshuffling. C-to-U type editing of mitochondrial RNA is also identified in A. kona, along with the presence of DYW-type PPR proteins encoded in the nucleus. Mapping of gene presence/absence onto a consensus phylogeny reveals a sporadic pattern of gene loss and genome reorganization in Discoba.
Materials and Methods
Cell Culture and DNA Extraction
Acrasis kona ATCC strain MYA-3509 (formerly Acrasis rosea) (Brown, Silberman, et al. 2012) was grown on CM+ (Corn Meal Plus) agar, with streaked Saccharomyces cerevisiae as the food source. For DNA isolation, cells were grown in Spiegel’s liquid medium (Spiegel 1982) in 250-ml flasks and shaken on a rotary shaker (120 cycles/min) at room temperature. Cells were harvested in 50-ml corning tubes after 48 h at a cell density of approximately 1 × 105/ml. Cell suspensions were transferred to Petri dishes and left for at least 1 h to allow the A. kona cells to settle and attach to the bottom. Cells were then washed three times with 10 mM phosphate buffer to remove the pellets of flocculated yeast. Cells were harvested by centrifugation and the DNA was extracted using the Blood & Cell Culture DNA Kit (Qiagen).
mtDNA Sequencing
A large portion of the A. kona mtDNA sequence was recovered from 454 shot-gun sequencing of total A. kona DNA (Fu C-J, Sheikh S, Baldauf SL, unpublished data). Four contigs of mtDNA sequence with size ranges from 3 to 17 kb were obtained by genomic assembly using Newbler v2.5 (Roche). These contigs were used for a baiting and iterative mapping approach using Illumina sequencing data to correct base-calling errors known to be associated with long single-nucleotide repeats in 454 reads with Mira (Hahn et al. 2013). Long range PCR was carried out using nested primers to cross gap regions using the LongAmp Taq PCR Kit (NEB) (supplementary table S1, Supplementary Material online). PCR products ranging from 2 to 7 kb were cloned using the CloneJET PCR Cloning Kit (Fermentas) (supplementary fig. S1, Supplementary Material online). Colonies containing inserts were sent for sequencing with ABI 3730 sequencer (Applied Biosystems) at Macrogen (Seoul, South Korea). Final gap closure was accomplished using a primer walking strategy.
RNA Extraction and Transcriptome Sequencing
RNA extraction and transcriptome sequencing was conducted as described in He et al. (2014). Briefly, cells were grown in Spiegel’s liquid medium and total RNA was extracted using TRI Reagent LS (Sigma-Aldrich). Poly (A)+ RNA was purified from 80 µg of total RNA using PolyAtract mRNA Isolation Systems (Promega) and sent for sequencing on a 454 GS FLX+ Titanium platform at Macrogen. Transcripts were assembled separately using the programs Newbler (v2.5, Roche) and Mira (v3.4) after removal of adapter sequences, and the results were combined using the program CAP3 (Huang and Madan 1999).
Genome Annotation
ORFs were annotated using BLASTp and PSI-BLASTp searches of the National Center for Biotechnology Information (NCBI) nr database. For ORFs lacking significant hits (E value cutoff = 1e−10), the more sensitive HHpred method, which uses profile Hidden Markov Models (HMMs), was used to search against all the databases provided on its web server (http://toolkit.tuebingen.mpg.de/hhpred/, last accessed September 1, 2014) (Söding 2005). The validity of hits from bacteria or viruses was checked by positional conservation patterns based on multiple alignments from the Conserved Domain Database at NCBI where available. Structural RNAs and potential introns were predicted using the automated gene annotation tool MFannot (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl, last accessed September 1, 2014) and the warnings in the output (e.g., alternative translation initiation sites, gene fusions, and exon–intron boundaries) were manually checked. Predicted boundaries of small and large ribosomal subunit RNA genes were verified in alignment with sequences from both Naegleria and jakobid mtDNAs. Transfer RNA (tRNA) genes were identified using tRNAscan-SE v1.23 (Lowe and Eddy 1997). Secondary structure of the largest noncoding mtDNA region was inferred with the MFold web server (Zuker 2003) using default settings and drawn with VARNA (Darty et al. 2009). Approximate tandem repeats were identified with tandem repeats finder (Benson 1999). The genome map was illustrated using DNAPlotter (Carver et al. 2009), followed by manual adjustment.
Gene Synteny
The mtDNAs of discobid species listed in table 1 were used for reciprocal BLASTn and tBLASTx searches to identify regions of similarity, insertions, and rearrangements. Artemis (Rutherford et al. 2000) and Artemis Comparison Tool (ACT) (Carver et al. 2005) were used to interactively visualize the genomic regions of interest. A cutoff score of 40 was used to determine the presence/absence of gene synteny blocks. The gene order of the ribosomal protein (r-protein) synteny block between representative alpha proteobacteria and discobid species was identified by reciprocal BLASTp search with a threshold of 1e−10 and visualized with Circoletto (Darzentas 2010).
Table 1.
General Features of Acrasis kona and Other Discoba mtDNAs
| Speciesa | Size (bp) | AT Content (%) |
AT Skew | GC Skew | Size Portion (%) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| PCGsb | RNAc | Noncoding | Overall | PCGs | RNA | Noncoding | ||||
| Heterolobosea | ||||||||||
| Acrasis kona | 51,458 | 84.0 | 72.4 | 89.8 | 83.3 | 0.102 | 0.354 | 82.5 | 10.7 | 6.8 |
| Naegleria gruberi | 49,843 | 79.3 | 67.1 | 85.0 | 77.8 | −0.087 | 0.170 | 81.2 | 11.9 | 6.9 |
| Naegleria fowleri | 49,531 | 75.9 | 60.5 | 83.2 | 74.8 | −0.079 | 0.196 | 79.9 | 11.6 | 8.5 |
| Jakobida | ||||||||||
| Andalucia godoyi | 67,656 | 64.3 | 51.3 | 69.4 | 63.7 | −0.012 | 0.015 | 81.0 | 10.2 | 8.8 |
| Histiona aroides | 70,177 | 64.7 | 52.5 | 67.2 | 64.6 | −0.308 | 0.047 | 81.2 | 9.7 | 9.1 |
| Jakoba bahamiensis | 65,327 | 68.3 | 54.0 | 76.8 | 67.8 | −0.102 | 0.138 | 82.1 | 10.1 | 7.8 |
| Jakoba liberad | 100,252 | 67.8 | 55.5 | 72.1 | 68.0 | 0.041 | −0.017 | 72.0 | 7.0 | 21.0 |
| Reclinomonas americana | 69,586 | 73.7 | 55.3 | 83.2 | 73.2 | −0.023 | 0.125 | 80.2 | 9.8 | 10.0 |
| Seculamonas ecuadoriensis | 69,158 | 68.2 | 53.9 | 76.8 | 68.1 | 0.017 | −0.011 | 77.8 | 9.7 | 12.5 |
| Tsukubamonadida | ||||||||||
| Tsukubamonas globosa | 48,463 | 67.4 | 56.5 | 70.6 | 66.2 | −0.136 | 0.100 | 76.8 | 13.7 | 9.5 |
aGenBank accession number: A. kona KJ679272; N. gruberi AF288092; N. fowleri JX174181; A. godoyi KC353352; H. aroides KC353353; J. bahamiensis KC353354; J. libera KC353355; R. americana KC353356; S. ecuadoriensis KC353359; T. globosa AB854048.
bPutative coding regions including annotated protein-coding genes and unknown ORFs.
cHeterolobosea and Tsukubamonadida (rRNA and tRNA); Jakobida (rRNA, tRNA, RNase P-RNA, and tmRNA).
dmtDNA is linear.
Codon Usage Analysis
For codon usage bias analysis, the values of expected effective number of codons (ENC or Nc) from GC content at synonymous third codon position (GC3s) under H0 (null hypothesis, i.e., no selection) were calculated according to the equation of Wright (1990): Nc = 2 + S + {29/[S2 + (1 − S)2]} (S denotes GC3s). If a given gene is only subject to G + C composition mutation constraint, it will lie just on the standard curve, whereas other kinds of selection and/or mutation pressure will cause values to lie above or below the curve (Fu et al. 2009). We also used its variant Nc′, which accounts for background nucleotide composition, to quantify bias in codon usage for individual genes in different mtDNAs using ENCprime (Novembre 2002). The measurement of codon deviation coefficient (CDC) was performed to investigate the potential influence of nucleotide positional heterogeneity on codon usage bias (Zhang et al. 2012), using the Composition Analysis Toolkit (CAT) with a statistical test of bootstrap resampling (10,000 replicates) under the default settings. GC content at first, second, and third codon positions (GC1, GC2, and GC3) was calculated for all annotated and unidentified ORFs with over 100 codons using codonW (codonw.sourceforge.net/) and visualized with GC Frame plot (watson.nih.go.jp/∼jun/cgi-bin/frameplot.pl). The two-sided Wilcoxon rank sum test was used to check the distribution of differences for the values of Nc′ and GC3s between mtDNAs. All the statistical analyses were carried out in R (www.R-project.org, last accessed September 1, 2014).
Amino Acid Compositional Homogeneity
For posterior predictive tests of compositional heterogeneity using PhyloBayes 3.2 (Lartillot et al. 2009), we used a concatenated protein data set consisting of 19 mtDNA-encoded proteins for 54 taxa (35 eukaryotes + 19 proteobacteria) including a total of 5,791 aligned amino acid positions (Burger et al. 2013). We tested the full data set and also recoded the amino acid data into the six Dayhoff groups (AGPST, C, DENQ, FWY, HKR, and ILMV) that tend to replace one another (Dayhoff et al. 1978). The tests use the default CAT model and run for 10,000 cycles, with first 5,000 cycles discarded as burn-in. The z score was used as a measure of the compositional deviation of individual taxa between the taxon-specific and global empirical frequencies over the 20 amino acids.
Identification of Nuclear-Encoded Mitochondrial Protein Genes
Protein-coding genes uniquely missing from A. kona mtDNA relative to other discobids were searched for in the A. kona draft nuclear genome by tBLASTn using protein sequences from the Naegleria and jakobid mtDNA as queries. All hits with an E value < 1e−10 were examined by protein multiple sequence alignment with corresponding Naegleria and jakobid sequences. Assembly coverage plots of A. kona nuclear contigs were checked for the corresponding loci of all predicted nuclear-encoded mitochondrial (NcMt) gene regions using Tablet (Milne et al. 2013). Transcriptional activity of the putative NcMt genes was checked by direct mapping of the 454 transcriptome reads by Newbler (v2.5, Roche) and by BLASTn against the assembled mRNA transcript sequences. Mitochondrial transit peptide sequences and the N-terminal cleavage sites were predicted using TargetP (Emanuelsson et al. 2007), Predotar (Small et al. 2004), and Mitoprot (Claros and Vincens 1996).
C-to-U RNA Editing Site Prediction and cDNA Synthesis
C-to-U type RNA editing sites in A. kona mtDNA were predicted using PREPACT (Lenz and Knoop 2013) (last accessed May 15, 2014). The probability of each candidate editing site was calculated by the percentage of the overlapping predictions against all the references from the output “commons.” Multiple alignment of the gene sequences containing the candidate sites was further checked. Oligonucleotide primer pairs were designed to flank the coding regions of four A. kona mitochondrial genes (nad1, atp6, cob, and cox3) with strong candidate sites (supplementary table S1, Supplementary Material online). For cDNA synthesis, total RNA was extracted and treated with DNase (Thermo). First strand cDNA was synthesized using the Phusion RT-PCR Kit (Thermo) with hexanucleotide random primer mix. PCR amplification of both mitochondrial genomic sequence and cDNA products was performed using the Phusion High-Fidelity DNA Polymerase (Thermo). PCR amplicons were cleaned with ExoSap-IT (GE Healthcare) and sent for direct sequencing.
Identification of DYW-Type PPR Proteins
Known DYW-type PPR protein sequences in N. gruberi (11) and Physcomitrella patens (10) (Knoop and Rüdinger 2010) were used as queries to search against the A. kona nuclear contigs, either with full length protein sequences (including variable length PLS repeat domains) or using only the conserved carboxy-terminal E/E+/DYW domain as query. All candidate proteins found were screened for the possible presence of PPR motifs using TPRpred (Karpenahalli et al. 2007). Homologous sequences were obtained by taxon-limited BLASTp searches of the NCBI database against all major groups of early branching land plants (Liverworts, Mosses, Hornworts, and Lycophytes) and additional taxa based on (Iyer et al. 2011; Schallenberg-Rüdinger et al. 2013), specifically Heterolobosea (N. gruberi), Amoebozoa (Acanthamoeba castellanii, Physarum polycephalum), Metazoa (Adineta riccia, Philodina roseola), Fungi (Laccaria bicolor), Charophyta (Nitella hyaline), and Malawimonas jakobiformis. Only sequences containing full E/E+/DYW domains and/or with conservative key amino acid positions were used in subsequent analyses. Sequence logos were generated using WEBLOGO (Crooks et al. 2004).
Phylogenetic Analyses
For putative A. kona NcMt genes, single gene trees were generated from inferred amino acid sequences aligned with MAFFT v7 (Katoh and Standley 2013). For the phylogeny of discobids, a concatenated data set of 24 mitochondrial proteins (Atp1, 3, 6, 8, 9, Cox1, 2, 3, 11, Cob, Nad1, 2, 3, 4, 4L, 5, 6, 7, 8, 9, 10, 11, Sdh2, and TufA) was used from taxa with complete mtDNA sequences. Conserved blocks were extracted using Gblocks with relaxed parameters (Castresana 2000). Multiple sequence alignment files are available upon request. Bayesian analysis was performed on NcMt proteins with MrBayes v3.2.2 (Ronquist et al. 2012) using a mixture of amino acid models. Searches consisted of two sets of four chains run over 1 million generations, discarding a burn-in of 25%. Bayesian analysis of discobid phylogeny and DYW-type PPR protein was performed with PhyloBayes MPI 1.4f (Lartillot et al. 2013), using the CAT + Gamma model and the predefined WLSR5 profile model (Wang et al. 2008), respectively, with constant sites removed (-dc). Analyses were run for at least 15,000 cycles (Max diff < 0.20), with the first 5,000 cycles discarded as burn-in. Maximum-likelihood analysis was conducted with RAxML v7.3.3 (Stamatakis et al. 2012) using the PROTGAMMALG model and the fast bootstrapping option (1,000 replicates). All phylogenetic analyses were run on the CIPRES Science Gateway (www.phylo.org, last accessed September 1, 2014).
Results
General Features, Gene Content, and Genome Organization in A. kona mtDNA
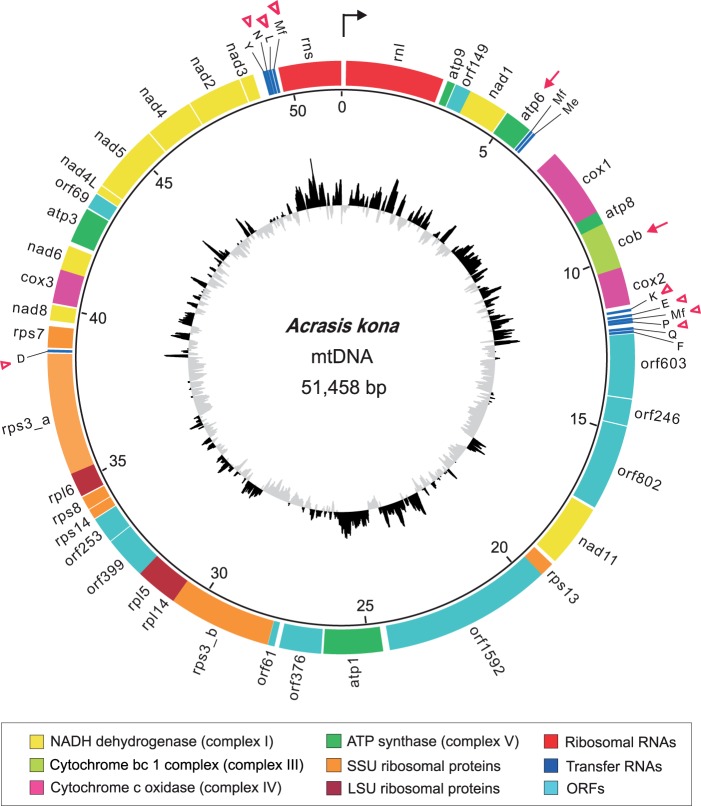
The A. kona mitochondrial genome was assembled into a single circular-mapping molecule with a size of 51,458 bp (fig. 1). Its overall A + T (AT) content is 83.3%, higher than those of the two Naegleria mtDNAs (74.8–77.8%), the only other sequenced heterolobosean mtDNAs. In fact, A. kona mtDNA has the highest AT content among known mtDNAs of free-living discobids for noncoding regions (89.8% AT), protein-coding genes (84% AT), and structural RNA genes (72.4% AT) (table 1). Despite this extreme AT-richness, A. kona mtDNA is predicted to consist of 93.2% coding regions, placing it among the most compact discobid mtDNAs, along with N. gruberi (93.1%) and Jakoba bahamiensis (92.2%), the most compact jakobid mtDNA (table 1). The largest noncoding region (755 bp) of A. kona mtDNA also contains its largest repetitive region, which exhibits extensive predicted secondary structure (supplementary fig. S2, Supplementary Material online).
Fig. 1.—
The mitochondrial genome of Acrasis kona. A circular map of the A. kona mtDNA is shown, drawn to scale as indicated by the inner circle and with coordinates in kilobases. The outermost track shows predicted genes, color-coded by functional category as indicated in the box below the map. The innermost circle shows GC content above (black) or below (gray) the genome average. Protein-coding genes with identified C-to-U type RNA editing are indicated with arrows. tRNA genes with predicted mismatches at the first three positions of the acceptor stem are indicated with triangles.
Despite its high predicted coding capacity, the number of protein-coding sequences with known functions in A. kona mtDNA is markedly less than in Naegleria, as are the number of predicted tRNA genes (fig. 2). Only 26 protein-coding genes and 11 different tRNA genes can be identified in A. kona mtDNA, versus 42 protein-coding genes and 20 tRNAs in both examined Naegleria mtDNAs and 41 protein-coding genes and 24 tRNAs in Tsukubamonas (fig. 2). We compared the sequence divergence of oxidative phosphorylation (OXPHOS) pathway genes in representative discobid mtDNAs (supplementary table S2, Supplementary Material online), as these are nearly universal among functional mitochondria and generally well conserved (Szklarczyk and Huynen 2010). The two Naegleria species have an overall divergence of 18.2% for these proteins at the amino acid level, which is similar to that between the two most closely related jakobids, Histiona aroides and Reclinomonas americana (16.2%). For comparison, there is 59.0% overall divergence between OXPHOS genes shared by Acrasis and Naegleria, reflecting their more distant relationship.
Fig. 2.—
A comparison of mtDNA gene contents among representative lineages of Discoba. Taxa names are shaded in purple (Heterolobosea), green (Tsukubamonadida), and yellow (Jakobida). Genes are color-coded according to the key in the box at the bottom right. mtDNA-encoded proteins exclusively found in Jakobida are indicated according to Burger et al. (2013). Nuclear-encoded mitochondrial (NcMt) proteins of two heterolobosean species (Acrasis kona and Naegleria gruberi) were identified using jakobid mtDNA genes as query (tBLASTn, E value < 1 e−10).
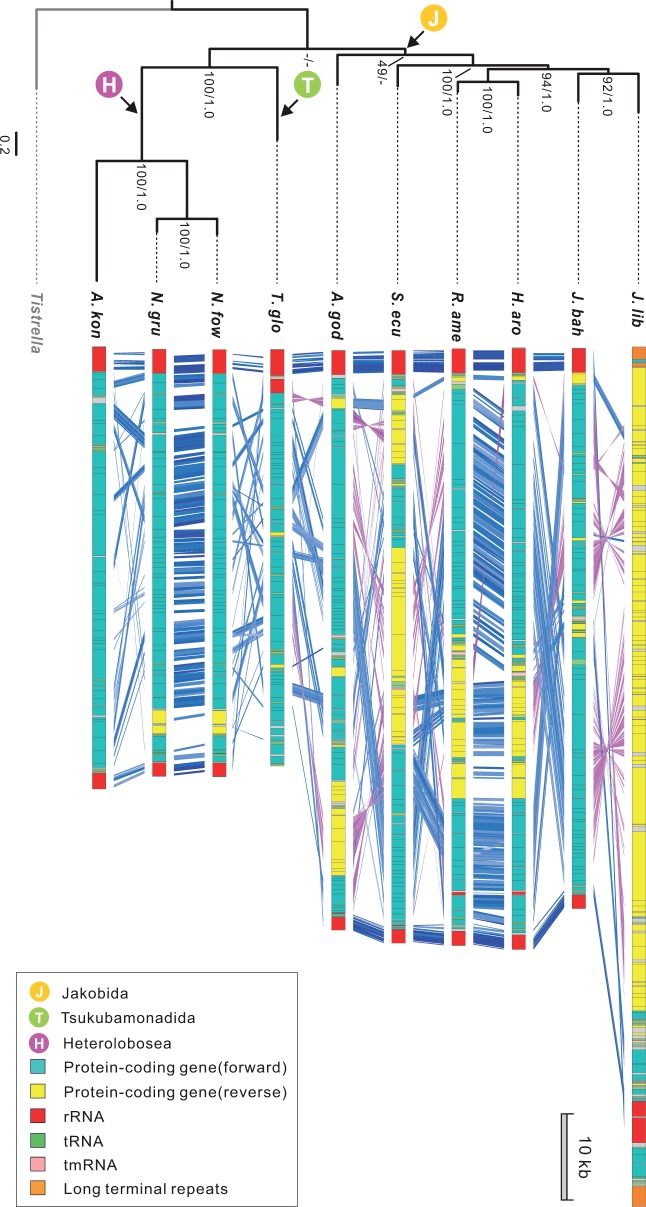
A mapping of the overall genome synteny among discobid species onto a consensus phylogeny of the taxa shows that the mtDNAs of the two closely related Naegleria species have a highly conserved gene order. In contrast, very little synteny is observed between Acrasis and Naegleria mtDNA (fig. 3). Good overall conservation of synteny is also seen between terminal clades of jakobids, although considerable gene transposition and inversion are found between the mtDNAs of more distantly related jakobids, especially between the earliest branches of the clade (fig. 3). One exception to this is four genes encoding NADH dehydrogenase subunits (nad4L, nad5, nad4, and nad2) (fig. 1). These share the same gene order in Acrasis, Tsukubamonas and all six jakobids, although they are dispersed in the Naegleria mtDNAs.
Fig. 3.—
A mapping of overall mtDNA gene synteny between different lineages of Discoba (Heterolobosea, Tsukubamonadida, and Jakobida) onto a consensus phylogeny. The tree was based on a concatenated data set of 24 mitochondrial proteins (7,313 aligned amino acid positions), with the α-proteobacterium Tistrella mobilis (NC_017956) as outgroup. Numbers indicate maximum-likelihood bootstrap values with RAxML (Stamatakis et al. 2012) and posterior probabilities with PhyloBayes (Lartillot et al. 2013). Genes are color-coded by functional category as indicated in the box. Genes in the forward versus reverse order are indicated in blue and purple lines, respectively, as generated by ACT (Carver et al. 2005) with a cut off value of 40.
A Highly Interrupted Ribosomal Protein Gene Cluster
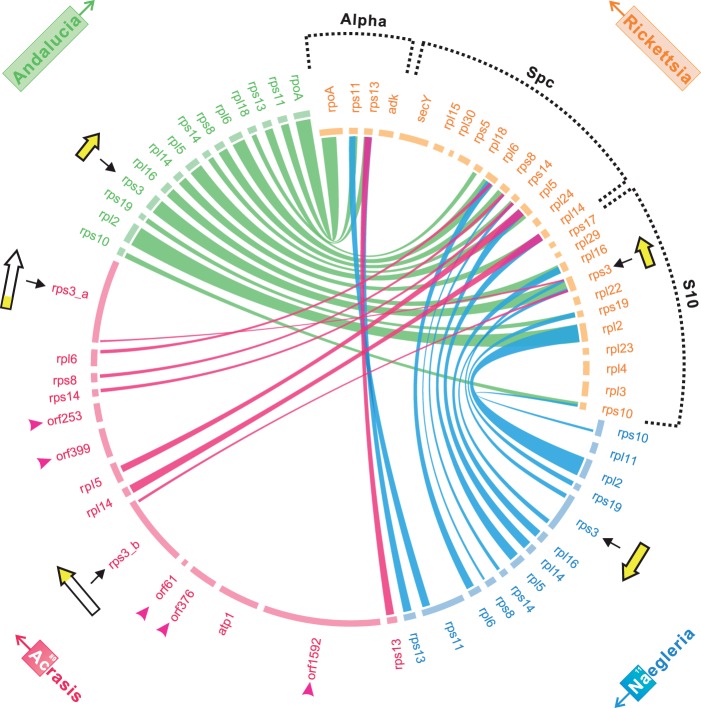
Of special interest is a highly conserved gene cluster recently identified in all jakobid mtDNAs, which corresponds to a large synteny block of r-protein operons (Burger et al. 2013). The order of these in the free-living α-proteobacterium Tistrella mobilis is (L11–L10–Beta–Str–S10–Spc–Alpha) (supplementary fig. S3, Supplementary Material online), which is thought to represent the ancestral organization of these operons in bacterial genomes (Yang and Sze 2008; Bratlie et al. 2010). We compared the arrangement of r-protein genes in heterolobosean and jakobid mtDNAs with those of close α-proteobacterial relatives, focusing particularly on Rickettsia where the L11–L10–Beta operons are separated from the Str–S10–Spc–Alpha operons but gene content and orders are well conserved within operons [supplementary fig. S3, Supplementary Material online; unlike (Burger et al. 2013)].
Gene order in the three contiguous r-protein operons—S10, Spc, and Alpha—is well conserved in Naegleria compared with Tsukubamonas, all jakobids, and α-proteobacteria (fig. 4). However, there are many fewer r-protein genes in A. kona mtDNA compared with Naegleria (8 vs. 17, respectively), and the only detectable gene synteny here in A. kona is in the Spc operon. The A. kona cluster further appears to be disrupted by the insertion of several putative protein-coding sequences in this synteny block, including atp1 and multiple ORFs of unknown function (fig. 4).
Fig. 4.—
A comparison of mtDNA gene order for the largest r-protein synteny block. The same block is compared with the generally contiguous S10, spectinomycin (Spc) and Alpha operons of α-proteobacteria as determined by BLASTp (E value < 1 e−10) against Rickettsia prowazekii genome (NC_000963). The locations of the rps3 genes within each synteny cluster are indicated with schematic models. The unknown ORFs in Acrasis kona are indicated with fuchsia triangles.
Interestingly, in A. kona mtDNA the five genes identified in the Spc operon are flanked on both sides by genes showing similarity to the rps3 gene, which is normally located inside the S10 operon (fig. 4). In A. kona, the gene is not only split, but the two halves designated as rps3_a and rps3_b are transposed relative to each other, separated by 4,588 bp that contains the five r-protein genes in the Spc operon plus two ORFs, and both reading frames are substantially extended (supplementary fig. S4, Supplementary Material online). Notably, the split occurs at roughly the same position as an approximately 300 aa insertion in Naegleria rps3. Together the two A. kona fragments encode a protein (2,099 aa) much larger than other known rps3 proteins (200–250 aa), and only the first 86 aa of rps3_a (1,133 aa) and the final 93 aa of rps3_b (966 aa) map to the predicted Naegleria rps3, at the N- and C-termini, respectively. GC content is also heterogeneous across the border between the conserved and extension portion of both fragments (supplementary fig. S4, Supplementary Material online). Moreover, the two genes flanking rps3 in the jakobid and Naegleria mtDNAs, rpl16 and rps19, both appear to be transferred to the nucleus in A. kona (see discussion).
Acquisition of Novel ORFs
Despite its lower gene content, the A. kona mtDNA genome (51.5 kb) is roughly the same size as those of Naegleria (49.5–49.8 kb) and Tsukubamonas (48.6 kb). This is due to the presence of ten predicted ORFs in A. kona mtDNA, constituting 26.5% of the genome and potentially encoding proteins ranging in size from 69 to 1,592 amino acids (supplementary table S3, Supplementary Material online). All ten ORFs occur on the sense DNA strand and in the same transcriptional orientation as the rest of the coding content of the genome (fig. 1). No significant sequence similarity (tBLASTx, E value threshold 1 e−10) was detected between the A. kona ORFs and the four ORFs in Naegleria mtDNA or any of the ORFs found in other discobid mtDNAs. The HMM search of A. kona ORFs for domain similarities against multiple databases (Pfam, SCOP, and InterPro) also did not generate significant hits (E value threshold 1 e−3).
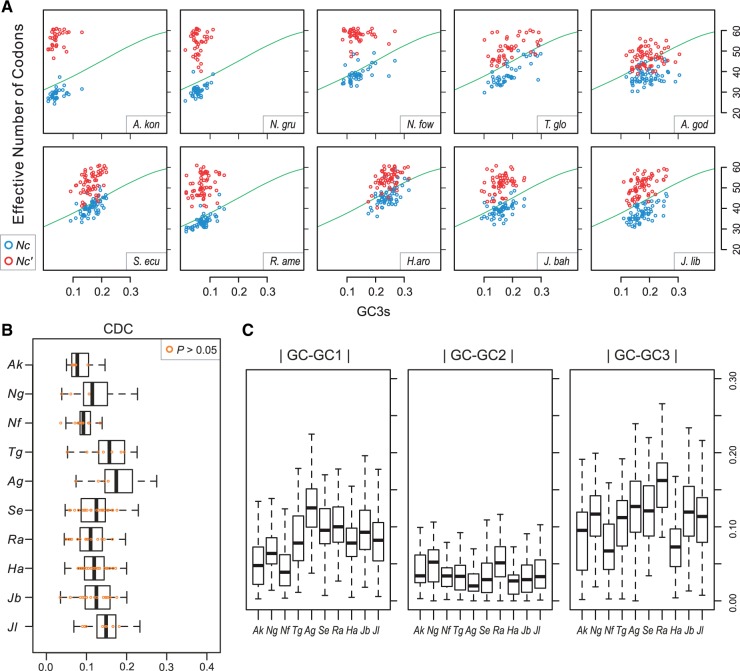
Codon usage bias analysis of the novel ORFs could show whether the putative genes have been exposed to the same mutation pressure as the more typical mitochondrial genes (e.g., OXPHOS pathway), and thus provide an indication of when these novel genes were acquired. A plot of Nc versus GC3s shows that values for all A. kona genes have generally small distance deviations from the standard curve (fig. 5A). When corrected for background nucleotide composition, the Nc′ values for most A. kona genes range from 50 to 61(overall 54.10) (fig. 5A). The calculated overall value of CDC of A. kona genes (0.091 ± 0.029) was also shown to be the lowest among all discobid mtDNAs (fig. 5B). Thus, the measurement of Nc, Nc′, and CDC all suggest no strong observed codon usage bias throughout A. kona mtDNA, likely reflecting a weak selection pressure on this genome despite its overall high AT content. In fact, variation in the heterogeneities of positional GC contents, particularly at first and third codon positions appears to be generally consistent with overall estimated codon usage bias in protein-coding sequences throughout discobid mtDNAs (fig. 5C).
Fig. 5.—
Codon usage bias and nucleotide positional heterogeneity of mitochondrial protein-coding sequences in the mtDNAs of Discoba. (A) The values of Nc (Wright 1990) and its variant (Nc′) (Novembre 2002) are plotted against GC3s. A line of standard curve based on the equation of Wright (1990) is superimposed on the graph. (B) Values of codon deviation coefficient (CDC) (Zhang et al. 2012) range from 0 (no bias) to 1(maximum bias). Genes with a CDC value of nonstatistical significance (P > 0.05) are indicated in orange circles. (C) Heterogeneity of positional GC content is represented by absolute differences between overall GC content and positional GC content at first, second, and third codon positions (GC1, GC2, and GC3). Ak, Acrasis kona; Ng, Naegleria gruberi; Nf, Naegleria fowleri; Tg, Tsukubamonas globosa; Ag, Andalucia godoyi; Ha, Histiona aroides; Jb, Jakoba bahamiensis; Jl, Jakoba libera; Ra, Reclinomonas americana; Se, Seculamonas ecuadoriensis.
The codon position-specific GC content of the A. kona ORFs is generally consistent with the rest of the protein-coding genes in its mtDNA (supplementary table S3, Supplementary Material online), and no significant difference was detected for Nc′ or GC3s between the protein-coding sequences and the unknown ORFs in A. kona (Wilcoxon rank sum test, P = 0.483 and 0.263, respectively). This suggests that these ORFs have resided in the A. kona mtDNA for a long enough time to adjust the base composition pattern to the strong AT mutation pressure. However, the largest ORF (ORF1592) has the highest GC content value at third codon position of all mitochondrial genes (0.163), which is about 2- to 3-fold higher than most other genes. That is largely due to a number of repeat elements in ORF1592, including two tandem repeats with a periodicity of 9 and 18 bp and heterogeneous GC3 content (supplementary fig. S2, Supplementary Material online). Meanwhile, ORF603, ORF246, and ORF802 tend to be lower in GC at both first and second codon positions than the typical mitochondrial genes, perhaps indicating that they evolve under lower selective constraints than the other genes (supplementary table S3, Supplementary Material online).
To examine whether lowered selective constraints have resulted in a more extreme amino acid bias in A. kona mtDNA, we examined the amino acid compositional heterogeneity for a set of 19 mitochondrial proteins (mainly involved in OXPHOS pathway) among a broad sampling of taxa (54 species). The compositional deviation for A. kona is among the highest detected of all taxa (supplementary table S4, Supplementary Material online), indicating a substantial violation of homogeneity (Lartillot et al. 2009). When these protein sequences were recoded using the six Dayhoff common amino acid substitution groups (Dayhoff et al. 1978), the z scores are decreased for most examined individual taxa except for Acrasis or Naegleria or for a scattered sampling of other taxa, particularly some amoebozoan species (Acantamoeba, Dictyostelium, and Hartmannella) (supplementary table S4, Supplementary Material online). The presence of biased amino acid composition in the OXPHOS proteins of A. kona mtDNA could possibly be linked to several attributes mentioned above, that is, a combined outcome of extreme AT content, high coding sequence divergence, and a loose constraint of selection pressure.
Functional Gene Transfer to the Nucleus
Among the 16 annotated mitochondrial protein-coding genes that are missing from A. kona mtDNA relative to Naegleria, 11 were detected as full predicted ORFs on A. kona nuclear contigs. The corresponding loci of each NcMt gene candidate on A. kona nuclear contigs were checked by coverage plots. Consistent homogeneous coverage patterns were shown for all these predicted genes, along with their flanking regions (supplementary fig. S5, Supplementary Material online). These 11 putative NcMt proteins include seven r-proteins, three OXPHOS proteins (nad7, nad9, and sdh2), and one protein involved in protein maturation (cox11) (supplementary table S5, Supplementary Material online). The missing genes in A. kona mtDNA include four from the S10 operon (rps10, rps19, rpl2, and rpl16), all of which are identified in A. kona nuclear contigs.
Phylogenetic trees for 10 of the 11 A. kona putative NcMt genes show them to be most closely related to their Naegleria mtDNA homologs (supplementary fig. S6, Supplementary Material online). This is despite the short lengths of many of these proteins (<130 aligned amino acid positions), which generally makes it difficult to get a strong phylogenetic signal. All of the 11 A. kona NcMt genes except rps2 are strongly predicted to carry a mitochondrial targeting signal. Moreover, 9 of the 11 A. kona NcMt genes are also predicted to encode N-terminal extensions relative to their counterparts in Naegleria mtDNA, potentially indicating nucleus-derived mitochondrial transit peptides ranging in size from 7 to 61 amino acids (supplementary table S5, Supplementary Material online). However, no sequence similarity was detected among these predicted transit peptides at the nucleotide or amino acid level. Transcriptome data also show that all the 11 A. kona putative NcMt genes are actively transcribed into polyA RNA (data not shown). Thus it appears that these A. kona putative NcMt genes represent functional gene transfer from mtDNA to the nucleus, sometime since the last common ancestor (LCA) of Acrasis and Naegleria.
C-to-U Type RNA Editing
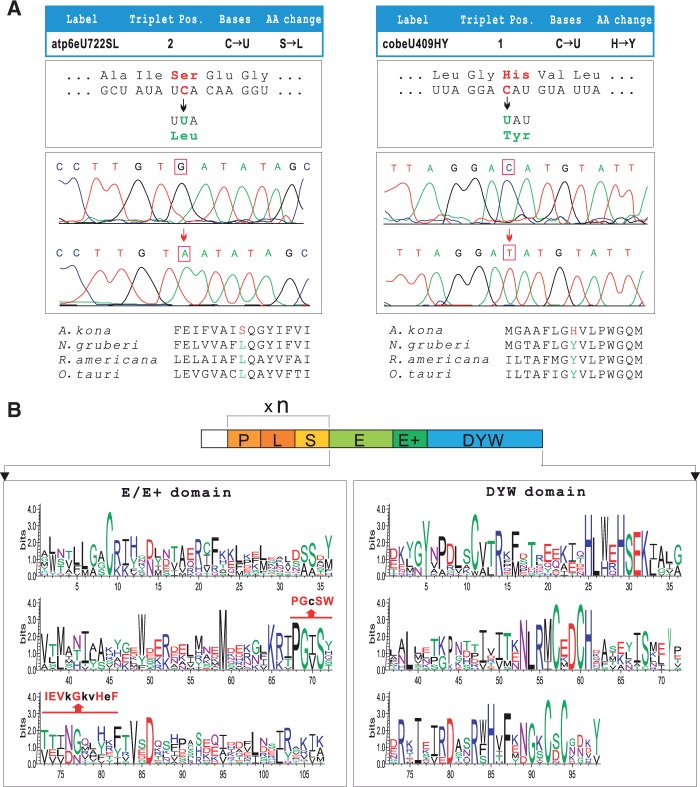
Sequence comparisons suggest the presence of six strong candidate sites of C-to-U RNA editing in four A. kona mtDNA genes—nad1, atp6, cob, and cox3. Editing was checked for all six sites by sequencing PCR products of the four genes from both genomic and cDNA templates. These experiments confirmed two of the predicted C-to-U editing sites, one each in the atp6 and cob genes (atp6eU722SL and cobeU409HY) (fig. 6A), but rejected the remaining four.
Fig. 6.—
C-to-U mitochondrial RNA editing and the DYW-type PPR proteins in the Acrasis kona genome. (A) Top: Two RNA editing sites in Acrasis mtDNA-encoded genes (atp6eU722SL and cobeU409HY). Middle: The PCR and RT-PCR products from genomic and cDNA templates. The Atp6 gene was sequenced using the antisense primer. Bottom: Conserved amino acid codons illustrated by alignments including Naegleria gruberi (Heterolobosea), Reclinomonas americana (Jakobida), and Ostreococcus tauri (green alga). (B) The schematic motif structure and plot of sequence conservation for the Acrasis DYW-type PPR proteins. A highly conserved 15 amino acid motif (PG box) identified in the land plants is indicated above the positions.
We searched the A. kona nuclear contigs for potential homologs of the DYW-type PPR proteins, which have been postulated as the key factors in C-to-U RNA editing in plant organelles (Salone et al. 2007). This identified 12 predicted ORFs with a recognizable E/E+/DYW domain at their carboxy-terminus. These ORFs were all further predicted to encode multiple PLS repeat domains by the program TPRpred (Karpenahalli et al. 2007). A highly conserved 15 amino acid motif or “PG box” (PGxSWIEVxGxxHxF) that bridges the E and E+ domain, previously identified in land plant DYW-type PPR proteins (Okuda et al. 2007) is also present in the 12 A. kona ORFs (fig. 6B).
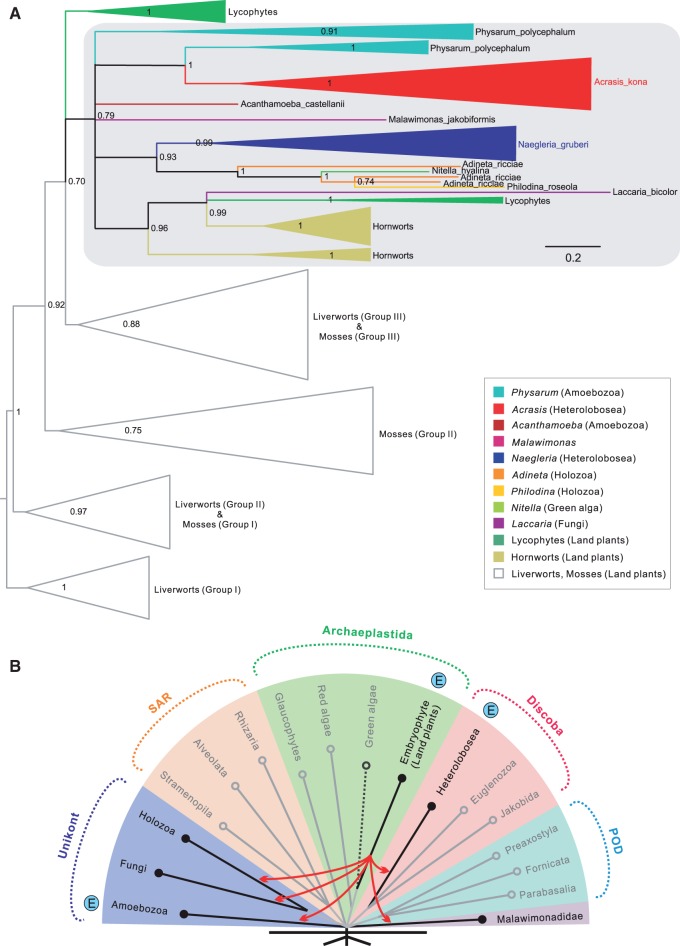
DYW-type PPR proteins have been identified in almost every major group of land plants except for marchantiid liverworts (Rüdinger et al. 2008), as well as in additional scattered taxa across the eukaryote tree of life (Knoop and Rüdinger 2010; Iyer et al. 2011; Schallenberg-Rüdinger et al. 2013). Phylogeny of the DYW-type PPR proteins further suggests that the genes encoding these proteins most probably have spread among eukaryotes through horizontal gene transfer (HGT) from early branching land plants (fig. 7A and B). Where found these sequences show a general trend toward lineage-specific expansion and diversification, including within Acrasis and Naegleria (supplementary fig. S7, Supplementary Material online). Interestingly, Acrasis DYW-type PPR proteins show a strong phylogenetic affinity for the distant-related amoebozoan Physarum (1.0 Bayesian posterior probability, biPP), whereas the homologues of Naegleria group together with the rotifer clade (0.93 biPP) (fig. 7A). This suggests possible multiple independent acquisition of the plant-type RNA editing factors in these two heterolobosean lineages.
Fig. 7.—
Phylogeny of DYW-type PPR proteins in the eukaryote species. (A) Bayesian phylogeny of the E/E+/DYW domain of the DYW-type PPR proteins. Taxa include a broad sampling of early branching land plants (Liverworts, Mosses, Hornworts, and Lycophytes), and those outside land plants (Heterolobosea [Acrasis kona, Naegleria gruberi], Amoebozoa [Acanthamoeba castellanii, Physarum polycephalum], Metazoa [Adineta ricciae, Philodina roseola], Fungi [Laccaria bicolor], Charophyta [Nitella hyaline], Malawimonas jakobiformis). Values correspond to posterior probabilities; all nodes with posterior probability less than 0.7 are collapsed. Taxon labels are color-coded according to the key at the bottom right. The strong phylogenetic affinity of the only known green algal sequence (N. hyaline) for rotifers may indicate possibly transcriptomic data contamination as reported in (Laurin-Lemay et al. 2012). A detailed list of taxa is shown in supplementary figure S7, Supplementary Material online. (B) A schematic tree showing the phylogenetic distribution of DYW-type PPR proteins in the major eukaryote lineages. Lineages with experimental evidence of C-to-U type RNA editing are indicated with “E.” Arrows indicate the probable HGT directions from land plants to the other eukaryotes. Dotted line indicates the doubtful presence of DYW-type PPR proteins in N. hyaline.
Discussion
Gene Loss and Genome Rearrangement in A. kona mtDNA
A complete sequence of the A. kona mtDNA shows that it is roughly the same size as the mtDNA of its closest sequenced relatives (Naegleria spp.), but has a remarkably different organization and gene content (figs. 2 and 3). Sixteen genes of known function, corresponding to nearly 40% of protein-coding genes in Naegleria are missing from A. kona mtDNA. Eleven of these missing genes are found on A. kona nuclear contigs, and phylogenetic analyses suggest that at least 10 of these 11 putative NcMt genes arose by direct functional transfer of mtDNA to the nucleus (Timmis et al. 2004). Seven of the 11 putative transfers involve r-proteins, consistent with evidence that r-protein genes tend to be lost from mtDNA more often than respiratory genes (Adams and Palmer 2003). It thus would be of interest to look for the preinsertion nuclear loci where such functional transfer has occurred upon the completion of the annotation of A. kona draft nuclear genome. The translation products of five remaining missing genes (rps4, rps11, ccmF, ccmC, and tatC) in A. kona mtDNA are usually present in the mitochondrial proteomes of free-living species (Gray et al. 2004). Thus there may have been additional gene transfers in Acrasis that may be difficult todetect due to the small size and/or low sequence conservation of the missing genes, possibly further complicated by fragmentation due to intron insertion within the nuclear genome.
The general pattern of genome degradation in A. kona mtDNA also includes loss of nearly half (9 of 20) of its tRNA genes relative to Naegleria mtDNA. This implies the need for extensive import of tRNAs from the cytosol (Salinas et al. 2008; Lithgow and Schneider 2010). In addition, mismatches were identified in the first 1–3 bp of the amino acid acceptor stem in 8 of the 11 remaining Acrasis tRNAs (fig. 1), as well as in 8 of the 20 predicted tRNAs in Naegleria mtDNA (data not shown). Such tRNAs would require editing in order to create the standard Watson–Crick base pairing necessary for functional mature tRNAs, thus a second RNA editing system (e.g., mitochondrial 5′-tRNA editing; Jackman et al. 2012) would probably be required in addition to the mRNA editing system (see above). The Acrasis tRNAs are otherwise well conserved in sequences, suggesting that they are not simply pseudogenes functionally replaced by additional tRNA imported from the cytosol.
Transfer to the nucleus of at least nine r-proteins in A. kona also coincides with massive rearrangement of its r-protein operons relative to the bacterial-like organization of these operons in Naegleria, Tsukubamonas, and all six examined jakobids (fig. 4). Gene order is further disrupted by the insertion of novel ORFs, as well as splitting and extension of the A. kona rps3 gene. Notably, rps3 is flanked in jakobid and Naegleria mtDNAs by two genes that are transferred to the nucleus in A. kona (rpl16 and rps19), allowing for the possibility that these phenomena may be linked. Splitting of the rps3 genes has previously been documented in a number of eukaryotes, often accompanied by insertion of additional functional domains or long peptides (Swart et al. 2012).
Mode and Tempo of Genome Evolution in Discoba
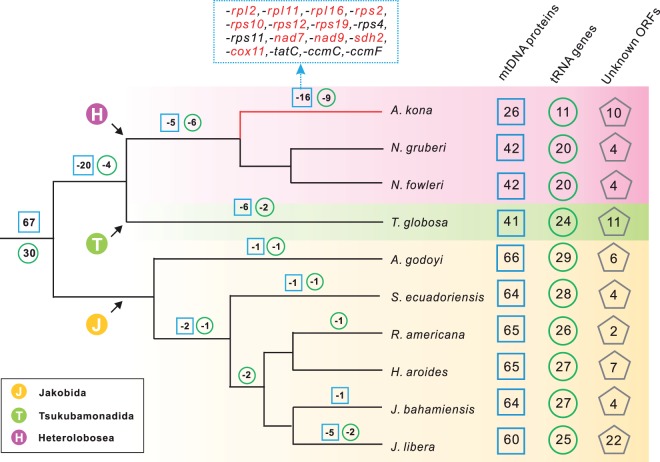
A mapping of mtDNA gene content onto a consensus phylogeny reveals a sporadic pattern of relative stasis and rampant gene loss in Discoba (fig. 8). Altogether 67 protein-coding genes are widely distributed in jakobid mtDNA (Burger et al. 2013) and therefore presumably present in the LCA of Discoba. Assuming that these mitochondrial genes were directly inherited from the mtDNA of the eukaryote LCA and that Tsukubamonas is more closely related to Heterolobosea than to Jakobida (Kamikawa et al. 2014), this implies a loss of 20 protein-coding genes between the LCA of Discoba and the LCA of Heterolobosea + Tsukubamonadida (fig. 8). Following the latter split, the Tsukubamonas lineage lost only six additional protein-coding genes, similar to the ancestral lineage of Acrasis + Naegleria, which lost five protein-coding genes. However, within the clade of Heterolobosea, gene loss appears to have halted entirely in the lineage leading to Naegleria, whereas the lineage leading to Acrasis appears to have continued the process of endosymbiotic gene transfer at the same or even an accelerated pace (fig. 8). Thus a sporadic pattern of evolutionary stasis and accelerated mtDNA gene loss is observed in Heterolobosea, a phenomenon rare for free-living organisms within other major groups of eukaryotes (Lang et al. 1999; Adams and Palmer 2003), in this case beginning with a much more gene-rich mtDNA ancestor.
Fig. 8.—
A mapping of mtDNA gene content onto a schematic phylogeny of Discoba, excluding Euglenozoa. Evolutionary loss of mtDNA gene content is indicated assuming the most parsimonious steps. Genes colored in red were identified on the Acrasis kona nuclear contigs (tBLASTn, E value < 1e−10).
The Discoba also show a preponderance of unknown ORFs, ranging from 2 to 22, most of which are not shared by any two mtDNAs. Thus, gain and loss of ORFs appears to be a very dynamic process in Discoba (fig. 8). However, these ORFs still constitute a small fraction of the potential protein-coding capacity of these mtDNAs, with the exception of A. kona, where ORFs constitute over a quarter (26.5%) of the genome, most of which are predicted to encode proteins over 200 amino acids in size. Several things suggest that the novel ORFs in A. kona mtDNA are probably functional genes. The ORFs exhibit an overall similar pattern of codon usage as the protein-coding sequences in the A. kona mtDNA, indicating that whatever their origin is, these ORFs have resided in the A. kona mtDNA for quite some time and have homogeneous base composition patterns as the rest of genome. Some of the ORFs also show faster evolution at third codon positions (supplementary table S3, Supplementary Material online). Transcripts corresponding to at least one ORF (ORF1592) were obtained by RT-PCR (data not shown). Investigating the potential significance of these ORFs in Acrasis will require sequencing and functional analysis of additional mtDNAs from acrasids and their close relatives.
C-to-U Type of RNA Editing in Heterolobosea mtDNAs through Ancient HGT
Different RNA editing systems are found across eukaryotes, particularly in organelles (Knoop 2011; Gray 2012). DYW-type PPR proteins are postulated as the key specificity determinants in C-to-U type editing (Salone et al. 2007; Zehrmann et al. 2009; Hayes et al. 2013), and co-occurrence of the DYW domain and organelle RNA editing is well documented in land plants (Rüdinger et al. 2012). However, the 12 putative DYW-type PPR proteins found in the A. kona nuclear genome far exceed the two confirmed C-to-U editing sites in its mtDNA, although additional editing sites may exist among the unknown ORFs that are not readily predicted. Likewise, only two C-to-U RNA editing sites were identified by extensive transcriptome analysis in N. gruberi (Rüdinger et al. 2011) despite the presence of 11 putative DYW-type PPR proteins in its nuclear genome (Knoop and Rüdinger 2010). Low RNA-editing activity in these mtDNAs could possibly have resulted from their extremely low GC content (table 1) (Jobson and Qiu 2008). Thus the excess of DYW-type PPR proteins suggests that they may play other roles, such as organellar endonucleolytic cleavage (Okuda et al. 2009) or transcript splicing (Ichinose et al. 2012) or have cytoplasmic activities. The genes targeted for C-to-U RNA editing differ between A. kona and N. gruberi, indicating that target sites are not highly conserved, which is also seen in land plants where it is presumed to be at least partly due to RNA-mediated gene conversion (Sloan et al. 2010).
The C-to-U editing type identified here in A. kona mtDNA is mostly restricted to land plant organelles where it is widespread (Fujii and Small 2011). DYW-type PPR proteins outside of land plants are intriguing. These proteins were recently identified in a scattering of species from across eukaryotes (Knoop and Rüdinger 2010; Iyer et al. 2011; Schallenberg-Rüdinger et al. 2013), mostly in species noted for their high levels of horizontally acquired genes, for example, Acanthamoeba (Clarke et al. 2013), Naegleria (Fritz-Laylin et al. 2010), Physarum (Watkins and Gray 2008), and Adineta (Gladyshev et al. 2008). Phylogenetic signals for possible origins of these HGTs are weak as the proteins are not well conserved in sequence. Nonetheless, there is strong support for lineage-specific expansion of these proteins in A. kona and N. gruberi. These two gene families also do not appear to group together but rather the DYW-type PPR proteins of Acrasis and Naegleria group strongly with Physarum and rotifer, respectively, suggesting that the two Heteroloboseans are either the source or the recipients of multiple independent HGT events. At least three strong candidate sites for C-to-U RNA editing were also predicted among protein-coding sequences in Tsukubamonas mtDNA (data not shown), which still needs further experimental proof. Nevertheless, it suggests that ancient HGT of DYW-type PPR proteins within certain discobid lineages might be more frequent than currently observed.
Concluding Remarks
The A. kona mitochondrial genome shows a number of unusual phenomena that may or may not be linked. The genome appears to be extreme in many different aspects: It has lost over 40% of its annotated coding capacity, massively rearranged its genome, become extremely AT rich and acquired ten novel ORFs that constitute over a quarter of the total mtDNA. This is in striking contrast to its sister lineage, Naegleria spp., whose mtDNA appears to closely resemble that of the Acrasis + Naegleria LCA. This considerably narrows the time frame within which these changes occurred. Thus, it should be possible to dissect some of these dynamic processes by examining additional heterolobosean mtDNAs.
Supplementary Material
Supplementary tables S1–S5 and figures S1–S7 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Matthew W. Brown (Mississippi State University) and Frederick W. Spiegel (University of Arkansas) for kindly providing the Acrasis kona culture, Gertraud Burger (Université de Montréal) for access to jakobid mtDNA data prior to public release, Franz B. Lang (Université de Montréal) for providing the concatenated data set of mitochondrial proteins, John Novembre (University of Chicago) for providing the ENCprime program, Ding He and Anders Larsson (Uppsala University) for valuable discussions, and anonymous reviewers for their insightful comments on an earlier version of the manuscript. This work was supported by grants from National Natural Science Foundation of China (No.30970424 to C.-J.F.), Knowledge Innovation Program of CAS (No. KSCX2-EW-G-6-4 to M.W.), and Swedish Research Council (Vetenskapsrådet Linnéstöd to S.L.B. and S.G.E.A.). C.-J.F., S.G.E.A., and S.L.B. conceived and designed the study; C.-J.F. and M.W. conducted the massive parallel sequencing; C.-J.F. performed all the other experiments and analyzed the data; S.S. participated in genome annotation and data analysis; C.-J.F., S.G.E.A., and S.L.B. wrote the paper. S.G.E.A. and S.L.B. are joint senior authors on this work.
Literature Cited
- Adams KL, Palmer JD. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol. 2003;29:380–395. doi: 10.1016/s1055-7903(03)00194-5. [DOI] [PubMed] [Google Scholar]
- Adl SM, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratlie MS, Johansen J, Drabløs F. Relationship between operon preference and functional properties of persistent genes in bacterial genomes. BMC Genomics. 2010;11:71. doi: 10.1186/1471-2164-11-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MW, Kolisko M, Silberman JD, Roger AJ. Aggregative multicellularity evolved independently in the eukaryotic supergroup Rhizaria. Curr Biol. 2012;22:1123–1127. doi: 10.1016/j.cub.2012.04.021. [DOI] [PubMed] [Google Scholar]
- Brown MW, Silberman JD, Spiegel FW. A contemporary evaluation of the acrasids (Acrasidae, Heterolobosea, Excavata) Eur J Protistol. 2012;48:103–123. doi: 10.1016/j.ejop.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Burger G, Gray MW, Forget L, Lang BF. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol. 2013;5:418–438. doi: 10.1093/gbe/evt008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver TJ, et al. ACT: the Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Nikolaev S. The zooflagellates Stephanopogon and Percolomonas are a clade (class Percolatea: Phylum Percolozoa) J Eukaryot Microbiol. 2008;55:501–509. doi: 10.1111/j.1550-7408.2008.00356.x. [DOI] [PubMed] [Google Scholar]
- Clarke M, et al. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 2013;14:R11. doi: 10.1186/gb-2013-14-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darty K, Denise A, Ponty Y. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzentas N. Circoletto: visualizing sequence similarity with Circos. Bioinformatics. 2010;26:2620–2621. doi: 10.1093/bioinformatics/btq484. [DOI] [PubMed] [Google Scholar]
- Dayhoff MO, Schwartz RM, Orcutt BC. A model of evolutionary change in proteins. In: Dayhoff MO, editor. Atlas of protein sequence and structure. Washington (DC): National Biomedical Research Foundation; 1978. pp. 345–352. [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Flegontov P, Gray MW, Burger G, Lukeš J. Gene fragmentation: a key to mitochondrial genome evolution in Euglenozoa? Curr Genet. 2011;57:225–232. doi: 10.1007/s00294-011-0340-8. [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin LK, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Ginger ML, Walsh C, Dawson SC, Fulton C. The Naegleria genome: a free-living microbial eukaryote lends unique insights into core eukaryotic cell biology. Res Microbiol. 2011;162:607–618. doi: 10.1016/j.resmic.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Xiong J, Miao W. Genome-wide identification and characterization of cytochrome P450 monooxygenase genes in the ciliate Tetrahymena thermophila. BMC Genomics. 2009;10:208. doi: 10.1186/1471-2164-10-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Small I. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 2011;191:37–47. doi: 10.1111/j.1469-8137.2011.03746.x. [DOI] [PubMed] [Google Scholar]
- Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008;320:1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- Gray MW. Evolutionary origin of RNA editing. Biochemistry. 2012;51:5235–5242. doi: 10.1021/bi300419r. [DOI] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Burger G. Mitochondria of protists. Annu Rev Genet. 2004;38:477–524. doi: 10.1146/annurev.genet.37.110801.142526. [DOI] [PubMed] [Google Scholar]
- Hahn C, Bachmann L, Chevreux B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 2013;41:e129. doi: 10.1093/nar/gkt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduk S, Ochsenreiter T. RNA editing in kinetoplastids. RNA Biol. 2010;7:229–236. doi: 10.4161/rna.7.2.11393. [DOI] [PubMed] [Google Scholar]
- Harding T, et al. Amoeba stages in the deepest branching heteroloboseans, including Pharyngomonas: evolutionary and systematic implications. Protist. 2013;164:272–286. doi: 10.1016/j.protis.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Hayes ML, Giang K, Berhane B, Mulligan RM. Identification of two pentatricopeptide repeat genes required for RNA editing and zinc binding by c-terminal cytidine deaminase-like domains. J Biol Chem. 2013;288:36519–36529. doi: 10.1074/jbc.M113.485755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D, Fiz-Palacios O, Fu CJ, Tsai CC, Baldauf SL. An alternative root for the eukaryote tree of life. Curr Biol. 2014;24:465–470. doi: 10.1016/j.cub.2014.01.036. [DOI] [PubMed] [Google Scholar]
- Herman EK, et al. The mitochondrial genome and a 60-kb nuclear DNA segment from Naegleria fowleri, the causative agent of primary amoebic meningoencephalitis. J Eukaryot Microbiol. 2013;60:179–191. doi: 10.1111/jeu.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose M, Tasaki E, Sugita C, Sugita M. A PPR-DYW protein is required for splicing of a group II intron of cox1 pre-mRNA in Physcomitrella patens. Plant J. 2012;70:271–278. doi: 10.1111/j.1365-313X.2011.04869.x. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Zhang D, Rogozin IB, Aravind L. Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic Acids Res. 2011;39:9473–9497. doi: 10.1093/nar/gkr691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Gott JM, Gray MW. Doing it in reverse: 3′-to-5′ polymerization by the Thg1 superfamily. RNA. 2012;18:886–899. doi: 10.1261/rna.032300.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobson RW, Qiu YL. Did RNA editing in plant organellar genomes originate under natural selection or through genetic drift? Biol Direct. 2008;3:43. doi: 10.1186/1745-6150-3-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikawa R, et al. Gene content evolution in discobid mitochondria deduced from the phylogenetic position and complete mitochondrial genome of Tsukubamonas globosa. Genome Biol Evol. 2014;6:306–315. doi: 10.1093/gbe/evu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpenahalli MR, Lupas AN, Söding J. TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics. 2007;8:2. doi: 10.1186/1471-2105-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiethega GN, Turcotte M, Burger G. Evolutionarily conserved cox1 trans-splicing without cis-motifs. Mol Biol Evol. 2011;28:2425–2428. doi: 10.1093/molbev/msr075. [DOI] [PubMed] [Google Scholar]
- Knoop V. When you can’t trust the DNA: RNA editing changes transcript sequences. Cell Mol Life Sci. 2011;68:567–586. doi: 10.1007/s00018-010-0538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop V, Rüdinger M. DYW-type PPR proteins in a heterolobosean protist: plant RNA editing factors involved in an ancient horizontal gene transfer? FEBS Lett. 2010;584:4287–4291. doi: 10.1016/j.febslet.2010.09.041. [DOI] [PubMed] [Google Scholar]
- Lang BF, et al. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 2013;62:611–615. doi: 10.1093/sysbio/syt022. [DOI] [PubMed] [Google Scholar]
- Laurin-Lemay S, Brinkmann H, Philippe H. Origin of land plants revisited in the light of sequence contamination and missing data. Curr Biol. 2012;22:R593–R594. doi: 10.1016/j.cub.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Lenz H, Knoop V. PREPACT 2.0: predicting C-to-U and U-to-C RNA editing in organelle genome sequences with multiple references and curated RNA editing annotation. Bioinform Biol Insights. 2013;7:1–19. doi: 10.4137/BBI.S11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow T, Schneider A. Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes. Philos Trans R Soc Lond B Biol Sci. 2010;365:799–817. doi: 10.1098/rstb.2009.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukeš J, Hashimi H, Zíková A. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr Genet. 2005;48:277–299. doi: 10.1007/s00294-005-0027-0. [DOI] [PubMed] [Google Scholar]
- Lurin C, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I, et al. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 2013;14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- Moreira S, Breton S, Burger G. Unscrambling genetic information at the RNA level. Wiley Interdiscip Rev RNA. 2012;3:213–228. doi: 10.1002/wrna.1106. [DOI] [PubMed] [Google Scholar]
- Novembre JA. Accounting for background nucleotide composition when measuring codon usage bias. Mol Biol Evol. 2002;19:1390–1394. doi: 10.1093/oxfordjournals.molbev.a004201. [DOI] [PubMed] [Google Scholar]
- Okuda K, et al. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell. 2009;21:146–156. doi: 10.1105/tpc.108.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Myouga F, Motohashi R, Shinozaki K, Shikanai T. Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc Natl Acad Sci U S A. 2007;104:8178–8183. doi: 10.1073/pnas.0700865104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive LS. The Mycetozoa: a revised classification. Bot Rev. 1970;36:59–89. [Google Scholar]
- Page FC, Blanton RL. The Heterolobosea (Sarcodina, Rhizopoda), a new class uniting the Schizopyrenida and the Acrasidae (Acrasida) Protistologica. 1985;21:121–132. [Google Scholar]
- Pánek T, Silberman JD, Yubuki N, Leander BS, Cepicka I. Diversity, evolution and molecular systematics of the Psalteriomonadidae, the main lineage of anaerobic/microaerophilic heteroloboseans (Excavata: Discoba) Protist. 2012;163:807–831. doi: 10.1016/j.protis.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Park JS, Simpson AGB. Characterization of Pharyngomonas kirbyi (= “Macropharyngomonas halophila” nomen nudum), a very deep-branching, obligately halophilic heterolobosean flagellate. Protist. 2011;162:691–709. doi: 10.1016/j.protis.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Ronquist F, et al. Mrbayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdinger M, Fritz-Laylin L, Polsakiewicz M, Knoop V. Plant-type mitochondrial RNA editing in the protist Naegleria gruberi. RNA. 2011;17:2058–2062. doi: 10.1261/rna.02962911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdinger M, Polsakiewicz M, Knoop V. Organellar RNA editing and plant-specific extensions of pentatricopeptide repeat proteins in jungermanniid but not in marchantiid liverworts. Mol Biol Evol. 2008;25:1405–1414. doi: 10.1093/molbev/msn084. [DOI] [PubMed] [Google Scholar]
- Rüdinger M, Volkmar U, Lenz H, Groth-Malonek M, Knoop V. Nuclear DYW-type PPR gene families diversify with increasing RNA editing frequencies in liverwort and moss mitochondria. J Mol Evol. 2012;74:37–51. doi: 10.1007/s00239-012-9486-3. [DOI] [PubMed] [Google Scholar]
- Rutherford K, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Salinas T, Duchêne AM, Maréchal-Drouard L. Recent advances in tRNA mitochondrial import. Trends Biochem Sci. 2008;33:320–329. doi: 10.1016/j.tibs.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Salone V, et al. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 2007;581:4132–4138. doi: 10.1016/j.febslet.2007.07.075. [DOI] [PubMed] [Google Scholar]
- Schallenberg-Rüdinger M, Lenz H, Polsakiewicz M, Gott JM, Knoop V. A survey of PPR proteins identifies DYW domains like those of land plant RNA editing factors in diverse eukaryotes. RNA Biol. 2013;10:1343–1350. doi: 10.4161/rna.25755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AGB, Inagaki Y, Roger AJ. Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of “primitive” eukaryotes. Mol Biol Evol. 2006;23:615–625. doi: 10.1093/molbev/msj068. [DOI] [PubMed] [Google Scholar]
- Sloan DB, MacQueen AH, Alverson AJ, Palmer JD, Taylor DR. Extensive loss of RNA editing sites in rapidly evolving silene mitochondrial genomes: selection vs. retroprocessing as the driving force. Genetics. 2010;185:1369–1380. doi: 10.1534/genetics.110.118000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Spiegel FW. The ultrastructure of the trophic cells of the protostelid Planoprotostelium aurantium. Protoplasma. 1982;113:165–177. [Google Scholar]
- Stamatakis A, et al. RAxML-Light: a tool for computing terabyte phylogenies. Bioinformatics. 2012;28:2064–2066. doi: 10.1093/bioinformatics/bts309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart EC, et al. The Oxytricha trifallax mitochondrial genome. Genome Biol Evol. 2012;4:136–154. doi: 10.1093/gbe/evr136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk R, Huynen MA. Mosaic origin of the mitochondrial proteome. Proteomics. 2010;10:4012–4024. doi: 10.1002/pmic.201000329. [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliff MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- Visvesvara GS, Moura H, Schuster FL. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol Med Microbiol. 2007;50:1–26. doi: 10.1111/j.1574-695X.2007.00232.x. [DOI] [PubMed] [Google Scholar]
- Wang HC, Li K, Susko E, Roger AJ. A class frequency mixture model that adjusts for site-specific amino acid frequencies and improves inference of protein phylogeny. BMC Evol Biol. 2008;8:331. doi: 10.1186/1471-2148-8-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins RF, Gray MW. Sampling gene diversity across the supergroup Amoebozoa: large EST data sets from Acanthamoeba castellanii, Hartmannella vermiformis, Physarum polycephalum, Hyperamoeba dachnaya and Hyperamoeba sp. Protist. 2008;159:269–281. doi: 10.1016/j.protis.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Wright F. The “effective number of codons” used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Yagi Y, et al. Pentatricopeptide repeat proteins involved in plant organellar RNA editing. RNA Biol. 2013;10:1236–1242. doi: 10.4161/rna.24908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Sze SH. Large-scale analysis of gene clustering in bacteria. Genome Res. 2008;18:949–956. doi: 10.1101/gr.072322.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yubuki N, Leander BS. Ultrastructure and molecular phylogeny of Stephanopogon minuta: an enigmatic microeukaryote from marine interstitial environments. Eur J Protistol. 2008;44:241–253. doi: 10.1016/j.ejop.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Zehrmann A, Verbitskiy D, Van Der Merwe JA, Brennicke A, Takenaka M. A DYW domain-containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell. 2009;21:558–567. doi: 10.1105/tpc.108.064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, et al. Codon Deviation Coefficient: a novel measure for estimating codon usage bias and its statistical significance. BMC Bioinformatics. 2012;13:43. doi: 10.1186/1471-2105-13-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.