Abstract
Tubulins belong to the most abundant proteins in eukaryotes providing the backbone for many cellular substructures like the mitotic and meiotic spindles, the intracellular cytoskeletal network, and the axonemes of cilia and flagella. Homologs have even been reported for archaea and bacteria. However, a taxonomically broad and whole-genome-based analysis of the tubulin protein family has never been performed, and thus, the number of subfamilies, their taxonomic distribution, and the exact grouping of the supposed archaeal and bacterial homologs are unknown. Here, we present the analysis of 3,524 tubulins from 504 species. The tubulins formed six major subfamilies, α to ζ. Species of all major kingdoms of the eukaryotes encode members of these subfamilies implying that they must have already been present in the last common eukaryotic ancestor. The proposed archaeal homologs grouped together with the bacterial TubZ proteins as sister clade to the FtsZ proteins indicating that tubulins are unique to eukaryotes. Most species contained α- and/or β-tubulin gene duplicates resulting from recent branch- and species-specific duplication events. This shows that tubulins cannot be used for constructing species phylogenies without resolving their ortholog–paralog relationships. The many gene duplicates and also the independent loss of the δ-, ε-, or ζ-tubulins, which have been shown to be part of the triplet microtubules in basal bodies, suggest that tubulins can functionally substitute each other.
Keywords: tubulin, TubZ, artubulin, FtsZ, eukaryotic evolution, gene duplication
Introduction
Tubulins belong to the most abundant proteins in eukaryotes and have therefore been used in dozens of studies aiming at determining species phylogenies (see e.g., Brown et al. 2009; Gong et al. 2010; Kurtzman 2011; Yi et al. 2012; Walker et al. 2012). Tubulins play critical roles in many cellular processes like the segregation of chromosomes in the mitotic and meiotic spindles, cell motility, intracellular transport, and in the assembly and stability of cilia and flagella (Nogales 2001; Libusová and Dráber 2006). Together with the prokaryotic FtsZ proteins, the tubulins comprise a large superfamily of GTPases able to build linear polymers (Dyer 2009; Aylett et al. 2011). For almost 30 years it was assumed that the tubulin family consisted of only three subfamilies (α, β, and γ) present in every eukaryote that has been studied (Oakley 2000). However, in the last 15 years, the tubulin superfamily expanded rapidly with the identification of further eukaryotic tubulin subfamilies (δ to κ), which were reported to be restricted to certain lineages or species. Some of those newfound subfamilies are suggested to be linked to specific subcellular structures like the δ-, ε-, and η-tubulins, which were proposed to be connected with the triplet microtubules of basal bodies underlying ciliary axonomes (Garreau de Loubresse et al. 2001; Ross et al. 2013). More surprisingly, supposed bacterial and archaeal homologs were discovered. For instance, two bacterial tubulin homologs, BtubA and BtubB, were found in the genus Prosthecobacter. They have most probably been derived by horizontal gene transfer (Jenkins et al. 2002; Schlieper et al. 2005) because their taxonomic distribution in prokaryotes is very narrow. In archaea of the genus Candidatus Nitrosoarchaeum, so-called “artubulins” have recently been described as bona fide tubulins implying an origin of these key components of eukaryotic cells in archaea (Yutin and Koonin 2012). In addition to FtsZ proteins, which, assemble into the cytokinetic ring essential for cell division and are present in most bacteria and many archaea (Huang et al. 2013), some bacteria encode a further polymer-forming FtsZ/tubulin superfamily homolog, called TubZ (Larsen et al. 2007; Aylett et al. 2010). It is tempting to speculate that the superfamily will expand even further with more genomes being sequenced.
Although tubulins are widely used in phylogenetic studies, a taxonomically broad and whole-genome-level-based analysis of the tubulin protein family has never been performed. A recent review provides a short overview of tubulin groups in major eukaryotic branches without, however, resolving subfamilies and duplicates (Wickstead and Gull 2011). It is well known for many protein families that most species not only contain different subfamily members but also often multiple copies of each subfamily. Reasons for multiple copies are branch- and species-specific duplications as well as whole-genome duplications, which have been discovered in vertebrates (Steinke et al. 2006; Van de Peer et al. 2010), yeasts (Wolfe and Shields 1997), and plants (Mühlhausen and Kollmar 2013) in recent years. Therefore, it is surprising to see species phylogenies based on or supported by tubulin sequences without the attempt to resolve the ortholog–paralog relationships within the data. Here, we performed a global analysis of more than 500 species from all eukaryotic kingdoms and from many closely related species to reveal the common set of subfamily members in the last common ancestor of the eukaryotes. Furthermore, we investigated their relation to the proposed archaeal (artubulins [Yutin and Koonin 2012]) and bacterial (BtubA/B [Schlieper et al. 2005]) tubulin homologs, their applicability in phylogenetic studies, and potential links of subfamilies and subtypes to cellular structures and functions.
Materials and Methods
Identification and Annotation of the Tubulin Genes
Tubulin genes have been identified in iterated TBLASTN searches of the completed or almost completed genomes of 504 species starting with the protein sequence of yeast Saccharomyces cerevisiae αTub. The respective genomic regions were submitted to AUGUSTUS (Stanke and Morgenstern 2005) to obtain gene predictions. However, feature sets are only available for a few species. Therefore, all hits were subsequently manually analyzed at the genomic DNA level. When necessary, gene predictions were corrected by comparison with the other tubulins included in the multiple sequence alignment. Where possible, expressed sequence tag (EST) data have been analyzed to help in the annotation process. In the last years, genome sequencing efforts have been extended from sequencing species from new branches to sequencing closely related organisms. Here, these species include, for example, seven ant species, 12 Drosophila species, and dozen mammals. Protein sequences from these closely related species have been obtained by using the cross-species functionality of WebScipio (Hatje et al. 2011, 2013). Nevertheless, also for all these genomes, TBLASTN searches have been performed. With this strategy, we wanted to ensure that we would not miss more divergent tubulin homologs, which might have been derived by species-specific inventions or duplications.
Some of the genes contain alternative splice forms. The different splice forms were not considered independently in the analysis. Instead, the same splice forms were taken for homologous tubulins. All sequence-related data (names, corresponding species, GenBank ID's, alternative names, corresponding publications, domain predictions, and sequences) and references to genome sequencing centers are available through the CyMoBase (www.cymobase.org, last accessed September 5, 2014) (Odronitz and Kollmar 2006).
Generating the Multiple Sequence Alignment
The tubulin sequence alignment in its current stage was created over years of assembling tubulin sequences. The initial alignment was created based on a few full-length sequences obtained from GenBank. Further sequences were added to this alignment by first aligning every newly predicted sequence to its supposed closest relative using ClustalW (Chenna et al. 2003) and subsequently adding this “aligned” sequence to the multiple sequence alignment. During the subsequent sequence validation process, the obtained alignment was manually adjusted by removing wrongly predicted sequence regions and by filling gaps. Still, in those sequences derived from low-coverage genomes, many gaps remained. To maintain the integrity of exons preceded or followed by gaps, gaps reflecting missing parts of the supposed protein sequences were added to the multiple sequence alignment. The alignment of the tubulins can be obtained from CyMoBase (www.cymobase.org) (Odronitz and Kollmar 2006) and supplementary data S1, Supplementary Material online. CyMoBase also offers a BLAST service that can be used for fast subfamily assignment and ortholog identification.
Comparison of the Sequence Identities and Similarities
Sequences designated “Fragment,” “Partial,” or “Pseudogene” were removed from the multiple sequence alignment. Sequence identity matrices (2D-matrix tables containing sequence identities scores for each pair of sequences) were calculated for each alignment using the method implemented in BioEdit (Tom Hall, http://www.mbio.ncsu.edu/bioedit/bioedit.html, last accessed September 5, 2014). Shortly, the reported numbers represent the ratio of identities to the length of the longer of the two sequences after positions where both sequences contain a gap are removed. Sequence similarity matrices were calculated with MatGAT (Campanella et al. 2003) using the BLOSUM62 substitution matrix and setting the gap opening and extending penalties to 12 and 2, respectively.
Preparation of the Data Sets for the Phylogenetic Analyses
The alignment of the 3,527 tubulins was treated with CD-Hit v.4.5.4 (Li and Godzik 2006) and gblocks v.0.91b (Talavera and Castresana 2007) to generate data sets with less redundancy and smaller blocks. First, sequences designated Fragment, Partial, or Pseudogene were removed from the multiple sequence alignment resulting in the data set called “100” (all 3,286 complete tubulin sequences, including archaeal and bacterial “tubulin” genes, accounting for 2,633 alignment positions). Further data sets were produced with CD-Hit applying similarity thresholds of 98% (1,998 sequences) to 70% (377 sequences). For all data sets, the number of alignment positions was reduced with gblocks applying least stringent selection criteria. The parameters were as follows: 1) The minimum number of sequences for a conserved position and the minimum of sequences for a flank position were set to the minimum (e.g. half the number of sequences plus one). 2) The maximum number of contiguous nonconserved positions was set to 8 and the minimum length of a block was set to 5. 3) The parameter for the allowed gap position was set to “with half” meaning that only positions within 50% or more of the sequences having a gap are treated as gap positions. The data sets accordingly contain 308 (“100”) to 175 positions (“70”).
Computing and Visualizing Phylogenetic Trees
Phylogenetic trees were generated for all data sets using the neighbor joining (NJ) and the maximum likelihood (ML) method. 1) ClustalW v.2.0.10 (Chenna et al. 2003) was used to calculate unrooted trees with the NJ method. For each data set, bootstrapping with 1,000 replicates was performed. Trees were corrected for multiple substitutions. 2) ML analyses with estimated proportion of invariable sites and resampling (1,000 replicates) were performed with FastTree v.2 (Price et al. 2010). ProtTest v.3.2 (Darriba et al. 2011) was used to determine the more appropriate of the two amino acid substitution models available within FastTree, Jones, Taylor, and Thorton (JTT) matrix (Jones et al. 1992), and Whelan and Goldman (WAG) model (Whelan and Goldman 2001). Within ProtTest, the tree topology was calculated with the BioNJ algorithm and both the branch lengths and the model of protein evolution were optimized simultaneously. The Akaike information criterion identified the WAG + Γ to be the best model. 3) Posterior probabilities were generated for the CD-Hit 70% data sets with and without applying gblocks using MrBayes v3.1.2 (Ronquist and Huelsenbeck 2003). Two independent runs each with 20,000,000 (gblocks reduced data set) and 5,000,000 generations (data set without gblocks applied), four chains, and a random starting tree were computed using the mixed amino acid option. MrBayes used the BLOSUM and WAG model for data set with and without gblocks applied, respectively. Trees were sampled every 1,000th generation and the first 25% of the trees were discarded as “burn-in” before generating a consensus tree. Phylogenetic trees were visualized with FigTree (http://tree.bio.ed.ac.uk/software/figtree/, last accessed September 5, 2014).
Results
Gene Identification and Annotation
Important note: For simplicity, file format handling, software requirements, and database compatibility, we use numbers instead of Greek letters for naming specific tubulins throughout the manuscript, the supplementary material, Supplementary Material online, and within CyMoBase (Odronitz and Kollmar 2006). Thus, Tub1 is used equivalent to α-tubulin or Tub3 equivalent to γ-tubulin.
All known tubulins have conserved sequences of very similar length. In addition, most α- and β-tubulin genes have long contiguous sequences (not interrupted by introns) resulting in long BLAST hits with high E values. Therefore, the identification of α-, β-, and γ-tubulins was straightforward. We mainly used the yeast Saccharomyces cerevisiae α-tubulin as starting sequence in TBLASTN searches in all genome assemblies. For the identification of the more divergent δ-, ε-, and ζ-tubulins, we also used members of these subtypes in the searches, especially in all those genomes where these subtypes seemed to be absent. The many available full-length cDNA sequences of α-, β-, and γ-tubulins helped in getting a well validated initial alignment of several hundred tubulin sequences. In contrast to the core part of the tubulins, the sequences at the N- and C-termini were difficult to assemble but could be resolved by manual inspection of the genomic DNA sequences and comparison within the sequence alignment. In particular, species of the Fungi kingdom contain several consecutive very short exons of 10–15 bp coding for the N-termini of the α-tubulins (for an example see Hatje et al. 2013). Even the yeasts including S. cerevisiae, which are known to contain only a few introns in their genomes, have at least one intron at the N-termini of their α-tubulin genes. In addition, in many cases, the C-termini of the α- and β-tubulins are encoded in separate exons. Because these exons code for the E-hook, which is the most divergent part of the tubulin sequences and in general of low complexity, they are missed in most gene predictions and could only be identified by comparative analysis. Searching different genome assemblies often revealed additional tubulin homologs. A striking example is the identification of two highly conserved tubulin genes, which are absent in the latest (and also the earlier) Bos taurus reference genome assembly (Elsik et al. 2009) but present in the alternative genome assembly (Zimin et al. 2009). We did not use any gene prediction data sets in our searches because we found out that not even all the human tubulins are included in the human RefSeq data set (see below). The quality and completeness of the gene prediction data sets of the other species are expected to be considerably worse. We surely would have missed many tubulins not only in the human but also in the other species’ genomes if we had analyzed gene prediction data sets instead of the genome assemblies directly. In addition to manually assembling all sequences, the multiple sequence alignment of the tubulins had been created and was maintained and improved manually (supplementary data S1, Supplementary Material online).
Sequences of which small parts were missing due to gaps in the genome assemblies (up to 5% of the supposed full-length sequence) were termed “Partials.” Sequences of which more than 5% were missing due to genome assembly gaps or incomplete EST data but which are otherwise unambiguous orthologs or paralogs were termed “Fragments.” “Partials” and “Fragments” are important to denote the presence of the tubulins in the respective species but were removed in the phylogenetic analyses. Tubulin genes were termed pseudogenes if they consisted of single pseudocoding exons that contained deletions and insertions (which would lead to frame shifts in the translations), in-frame stop codons, and/or missed considerable parts of a “normal” full-length tubulin. Although this procedure was reliable for annotating human pseudogenes, presuming that the human reference genome assembly is almost complete and does not contain any sequencing and assembly errors, annotating pseudogenes in other species was often more difficult. For example, several of the low-coverage fungi genome assemblies are of lower quality, and it would not be surprising to observe sequencing and assembly errors within coding regions. In these cases, we did not annotate the respective tubulin genes as pseudogenes. In several mammalian genomes, there are tubulin (pseudo)genes consisting of single or multiple exons without any reading-frame interrupting insertions/deletions/mutations. When comparing these (pseudo)genes to their closest homologs, the (pseudo)genes contain mutations leading to amino acid substitutions that are completely unlikely for the respective tubulin subtype, like a mutation of a 100% conserved arginine to a tryptophan. For example, this particular mutation is present twice in the manatee Trichechus manatus α-tubulin Tub1E pseudogene. These tubulins were thus also annotated as pseudogenes.
In total, the tubulin data set contains 3,524 sequences from 504 organisms (table 1, supplementary data S1, Supplementary Material online). In total, 3,353 sequences are complete, and an additional 64 sequences are partially complete. For plotting the presence or absence of tubulins across the tree of eukaryotes, we only included those species whose genomes have been sequenced with high coverage and which provided reliable data in many other studies (Odronitz and Kollmar 2007; Odronitz et al. 2009; Eckert et al. 2011; Kollmar et al. 2012). Nevertheless, low-coverage genomes have also been analyzed because every single piece of sequence could be very important to resolve ambiguous regions in related species or to clarify phylogenetic questions. For example, we analyzed the incomplete genome of the tammar wallaby Macropus eugenii to reveal whether ζ-tubulins are present in all Metatheria.
Table 1.
Data Statistics
| Total | α | β | γ | δ | ε | ζ | |
|---|---|---|---|---|---|---|---|
| Sequence | |||||||
| Total | 3,524 | 1,420 | 1,313 | 490 | 121 | 131 | 49 |
| Pseudogenes | 131 | 73 | 54 | 3 | 0 | 1 | 0 |
| Completeness | |||||||
| Complete | 3,353 | 1,363 | 1,239 | 470 | 117 | 120 | 43 |
| Partials | 64 | 14 | 31 | 8 | 1 | 8 | 2 |
| Fragments | 107 | 43 | 43 | 12 | 3 | 2 | 4 |
| Species | |||||||
| Total | 504 | 438 | 436 | 428 | 120 | 130 | 49 |
| Sequences in taxa | |||||||
| Metazoa | 1,479 | 639 | 532 | 121 | 80 | 83 | 24 |
| Fungi | 1,093 | 437 | 387 | 265 | 1 | 3 | 0 |
| Apusozoa | 6 | 1 | 1 | 1 | 1 | 1 | 1 |
| Amoebozoa | 46 | 21 | 14 | 11 | 0 | 0 | 0 |
| SAR | 294 | 110 | 86 | 36 | 25 | 26 | 11 |
| Cryptophyta | 17 | 5 | 6 | 5 | 0 | 0 | 1 |
| Haptophyta | 18 | 8 | 6 | 2 | 0 | 1 | 1 |
| Excavata | 138 | 48 | 53 | 8 | 10 | 10 | 9 |
| Viridiplantae | 381 | 135 | 204 | 32 | 3 | 5 | 2 |
Classification
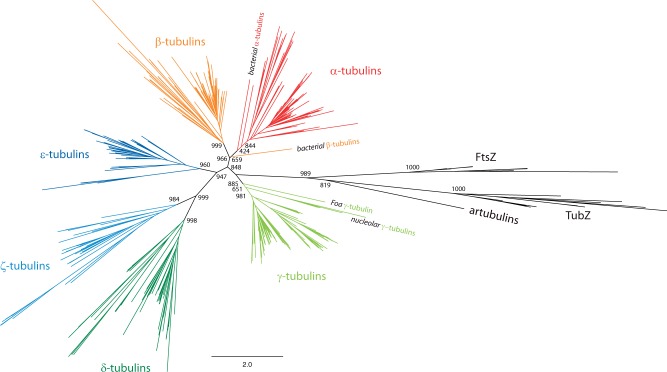
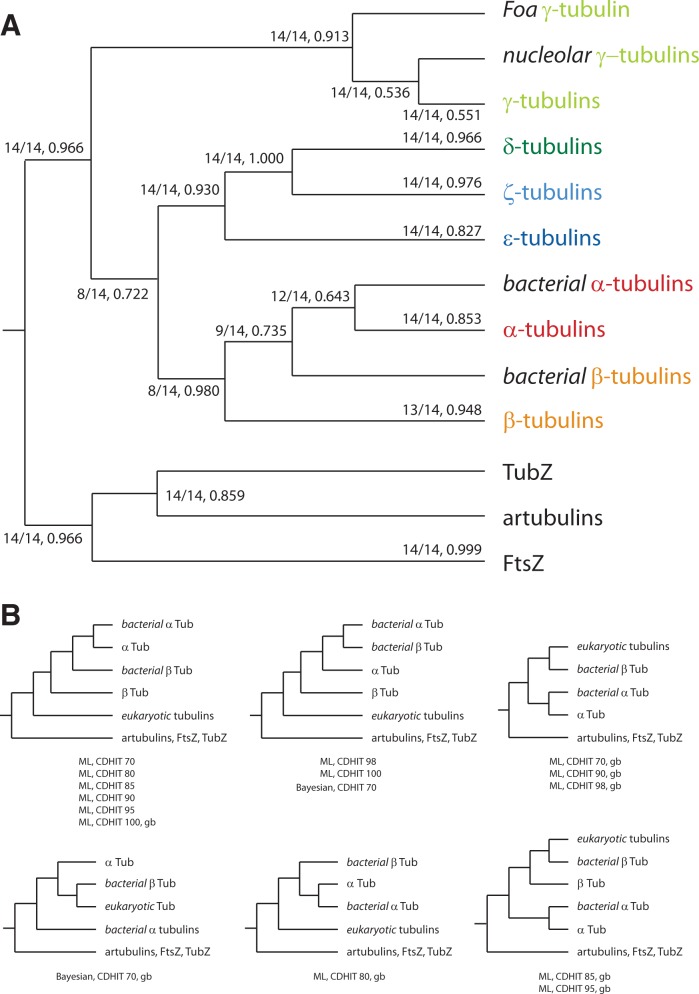
To infer the phylogenetic relationship of the major tubulin subfamilies and the relation of the many gene duplicates, we reconstructed phylogenetic trees using ML, NJ, and Bayesian methods. In these trees, we also included the bacterial tubulins from Prosthecobacter species (Jenkins et al. 2002), the artubulins (Yutin and Koonin 2012), several TubZ proteins (Larsen et al. 2007; Aylett et al. 2010), and some FtsZ homologs (Dyer 2009), which were intended to be used as outgroup. The trees were generated on full and reduced data sets, in which redundant sequences, divergent regions, and unique positions were removed at various stringencies (supplementary table S1, Supplementary Material online). Fragments, Partials, and pseudogenes were excluded. The resulting tree topologies were almost identical showing strong support for six major classes of eukaryotic tubulins in all trees (fig. 1). For example, within the ML tree based on a reduced data set (sequence similarity threshold of 70%), four subtypes are supported by a bootstrap value higher than 96% (fig. 1 and supplementary fig. S1, Supplementary Material online), and the α- and γ-tubulin groups are supported by bootstrap values of 84% and 89%, respectively. We defined six classes by their bootstrap support values of the respective class-forming nodes and by the presence of class members from all major eukaryotic kingdoms. This way, for example, δ-tubulins are separated from ζ-tubulins although both always form a highly supported common group. Species from all eukaryotic kingdoms encode both subfamily members (table 1); the alternative to defining δ- and ζ-tubulins as distinct classes would be to define both as subtypes of a superclass with both subtypes already present in the last common ancestor of the eukaryotes. The tubulins previously termed “η” group to the ζ-tubulins from Trypanosoma and Leishmania species. We suggest naming these tubulins ζ-tubulins to not interrupt the alphabetic naming. The Paramecium tetraurelia tubulins, which had previously been proposed to form distinct new classes of “θ”- and “ι”-tubulins (Libusová and Dráber 2006), group together with Tetrahymena thermophila, Oxytricha trifallax, and Naegleria gruberi tubulins at the base of the β-tubulins (supplementary fig. S1, Supplementary Material online). Because the taxonomic sampling of this group is limited to Ciliates and Naegleria, and because this group of sequences in all trees groups to the β-tubulins, we suggest classifying these also as β-tubulins and not as separate classes. Similarly, the tubulin previously termed “κ” always groups to the α-tubulins (supplementary fig. S1, Supplementary Material online) and was renamed accordingly. In addition, the consistent grouping of the bacterial α- and β-tubulins at the base of their eukaryotic orthologs (fig. 2A) supports the new classification of the “i”- to “κ”-tubulins.
Fig. 1.—
Phylogenetic tree of the tubulin protein family. Unrooted ML topology generated under the WAG + Γ model in FastTree showing branch lengths for 75 α-, 69 β-, 84 γ-, 50 δ-, 45 ε-, 32 ζ-tubulins, 2 bacterial tubulins, 1 “artubulin,” 11 bacterial FtsZ, and 8 bacterial TubZ proteins. CD-Hit (70% identity) was used to obtain a representative data set for subfamily classification and visualization. Support for the major branchings indicating the grouping of the tubulins and FtsZ family members into different subtypes is given as likelihood bootstraps (FastTree). The scale bar corresponds to estimated amino acid substitutions per site.
Fig. 2.—
Schematic tree of the tubulin subfamilies. (A) Schematic consensus tree from 14 trees reconstructed with the ML method and based on full and reduced data sets, in which redundant sequences, divergent regions, and unique positions were removed at various stringency levels. The first number at branches denotes the number of trees supporting the respective branch followed by the median of the support values (see supplementary table S2, Supplementary Material online, for more details). (B) The small trees show the alternative topologies for the branching of the bacterial tubulins. CDHIT, application of CD-Hit with the given similarity threshold; gb, use of gblocks.
The relative grouping of the major subtypes is conserved in almost all trees with the δ- and ζ-tubulins forming a group closest related to the ε-tubulins, and the α- and β-tubulins forming a supergroup. From the set of ML trees, we reconstructed a most parsimonious consensus tree showing the occurrence and average support for each branching (fig. 2A and supplementary table S2, Supplementary Material online). In the NJ trees, the nucleolar γ-tubulins and the Piroplasmida (Theileria and Babesia species) ε-tubulins do not group to the other γ- and ε-tubulins. The other eukaryotic tubulins group together as in the ML trees (supplementary table S2, Supplementary Material online). In contrast to the conserved topology of the eukaryotic tubulin subgroups, the bacterial tubulins branch differently in 50% of the ML trees (fig. 2B). In the trees reconstructed on full-length alignments, both the α- and β- bacterial tubulins group to the eukaryotic α- and β-tubulins, whereas in the trees reconstructed from the gblock-reduced alignments, the bacterial tubulins also group outside all eukaryotic tubulins and many other topologies. However, in 86% of the trees, the bacterial α-tubulins group closest to the eukaryotic α-tubulins.
Human Tubulins and Their Vertebrate Orthologs
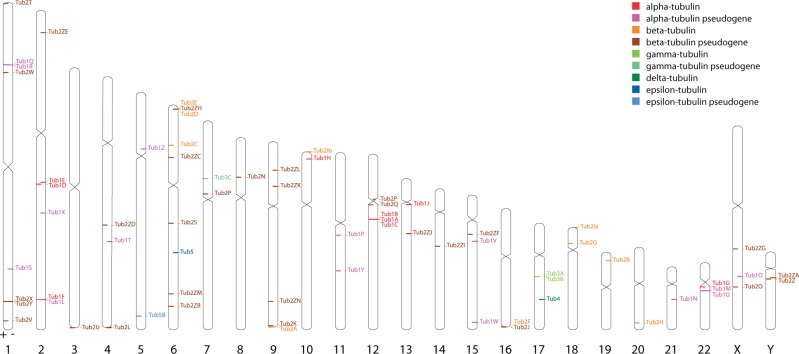
The human genome contains 23 tubulin genes and at least 48 pseudogenes (fig. 3, supplementary figs. S3–S6 and table S3, Supplementary Material online). We identified and assembled all fragments of tubulin genes with (pseudo-)coding regions of at least 150 amino acids. Pseudogenes have been identified for all tubulin subfamilies except δ-tubulin. The tubulins and pseudotubulins are spread over all chromosomes, and there is no specific enrichment in any chromosomal region (fig. 3). Noteworthy, many of the β-tubulin genes are located in telomere regions, which might have hindered their identification and characterization, so far. For example, the β-tubulin Tub2Ib gene has not been identified at all yet and is therefore not included in the RefSeq data set or the UCSC Genome browser. α-tubulins are never clustered together with β-tubulins as has been found for example for the Kinetoplastid tubulins (Jackson et al. 2006). However, there are several clusters of class-specific tandem gene duplications like the cluster of three α-tubulins on chromosome 12, the cluster of two β-tubulins on chromosome 6, and the cluster of γ-tubulins on chromosome 17 (fig. 3, supplementary fig. S7, Supplementary Material online).
Fig. 3.—
Chromosomal location of human tubulin genes. The human tubulin genes and pseudogenes are distributed over all chromosomes. Some genes appear in clusters of tandemly arranged gene duplicates like the α-tubulins Tub1A, Tub1B, and Tub1C on chromosome 12, the β-tubulins Tub2D and Tub2E on chromosome 2, and the γ-tubulins Tub3A and Tub3B on chromosome 17 (see also supplementary fig. S6, Supplementary Material online). The ideogram was produced with Idiographica based on the human hg19 chromosome assembly (Kin and Ono 2007).
It had already been noted that most of the human, mouse, and rat α-tubulins are conserved between these species and should therefore get the same name (Khodiyar et al. 2007). Orthology had been assigned based on phylogenetic grouping of the respective cDNA sequences and synteny of the genomic regions. To determine the conservation across all vertebrates, we analyzed the genomes of 22 species (supplementary tables S4 and S5, Supplementary Material online). In contrast to the other vertebrates, all mammals contain two γ-tubulins in a cluster of tandemly arrayed genes implying that the γ-tubulin duplication happened in their last common ancestor. Only in the rat genome, a second γ-tubulin tandem gene duplicate could be found (supplementary fig. S7, Supplementary Material online). The fish α- and β-tubulins are already too divergent to reliably group them with the mammalian homologs. From the amphibians and the sauropsids, only the α-tubulins could unambiguously be assigned to mammalian subtypes (supplementary tables S4 and S5, Supplementary Material online). Subtype grouping was done by inspecting the phylogenetic trees and by comparing the sequences and gene structures. The most prominent sequence differences are in the C-terminal E-hooks (supplementary fig. S8, Supplementary Material online). In fact, these could exclusively be used for classification. Comparing the synteny at the respective genomic regions could also be used as additional hint for classification. However, synteny breaks over time, and it can hardly be distinguished, whether the synteny got lost at a tubulin gene locus or whether the respective tubulin gene has been moved from its original location. Examples are the α-tubulin Tub1D and Tub1E genes (called tubulin α 3A and α 3B in mouse, α 3D and α 3E in human [Khodiyar et al. 2007]) that have been given different names in mouse and human because their respective genomic regions are not syntenic. The distance between the loci of the human paralogs is about 1.3 Mb, the respective distance between mouse loci is more than 20 Mb. However, in many mammals like the elephant Loxodonta africana and the guinea pig Cavia porcellus, the same orthologs are closely located in a cluster of tandemly arrayed gene duplicates (supplementary fig. S7, Supplementary Material online). In other mammals like the Florida manatee T. manatus, the cluster of closely located genes is still present although one of the genes has turned into a pseudogene. Interestingly, the two paralogs are always on opposite strands and on the same chromosome independently of their proximity (supplementary fig. S9, Supplementary Material online). This implies that the paralogs originated by duplication in the last common ancestor of the mammals as a cluster of closely located genes. Furthermore, the respective region was strongly involved in major genomic rearrangements leading to different arrangements/locations of the orthologs in extant species. However, the sequences remained 100% conserved within and across species. In contrast, the cluster of the α-tubulins Tub1A, Tub1B, and Tub1C (supplementary fig. S7, Supplementary Material online) has been retained in all mammals, in frog, and in the anole lizard but has completely been lost in birds (supplementary table S4, Supplementary Material online).
Some mammals have lost specific tubulins, either completely or by turning them into pseudogenes, like the α-tubulin Tub1H (called “tubulin, α-like 3” in human [Khodiyar et al. 2007]) pseudogenes in the elephant and manatee, and the missing Tub1H gene in the guinea pig and the opossum (supplementary table S4, Supplementary Material online). Some species have additional duplications like the Tub1C duplication in cow (supplementary fig. S7, Supplementary Material online). Interestingly, many species also encode α-tubulin subtypes that have been lost in human and mouse. Thus, the opossum, dog, squirrel, and ferret contain a Tub1I gene, which is not a clear paralog of any of the other α-tubulins but is also present in frog, the anole lizard, and birds (supplementary table S4, Supplementary Material online) indicating an origin at least in the ancient Sarcopterygii. In addition, many mammals contain a Tub1K α-tubulin gene, which is also present in frogs and the anole lizard. In all species, this Tub1K gene is located in a cluster on opposite strands together with the Tub1F gene (supplementary fig. S7, Supplementary Material online). At the same location in humans is the Tub1L α-tubulin pseudogene, which, however, diverged so far that unambiguous subtype sequence similarity could not be inferred. In mouse, the Tub1F gene is the only α-tubulin gene on chromosome 1, and there are no pseudogenes as well, implying that the Tub1K paralog completely disappeared already. The guinea pig contains a unique species-specific cluster of two additional α-tubulins. The frog, anole lizard, and birds have additional duplicates for several of their genes (supplementary fig. S7 and tables S4 and S5, Supplementary Material online). This demonstrates that mammals, amphibians, and sauropsids have branch-specific α-tubulin gene duplications, the Tub1D/Tub1E cluster in mammals, cluster of Tub1F and Tub11G duplicates in sauropsids, and clusters of Tub1H and Tub1K duplicates in frogs.
The β-tubulin subfamily developed less dynamically than the α-tubulins within the vertebrates. Except for single gene losses in a few species, all mammals share the same set of eight β-tubulins including the tandemly arrayed cluster of the β-tubulins Tub2D and Tub2E (named TUBB2B and TUBB2A in human, respectively; supplementary fig. S7, Supplementary Material online). Primates have an additional β-tubulin subtype, Tub2I. The squirrel Spermophilus tridecemlineatus contains three unique β-tubulins, Tub2J, Tub2K, and Tub2L, of which the first two are arranged in a cluster (supplementary fig. S7, Supplementary Material online).
Alpha- and Beta-Tubulins
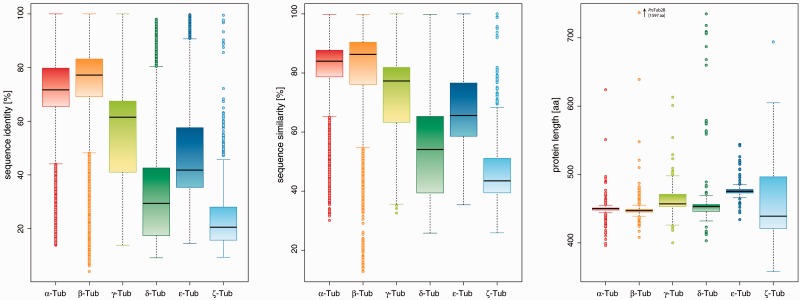
Among the tubulin family, the α- and β-tubulins comprise the largest groups. They are strongly conserved in sequence and protein lengths (fig. 4) with the following exceptions. The Chytridiomycota and Neocallimastigomycota each encode a β-tubulin variant containing a C-terminal extension of up to 1,100 residues. The sequences of these C-terminal extensions are of low complexity and not conserved across species but are supported by conserved gene structures. Other α-tubulin subclasses lost the E-hook, the acidic C-terminus of α- and β-tubulins. For example, the mammalian α-tubulin Tub1H orthologs have lost the E-hook, which is still present in the Tub1H orthologs of birds, frog, and the anole lizard (supplementary fig. S8, Supplementary Material online), implying that the loss must have happened in the last common mammalian ancestor. Other species with E-hook-less α-tubulins are the Babesiae/Theileriae, the Entamoebae, insects, Ciliates, and Naegleria. Four of the very divergent Paramecium β-tubulins do not contain a P-loop sequence anymore suggesting these tubulins function as structural building block and not in tubulin polymerization. The α- and β-tubulins are often clustered in the genome. Although the Trypanosoma species seem to be the only organisms with clusters of tandemly arrayed α- and β-tubulins (Jackson et al. 2006), only clusters of either α- or β-tubulins have been found in other species like the vertebrates (see above) or insects (supplementary tables S6 and S7, Supplementary Material online).
Fig. 4.—
Sequence conservation in tubulins. Box plots of the sequence identities (left) and similarities (middle) of all complete bacterial and eukaryotic tubulins, excluding pseudogenes. On the right, box plots of the protein lengths are shown.
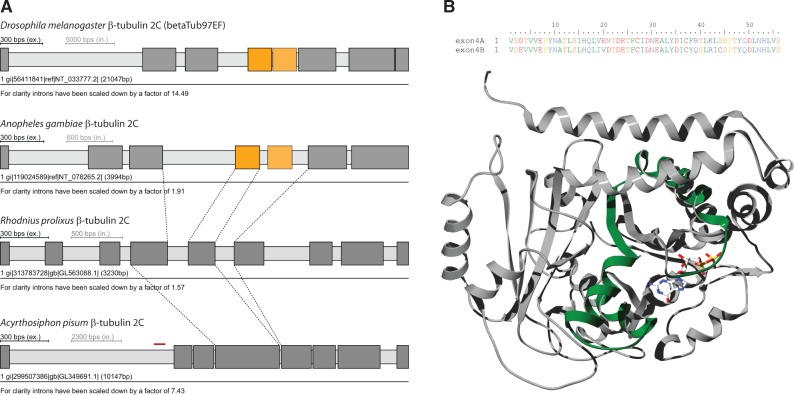
Because of their strong sequence and length conservation, alternative splice variants for α- and β-tubulins seemed very unlikely. However, by using a recently developed software to predict mutually exclusive spliced exons (MXEs) based on reading frame and splice site conservation, sequence similarity, and exon length constrains (Pillmann et al. 2011), we identified a cluster of mutually exclusive spliced exon candidates in the Drosophila melanogaster betaTub97EF gene (Hatje and Kollmar 2013). The betaTub97EF gene belongs to a subgroup of β-tubulins present in all Diptera with further orthologs in the Paraneoptera Rhodnius prolixus and Acyrthosiphon pisum (fig. 5A) implying that a β-tubulin of this subgroup must have already been present in the last common ancestor of the Neoptera. Most of the intron positions are shared between the Diptera and Paraneoptera homologs. The exon, which is part of the cluster of MXEs conserved in all Diptera, is still present in Rhodnius, whereas it is fused to the respective 5’-exon in Acyrthosiphon (fig. 5A). The two MXEs of the Drosophila betaTub97EF gene have a sequence identity of 76.7% at the protein level and code for a central part of the β-tubulin structure (fig. 5B), excluding that these exons could be spliced as differentially included exons as they had been annotated in the Drosophila Flybase r5.36 release.
Fig. 5.—
The mutually exclusive spliced insect β-tubulin 2C genes. (A) The Diptera encode β-tubulins containing a cluster of mutually exclusive spliced exons (MXEs). This cluster most probably appeared by exon duplication in the ancestor of the Diptera, because the gene structures are conserved in other insects that diverged prior to the emergence of the Diptera. Exons and introns are represented as dark- and light-gray bars, respectively; MXEs are shown in color. The opacity of the color of the 3’ of the alternative exons corresponds to the alignment score of the alternative exon to the original one (5’-exon). (B) The structural region covered by the MXEs of the Drosophila gene is shown mapped onto the crystal structure of β-tubulin from sheep brain (PDB-ID: 3RYC) (Nawrotek et al. 2011).
Gamma-, Delta-, Epsilon-, and Zeta-Tubulins
Like α- and β-tubulins, γ-tubulins are ubiquitous, as has already been speculated based on limited data almost 15 years ago (Oakley 2000). We did not find a single species that misses the γ-tubulin, although γ-tubulins can be very divergent (e.g. the Fonticula alba Tub3). About half of the species contain a single γ-tubulin gene, whereas two paralogs are found in mammals, Diptera, many Basidiomycotes, Rhizopus fungi, Bacillariophyta, and Haptophyta, and up to three paralogs in flowering plants. So far, δ-, ε-, and “η”-tubulins were reported to be restricted to certain lineages, mainly protists (Breviario et al. 2013). A so-called ζ-tubulin has only been identified in Trypanosoma species (Vaughan et al. 2000). In our exhaustive search, δ-, ε- and ζ-tubulins have been identified in all major kingdoms of the eukaryotes (table 1). However, in contrast to the ubiquitous α-, β-, and γ-tubulins, the δ-, ε-, and ζ-tubulins have been lost independently in many branches and extant species implying that they do not perform essential functions in most cells. For example, all Dikarya (Ascomycota and Basidiomycota) and seed plants (Spermatophyta) do not contain any δ-, ε-, and ζ-tubulins. Examples for a more recent loss of the δ-, ε-, and ζ-tubulins are the Diptera, which include the Drosophila and mosquitoes (fig. 6). Although species contain the ε-tubulin independent of the δ- and ζ-tubulins, the presence of the ζ-tubulin seems to be coupled to the presence of the δ-tubulin, with the exceptions of Bigelowiella natans, Emiliania huxleyi, and Guillardia theta. Homo sapiens and Mus musculus do not contain a ζ-tubulin. By analyzing many of the available mammalian genome assemblies, we could reveal that ζ-tubulins are present in all sequenced Metatheria (Monodelphis domestica, M. eugenii, and Sarcophilus harrisii) but absent in Eutherians (fig. 6). Also in contrast to α-, β-, and γ-tubulins, none of the analyzed species contains duplicated δ-, ε-, and ζ-tubulins. Furthermore, several alternative splice variants for the mammalian δ-tubulin genes can be identified in the cDNA/EST databases, although most are probably pseudoisoforms resulting in nonfunctional proteins (supplementary fig. S10, Supplementary Material online).
Fig. 6.—
Schematic tree of analyzed metazoans and their tubulins. Abstract representation of the phylogenetic tree of the Metazoa constructed by analyzing data from Wägele and Bartolomaeus (2014). Branches are shown at which changes in the presence of the δ-, ε-, and ζ-tubulins happened. At each leaf, one representative species of the branch is printed. Branch lengths are arbitrary. White boxes illustrate the loss of the respective tubulin subfamily member in the branch or species.
Discussion
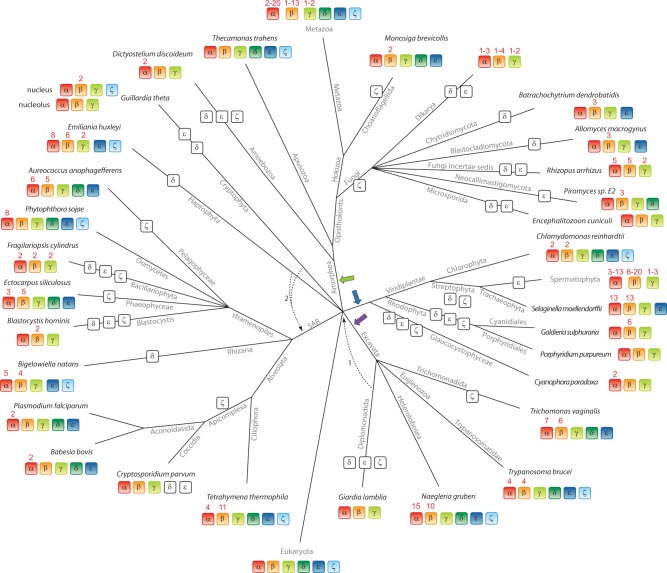
The consensus of the generated phylogenetic trees, based on different tree reconstruction methods and varying data sets, shows six major eukaryotic tubulin subfamilies. Members of all subfamilies are present in all major kingdoms of the eukaryotes implying that an ancestor of each subfamily must have been present in the last common ancestor of the eukaryotes (fig. 7). According to these trees, the previously named “η”-tubulins group together with the ζ-tubulins, and the “θ”-, “ι”-, and “κ”-tubulins, which have only been identified in the Ciliate P. tetraurelia so far, group together with the other eukaryotic β- and α-tubulins. The latter grouping has already been shown in other studies (Dutcher 2003; Yutin and Koonin 2012), but the respective tubulins have not been renamed yet. The grouping of the “η”-tubulins to the ζ-tubulins has probably not been recognized so far because of the very limited number of sequences used in the analysis (Dutcher 2003) or because either one or the other group has not been included in the study (Vaughan et al. 2000; Libusová and Dráber 2006; Yutin and Koonin 2012). Thus, our analysis leads to a consolidation of the eukaryotic tubulin family. The broad taxonomic sampling of the data across the eukaryotic tree suggests that the eukaryotic tubulin family is now complete with six subfamilies.
Fig. 7.—
Evolution of the tubulin protein family across eukaryotes. The tree has been reconstructed by evaluating recent literature (Parfrey et al. 2010; Adl et al. 2012; He et al. 2014) for those eukaryotic branches that have been included in this study. However, especially the grouping of taxa that emerged close to the origin of the eukaryotes remains highly debated. Therefore, alternative branchings are also indicated in the tree. The phylogeny of the supposed supergroup Excavata is the least understood because only a few species of this branch have been completely sequenced so far. Although the grouping of the Heterolobosea, Trichomonada, and Euglenozoa into the Excavata is found in most analyses, the grouping of the Diplomonadida as separate phylum or as part of the Excavata is still debated (arrow 1; Simpson et al. 2006). The placement of the Haptophyceae and Cryptophyta to the SAR (arrow 2; Nozaki et al. 2009; Keeling 2009) is supported by some studies although most analyses are in contrast (Hampl et al. 2009; Parfrey et al. 2010). At each leaf of the tree, one representative species of the branch is printed. Branch lengths are arbitrary. The tree illustrates the presence (colored boxes) and absence (white boxes) of each tubulin subfamily in the corresponding species. The LECA must have already contained one member of each of the six subfamilies, as indicated, independent of whether the root of the eukaryotes is placed at the base of six eukaryotic supergroups as shown or whether the root is placed at alternative positions like the unikont–bikont split (green arrow; unikonts/Amorphea versus all other eukaryotes) or the photosynthetic–nonphotosynthetic split (blue arrow), or between Excavata and Neozoa (all other eukaryotes; purple arrow) (He et al. 2014). Numbers above tubulins indicate the number or range of duplicates within the species or taxon, respectively.
A recent analysis has proposed an expansion of the tubulin family into the archaea kingdom, and the respective homologs have been named “artubulins” accordingly (Yutin and Koonin 2012). These artubulins have been denoted tubulins because the best BLAST hits turned out to be γ-tubulins, because their sequences could be aligned with tubulins, and because they grouped between FtsZ and eukaryotic tubulins in the constructed phylogenetic tree (Yutin and Koonin 2012). The placing of the root of the phylogenetic tree between FtsZ and the artubulins turned the eukaryotic tubulins to a sister group of the artubulins. This was justified by the argument that alternative scenarios such as rooting the tree by artubulins would imply an ancient duplication followed by a massive loss of artubulins in all bacteria and archaea, which would be highly nonparsimonious (Yutin and Koonin 2012). However, in the proposed parsimonious scenario, the artubulins must have similarly been lost in all archaea except for the two Nitrosarchaea. The argumentation stands and falls with denoting the artubulins tubulin homologs. The artubulins could have also evolved by a Nitrosarchaea-specific duplication of the FtsZ gene with subsequent substantial mutations turning them FtsZ-like. Here, we have not only included FtsZ proteins in the analysis but also bacterial TubZ proteins, which are known to be FtsZ-like. The artubulins grouped to these TubZ proteins in all our phylogenetic trees forming together a sister group to the FtsZ proteins (figs. 1 and 2). This suggests that the artubulins are either TubZ homologs or the founding members of another FtsZ subfamily. Hence, the artubulins cannot be regarded as tubulins or tubulin homologs. They should be renamed because their present naming implies a common ancient origin with tubulins, which is not supported by the data.
Another, not finally resolved question was the placing of the bacterial tubulin homologs from Prosthecobacter species (Schlieper et al. 2005; Sontag et al. 2009). Some proposed mosaic sequences with intertwining features from both α- and β-tubulins indicating an origin prior to the split of the eukaryotic α- and β-tubulins (Martin-Galiano et al. 2011; Yutin and Koonin 2012). Other studies, however, did not find connections to any particular modern tubulin subfamily and suggested that bacterial tubulins were derived from an ancestor of the entire tubulin superfamily (Pilhofer et al. 2011). In most of our trees, the bacterial tubulins group to the eukaryotic α- and β-tubulins albeit with different topologies (fig. 2). In 86% of the trees, the same set of bacterial tubulins groups as sister group to the eukaryotic α-tubulins. Therefore, we termed this clade bacterial α-tubulins and correspondingly the other group bacterial β-tubulins although their placement in the trees varies. We did not observe any tree, in which all bacterial tubulins form a clade grouping sister to the combined eukaryotic α- and β-tubulins. This has been found by others (Yutin and Koonin 2012) and would imply a horizontal gene transfer prior to the split of the eukaryotic α- and β-tubulins. According to our data, the most probable scenario suggests a horizontal gene transfer after the split of the eukaryotic α- and β-tubulins but before early eukaryotic diversification started. Although the bacterial α-tubulins consistently group to the eukaryotic α-tubulins, the bacterial β-tubulins diverged so far that an unambiguous relationship to the eukaryotic β-tubulins cannot be inferred anymore.
Because of their ubiquitous distribution in eukaryotes, α- and β-tubulins are often used in phylogenetic studies aiming to reveal taxonomic relationships. However, of the 504 species analyzed here, 85% contain either duplicated α- and/or duplicated β-tubulins (supplementary fig. S2, Supplementary Material online). Duplications are not linked to major early eukaryotic branching events (fig. 7) and not even to the many well-known whole-genome duplication events but happened in recent branchings and are often species specific. This counts for all major kingdoms of the eukaryotes. In contrast, other protein families retained major duplication events like the plant myosins that can be grouped according to the dozens of whole-genome duplications (Mühlhausen and Kollmar 2013). A similar grouping is not possible for plant tubulins. Even very closely related plants contain completely different numbers and subtypes of α- and β-tubulins (supplementary fig. S2, Supplementary Material online). Similarly, the 56 analyzed Basidiomycotes have various numbers of α-, β and γ-tubulins, and if duplicates are present, these do not always belong to the same subclass. As species phylogenies are rarely based on whole-genome data but on sequencing single genes, it is highly probable that all tubulin-based trees—at least in part—do not represent the true species phylogeny.
α-, β-, and γ-tubulins are ubiquitous, and thus, their main functions should be conserved throughout the eukaryotes. Fine-tuned functions should therefore mainly result from the many possible posttranslational modifications and the sequence differences between duplicates. Because of the many independent duplications and multiplications of either or both the α- and β-tubulins functional specialization of a certain subclass for cytoskeletal, axonemal, A-, B-, or C-tubules, or any other distinct microtubule substructure cannot be inferred, as has been suggested by others. The almost identical tubulin duplicates in many species also seem to contradict the multitubulin hypothesis, firstly proposed by Fulton and Simpson in 1976 (Fulton and Simpson 1976) stating that each tubulin protein contributes to distinct microtubule structures. Similarly, many species such as Giardia lamblia, the Apicomplexa species, and the Apusozoa Thecamonas trahens contain only single α- and β-tubulin genes, but distinct microtubule substructures showing that these can be build without tubulin diversity. Also, there is no obvious evidence for the concerted evolution of α- and β-tubulins as has been suggested for some insect tubulins (Nielsen et al. 2010). Almost all species with α- and β-tubulin duplicates have different numbers of their α- and β-tubulins with sometimes striking differences such as 4 and 11 α- and β-tubulins, respectively, in Tetrahymena species, 20 and 7 α- and β-tubulins, respectively, in Nematostella vectensis, and 8 and 20 α- and β-tubulins, respectively, in Populus trichocarpa (supplementary fig. S2, Supplementary Material online). These numbers suggest that within species every α-tubulin should be compatible with every β-tubulin. Therefore, the tubulin duplicates were also not derived by the classical model of duplication and divergence (Ohno 1970), which states that a duplicated gene will only be retained if it developed a totally new function (neofunctionalization). Rather, the cellular abundance of the tubulins tolerates multiple gene copies without enforcing mutations toward subfunctionalizations and transcription rate reductions. It is highly unlikely that the last common ancestor of the eukaryotes already contained duplicates of tubulin subfamilies as these would have been apparent in eukaryote-wide distinct subgroups. Thus, all the distinct microtubule substructures present in the last common eukaryotic ancestor were built by single α- and β-tubulins, whereas subfunctionalization by gene duplication is a more recent process. The many differences in α- and β-tubulin gene inventories in vertebrates, insects, and plants (supplementary tables S4–S7, Supplementary Material online) show that this process is branch- and species specific, that it is still ongoing, and that it happened independently in almost all eukaryotic branches.
δ-, ε-, and ζ-(“η”-) tubulins have been reported to be important or essential for basal bodies stability and assembly in Paramecium and Tetrahymena, respectively (Garreau de Loubresse et al. 2001; Dutcher 2003; Libusová and Dráber 2006). According to our data, none of the three tubulin subfamilies seems to be essential for basal bodies because the presence of cilia or flagella does not correlate with the presence of δ-, ε-, and ζ-tubulin genes in the respective species (fig. 7). On the other hand, δ-, ε-, and ζ tubulins are only present in eukaryotes that have cilia/basal bodies, indicating that their function is only related to the function of cilia. The ε-tubulin, for example, is absent in the beetle Tribolium castaneum, in fungi of the Neocallimastigomycota clade and in G. theta, and the δ-tubulin is missed in the Trachaeophyta plants, in Chytridiomycota and in Blastocladiomycota fungi, the Haptophyte E. huxleyi, and the Rhizaria B. natans. Species having cilia but no δ-, ε-, and ζ-tubulins are the protozoan parasite Giardia lamblia, the leech Helobdella robusta, and the Diptera (figs. 6 and 7). Because many of the species having cilia contain the δ- or the ε-tubulin, these subtypes might functionally substitute each other. The ζ-tubulin is the least distributed and conserved tubulin subfamily and might be important for fine-tuning functions that are otherwise performed by one or a combination of the δ- and ε-tubulins. Because the last common ancestor of the eukaryotes contained δ-, ε-, and ζ-tubulins and the presence of these tubulins is correlated to cilia/basal bodies in extant species, it is highly likely that the ancestral function of the δ-, ε-, and ζ-tubulins was related to basal bodies.
Data Access
The alignment of the tubulins can be obtained from CyMoBase (www.cymobase.org) and supplementary data S1, Supplementary Material online.
Supplementary Material
Supplementary figures S1–S10, tables S1–S7, and data S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
P.F. and M.K. designed the study and analyzed the data. P.F., S.M., S.D., J.H., A.Z., and M.K. annotated tubulin genes. S.M. performed phylogenetic computations. T.C. participated in data interpretation. M.K. wrote the manuscript. All authors read and approved the final manuscript. The authors thank Prof. Christian Griesinger for his continuous generous support and the reviewers for their insightful comments. This work was supported by grants KO 2251/3-1, KO 2251/3-2, and KO 2251/3-3 of the Deutsche Forschungsgemeinschaft to M.K., a Synaptic Systems fellowship to S.M., and by grant I80798 of the VolkswagenStiftung to M.K.
Literature Cited
- Adl SM, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylett CHS, Löwe J, Amos LA. New insights into the mechanisms ofcytomotive actin and tubulin filaments. Int Rev Cell Mol Biol. 2011;292:1–71. doi: 10.1016/B978-0-12-386033-0.00001-3. [DOI] [PubMed] [Google Scholar]
- Aylett CHS, Wang Q, Michie KA, Amos LA, Löwe J. Filament structure of bacterial tubulin homologue TubZ. Proc Natl Acad Sci U S A. 2010;107:19766–19771. doi: 10.1073/pnas.1010176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breviario D, Gianì S, Morello L. Multiple tubulins: evolutionary aspects and biological implications. Plant J Cell Mol Biol. 2013;75:202–218. doi: 10.1111/tpj.12243. [DOI] [PubMed] [Google Scholar]
- Brown MW, Spiegel FW, Silberman JD. Phylogeny of the “forgotten” cellular slime mold, Fonticula alba, reveals a key evolutionary branch within Opisthokonta. Mol Biol Evol. 2009;26:2699–2709. doi: 10.1093/molbev/msp185. [DOI] [PubMed] [Google Scholar]
- Campanella JJ, Bitincka L, Smalley J. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics. 2003;4:29. doi: 10.1186/1471-2105-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK. Long-lost relatives reappear: identification of new members of the tubulin superfamily. Curr Opin Microbiol. 2003;6:634–640. doi: 10.1016/j.mib.2003.10.016. [DOI] [PubMed] [Google Scholar]
- Dyer N. Tubulin and its prokaryotic homologue FtsZ: a structural and functional comparison. Sci Prog. 2009;92:113–137. doi: 10.3184/003685009X461431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert C, Hammesfahr B, Kollmar M. A holistic phylogeny of the coronin gene family reveals an ancient origin of the tandem-coronin, defines a new subfamily, and predicts protein function. BMC Evol Biol. 2011;11:268. doi: 10.1186/1471-2148-11-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsik CG, et al. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. 2009;324:522–528. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton C, Simpson PA. Selective synthesis and utilisation of flagellar tubulin. The multi-tubulin hypothesis. In: Goldman R, Pollard T, Rosenbaum J, editors. Cell motility. Vol. 3. New York: Cold Spring Harbor Laboratory Press; 1976. pp. 987–1005. [Google Scholar]
- Garreau de Loubresse N, Ruiz F, Beisson J, Klotz C. Role of delta-tubulin and the C-tubule in assembly of Paramecium basal bodies. BMC Cell Biol. 2001;2:4. doi: 10.1186/1471-2121-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, et al. Alpha-tubulin and small subunit rRNA phylogenies of peritrichs are congruent and do not support the clustering of mobilids and sessilids (Ciliophora, Oligohymenophorea) J Eukaryot Microbiol. 2010;57:265–272. doi: 10.1111/j.1550-7408.2010.00472.x. [DOI] [PubMed] [Google Scholar]
- Hampl V, et al. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci U S A. 2009;106:3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatje K, Hammesfahr B, Kollmar M. WebScipio: reconstructing alternative splice variants of eukaryotic proteins. Nucleic Acids Res. 2013;41:W504–W509. doi: 10.1093/nar/gkt398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatje K, Kollmar M. Expansion of the mutually exclusive spliced exome in Drosophila. Nat Commun. 2013;4:2460. doi: 10.1038/ncomms3460. [DOI] [PubMed] [Google Scholar]
- Hatje K, et al. Cross-species protein sequence and gene structure prediction with fine-tuned Webscipio 2.0 and Scipio. BMC Res Notes. 2011;4:265. doi: 10.1186/1756-0500-4-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D, et al. An alternative root for the eukaryote tree of life. Curr Biol. 2014;24:465–470. doi: 10.1016/j.cub.2014.01.036. [DOI] [PubMed] [Google Scholar]
- Huang K-H, Durand-Heredia J, Janakiraman A. FtsZ ring stability: of bundles, tubules, crosslinks, and curves. J Bacteriol. 2013;195:1859–1868. doi: 10.1128/JB.02157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AP, Vaughan S, Gull K. Evolution of tubulin gene arrays in Trypanosomatid parasites: genomic restructuring in Leishmania. BMC Genomics. 2006;7:261. doi: 10.1186/1471-2164-7-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins C, et al. Genes for the cytoskeletal protein tubulin in the bacterial genus Prosthecobacter. Proc Natl Acad Sci U S A. 2002;99:17049–17054. doi: 10.1073/pnas.012516899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. Chromalveolates and the evolution of plastids by secondary endosymbiosis. J Eukaryot Microbiol. 2009;56:1–8. doi: 10.1111/j.1550-7408.2008.00371.x. [DOI] [PubMed] [Google Scholar]
- Khodiyar VK, et al. A revised nomenclature for the human and rodent α-tubulin gene family. Genomics. 2007;90:285–289. doi: 10.1016/j.ygeno.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Kin T, Ono Y. Idiographica: a general-purpose web application to build idiograms on-demand for human, mouse and rat. Bioinformatics. 2007;23:2945–2946. doi: 10.1093/bioinformatics/btm455. [DOI] [PubMed] [Google Scholar]
- Kollmar M, Lbik D, Enge S. Evolution of the eukaryotic ARP2/3 activators of the WASP family: WASP, WAVE, WASH, and WHAMM, and the proposed new family members WAWH and WAML. BMC Res Notes. 2012;5:88. doi: 10.1186/1756-0500-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman CP. Phylogeny of the ascomycetous yeasts and the renaming of Pichia anomala to Wickerhamomyces anomalus. Antonie Van Leeuwenhoek. 2011;99:13–23. doi: 10.1007/s10482-010-9505-6. [DOI] [PubMed] [Google Scholar]
- Larsen RA, et al. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 2007;21:1340–1352. doi: 10.1101/gad.1546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Libusová L, Dráber P. Multiple tubulin forms in ciliated protozoan Tetrahymena and Paramecium species. Protoplasma. 2006;227:65–76. doi: 10.1007/s00709-005-0152-0. [DOI] [PubMed] [Google Scholar]
- Martin-Galiano AJ, et al. Bacterial tubulin distinct loop sequences and primitive assembly properties support its origin from a eukaryotic tubulin ancestor. J Biol Chem. 2011;286:19789–19803. doi: 10.1074/jbc.M111.230094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlhausen S, Kollmar M. Whole genome duplication events in plant evolution reconstructed and predicted using myosin motor proteins. BMC Evol Biol. 2013;13:202. doi: 10.1186/1471-2148-13-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrotek A, Knossow M, Gigant B. The determinants that govern microtubule assembly from the atomic structure of GTP-tubulin. J Mol Biol. 2011;412:35–42. doi: 10.1016/j.jmb.2011.07.029. [DOI] [PubMed] [Google Scholar]
- Nielsen MG, Gadagkar SR, Gutzwiller L. Tubulin evolution in insects: gene duplication and subfunctionalization provide specialized isoforms in a functionally constrained gene family. BMC Evol Biol. 2010;10:113. doi: 10.1186/1471-2148-10-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E. Structural insight into microtubule function. Annu Rev Biophys Biomol Struct. 2001;30:397–420. doi: 10.1146/annurev.biophys.30.1.397. [DOI] [PubMed] [Google Scholar]
- Nozaki H, et al. Phylogenetic positions of Glaucophyta, green plants(Archaeplastida) and Haptophyta (Chromalveolata) as deduced from slowly evolving nuclear genes. Mol Phylogenet Evol. 2009;53:872–880. doi: 10.1016/j.ympev.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Oakley BR. An abundance of tubulins. Trends Cell Biol. 2000;10:537–542. doi: 10.1016/s0962-8924(00)01857-2. [DOI] [PubMed] [Google Scholar]
- Odronitz F, Becker S, Kollmar M. Reconstructing the phylogeny of 21 completely sequenced arthropod species based on their motor proteins. BMC Genomics. 2009;10:173. doi: 10.1186/1471-2164-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. Pfarao: a web application for protein family analysis customized for cytoskeletal and motor proteins (CyMoBase) BMC Genomics. 2006;7:300. doi: 10.1186/1471-2164-7-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. New York: Springer-Verlag; 1970. Evolution by gene duplication. [Google Scholar]
- Parfrey LW, et al. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst Biol. 2010;59:518–533. doi: 10.1093/sysbio/syq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilhofer M, Ladinsky MS, McDowall AW, Petroni G, Jensen GJ. Microtubules in bacteria: ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton. PLoS Biol. 2011;9:e1001213. doi: 10.1371/journal.pbio.1001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillmann H, Hatje K, Odronitz F, Hammesfahr B, Kollmar M. Predicting mutually exclusive spliced exons based on exon length, splice site and reading frame conservation, and exon sequence homology. BMC Bioinformatics. 2011;12:270. doi: 10.1186/1471-2105-12-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ross I, Clarissa C, Giddings TH, Jr, Winey M. ε-tubulin is essential in Tetrahymena thermophila for the assembly and stability of basal bodies. J Cell Sci. 2013;126:3441–3451. doi: 10.1242/jcs.128694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieper D, Oliva MA, Andreu JM, Löwe J. Structure of bacterial tubulin BtubA/B: evidence for horizontal gene transfer. Proc Natl Acad Sci U S A. 2005;102:9170–9175. doi: 10.1073/pnas.0502859102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AGB, Inagaki Y, Roger AJ. Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of “primitive” eukaryotes. Mol Biol Evol. 2006;23:615–625. doi: 10.1093/molbev/msj068. [DOI] [PubMed] [Google Scholar]
- Sontag CA, Sage H, Erickson HP. BtubA-BtubB heterodimer is an essential intermediate in protofilament assembly. PLoS One. 2009;4:e7253. doi: 10.1371/journal.pone.0007253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33:W465–W467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinke D, Hoegg S, Brinkmann H, Meyer A. Three rounds (1R/2R/3R) of genome duplications and the evolution of the glycolytic pathway in vertebrates. BMC Biol. 2006;4:16. doi: 10.1186/1741-7007-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Maere S, Meyer A. 2R or not 2R is not the question anymore. Nat Rev Genet. 2010;11:166. doi: 10.1038/nrg2600-c2. [DOI] [PubMed] [Google Scholar]
- Vaughan S, et al. New tubulins in protozoal parasites. Curr Biol. 2000;10:R258–R259. doi: 10.1016/s0960-9822(00)00414-0. [DOI] [PubMed] [Google Scholar]
- Wägele JW, Bartolomaeus T. Berlin (Germany): De Gruyter; 2014. Deep metazoan phylogeny: the backbone of the tree of life, new insights from analyses of molecules, morphology, and theory of data analysis. [cited 2014 Aug 18] Available from: http://www.degruyter.com/view/product/181254. [Google Scholar]
- Walker DM, Castlebury LA, Rossman AY, White JF. New molecular markers for fungal phylogenetics: two genes for species-level systematics in the Sordariomycetes (Ascomycota) Mol Phylogenet Evol. 2012;64:500–512. doi: 10.1016/j.ympev.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Wickstead B, Gull K. The evolution of the cytoskeleton. J Cell Biol. 2011;194:513–525. doi: 10.1083/jcb.201102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Yi Z, Katz LA, Song W. Assessing whether alpha-tubulin sequences are suitable for phylogenetic reconstruction of Ciliophora with insights into its evolution in euplotids. PloS One. 2012;7:e40635. doi: 10.1371/journal.pone.0040635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Koonin EV. Archaeal origin of tubulin. Biol Direct. 2012;7:10. doi: 10.1186/1745-6150-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, et al. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 2009;10:R42. doi: 10.1186/gb-2009-10-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.