Abstract
Screening of a deeply sequenced transcriptome using Illumina sequencing as well as the genome of the tardigrade Hypsibius dujardini revealed a set of five opsin genes. To clarify the phylogenetic position of these genes and to elucidate the evolutionary history of opsins in Panarthropoda (Onychophora + Tardigrada + Arthropoda), we reconstructed the phylogeny of broadly sampled metazoan opsin genes using maximum likelihood and Bayesian inference methods in conjunction with carefully selected substitution models. According to our findings, the opsin repertoire of H. dujardini comprises representatives of all three major bilaterian opsin clades, including one r-opsin, three c-opsins, and a Group 4 opsin (neuropsin/opsin-5). The identification of the tardigrade ortholog of neuropsin/opsin-5 is the first record of this opsin type in a protostome, but our screening of available metazoan genomes revealed that it is also present in other protostomes. Our opsin phylogeny further suggests that two r-opsins, including an “arthropsin,” were present in the last common ancestor of Panarthropoda. Although both r-opsin lineages were retained in Onychophora and Arthropoda, the arthropsin was lost in Tardigrada. The single (most likely visual) r-opsin found in H. dujardini supports the hypothesis of monochromatic vision in the panarthropod ancestor, whereas two duplications of the ancestral panarthropod c-opsin have led to three c-opsins in tardigrades. Although the early-branching nodes are unstable within the metazoans, our findings suggest that the last common ancestor of Bilateria possessed six opsins: Two r-opsins, one c-opsin, and three Group 4 opsins, one of which (Go opsin) was lost in the ecdysozoan lineage.
Keywords: opsin evolution, vision, transcriptomics, Tardigrada, Panarthropoda
Introduction
Opsins are light-sensitive proteins used for photoreception. These proteins function as G protein-coupled receptors (GPCRs) that trigger phototransduction cascades associated with animal vision and circadian clocks (e.g., Arendt et al. 2004; Velarde et al. 2005; Rubin et al. 2006; review Hankins et al. 2008; review Shichida and Matsuyama 2009; review Fain et al. 2010). Previous studies had unveiled that the last common ancestor of Bilateria possessed representatives of three major opsin clades, including ciliary [=c-opsins], rhabdomeric [=r-opsins], and Group 4 opsins (review Porter et al. 2012). Among these three clades, only the c-opsins and r-opsins have been known to be involved in vision (e.g., Koyanagi et al. 2008; Land and Nilsson 2012; Koyanagi and Terakita 2013). Independent diversification of c-opsins in vertebrates and r-opsins in arthropods has led to convergent evolution of color vision in these animals (review Shichida and Imai 1998).
Within Panarthropoda (Onychophora + Tardigrada +Arthropoda), color vision has been confirmed only in arthropods, whereas onychophorans most likely show monochromatic vision due to the presence of a single visual r-opsin, dubbed onychopsin (Hering et al. 2012). Moreover, although all three major bilaterian opsin clades are represented in the arthropod lineage, Group 4 opsins are absent from onychophorans (Eriksson et al. 2013). However, because the corresponding information is completely missing from tardigrades, the opsin repertoire in the last common ancestor of Panarthropoda remains unknown.
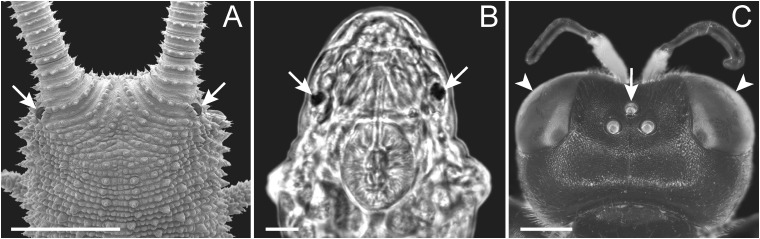
Tardigrades typically possess a pair of simple, ocellus-like eyes (Kristensen 1983; Dewel et al. 1993; Greven 2007)—a situation which is similar to that in onychophorans but different from that in arthropods, which show two types of visual organs: The median ocelli and the compound eyes (fig. 1A–C). Although the onychophoran eyes have been homologized with the median ocelli of arthropods (Mayer 2006), the homology of the tardigrade eyes remains obscure (Greven 2007). Experimental data revealed that tardigrades clearly respond to light (Marcus 1929; Baumann 1961; Ramazzotti and Maucci 1983; Beasley 2001), but beyond this nothing is known about the photoreceptive system in these animals.
Fig. 1.—
Visual organs in representatives of Panarthropoda. Although most onychophorans and tardigrades have a pair of simple, ocellus-like eyes (arrows in A and B), arthropods usually show two types of visual organs: The compound eyes (arrowheads in C) and the median ocelli (arrow in C). (A) Scanning electron micrograph of the head of the onychophoran Principapillatus hitoyensis in dorsal view. Scale bar: 1 mm. (B) Light micrograph of the head of the tardigrade Hypsibius dujardini in dorsal view. Scale bar: 10 µm. (C) Stereomicrograph of the head of the hymenopteran Ectemnius cavifrons in dorsal view. Scale bar: 1 mm.
The major objective of our study is therefore to analyze the opsin repertoire in a tardigrade to clarify the following questions: 1) Do tardigrades show only a single r-opsin as in onychophorans (Hering et al. 2012), or is there evidence for multiple visual pigments as in arthropods (e.g., review Briscoe and Chittka 2001; review Marshall et al. 2007; Henze et al. 2012)? 2) Did the last common ancestor of Onychophora, Tardigrada, and Arthropoda have monochromatic vision? 3) How many orthologs of c-opsins and Group 4 opsins have been retained in the tardigrade lineage? 4) Did losses and duplications of opsin genes occur in tardigrades and, if so, how many? and 5) What was the opsin composition in the last common ancestor of Panarthropoda and Bilateria?
To answer these questions, we sequenced and analyzed the transcriptome of the eutardigrade Hypsibius dujardini using an Illumina-based sequencing approach. In addition, we screened various metazoan genomes, including the recently released genome of H. dujardini, and reconstructed the phylogeny of broadly sampled metazoan opsin genes, which allowed us to firmly place the tardigrade sequences in the bilaterian opsin tree.
Materials and Methods
Specimens, Library Preparation, Sequencing, and Assembly
Specimens of Hypsibius dujardini (Doyère, 1840) (Eutardigrada, Hypsibiidae) were obtained commercially from Sciento (Manchester, UK). Several hundred specimens were used to extract total RNA using TRIzol reagent (Invitrogen, Carlsbad, CA) and RNeasy MinElute Cleanup Kit (Qiagen, Hilden, Germany) according to the manufacturers’ protocols. Library preparation for double indexing (Meyer and Kircher 2010; Kircher et al. 2012), 76 cycles paired-end sequencing on an Illumina Genome Analyzer IIx (San Diego, CA), and postsequencing processing (adapter trimming, removal of reads with falsely paired indices and filtering of reads at three different levels of stringency) were performed as described by Hering et al. (2012). Each of the three obtained data sets (Filter15, Filter25, and Filter30) was then assembled de novo using two different software packages to assess the occurrence of opsin transcripts in a broader methodological framework: CLC Genomics Workbench v5.1 (CLC bio, Århus, Denmark), and IDBA-Tran v1.1.0 (Peng et al. 2013). The IDBA-Tran assemblies were done twice, allowing for the retention of one or three isoforms of a transcript, respectively, using the -num_isoform option (additional assembly parameters and statistics; supplementary table S1, Supplementary Material online).
Obtaining Opsin Sequences from the Transcriptome of H. dujardini and Publicly Available Metazoan Genomes
To obtain the sequences of putative opsin genes from the transcriptome of H. dujardini, BLAST v2.2.27+ (Altschul et al. 1997) and HMMER v3.1b1 (http://hmmer.org/, last accessed September 10, 2014; Eddy 1998) were used in custom Perl scripts on nine assemblies in total as described by Hering et al. (2012) with the following modifications: For the tBLASTn/BLASTP searches, 16 opsin sequences from all major opsin groups were used as bait sequences with an E value of 1e-5 as a threshold (for accession numbers of all query sequences, see supplementary table S2, Supplementary Material online). For the HMMER search, the same value was used, and the search was performed by applying previously built HMMER profiles (Hering et al. 2012). In total, 1,634 nonredundant contigs were obtained as putative opsin genes and used as candidates in further analyses. For reciprocal BLAST searches against the nr database of GenBank, 530 nonredundant contigs from a BLASTP search with the E value 1e-10 and a HMMER search with the E value 1e-20 were used and every best hit was stored (101 hits after the removal of redundant sequences). Furthermore, we mined the publicly available genomes of the annelids Capitella teleta and Helobdella robusta, and the mollusks Lottia gigantea (http://genome.jgi.doe.gov/, last accessed September 10, 2014) and Crassostrea gigas (http://gigadb.org/, last accessed September 10, 2014) to enrich our metazoan opsin data set by using BLAST searches. In addition, we screened the genomes of the aphid Acyrthosiphon pisum (http://www.aphidbase.com/, last accessed September 10, 2014) and the water flea Daphnia pulex (http://genome.jgi.doe.gov/, last accessed September 10, 2014) to identify putative orthologs of the new opsin-5 gene (vertebrate opsin-5/neuropsin-like gene) from H. dujardini. All identified putative opsin genes from these genomes were checked for the presence of lysine at the retinal-binding site corresponding to the K296 position of bovine rhodopsin (Palczewski et al. 2000). Due to the uncertain placement of one of the opsins of the ctenophore Mnemiopsis leidyi (MleiOpsin3; see Schnitzler et al. 2012) and an unusual insertion downstream of the predicted retinal-binding site corresponding to the K296 position of bovine rhodopsin, it is unclear whether or not MleiOpsin3 is a functional opsin gene. We therefore decided to exclude MleiOpsin3 before our analyses (supplementary fig. S8, Supplementary Material online).
Computational Preanalyses
To decide whether or not a contig identified by BLAST/HMMER is a potential opsin gene, several maximum-likelihood (ML) analyses were performed to sort out nonopsin contigs. Therefore, we aligned all query sequences for the BLAST searches, and the opsin sequences used to build the HMMER profiles to an opsin master alignment using the web server of MAFFT version 7 (Katoh and Standley 2013). Thereafter, we extended this alignment with the 101 unique best hits obtained by the reciprocal BLAST search using the -add option in MAFFT. In the next step, the initially obtained 1,634 candidate contigs were split into batches of 150 sequences and each batch was also added to the master alignment using MAFFT (-add option) resulting in 11 separate alignments. Each of the 11 alignments was masked using ALISCORE v2.2 (Misof and Misof 2009; Kück et al. 2010) and ALICUT v2.3 (http://www.utilities.zfmk.de, last accessed September 10, 2014) to exclude randomly aligned sections before tree reconstruction (sliding window size 64, comparing 10,000 random pairs). After running 11 independent ML analyses to obtain the best tree for each data set using RAxML v7.5.8 (Stamatakis 2006) with the PROTGAMMAAUTO option for automatic selection of the best-fitting substitution model (10x LG+G, 1x MTZOA+G), a total of 43 in-group opsin transcripts from all assemblies were curated manually using BioEdit v7.0.9 (Hall 1999) and CLC Main Workbench v6.8.4 (CLC bio, Århus, Denmark). These transcripts yielded five different opsin genes (Hd-r-opsin, Hd-c-opsin1, Hd-c-opsin2, Hd-c-opsin3, and Hd-neuropsin) for the tardigrade H. dujardini, named after the clade in which they occur.
Cloning and Rapid Amplification of cDNA Ends
Rapid amplification of cDNA ends (RACE) was performed using SMARTer RACE cDNA Amplification Kit (Clontech Laboratories, Inc., Mountain View, CA) according to the manufacturer’s protocol for a putative c-opsin (Hd-c-opsin3) and putatively new Hd-neuropsin of H. dujardini, due to the short length of the corresponding contigs obtained from our transcriptome data. The fragments of all five identified opsin genes of H. dujardini were cloned from cDNA using the pGEM-T Vector System (Promega, Madison, WI) and verified by Sanger sequencing (GATC Biotech, Konstanz, Germany) using a standard M13 amplification. The sequences of the tardigrade opsin genes (Hd-r-opsin, Hd-c-opsin1, Hd-c-opsin2, Hd-c-opsin3, and Hd-neuropsin) were submitted to GenBank under the accession numbers KM086335–KM086339.
Final Sequence Alignment and Masking
The final data set comprised 401 metazoan opsin and other closely related GPCRs, such as receptors for somatostatin, allatostatin, dopamine, octopamine, and melatonin as outgroups, selected according to the reconstructed trees from our preanalyses. Before the final alignment, a Pfam v27.0 domain search (Punta et al. 2012) was performed and all sequences were trimmed manually at ±20 amino acids up- and downstream from the predicted seven-transmembrane domain (7tm_1; PF00001). This was done to allow for a more accurate identification of homologous positions during the alignment step by removing poorly alignable regions a priori, such as the unconserved regions flanking the actual domain. The alignment was done using MAFFT version 7 with the most accurate option L-INS-i and default parameters. The software Noisy rel. 1.5.12 (Dress et al. 2008) was used to mask the alignment by removing homoplastic and random-like positions (-cutoff = 0.8, -seqtype = P, -shuffles = 20,000).
Model Choice and Cross-Validation
Several analyses were performed to obtain the best-fitting substitution model for the phylogenetic analyses of the final data set. First, ProtTest v3.3 (Darriba et al. 2011) revealed LG+G+F as the best-fitting model according to the Akaike information criterion (AIC; Akaike 1974), Bayesian Information Criterion (Schwarz 1978), corrected AIC (Sugiura 1978; Hurvich and Tsai 1989), and Decision Theory Criterion (Minin et al. 2003). Second, a 10-fold cross-validation with ten replicates for each chosen model (LG, GTR, CAT-LG, CAT-GTR, C20-LG, C20-GTR, C30-LG, C30-GTR, C40-LG, C40-GTR, C50-LG, C50-GTR, C60-LG, C60-GTR, WLSR5-LG, WLSR5-GTR) was performed for comparison using the multicore version of PhyloBayes MPI v1.4f (Lartillot et al. 2009). Therefore, the original alignment was split randomly ten times into a learning set (9/10 of the initial data set) and a test set (1/10 of the initial data set). Markov chains were run for 1,500 generations on each learning set (160 chains in total) of the models for comparison. For each of the replicates, the cross-validated likelihoods were calculated under the test set, averaged over the posterior distribution of the learning set, discarding the first 500 sampled points as burn-in and using the remaining 1,000 generations. Finally, the cross-validation log-likelihood scores per model were averaged over the ten replicates and used to rank the fit to the initial data set. According to this, the site-heterogeneous CAT-GTR model fitted the data best in eight out of the ten replicates. As the best non site-heterogeneous model, the data set -specific GTR model fitted the data better than the empirical LG model (supplementary table S3, Supplementary Material online). We therefore decided to use the GTR model for the final ML analyses using RAxML (due to the lack of site-heterogeneous models) and CAT-GTR to conduct Bayesian inference (BI) analyses with PhyloBayes.
ML and BI Analyses
The best ML tree was obtained running 100 independent inferences on the final data set with GAMMA correction of the final tree using the Pthreads-AVX version of RAxML v7.7.8 (fig. 2 and supplementary fig. S1, Supplementary Material online). A data set -specific GTR substitution matrix was estimated during a prior single run and then provided as substitution matrix for the above-mentioned run. Bootstrap support values (BS) were calculated using the rapid bootstrapping algorithm implemented in RAxML from 1,000 pseudoreplicates on the original alignment. To check whether or not a sufficient number of replicates has been generated (Pattengale et al. 2009), bootstrap convergence was assessed a posteriori according to the weighted Robinson–Foulds distance criterion using the –I autoMRE option of RAxML (supplementary fig. S1, Supplementary Material online). BI analysis was performed using the multicore version of PhyloBayes MPI v1.4f under the site-heterogeneous CAT-GTR model, which is the best-fitting model according to the cross-validation test. Two Markov chains were run independently for 60,000 generations each. Convergence of the chains was assessed using bpcomp and tracecomp statistics of PhyloBayes and Tracer v1.5.0 (Rambaut and Drummond 2009). Therefore, bpcomp and tracecomp were run multiple times. The burn-in was increased by 1,000 every iteration (sampling every tenth tree), beginning with a burn-in of 1,000, and finishing with a burn-in of 59,000. The obtained statistics and the log-likelihood traces of the runs were summarized and used for reliable assessment of chain convergence as described in the PhyloBayes manual, in dependency on the burn-in (summary statistics and parameter for chain convergence; supplementary fig. S9, Supplementary Material online). Accordingly, the first 21,000 trees of each chain were discarded and every second tree thereafter was used to calculate the 50% majority rule consensus tree with posterior probability support values (supplementary fig. S2, Supplementary Material online).
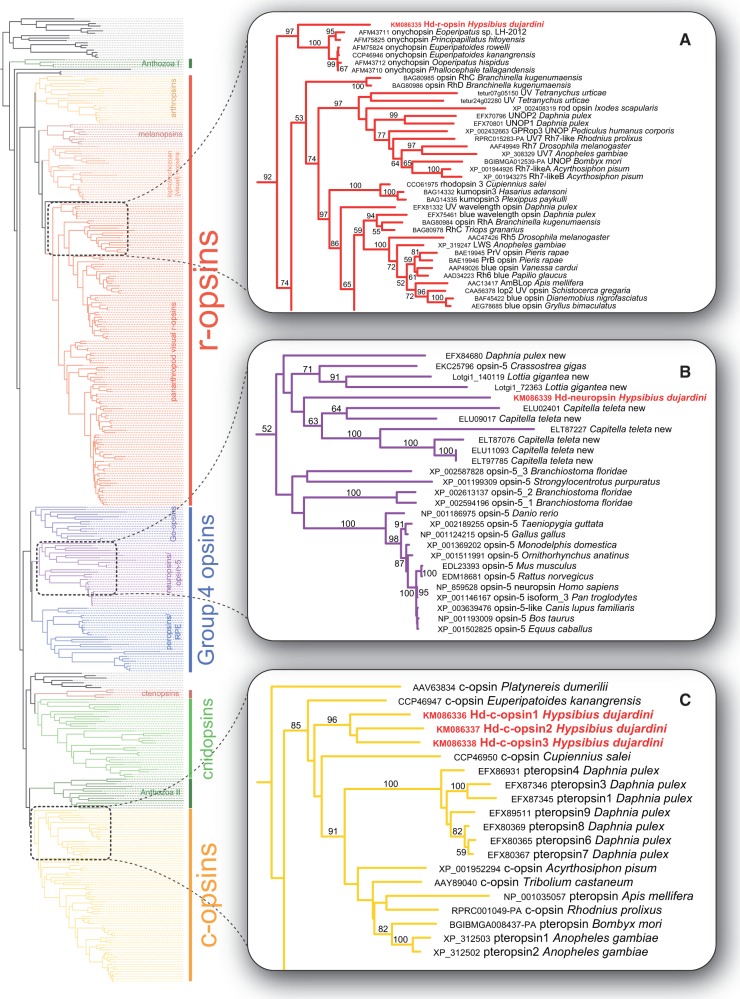
Fig. 2.—
Simplified best obtained ML tree of the full metazoan opsin gene data set and the placement of the five opsin genes from the tardigrade Hypsibius dujardini (highlighted in red). See supplementary figure S1, Supplementary Material online, for the uncondensed tree. (A) Detail view of the bilaterian r-opsin clade, in which Hd-r-opsin of H. dujardini occurs as the sister group to the onychophoran onychopsin clade. (B) Detail view of the neuropsin/opsin-5 subgroup of Group 4 opsins, illustrating the placement of Hd-neuropsin of H. dujardini among the neuropsins from other protostomes. (C) Detail view of the bilaterian c-opsin clade, in which Hd-c-opsin1, Hd-c-opsin2 and Hd-c-opsin3 of H. dujardini are nested as a monophyletic group within the panarthropod c-opsin clade containing the arthropod pteropsins.
Sensitivity Analyses Using Leaf Stability
To identify “unstable” taxa in the reconstructed phylogeny of opsins, especially among the “early-branching” clades, we calculated the leaf stability indices (LS; Thorley and Wilkinson 1999) for each branch using Phyutility v2.2.6 (Smith and Dunn 2008), with 1,000 trees derived from RAxML bootstrapping as input, and mapped these LS indices on the corresponding ML tree using iTOL v2.1 (Letunic and Bork 2011) (LS indices; supplementary fig. S1, Supplementary Material online). Following this, we excluded those taxa from the original data set with LS indices ≤0.50 and ≤0.60, respectively, except for the ctenophore opsins (LS = 0.57) due to their importance for our study. These pruned data sets were aligned, masked and reanalyzed separately using RAxML v7.7.8 (GTR+G) with 1,000 bootstrap pseudoreplicates, including the a posteriori Bootstrap convergence assessment as described above for the full data set (supplementary figs. S3 and S5, Supplementary Material online). In contrast to the BI on the full data set, two Markov chains for each pruned data set were run for 30,000 instead of 60,000 generations using PhyloBayes MPI v1.4f (CAT-GTR). After the assessment of chain convergence, as described for the full data set (supplementary figs. S10 and S11, Supplementary Material online), the first 18,000 trees (LS≤0.50 data set) and 16,000 trees, respectively (LS≤0.60 data set), were discarded as burn-in and every tree thereafter was used to calculate the 50% majority rule consensus tree with posterior probability support values (supplementary figs. S4 and S6, Supplementary Material online).
Results
Identification of Opsin Genes in the Transcriptome of the Tardigrade H. dujardini
The sequencing of the H. dujardini transcriptome library with an Illumina Genome Analyzer IIx yielded 68,214,238 filtered paired-end reads. Our screening pipeline to search for putative opsin candidates included three different filtering stringencies of the raw data (Filter15, Filter25, Filter30), two different assemblers (CLC, IDBA), and two different search algorithms (BLAST, HMMER) (see the Materials and Methods section for more details). Sorting out the false positives resulted in a total of 43 contigs belonging to five different opsin genes. The sequences of two of them had to be extended by RACE. We successfully cloned partial sequences of all opsin transcripts, which we named after the clade in which they occur in our phylogenetic analyses (Hd-r-opsin: 703, Hd-c-opsin1: 664, Hd-c-opsin2: 704, Hd-c-opsin3: 702, Hd-neuropsin: 605 nt).
To verify our results and to check for additional opsin genes, we also screened the recently released genome of H. dujardini (http://badger.bio.ed.ac.uk/H_dujardini/, last accessed September 10, 2014). With the exception of a second, slightly different predicted transcript from the genome (nHd.2.3.1.g15325), which is most similar to our Hd-neuropsin and therefore perhaps an isoform of it, we found no evidence for any additional opsin genes apart from those already obtained from our transcriptomic data.
Phylogenetic Analyses of Metazoan Opsin Genes and the Placement of H. dujardini opsins
In our ML analysis of the full metazoan opsin data set, all three major bilaterian opsin subgroups were recovered, including r-opsins, c-opsins, and Group 4 opsins sensu Porter et al. (2012) (fig. 2 and supplementary fig. S1, Supplementary Material online). The best ML tree shows a monophyletic clade of the bilaterian c-type opsins with a weak BS (BS < 50), containing vertebrate visual pigments, chordate brain opsins (pinopsins, parapinopsins, vertebrate ancient opsins, teleost multiple tissue opsins, and encephalopsins), arthropod brain opsins (pteropsins), a brain opsin from the annelid Platynereis dumerilii (see Arendt et al. 2004), and a c-type opsin from the onychophoran Euperipatoides kanangrensis (see Eriksson et al. 2013) (fig. 2C and supplementary fig. S1, Supplementary Material online). Notably, none of the opsin sequences obtained from the trochozoan genomes, including the annelids C. teleta and He. robusta, and the mollusks L. gigantea and Cr. gigas, fall into this clade.
Within the bilaterian c-opsin clade, three of the five obtained opsin genes from the tardigrade H. dujardini (Hd-c-opsin1, Hd-c-opsin2, and Hd-c-opsin3) emerge as the sister group to all known c-opsins/pteropsins from arthropods. The tardigrade and arthropod c-opsins/pteropsins in turn cluster with the onychophoran c-opsin, altogether forming a well-supported monophyletic clade (BS = 85) of panarthropod c-opsins/pteropsins (fig. 2C and supplementary fig. S1, Supplementary Material online). Our analysis further revealed a highly supported clade (BS = 99) consisting exclusively of the anthozoan opsin genes from Nematostella vectensis and Acropora digitifera (hereafter referred to as “Anthozoa II”) as the sister group to the bilaterian c-opsins (supplementary fig. S1, Supplementary Material online). However, this relationship exhibits a low BS value (BS < 50), and our LS analysis shows that the position of Anthozoa II is unstable in the metazoan opsin tree (LS = 0.47).
A group of several uncharacterized opsins from the genomes of L. gigantea and Cr. gigas, the sequence of Sp-opsin2 from the sea urchin Strongylocentrotus purpuratus (see Raible et al. 2006), and the c-type opsin from the brachiopod Terebratalia transversa (see Passamaneck et al. 2011) was recovered as sister to the clade of the “cnidopsins” sensu Plachetzki et al. (2007, 2010) (hereafter used for all cnidarian opsins to the exclusion of the anthozoan opsins I and II; fig 2 and supplementary fig. S1, Supplementary Material online) and the ctenophoran opsins (hereafter referred to as “ctenopsins”), also with low nodal support (BS < 50). This clade in turn occurs as the sister group to the clade [Anthozoa II + c-opsins], although this relationship shows only weak bootstrap and LS values (BS < 50; LS ≤ 0.59). Interestingly, the two ctenopsins from both ctenophoran species included in the analyses (M. leidyi and Pleurobrachia pileus) are monophyletic with maximum support value (BS = 100), suggesting that they are ctenophoran in-paralogs.
The second major clade of the bilaterian opsins—Group 4 opsins sensu Porter et al. (2012)—includes peropsins, RGR opsins, Go-opsins, and an assemblage of neuropsins and “opsin-5” genes. The sister group relationship of Group 4 opsins to all above-mentioned c-type opsins, cnidopsins, Anthozoa II opsins, ctenopsins, and related opsins is weakly supported in our analyses (BS < 50; see fig. 2 and supplementary fig. S1, Supplementary Material online). Surprisingly, one of our obtained putative opsin genes from the tardigrade H. dujardini appears together with other protostome taxa as the sister group to the deuterostome neuropsins/opsin-5 group (Tarttelin et al. 2003), both forming a monophyletic clade (BS = 52) within the Group 4 opsins (fig. 2B). Besides the tardigrade sequence, this protostome neuropsin clade consists of single putative neuropsin homologs of D. pulex and Cr. gigas, two homologs of L. gigantea and six homologs of C. teleta based on their screened genomes.
A monophyletic clade of r-type opsins, the third major bilaterian opsin group, was recovered as the sister group to the clade [c-opsins (including related opsins) + Anthozoa II + cnidopsins + ctenopsins + Group 4 opsins] with strong BS (BS = 85). Compared with the c-opsin and Group 4 opsin clades, the r-opsins occur as the best-supported and the earliest-branching clade. Moreover, the leaf stabilities of the r-opsins are by far the highest among all taxa included (LS ≥ 78), exceeded only by the outgroup taxa (supplementary fig. S1, Supplementary Material online). Within the r-opsin clade, the fifth opsin candidate from the tardigrade H. dujardini occurs as sister to the visual r-opsins of Onychophora (onychopsin genes sensu Hering et al. 2012), which together form a well-supported monophyletic clade (BS = 97) (fig. 2A and supplementary fig. S1, Supplementary Material online). The r-opsins of the Tardigrada + Onychophora clade emerge as the closest relatives to a highly diverse group of visual r-opsins of Arthropoda. These arthropod r-opsins are subdivided into two major subgroups that to some extent reflect their spectral sensitivity, that is, ultraviolet (UV) and short-wavelength sensitive versus medium- and long-wavelength sensitive opsins. The tardigrade r-opsin is unlikely to be a UV-sensitive visual pigment, as it has methionine (M) instead of lysine (K) at the position corresponding to G90 of the bovine rhodopsin (Palczewski et al. 2000), which has been shown to be responsible for UV tuning properties of arthropod opsins (Salcedo et al. 2003).
In addition to the panarthropod visual r-opsins, the rhabdomeric opsins are further subdivided into three monophyletic clades according to our analyses: 1) The lophotrochozoan (most likely visual) r-opsins, 2) the chordate nonvisual r-opsins (=melanopsins), and 3) the arthropsins (initially described by Colbourne et al. 2011) that form the earliest-branching subgroup. Most intriguingly, in addition to the eight arthropsin paralogs described from the crustacean D. pulex and the two putative arthropsins from the onychophoran E. kanangrensis and the spider Cupiennius salei (see Colbourne et al. 2011; Eriksson et al. 2013), we identified putative arthropsin sequences in the genomes of the pea aphid A. pisum as well as the annelid C. teleta and the mollusks L. gigantea and Cr. gigas. Moreover, even r-opsin4 from the annelid P. dumerilii (see Randel et al. 2013) and Amphiop6 from the lancelet Branchiostoma belcheri (see Koyanagi et al. 2002) occur as members of the arthropsin clade in our cladograms (supplementary fig. S1, Supplementary Material online).
The least stable taxa in our full data set ML analysis (LS ≤ 0.46) are the Sp-opsin5 sequences from two sea urchins (Raible et al. 2006; Lesser et al. 2011) and the Go-opsin2 sequence from the brachiopod T. transversa (see Passamaneck and Martindale 2013) that branch off at the base of the tree as well as a second clade of anthozoan-specific opsins (hereafter referred to as Anthozoa I opsins) containing two paralogs from the genome of N. vectensis and the acropsin3 sequences (Mason et al. 2012) from two Acropora species. Sp-opsin5 and its ortholog from Strongylocentrotus droebachiensis have been classified as rhabdomeric opsins (Lesser et al. 2011), but these opsins clearly cluster outside the r-opsin clade in our and Lesser et al.’s (2011) phylogenetic analyses. In contrast, according to our results, Sp-opsin4 from S. purpuratus is deeply nested within the r-opsin clade (supplementary fig. S1, Supplementary Material online).
The 50% majority rule consensus tree of our BI analysis of the full metazoan opsin data set revealed a similar topology to our ML analysis, except that the Anthozoa II clade is deeply nested within the c-opsins clade. However, as mentioned above, the Anthozoa II opsins show low LS indices (LS = 0.47) (supplementary fig. S2, Supplementary Material online). The most conspicuous deviation from the ML tree is the unresolved topology at the base of the tree, indicated by polytomous branches of the particular opsin clades (bilaterian c-opsins, r-opsins, Group 4 opsins, cnidopsins, ctenopsins, and Anthozoa I). Nevertheless, the placement of the tardigrade opsins in the BI analysis corresponds to that in the ML tree. The same holds true for the ML and BI analyses, from which we excluded the putatively unstable taxa with LS indices LS ≤ 0.50 and LS ≤ 0.60, respectively (supplementary figs. S3–S6, Supplementary Material online).
Discussion
Opsin Repertoire in the Tardigrade H. dujardini
Our transcriptomic analyses of the opsin repertoire in the tardigrade H. dujardini revealed a set of five opsin genes, including one r-opsin (Hd-r-opsin), three c-opsins (Hd-c-opsin1, Hd-c-opsin2, and Hd-c-opsin3), and a neuropsin/opsin-5 (Hd-neuropsin). We found essentially the same set of genes in the publicly available genome of this species, with the exception of an additional neuropsin/opsin-5 isoform. However, this isoform is unlikely to be a functional gene, as its expression could not be confirmed by the transcriptomic analyses and gene cloning. Moreover, we have noticed some inconsistencies between our cloned transcripts on the one hand and the genomic sequences and predicted transcripts on the other hand, in which short fragments from putatively expressed regions are either absent or repeated (supplementary fig. S7, Supplementary Material online). This suggests that either the genomic contigs and/or scaffolds were assembled incorrectly or, if the assembly was correct and the introns indeed contain such repeated exonic sequences, the actual splice sites were predicted incorrectly during the gene annotation process. It is therefore unclear whether the additional neuropsin/opsin-5 isoform is a real, albeit nonfunctional sequence, or an assembly artifact. Nevertheless, irrespective of these inconsistencies and minor differences in nucleotide composition (probably due to heterozygosity), the available genome sequences correspond well to our transcriptomic data, suggesting that H. dujardini has at least five functional opsin genes. According to our phylogenetic analyses, these genes cluster within the three major bilaterian opsin clades.
Evidence for Monochromatic Vision in the Last Common Ancestor of Panarthropoda
Typically, either r-opsins (as in arthropods) or c-opsins (as in vertebrates) are involved in animal vision (e.g., Arendt and Wittbrodt 2001; Arendt 2003; Vopalensky and Kozmik 2009; Land and Nilsson 2012). Given that Tardigrada is one of the closest arthropod relatives, one would expect that H. dujardini employs the single identified r-opsin as the sole visual pigment, suggesting monochromatic vision in this tardigrade species. However, in addition to numerous microvilli that may act as rhabdomeric photoreceptive structures, at least one ciliary cell has been reported from the eye of Milnesium tardigradum and Halobiotus crispae (see Kristensen 1983; Dewel et al. 1993; Greven 2007). Although the cilia described from Mi. tardigradum and Ha. crispae are unlikely to be involved in photoreception, a potential function of the three identified c-opsins in H. dujardini in visual photoreception and, hence, in color vision cannot be excluded as long as the corresponding gene expression data are unavailable. However, irrespective of the function of these genes, our phylogenetic analyses suggest that the three c-opsin genes have evolved by gene duplication in the tardigrade lineage (or a tardigrade subgroup) and that the last common ancestor of Panarthropoda possessed only one c-opsin gene (fig. 3A). This finding and the identification of a single r-opsin gene in H. dujardini are in line with the assumption of monochromatic vision in the last common ancestor of Onychophora, Tardigrada, and Arthropoda (Hering et al. 2012).
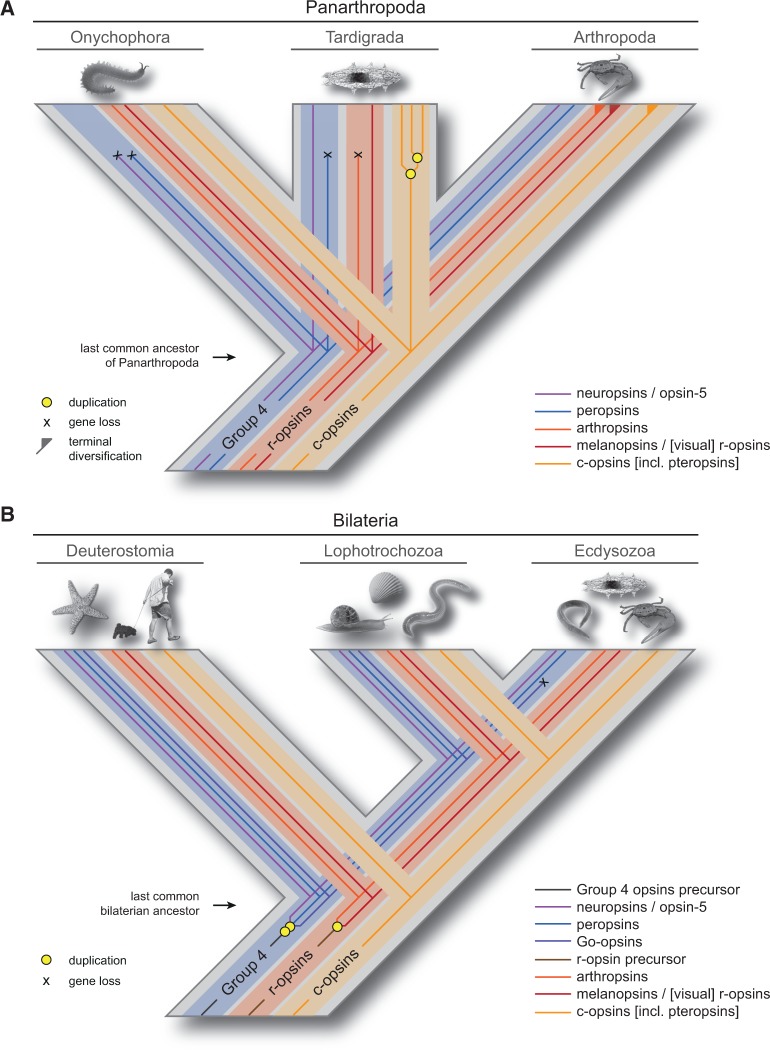
Fig. 3.—
Hypotheses on the evolutionary history of opsin genes in Panarthropoda and Bilateria based on our phylogenetic analyses of metazoan opsin sequences. (A) Scenario on opsin evolution in Panarthropoda. The trichotomy is due to the uncertain position of Tardigrada as the sister group of Arthropoda, Onychophora, or Onychophora plus Arthropoda (see Mayer et al. 2013 and references therein). According to this scenario, the last common ancestor of Panarthropoda possessed five opsin genes: One c-opsin, two r-opsins, and two Group 4 opsins. The three c-opsin genes of H. dujardini might have evolved by gene duplications either in the tardigrade lineage, or a tardigrade subgroup. (B) Scenario on opsin evolution in Bilateria, according to which the last common bilaterian ancestor possessed six opsin genes. Although the arthropsin and melanopsin/[visual] r-opsin lineages arose by a duplication of the r-opsin precursor, two additional duplications in the bilaterian Group 4 opsin clade gave rise to the neuropsin/opsin-5, peropsin, and Go-opsin lineages. Note that the Go-opsin gene was subsequently lost in the last common ancestor of Ecdysozoa.
Neuropsins Were Present in the Last Common Bilaterian Ancestor
Our phylogenetic analyses revealed that at least one opsin gene of H. dujardini belongs to the neuropsin/opsin-5 clade—a subgroup of the Group 4 opsins. To our knowledge, this is the first report of a neuropsin from a protostome, as this type of opsin so far has been known only from vertebrates (review Koyanagi and Terakita 2013). In addition to the tardigrade neuropsin, we identified up to six putative neuropsin homologs in the genomes of other protostomes, including the annelid C. teleta, the mollusks Cr. gigas and L. gigantea, and the crustacean D. pulex. The latter is most astonishing because of its well-characterized opsin repertoire of hitherto 46 identified opsin genes (Colbourne et al. 2011). The identification of neuropsin/opsin-5 members in representatives of Lophotrochozoa, Ecdysozoa, and Deuterostomia suggests an origin of this opsin lineage prior to the split Deuterostomia + Protostomia (fig. 3B). While the function of neuropsin/opsin-5 homologs in protostomes is unknown, the vertebrate Opsin-5 (Opn5) homolog acts as a UV-sensitive GPCR, which activates the Gi-type G proteins (Yamashita et al. 2010) and is expressed in various tissues, including the neural retina, deep brain, testis, and even the outer ear (Tarttelin et al. 2003; Nakane et al. 2010; Kojima et al. 2011; Yamashita et al. 2014). In light of our findings, it would be interesting to know whether the protostome neuropsin/opsin-5 homologs also have a function as UV-sensitive GPCRs or rather act as retinal photoisomerases, similar to the closely related peropsins (Koyanagi et al. 2002).
“Arthropsins” Are Not Restricted to Arthropods
Arthropsins were initially described from the genome of the crustacean D. pulex, in which eight members of this putatively novel opsin group clustered as an early-branching clade within the r-opsins (Colbourne et al. 2011). Interestingly, another early-branching r-opsin, Amphiop6—which was identified previously in the chordate B. belcheri and did not group with members of the melanopsin clade (Koyanagi et al. 2005)—was subsequently recovered as sister to the arthropsins (Hering et al. 2012). Moreover, Randel et al. (2013) identified “another stable r-opsin clade (Clade I) with both mollusk and annelid sequences,” the phylogenetic relationship of which remained unclear, as neither the Daphnia arthropsins nor Amphiop6 were included in their study. The results of our phylogenetic analyses now show that all of these sequences, including Randel et al.’s (2013) “Clade I” and additional members from other bilaterian taxa, form a monophyletic group, which thus includes representatives of Onychophora, Arthropoda, Lophotrochozoa, and Chordata. This implies that arthropsins are not restricted to arthropods but are the result of an ancient duplication of the r-opsin precursor in the last common bilaterian ancestor (fig. 3B). Although additional duplications have occurred in some lineages (e.g., in D. pulex; see Colbourne et al. 2011), an arthropsin homolog might have been lost in the tardigrade linage, as it is not found in the transcriptomic or genomic data from H. dujardini (fig. 3A).
Conclusions
To determine the position of the tardigrade opsins, we performed an extensive phylogenetic analysis of broadly sampled metazoan opsins, including newly identified members from the available metazoan genomes. Our results suggest that the last common bilaterian ancestor possessed six opsins belonging to the three major opsin clades (fig. 3B): 1) Two r-opsins (one of which was an arthropsin); 2) one c-opsin; and 3) three Group 4 opsins (including a neuropsin, a peropsin, and a Go opsin that was most likely lost in the ecdysozoan lineage). Because none of the opsins from the nonbilaterian taxa falls into any of these major clades, we suggest that a single duplication of the r-opsin precursor and two duplications within the Group 4 opsin clade have occurred within the bilaterian lineage (fig. 3B). Unfortunately, beyond this no unambiguous conclusion is possible on the origin and relationship of the three major bilaterian opsin clades, possibly due to the lack of sufficient phylogenetic signal to robustly resolve the deepest nodes of the opsin gene tree. This, in conjunction with the generally ambiguous placement of the nonbilaterian taxa, such as Cnidaria and Ctenophora within the metazoans (review Philippe et al. 2011), makes it a challenging task to draw any conclusions about prebilaterian opsin evolution.
Our analyses revealed three clades of cnidarian opsins: Anthozoa I, Anthozoa II, and cnidopsins (sensu Plachetzki et al. 2007). Although the Anthozoa I and II clades exclusively contain anthozoan sequences, the cnidopsins comprise representatives of Anthozoa, Cubozoa, and Hydrozoa, supporting the results of previous studies (Suga et al. 2008; Plachetzki et al. 2010; Porter et al. 2012). However, the placement of the cnidopsins differs among these studies, in which they form the sister group to various clades, including [r-opsins + Group 4 opsins] (Plachetzki et al. 2007), [r-opsins + c-opsins + Group 4 opsins] (Plachetzki et al. 2010), [Group 4 opsins] (Feuda et al. 2012), and [c-opsins] (Suga et al. 2008; Hering et al. 2012; Porter et al. 2012), depending on the underlying reconstruction method and the substitution model used. In our analyses, the cnidopsins generally form the sister group of the ctenophoran opsins (=ctenopsins), but the placement of the entire cnidopsins/ctenopsins clade is ambiguous and depends on the analysis parameters used. The same applies to the Arthropsin I and II clades, the position of which is unstable.
These discrepancies between the studies and the methods used are not surprising, given the old age of the major metazoan lineages, dating back to approximately 700 Ma, and the cladogenesis events that were highly compressed in time (e.g., Rokas et al. 2005). This might have led to a considerable accumulation of homoplasies and, hence, to an erosion of the phylogenetic signal in the molecular data. Therefore, currently it seems impossible to reconstruct with confidence the early-branching nodes and to reconcile a reliable scenario on the evolutionary history of the metazoan opsins based solely on opsin phylogeny. Nevertheless, our phylogenetic framework allows for the following conclusions on the evolution of opsins in panarthropods:
The last common ancestor of Panarthropoda most likely possessed a c-opsin, two r-opsins (an arthropsin and an additional [probably visual] r-opsin, which was not UV-sensitive), and two Group 4 opsins (a neuropsin and a peropsin)—a set that had been inherited from the last common ancestor of Ecdysozoa (fig. 3A and B).
This ancestral set of opsin genes was retained in the last common ancestor of Arthropoda (fig. 3A), although subsequent duplications and losses occurred in some arthropod lineages (e.g., Briscoe 2000; Colbourne et al. 2011; Porter et al. 2013).
The last common ancestor of Onychophora had retained a c-opsin and two r-opsins (arthropsin and onychopsin; see Hering et al. 2012; Eriksson et al. 2013) from the panarthropod ancestor, whereas the two Group 4 opsins (neuropsin and peropsin) were lost in the onychophoran lineage (fig. 3A). However, this hypothesis requires confirmation by genomic analyses.
The last common ancestor of Tardigrada (or the tardigrade subgroup containing H. dujardini) most likely possessed an r-opsin, three c-opsins, and a neuropsin, whereas the arthropsin and the peropsin were lost in the tardigrade lineage (fig. 3A).
Supplementary Material
Supplementary figures S1–S11 and tables S1–S3 are available at Genome Biology and Evolution online (http://gbe.oxfordjournals.org/).
Acknowledgments
The authors are thankful to Matthias Meyer, Birgit Nickel and Martin Kircher for assistance with library preparation and sequencing, Vladimir Gross for proofreading, and Sandy Richter and Christoph Bleidorn for comments on the manuscript. We acknowledge support from the German Research Foundation (DFG) and Leipzig University within the program of Open Access Publishing. This work was supported by a grant from the German Research Foundation (DFG; grant Ma 4147/3-1) to G.M., who is a Research Group Leader supported by the Emmy Noether Programme of the DFG.
Literature Cited
- Akaike H. A new look at the statistical model identification. IEEE T Automat Control. 1974;19:716–723. [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D. Evolution of eyes and photoreceptor cell types. Int J Dev Biol. 2003;47:563–571. [PubMed] [Google Scholar]
- Arendt D, Tessmar-Raible K, Snyman H, Dorresteijn AW, Wittbrodt J. Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science. 2004;306:869–871. doi: 10.1126/science.1099955. [DOI] [PubMed] [Google Scholar]
- Arendt D, Wittbrodt J. Reconstructing the eyes of Urbilateria. Philos Trans R Soc B Lond Biol Sci. 2001;356:1545–1563. doi: 10.1098/rstb.2001.0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann H. Der Lebenslauf von Hypsibius (H.) convergens Urbanowicz (Tardigrada) Zool Anz. 1961;167:362–381. [Google Scholar]
- Beasley CW. Photokinesis of Macrobiotus hufelandi (Tardigrada, Eutardigrada) Zool Anz. 2001;240:233–236. [Google Scholar]
- Briscoe AD. Six opsins from the butterfly Papilio glaucus: molecular phylogenetic evidence for paralogous origins of red-sensitive visual pigments in insects. J Mol Evol. 2000;51:110–121. doi: 10.1007/s002390010071. [DOI] [PubMed] [Google Scholar]
- Briscoe AD, Chittka L. The evolution of color vision in insects. Annu Rev Entomol. 2001;46:471–510. doi: 10.1146/annurev.ento.46.1.471. [DOI] [PubMed] [Google Scholar]
- Colbourne JK, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewel RA, Nelson DR, Dewel WC. Tardigrada. In: Harrison FW, Rice ME, editors. Microscopic anatomy of lnvertebrates: Onychophora, Chilopoda, and lesser Protostomata. New York: Wiley-Liss; 1993. pp. 143–183. [Google Scholar]
- Dress A, et al. Noisy: identification of problematic columns in multiple sequence alignments. Alg Mol Biol. 2008;3:7. doi: 10.1186/1748-7188-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Eriksson BJ, Fredman D, Steiner G, Schmid A. Characterisation and localisation of the opsin protein repertoire in the brain and retinas of a spider and an onychophoran. BMC Evol Biol. 2013;13:186. doi: 10.1186/1471-2148-13-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fain GL, Hardie R, Laughlin SB. Phototransduction and the evolution of photoreceptors. Curr Biol. 2010;20:R114–R124. doi: 10.1016/j.cub.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuda R, Hamilton SC, McInerney JO, Pisani D. Metazoan opsin evolution reveals a simple route to animal vision. Proc Natl Acad Sci U S A. 2012;109:18868–18872. doi: 10.1073/pnas.1204609109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greven H. Comments on the eyes of tardigrades. Arthropod Struct Dev. 2007;36:401–407. doi: 10.1016/j.asd.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hankins MW, Peirson SN, Foster RG. Melanopsin: an exciting photopigment. Trends Neurosci. 2008;31:27–36. doi: 10.1016/j.tins.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Henze M, Dannenhauer K, Kohler M, Labhart T, Gesemann M. Opsin evolution and expression in arthropod compound eyes and ocelli: insights from the cricket Gryllus bimaculatus. BMC Evol Biol. 2012;12:163. doi: 10.1186/1471-2148-12-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering L, et al. Opsins in Onychophora (velvet worms) suggest a single origin and subsequent diversification of visual pigments in arthropods. Mol Biol Evol. 2012;29:3451–3458. doi: 10.1093/molbev/mss148. [DOI] [PubMed] [Google Scholar]
- Hurvich CM, Tsai C-L. Regression and time series model selection in small samples. Biometrika. 1989;76:297–307. [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Sawyer S, Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012;40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima D, et al. UV-sensitive photoreceptor protein OPN5 in humans and mice. PLoS One. 2011;6:e26388. doi: 10.1371/journal.pone.0026388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi M, Kubokawa K, Tsukamoto H, Shichida Y, Terakita A. Cephalochordate melanopsin: evolutionary linkage between invertebrate visual cells and vertebrate photosensitive retinal ganglion cells. Curr Biol. 2005;15:1065–1069. doi: 10.1016/j.cub.2005.04.063. [DOI] [PubMed] [Google Scholar]
- Koyanagi M, Nagata T, Katoh K, Yamashita S, Tokunaga F. Molecular evolution of arthropod color vision deduced from multiple opsin genes of jumping spiders. J Mol Evol. 2008;66:130–137. doi: 10.1007/s00239-008-9065-9. [DOI] [PubMed] [Google Scholar]
- Koyanagi M, Terakita A. Diversity of animal opsin-based pigments and their optogenetic potential. Biochim Biophys Acta. 2013;1837:710–716. doi: 10.1016/j.bbabio.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Koyanagi M, Terakita A, Kubokawa K, Shichida Y. Amphioxus homologs of Go-coupled rhodopsin and peropsin having 11-cis- and all-trans-retinals as their chromophores. FEBS Lett. 2002;531:525–528. doi: 10.1016/s0014-5793(02)03616-5. [DOI] [PubMed] [Google Scholar]
- Kristensen RM. The first record of cyclomorphosis in Tardigrada based on a new genus and species from Arctic meiobenthos. J Zool Syst Evol Res. 1983;20:249–270. [Google Scholar]
- Kück P, et al. Parametric and non-parametric masking of randomness in sequence alignments can be improved and leads to better resolved trees. Front Zool. 2010;7:10. doi: 10.1186/1742-9994-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land MF, Nilsson D-E. Animal eyes. New York: Oxford University Press; 2012. [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- Lesser MP, Carleton KL, Böttger SA, Barry TM, Walker CW. Sea urchin tube feet are photosensory organs that express a rhabdomeric-like opsin and PAX6. Proc R Soc B Biol Sci. 2011;278:3371–3379. doi: 10.1098/rspb.2011.0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus E. Tardigrada. In: Bronn HG, editor. Klassen und ordnungen des tierreichs. Leipzig: Akademische Verlagsgesellschaft m.b.H; 1929. p. 608. [Google Scholar]
- Marshall J, Cronin TW, Kleinlogel S. Stomatopod eye structure and function: a review. Arthropod Struct Dev. 2007;36:420–448. doi: 10.1016/j.asd.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Mason B, et al. Evidence for multiple phototransduction pathways in a reef-building coral. PLoS One. 2012;7:e50371. doi: 10.1371/journal.pone.0050371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer G. Structure and development of onychophoran eyes: what is the ancestral visual organ in arthropods? Arthropod Struct Dev. 2006;35:231–245. doi: 10.1016/j.asd.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Mayer G, et al. Selective neuronal staining in tardigrades and onychophorans provides insights into the evolution of segmental ganglia in panarthropods. BMC Evol Biol. 2013;13:230. doi: 10.1186/1471-2148-13-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010 (6), doi:10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- Minin V, Abdo Z, Joyce P, Sullivan J. Performance-based selection of likelihood models for phylogeny estimation. Syst Biol. 2003;52:674–683. doi: 10.1080/10635150390235494. [DOI] [PubMed] [Google Scholar]
- Misof B, Misof K. A Monte Carlo approach successfully identifies randomness in multiple sequence alignments: a more objective means of data exclusion. Syst Biol. 2009;58:21–34. doi: 10.1093/sysbio/syp006. [DOI] [PubMed] [Google Scholar]
- Nakane Y, et al. A mammalian neural tissue opsin (Opsin 5) is a deep brain photoreceptor in birds. Proc Natl Acad Sci U S A. 2010;107:15264–15268. doi: 10.1073/pnas.1006393107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Passamaneck Y, Furchheim N, Hejnol A, Martindale M, Luter C. Ciliary photoreceptors in the cerebral eyes of a protostome larva. EvoDevo. 2011;2:6. doi: 10.1186/2041-9139-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passamaneck YJ, Martindale MQ. Evidence for a phototransduction cascade in an early brachiopod embryo. Integr Comp Biol. 2013;53:17–26. doi: 10.1093/icb/ict037. [DOI] [PubMed] [Google Scholar]
- Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? In: Batzoglou S, editor. Research in computational molecular biology. Berlin, Heidelberg: Springer; 2009. pp. 184–200. [DOI] [PubMed] [Google Scholar]
- Peng Y, et al. IDBA-tran: a more robust de novo de Bruijn graph assembler for transcriptomes with uneven expression levels. Bioinformatics. 2013;29:i326–i334. doi: 10.1093/bioinformatics/btt219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, et al. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 2011;9:e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachetzki DC, Degnan BM, Oakley TH. The origins of novel protein interactions during animal opsin evolution. PLoS One. 2007;2:e1054. doi: 10.1371/journal.pone.0001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachetzki DC, Fong CR, Oakley TH. The evolution of phototransduction from an ancestral cyclic nucleotide gated pathway. Proc R Soc B Biol Sci. 2010;277:1963–1969. doi: 10.1098/rspb.2009.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ML, et al. Shedding new light on opsin evolution. Proc R Soc B Biol Sci. 2012;279:3–14. doi: 10.1098/rspb.2011.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ML, et al. The evolution of complexity in the visual systems of stomatopods: insights from transcriptomics. Integr Comp Biol. 2013;53:39–49. doi: 10.1093/icb/ict060. [DOI] [PubMed] [Google Scholar]
- Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raible F, et al. Opsins and clusters of sensory G-protein-coupled receptors in the sea urchin genome. Dev Biol. 2006;300:461–475. doi: 10.1016/j.ydbio.2006.08.070. [DOI] [PubMed] [Google Scholar]
- Ramazzotti G, Maucci W. Verbania Pallanza: Memorie dell'Istituto Italiano di Idrobiologia Dott. Marco De Marchi; 1983. Il Phylum Tardigrada. [Google Scholar]
- Rambaut A, Drummond AJ. 2009. Tracer v1.5. [cited 2014 Jan 1]. Available from: http://tree.bio.ed.ac.uk/software/tracer/ [Google Scholar]
- Randel N, Bezares-Calderón LA, Gühmann M, Shahidi R, Jékely G. Expression dynamics and protein localization of rhabdomeric opsins in Platynereis larvae. Integr Comp Biol. 2013;53:7–16. doi: 10.1093/icb/ict046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Krüger D, Carroll SB. Animal evolution and the molecular signature of radiations compressed in time. Science. 2005;310:1933–1938. doi: 10.1126/science.1116759. [DOI] [PubMed] [Google Scholar]
- Rubin EB, et al. Molecular and phylogenetic analyses reveal mammalian-like clockwork in the honey bee (Apis mellifera) and shed new light on the molecular evolution of the circadian clock. Genome Res. 2006;16:1352–1365. doi: 10.1101/gr.5094806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo E, Zheng L, Phistry M, Bagg EE, Britt SG. Molecular basis for ultraviolet vision in invertebrates. J Neurosci. 2003;23:10873–10878. doi: 10.1523/JNEUROSCI.23-34-10873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzler C, et al. Genomic organization, evolution, and expression of photoprotein and opsin genes in Mnemiopsis leidyi: a new view of ctenophore photocytes. BMC Biol. 2012;10:107. doi: 10.1186/1741-7007-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G. Estimating the dimension of a model. Ann Stat. 1978;6:461–464. [Google Scholar]
- Shichida Y, Imai H. Visual pigment: G-protein-coupled receptor for light signals. Cell Mol Life Sci. 1998;54:1299–1315. doi: 10.1007/s000180050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichida Y, Matsuyama T. Evolution of opsins and phototransduction. Philos Trans R Soc B Lond Biol Sci. 2009;364:2881–2895. doi: 10.1098/rstb.2009.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Dunn CW. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics. 2008;24:715–716. doi: 10.1093/bioinformatics/btm619. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Suga H, Schmid V, Gehring WJ. Evolution and functional diversity of jellyfish opsins. Curr Biol. 2008;18:51–55. doi: 10.1016/j.cub.2007.11.059. [DOI] [PubMed] [Google Scholar]
- Sugiura N. Further analysis of the data by Akaike's information criterion and the finite corrections. Commun Stat Theory Methods. 1978;7:13–26. [Google Scholar]
- Tarttelin EE, Bellingham J, Hankins MW, Foster RG, Lucas RJ. Neuropsin (Opn5): a novel opsin identified in mammalian neural tissue. FEBS Lett. 2003;554:410–416. doi: 10.1016/s0014-5793(03)01212-2. [DOI] [PubMed] [Google Scholar]
- Thorley JL, Wilkinson M. Testing the phylogenetic stability of early tetrapods. J Theor Biol. 1999;200:343–344. doi: 10.1006/jtbi.1999.0999. [DOI] [PubMed] [Google Scholar]
- Velarde RA, Sauer CD, O. Walden KK, Fahrbach SE, Robertson HM. Pteropsin: a vertebrate-like non-visual opsin expressed in the honey bee brain. Insect Biochem Mol Biol. 2005;35:1367–1377. doi: 10.1016/j.ibmb.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Vopalensky P, Kozmik Z. Eye evolution: common use and independent recruitment of genetic components. Philos Trans R Soc B Lond Biol Sci. 2009;364:2819–2832. doi: 10.1098/rstb.2009.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, et al. Opn5 is a UV-sensitive bistable pigment that couples with Gi subtype of G protein. Proc Natl Acad Sci U S A. 2010;107:22084–22089. doi: 10.1073/pnas.1012498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, et al. Evolution of mammalian Opn5 as a specialized UV-absorbing pigment by a single amino acid mutation. J Biol Chem. 2014;289:3991–4000. doi: 10.1074/jbc.M113.514075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.