Abstract
Campylobacter is the most common bacterial cause of gastroenteritis in the European Union with over 200,000 laboratory-confirmed cases reported annually. This is the first study to describe findings related to comparative genomics analyses of the sequence type (ST)-677 clonal complex (CC), a Campylobacter jejuni lineage associated with bacteremia cases in humans. We performed whole-genome sequencing, using Illumina HiSeq sequencing technology, on five related ST-677 CC isolates from two chicken farms to identify microevolution taking place at the farms. Our further aim was to identify novel putative virulence determinants from the ST-677 CC genomes. For this purpose, clinical isolates of the same CC were included in comparative genomic analyses against well-known reference strains of C. jejuni. Overall, the ST-677 CC was recognized as a highly clonal lineage with relatively small differences between the genomes. Among the farm isolates differences were identified mainly in the lengths of the homopolymeric tracts in genes related to the capsule, lipo-oligosaccharide, and flagella. We identified genomic features shared with C. jejuni subsp. doylei, which has also been shown to be associated with bacteremia in humans. These included the degradation of the cytolethal distending toxin operon and similarities between the capsular polysaccharide biosynthesis loci. The phase-variable GDP-mannose 4,6-dehydratase (EC 4.2.1.47) (wcbK, CAMP1649), associated with the capsular polysaccharide biosynthesis locus, may play a central role in ST-677 CC conferring acid and serum resistance during different stages of infection. Homology-based searches revealed several additional novel features and characteristics, including two putative type Vb secretion systems and a novel restriction modification/methyltransferase gene cluster, putatively associated with pathogenesis and niche adaptation.
Keywords: Campylobacter jejuni, capsule, phase variation, serum resistance, niche adaptation, bacteremia
Introduction
Campylobacter is by far the most common bacterial cause of gastroenteritis in Finland, with an incidence of around 78/100,000 inhabitants (Kuusi and Nakari 2012). Campylobacter jejuni is the main species identified. In the European Union (EU), 214,268 human cases were reported in 2012 (EFSA and ECDC 2014). The estimated costs of campylobacteriosis to public health systems in the EU are around 2.4 billion euros per year (EFSA Panel on Biological Hazards [BIOHAZ] 2011). Campylobacteriosis occurs mostly as a self-limiting noncomplicated infection; however, in some instances the infection may lead to a more severe outcome, for example, bacteremia, and such postinfection complications as reactive arthritis and Guillain–Barré syndrome (Blaser 1997).
The two most prevalent clonal complexes (CCs), defined by multilocus sequence typing (MLST), in the past 10 years have been ST-45 and ST-21 CC, representing 44% and 19% of the clinical isolates among domestically acquired infections in Finland, respectively (de Haan et al. 2010). However, the third most common CC, the ST-677, occurred in nearly 12% of the isolates. ST-677 CC consists of a limited number of STs, mainly ST-677 and ST-794, which share only a few alleles with other CCs. In our previous study, ST-677 CC was found to be associated with patients drinking nonchlorinated water from a small water plant or water from natural sources (Kärenlampi et al. 2007). Later on, ST-677 CC has been associated with wild birds, rabbits, and environmental sources in northwest England (Kwan et al. 2008). However, it does not seem to be limited to any particular animal host, as it has been recovered from chicken, cattle and surface waters in Finland (de Haan et al. 2014) and from chicken, cattle, wild birds, wild rabbits, environmental water and diarrheic dogs from other parts of Europe (Kwan et al. 2008; Griekspoor et al. 2010; Kittl et al. 2013). In 2007, ST-677 CC was the second most common CC among chicken isolates in Finland (de Haan et al. 2010).
Interestingly, isolates of the ST-677 CC were found to be more common among patients requiring hospitalization and a longer stay in the hospital, suggesting a link with a more severe disease (Kärenlampi et al. 2007). Moreover, a recent study revealed that ST-677 CC was associated with bacteremia in Finnish patients (during 1998–2007) (Feodoroff et al. 2013). In that study, isolates representing the ST-677 CC were also shown to be significantly more resistant to normal human serum than all other isolates. Earlier, we demonstrated that ST-677 CC and ST-48 CC were associated with Penner serotype HS4 complex (Kärenlampi et al. 2007). Furthermore, C. jejuni HS4 strains have been shown to occur in higher proportions in blood isolates than in fecal isolates, suggesting that these strains are likely to be more serum-resistant and/or invasive (Skirrow et al. 1993). These results suggest that serum resistance and invasiveness are at least partly associated with the capsular polysaccharide (CPS), which is the major determinant in Penner serotyping of C. jejuni (Karlyshev et al. 2000). Other bacterial and host genetic determinants leading to bacteremia and sepsis are largely unknown.
To date, scant information exists on the genomic differences between epidemiologically linked C. jejuni isolates during natural infection cycles. This information is needed for use in genomic epidemiology when assessing criteria to include/exclude isolates in an outbreak investigation. To address these issues, we sequenced two sets of related strains representing the ST-677 CC to evaluate the genome microevolution taking place on a chicken farm and to identify genetic features and characteristics potentially explaining the more severe disease associated with ST-677 CC in human patients. To support our data, we included in the comparative analysis six whole-genome shotgun sequences of epidemiologically unrelated clinical isolates of ST-677 CC from Sweden and the United Kingdom, representing different isolation times and geographical areas.
Materials and Methods
Bacterial Strains
As part of a study aimed at deciphering the risk factors for campylobacter colonization at broiler chicken farms in Finland, fresh fecal or cloacal swabs from chicken and boot sock samples were collected simultaneously from the grounds inside and outside of chicken houses, including water samples from puddles on the grounds. Approximately, 100 farms that had previously produced Campylobacter-positive flocks were investigated. The C. jejuni isolates were subsequently typed by pulsed-field gel electrophoresis (PFGE). On five occasions, C. jejuni was detected both inside the house and from environmental samples. On two farms, the PFGE profiles of temporally associated isolates were the same and closely related with each other, and these isolates were selected for further studies.
The isolates from farm A (date of sampling: September 9, 2004) were from a fresh fecal swab inside the chicken house (5070, ST-677), a boot sock sample from inside the chicken house (5071, ST-677), and a sample from a rain water puddle outside the chicken house (5072, ST-677). On farm C (date of sampling: August 13, 2003), the isolates were from a cloacal swab (3515, ST-794) and an environmental boot sock sample collected from the grounds outside the chicken house (3516, ST-794).
Draft genomes retrieved from public databases and included for comparative studies comprised strain LMG9872 (ST-677, accession number NZ_AIPM00000000) isolated from the cerebrospinal fluid of a meningitis patient in Sweden in July 1979 (Norrby et al. 1980) and five genomes of clinical ST-677 CC isolates collected from 2010 to 2012 from gastroenteritis patients in the United Kingdom that were available in the Campylobacter PubMLST database (ST-677: OXC6332, OXC7095, OXC7345, OXC7358; ST-794: OXC5341).
Whole-Genome Sequencing and Assembly
DNA was prepared from single-colony isolates using the Wizard genomic DNA purification kit (Promega, Mannheim, Germany). The genome sequences were determined using a paired-end library and Illumina HiSeq sequencing technology with 100 cycles (Base-Clear BV, Leiden, The Netherlands). The sequence coverage for the C. jejuni genomes was >500-fold. The reads were filtered using the ConDeTri Perl script (Smeds and Kunstner 2011) using default settings and a minimum read length of 75 nucleotides. Only sequences passing the quality threshold in both paired reads were assembled into contigs using ABySS 1.3.5 (Simpson et al. 2009). In specific regions of interest, gaps in the genome of isolate 5070 were closed by traditional Sanger sequencing. The genome assembly was edited using gap5 (Staden et al. 2000; Bonfield and Whitwham 2010) of the Staden Package (Staden et al. 2000).
Whole-Genome MLST
Gene-by-gene population annotation and analysis for whole-genome MLST (wgMLST) was performed using Bacterial Isolate Genome Sequence Database (BIGSdb). Allelic profiles for all of the isolates were retrieved for each locus defined in the database. The NeighborNet algorithm was used to construct a distance matrix from the shared loci of the isolates, and the resulting phylogenetic network was visualized using SplitsTree4 (Huson and Bryant 2006).
Genome Annotation and Comparative Genomics Analyses
Gene calling and functional annotation were done using the Rapid Annotation using Subsystem Technology (RAST) web server (Aziz et al. 2008; Overbeek et al. 2014). Pairwise single-nucleotide polymorphism (SNP) analysis among the farm isolates was performed using the breseq pipeline with default settings (Barrick et al. 2009; Barrick and Lenski 2009). Sequence-based comparisons, genome browsing, and alignments of the synteny of genomic regions were performed using the SEED Viewer (Overbeek et al. 2005, 2014) with default parameters, Mauve (Darling et al. 2004, 2010), Artemis (Rutherford et al. 2000), ACT (Carver et al. 2005), and BRIG using BLAST+ BLASTn with the parameter –evalue equal to 0.001 (Alikhan et al. 2011). Novel open reading frames (ORFs) that were identified against reference C. jejuni genomes were further analyzed using the InterProScan (Quevillon et al. 2005) and by BLASTP searches (Altschul et al. 1990, 1997) against the nonredundant protein sequences (nr) database in GenBank (Benson et al. 2013).
Phylogenetic Analyses
Genealogy Reconstruction
The clonal genealogy of the strains based on draft genome sequences was estimated using a model-based approach to determine bacterial microevolution implemented in ClonalFrame (Didelot and Falush 2007). The contigs were aligned using progressive Mauve (Darling et al. 2010), and collinear blocks bigger than 500 bp were filtered using the perl script stripSubsetLCBs available in the ClonalOrigin package (Didelot et al. 2010). ClonalFrame was run with 10,000 burn-in iterations followed by 10,000 data collection iterations. The strict (100%) consensus tree represents combined data from three independent runs.
Evolution of the Phase-Variable Gene wcbK
All alleles of nontruncated C. jejuni wcbK (GDP-mannose 4,6-dehydratase, CAMP1649) were obtained from the BIGSdb (Jolley and Maiden 2010) implemented in the Campylobacter PubMLST website (http://pubmlst.org/campylobacter/, last accessed May 2014) and from several C. jejuni genomes available in GenBank. Alignment of wcbK sequences was first generated at the amino acid level using MAFFT-FFT-NS-I v. 7 (Katoh and Standley 2013), then back-translated to nucleotide sequence using the TranslatorX perl script (Abascal et al. 2010). A distance matrix was then calculated using MEGA5, and the genes were grouped based on nucleotide identity (>90%). One allele from each group was randomly selected. The evolutionary relationship of the selected alleles of the wcbK was reconstructed using Bayesian phylogenetic inference. Two independent analyses of four Markov chain Monte Carlo (MCMC) chains run for 10 million generations with a tree sample every 5,000 generations were conducted for each allele using MrBayes v. 3.2.1 (Ronquist et al. 2012). The substitution model space was sampled during Bayesian MCMC to avoid a priori model testing (Huelsenbeck et al. 2004). The number of discrete categories used to approximate the gamma distribution was set to 6.
Results
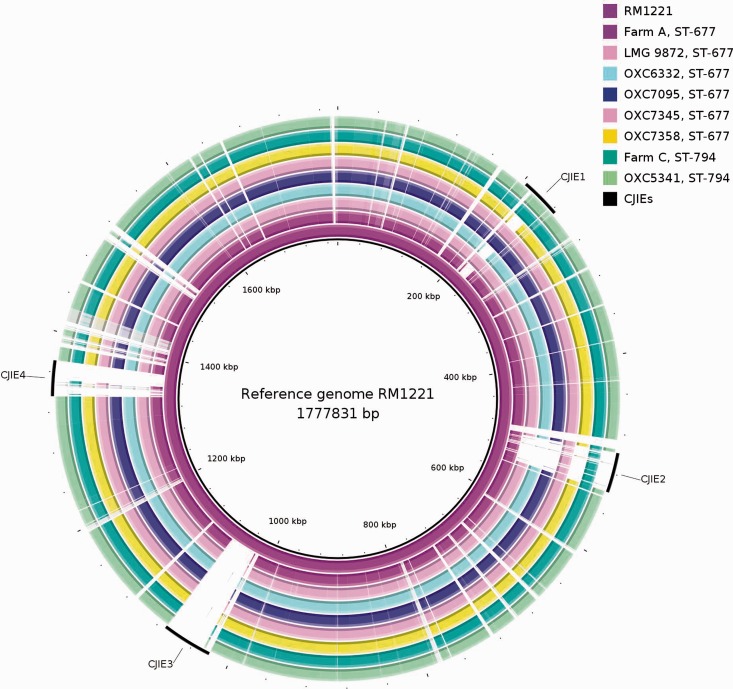
An overview of the C. jejuni genomes sequenced in this study is presented in table 1. Overall, the genome sequences were highly conserved (supplementary table S1, Supplementary Material online). There was only one gene missing or highly divergent between the different isolates from the same farm. The number of missing or highly divergent genes between all isolates included in this study (farm isolates and unassociated strains) was less than 115 and 50 ORFs for the ST-677 and ST-794 isolates, respectively. In contrast, the two STs differed by up to 113 ORFs. The main differences between the farm isolates and epidemiologically unrelated clinical isolates were associated with the C. jejuni-integrated elements (CJIEs) (fig. 1). Moreover, the ST-677 CC isolates were found to carry a degenerated CRISPR-Cas operon (supplementary fig. S1A, Supplementary Material online).
Table 1.
Overview of the Draft Genomic Data of the Campylobacter jejuni ST-677 CC Isolates Used in This Study
| Isolate | 5070 | 5071 | 5072 | 3515 | 3516 | LMG9872 | OXC5341 | OXC6332 | OXC7095 | OXC7345 | OXC7358 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Farm | A | A | A | C | C | — | — | — | — | — | — |
| MLST | 677 | 677 | 677 | 794 | 794 | 677 | 794 | 677 | 677 | 677 | 677 |
| CC | 677 | 677 | 677 | 677 | 677 | ||||||
| No. of contigs | 34 (15) | 36 | 36 | 32 | 34 | 91 | 42 | 40 | 37 | 37 | 37 |
| Genome size (Mbp) | 1.65 (1.65) | 1.65 | 1.65 | 1.66 | 1.66 | 1.62 | 1.63 | 1.64 | 1.63 | 1.66 | 1.64 |
| No. of CDSs | 1,734 (1,744) | 1,738 | 1,734 | 1,758 | 1,756 | 1,694 | 1,723 | 1,709 | 1,708 | 1,758 | 1,715 |
Note.—Strains sequenced in this study are indicated in italic font. Numbers in parentheses describe the data with inclusion of Sanger sequencing data used to close gaps in regions of interest.
Fig. 1.—
BRIG image highlighting the CJIEs using strain RM1221 as a reference. Upper and lower identity thresholds of 90% (dark color) and 70% (light color) were used for visualizing the difference between sequences. White areas correspond to sequences with less than 70% similarity (BLASTN) against the reference.
Whole-Genome MLST
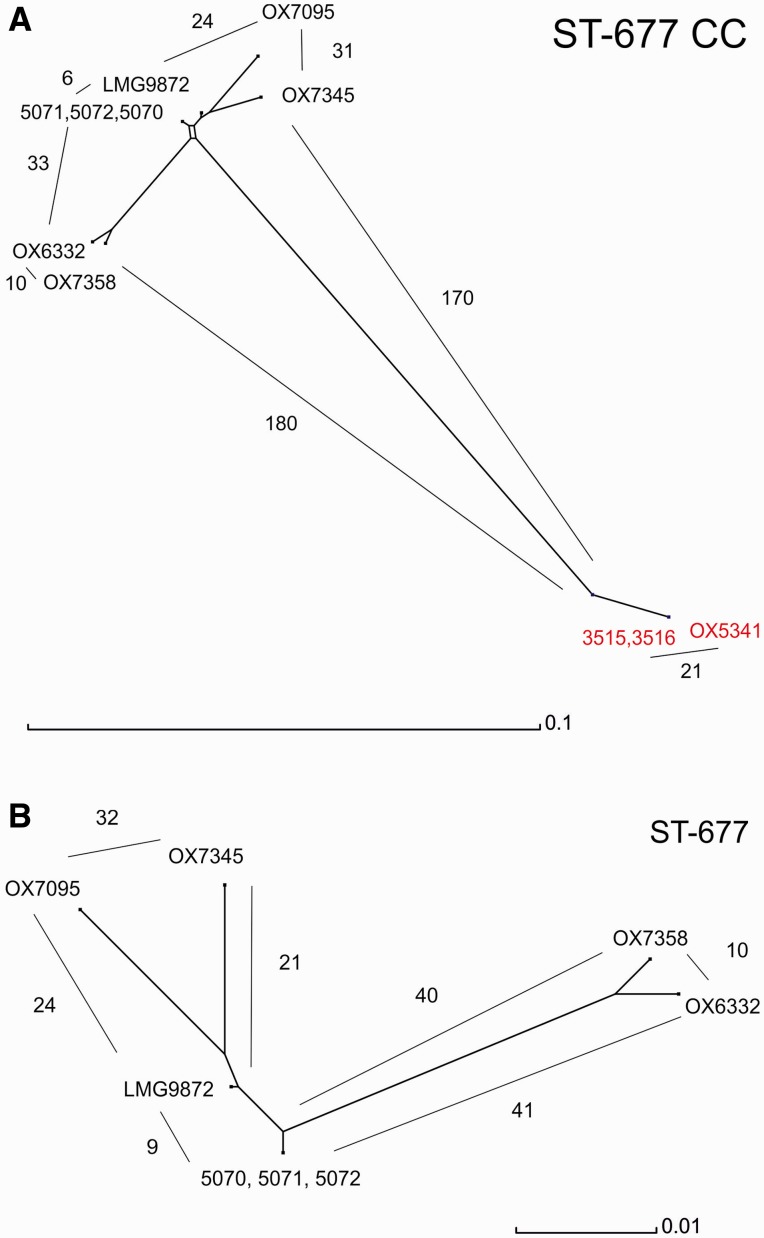
Figure 2A and B shows the NeighborNet networks of the ST-677 CC and ST-677 isolates, respectively, including the chicken farm isolates and clinical isolates from Sweden and the United Kingdom. Using the shared loci obtained by the Campylobacter BIGSdb query, no differences were identified within the three farm A (ST-677) and two farm C (ST-794) isolates (fig. 2A). The unrelated ST-677 CC isolates showed more than 20 allelic differences, except in the case of the Swedish clinical isolate LMG9872, which showed only nine allelic differences against the farm A isolates among the 1,319 shared loci (fig. 2B). The allelic differences between LMG9872 and farm A isolates are presented in detail in table 2. All of the observed differences were SNPs, and none of them occurred in homopolymeric tracts of contingency genes. The same two alleles occurred also among the same loci in the patient isolates from the United Kingdom, except in two cases (CAMP0952, mapA, and CAMP0999, cj1078), where all the clinical isolates represented the same allele type as indicated in table 2.
Fig. 2.—
NeighborNet phylogeny of the farm and clinical isolates. Networks were drawn from the shared loci of each set of isolates. (A) ST-677 CC (1,286 shared loci). ST-677 isolates are shown in black and ST-794 in red. (B) ST-677 (1,319 shared loci). Numbers of allelic differences between the strains are indicated by lines and Arabic numbers. Loci and alleles were defined using BIGSdb (http://pubmlst.org/campylobacter/, last accessed September 12, 2014).
Table 2.
Locus, Function, and Alleles in Which Differences Were Observed between the Campylobacter jejuni ST-677 Farm Isolates and the Clinical Isolate Obtained from a Meningitis Patient from Sweden (LMG9872) 25 Years Apart
| Locus | Function | Allelic Varianta |
Sequence Difference | |
|---|---|---|---|---|
| Farm Isolates | LMG9872 | |||
| CAMP0231 (cj0261c) | SAM-dependent methyltransferase | 61 | 18 | 173: G→A |
| CAMP0466 (alaS) | Alanyl-tRNA synthetase | 146 (OXC) | 40 | 1635: C→T |
| CAMP0530 (ribA) | GTP cyclohydrolase II | 94 | 30 | 897: T→A |
| CAMP0565 (cj0607) | Macrolide export ATP-binding/permease protein MacB | 140 | 37 | 697: C→T |
| CAMP0952 (mapA) | Outer-membrane lipoprotein MapA | 61 | 28 (OXC) | 295:A→G |
| CAMP0963 (cj1040c) | MFS transport protein | 19 | 25 | 850: G→A |
| CAMP0999 (cj1078) | Putative periplasmic protein | 10 | 2 (OXC) | 502: A→G |
| CAMP1131 (rbn) | Ribonuclease BN | 64 | 24 | 128: T→C |
| CAMP1193 (pyrH) | Uridylate kinase | 59 | 24 | 315: G→A |
aThe allele number is followed by OXC in parentheses when all clinical isolates obtained from the BIGSdb contained the same allele.
Genealogy Reconstruction
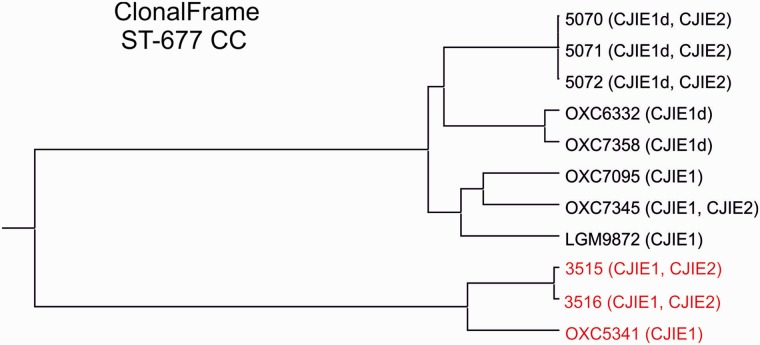
The ClonalFrame genealogy of the whole genomes of our chicken farm isolates and the clinical ST-677 CC isolates is shown in figure 3. This figure shows a similar clustering of the isolates as the wgMLST NeighborNet; however, the evolutionary distance between LMG9872 and the farm A isolates (5070, 5071, and 5072) is more pronounced. Variable genomic regions (not present in all isolates) were excluded from the ClonalFrame analysis, yet an association with the deletion within CJIE1 in the genomes was evident.
Fig. 3.—
ClonalFrame genealogy depicting the evolutionary relationship of the ST-677 CC isolates. In parentheses, the presence of CJIEs is indicated (“d” meaning large deletion in the CJIE1-like element). ST-677 isolates are shown in black and ST-794 in red.
CJIEs and Other Mobile Genetic Elements
The CJIE1-like element was found to be present in all studied ST-677 CC genomes. The CJIE1 was located in the same position in all ST-677 CC isolates (next to cj0661c, GTPase Era), but differently from RM1221, the reference strain for integrated elements in C. jejuni (Fouts et al. 2005). The gene content of CJIE1 was also highly conserved among the ST-677 CC, except for a deletion of 24 ORFs in ST-677 farm A isolates and one clinical isolate (OXC7358), consisting mainly of hypothetical proteins, phage baseplate proteins, and a phage tail protein. Compared with the CJIE1 in RM1221 (total 72 ORFs), 55 and 35 ORFs were missing (or highly divergent) in farm A and C genomes, respectively. Furthermore, the ST-677 CC isolates contained 18 (farm A) and 20 (farm C) ORFs absent in RM1221. The conserved parts in the CJIE1 contained among others a DNA adenine methylase and an endonuclease I precursor (EC 3.1.21.1).
The CJIE2-like element was missing completely in LMG9872 and other clinical isolates, except for OXC7345 (ST-677). Furthermore, in farm C (ST-794) isolates the CJIE2 was located in a different position (cj1359/cj1361c) than in ST-677 genomes, which contained the CJIE2 in the same position as RM1221 (cj0493/cj0494). In the CJIE2-like element, sequence similarities were also much lower (ranging from 49% to 100% amino acid sequence identities) than in the CJIE1 (98–100%), and differences in gene content were more evident. The conserved parts in all genomes contained, among others, an adenine-specific methyltransferase and a DNA/RNA nonspecific endonuclease. A putative secreted serine protease (homolog of cj1365c), located close to the integration site of the CJIE2-like element in farm C (ST-794) isolates, was found to be missing in all ST-677 CC isolates included in this study.
A small putative cryptic plasmid was identified in LMG9872 and one OXC isolate (OXC7095), showing sequence similarity and conserved synteny with cryptic plasmid pCC2228-2 of Campylobacter coli RM2228.
Genome Microevolution
Genes in which we identified differences using breseq among the C. jejuni ST-677 and ST-794 farm isolates are shown in table 3. The differences between the isolates among both STs were present mainly in the poly-G tracts of genes located in the lipo-oligosaccharide (LOS), flagellar glycosylation, and capsular polysaccharide gene loci. Only one SNP was observed in a noncontingency gene between the ST-794 isolates, leading to a synonymous substitution in a hypothetical protein located in CJIE1. The predicted length variations in the encoded protein sequences between the ON and OFF phases are also shown in table 3. All ST-677 CC strains included in our study were found to have a LOS class O biosynthetic locus according to the scheme presented previously (Parker et al. 2008). Sequence similarities with RM3423 (type strain for LOS class O, Penner HS27 reference strain) ranged from 90% to 99% (amino acid identity) in all ORFs. Homopolymeric tract length variation occurred in ORFs 23o and 25o, resulting in premature stop codons in all isolates. The predicted protein encoded by the truncated 23o transcript was not lacking any known protein domain and was nearly of the same size as the nontruncated version. However, in ORF 25o there was an additional formyl transferase, N-terminal domain (Pfam: PF00551), predicted in the encoded protein in the ON phase. Homology-based searches revealed that the flagellar glycosylation gene locus of ST-677 CC is highly similar to C. jejuni strain M1 (ST-137, ST-45 CC) (99–100% amino acid identity). The cjj5070_11410c (homolog of CAMP1228, cj1310c) was predicted to encode a serine protease (PANTHER protease family S9B,C dipeptidyl-peptidase IV-related domain, PTHR11731) in the ON phase.
Table 3.
Genomic Region, Locus Tag, and Putative Function of Genes and Intergenic Regions in Which Differences Were Observed According to breseq Analysis among the Campylobacter jejuni ST-677 CC Farm A (ST-677) and C (ST-794) Isolates, Respectively
| Isolation Place/Genomic Region | Locus Taga | Putative Function | In-Frame ORF polyG -Tract Length [aa ON/OFF] or Substitution | Isolate |
||||
|---|---|---|---|---|---|---|---|---|
| 5070 | 5071 | 5072 | 3515 | 3516 | ||||
| Farm A (ST-677) | ||||||||
| Capsule biosynthesis | cjj5070_05630 | FIG00470126: hypothetical protein | G(9) [400/48] | G(7) | G(8) | G(8) | ||
| cjj5070_05700c (CAMP1649) | GDP-mannose 4,6-dehydratase (EC 4.2.1.47) | G(9) [344/286] | G(11) | G(11) | G(9) | |||
| LOS biosynthesis | cjj5070_09800 | Unknown (LOS ORF 23o) | G(11) [147/138] | G(9) | G(10) | G(10)b | ||
| cjj5070_09820 | Hypothetical protein (LOS ORF 25o) | G(11) [427/156] | G(9) | G(10) | G(9) | |||
| Flagellin modification | cjj5070_11410c (CAMP1228, cj1310c) | FIG00469527: hypothetical protein | G(9) [403/204] | G(9) | G(8) | G(9) | ||
| Other | cjj5070_12060/12070 | Putative periplasmic protein/putative periplasmic protein | Intergenic | G(9) | G(9) | G(8) | ||
| Farm C (ST-794) | ||||||||
| CJIE1 | cjj5070_04520 | FIG00469983: hypothetical protein | A113A | GCG | GCA | |||
| LOS biosynthesis | cjj5070_09820 | Hypothetical protein (LOS ORF 25o) | G(11) [427/156] | G(9) | G(10) | |||
| Other | cjj5070_12060/12070 | Putative periplasmic protein/putative periplasmic protein | Intergenic | G(10) | G(9) | |||
aLocus tags of C. jejuni isolate 5070 (this study). The locus from BIGSdb when present is indicated in parentheses.
bG-tract length defined by Sanger sequencing.
Homopolymeric tract length variation was observed also among the clinical isolates from Sweden and the United Kingdom, with homologs of CJJ5070_09800 and CJJ5070_09820, involved in LOS biosynthesis, being in the OFF phase and homologs of CJJ5070_11410c and CJJ5070_05700c, located in the flagellar glycosylation locus and the capsular polysaccharide gene locus, variable between the ON/OFF phases. In addition to these sequence variations, predicted mutations compared against isolate 5070 occurred, ranging from 55 for LMG9872 to 451 for OXC7345 (all SNPs including the ones predicted in the CJIEs) in the breseq analysis (data not shown).
Capsular Polysaccharide Gene Locus (37,071 bp)
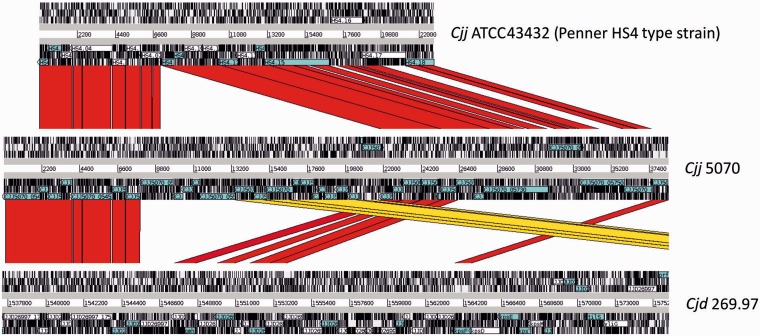
A discontinuous megablast search (BLASTN) of the whole capsule region starting from kpsC (cjj5070_05410c) and ending in kpsF (cjj5070_05770c) of strain 5070 was performed against the nr. The closest match was C. jejuni strain 32488 (ST-1460, ST-48 CC), showing a similar capsular polysaccharide gene locus (97% identity for full length) as our isolates. The second closest match was the Penner serotype HS4 type strain ATCC43432 (97–100% amino acid identities, range 92–100%), and the third one was C. jejuni subsp. doylei 269.97. Figure 4 shows the ACT comparison of the capsular polysaccharide biosynthesis loci of 5070, ATCC43432 and 269.97. Compared with C. jejuni ATCC43432, there is a large insertion in the 5070 capsule similar to a gene cluster found in C. jejuni subsp. doylei 269.97 located outside the capsular biosynthesis region defined by the kpsC and kpsF, resulting in one of the largest capsule encoding regions reported for C. jejuni.
Fig. 4.—
ACT comparison of the CPS gene cluster (from kpsC, cjj5070_05410c to kpsF, cjj5070_05770c) of strain 5070 against ATCC43432 (Penner HS4 type strain) and strain 269.97. Cjd, C. jejuni subspecies doylei; and Cjj, C. jejuni subsp. jejuni.
Evolution of the Phase-Variable Gene wcbK (CAMP1649)
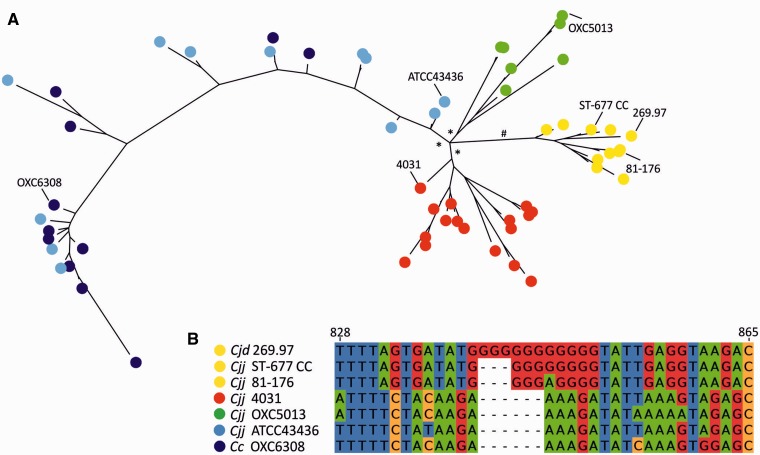
The capsule locus was found to contain a total of seven poly-G/C tracts of nine or more nucleotides in length. In the nearest published capsule, there were four poly-G/C tracts, two of which were present in the ST-677 CC genomes as well, and homopolymeric poly-G tract length variation was observed in one of them (table 3). Further analysis of this gene, the GDP-mannose 4,6-dehydratase (cjj5070_05700c, wcbK, CAMP1649), showed that 9 (or 12) Gs resulted in an in-frame ORF, whereas 11 Gs resulted in a premature stop codon and a truncated product likely to be unfunctional as the CATH superfamily (3.90.25.10) UDP-galactose 4-epimerase domain 1, present in the full-length protein encoded by the gene, was missing from the truncated version. We searched the Campylobacter BIGSdb for all C. jejuni alleles for this locus and made a phylogenetic tree based on a representative selection of the alleles (fig. 5A). The sequence alignment (fig. 5B) revealed that this gene was contained a poly-G tract only in one cluster of the C. jejuni isolates, and a homolog of this gene was completely missing from some C. jejuni strains, including NCTC11168, 81116 and M1. Supplementary table S2, Supplementary Material online, shows the distribution of the different alleles of CAMP1649 among the isolates available in the Campylobacter BIGSdb. From these data, it is evident that this locus has not evolved strictly in association with MLST lineages and that, for example, in ST-45 several different alleles are found and a large portion of the isolates (39%) lack this locus completely (supplementary table S3, Supplementary Material online). However, only one allele was found in the ST-677 CC in all isolates, and was absent from other STs/CCs, with varying lengths of the homopolymeric G-tract. In strain 81-176 (allele 47), the poly-G tract contained a single A inside the G-tract and is likely not phase-variable similar to allele 95 (fig. 5 and supplementary table S2, Supplementary Material online).
Fig. 5.—
Evolution of wcbK (CAMP1649) in a representative selection of C. jejuni and C. coli strains. (A) Bayesian phylogenetic tree based on a selection of isolates obtained from Campylobacter PubMLST and GenBank containing the CAMP1649 locus. Branches indicated by * are supported by more than 85%, and the branch indicated by # is supported by more than 90%. Positions of the strains selected for the sequence alignment (shown in panel B) are indicated by strain numbers in the phylogenetic tree. Dark blue circles represent C. coli isolates. (B) Homopolymeric G-tract was identified to be present only in one cluster of isolates (yellow clade). Cjd, C. jejuni subspecies doylei; Cjj, C. jejuni subsp. jejuni; and Cc, C. coli.
Other Putative Virulence-Associated Genes and Features in ST-677 CC
Cytolethal Distending Toxin Operon
The cytolethal distending toxin operon was highly degenerated in all ST-677 CC isolates, with cdtA and cdtB missing nearly completely and cdtC truncated and likely to be a pseudogene (supplementary fig. S1B, Supplementary Material online).
Iron Acquisition
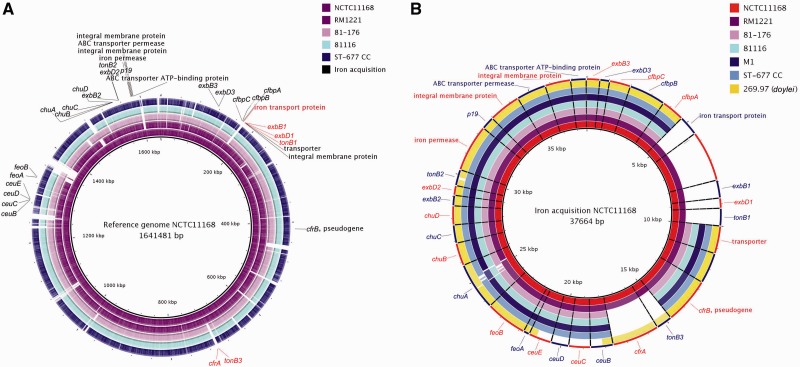
Similar to C. jejuni strain 81-176, the ferric enterobactin uptake receptor (cfrA) as well as the adjacent tonB3 gene and the tonB1 ferric siderophore transport system are missing in ST-677 CC (fig. 6A and B). Strain 81-176 has been shown to encode an alternative ferric enterobactin receptor cfrB (pseudogene in NCTC11168) (Xu et al. 2010), which we found to be present also in the ST-677 CC isolates (cjj5070_01700). Furthermore, chuA (CAMP1510), associated with hemin uptake in C. jejuni, was most variable between the reference strains and ST-677 CC isolates. Sequence variation accumulated in the same regions as for strain 81-176 (fig. 6B).
Fig. 6.—
BRIG images visualizing genes involved in iron uptake in C. jejuni using NCTC11168 as a reference. (A) Chromosomal location and distribution of selection of genes involved in iron uptake in reference strain NCTC11168. Genes found to be missing in the ST-677 CC are highlighted in red. (B) Detailed analysis of the sequence conservation in selected genes. White areas correspond to sequences with less than 70% similarity (BLASTN) against the reference.
IgA1/Serine Protease
We identified a conserved hypothetical protein (CJJ5070_17030) and a serine protease Pet putative autotransporter protein (CJJ5070_17040) (supplementary fig. S1C, Supplementary Material online), both coding sequences (CDSs) missing completely in strain NCTC11168 and highly fragmented in other C. jejuni genomes, yet conserved among the ST-677 CC isolates. In RM1221, the integration site of CJIE1 is located next to the site where the highly fragmented gene (homolog of cjj5070_17030) is found. InterProScan revealed that the hypothetical protein CJJ5070_17030 belongs to the Pfam family Peptidase_S6 (PF02395, MEROPS peptidase family S6), a group of serine peptidases, and contains an immunoglobulin A1 protease domain. The ORF annotated by RAST as serine protease Pet (CJJ5070_17040) contained an outer-membrane autotransporter barrel domain (IPR006315) and autotransporter beta-domain (IPR005546). This type of organization is typical for a two-partner secretion (TPS) pathway, also known as type Vb secretion system (T5bSS), with the passenger (TpsA) and translocator (TpsB) functions translated as separate polypeptide chains and the genes typically organized in an operon (Henderson et al. 2004).
Filamentous Hemagglutinin Domain Protein
We found two syntenic homologous gene clusters in different chromosomal locations (supplementary fig. S1D, Supplementary Material online) that contained a putative periplasmic protein, a filamentous hemagglutinin domain protein, and a putative hemolysin activation/secretion protein (CJJ5070_02700–CJJ5070_02740) or a hypothetical protein (CJJ5070_12170–CJJ5070_12210) with a polypeptide-transport-associated ShlB-type (IPR013686) domain of the TpsB transporter family (T5bSS). These genes were more conserved in the ST-677 CC genomes than their counterparts in several other C. jejuni genomes. In a recent study from Japan (Asakura et al. 2012), the filamentous hemagglutination domain protein (FHA) (CJJCF936_0827) was shown to affect host cell binding, and especially the N-terminus region was required for the increased binding affinity to heparan sulfate. This region was found to be well conserved among the two homologous proteins, even though the sequences overall were highly variable (<9% amino acid identity).
IceA1 Gene Cluster
Genes involved in methionine biosynthesis (O-acetylhomoserine sulfhydrylase, homolog of cj1727c, metB; homoserine O-succinyltransferase cj1726c, metA) were missing in the ST-677 CC genomes. Instead, there was an insertion of a novel gene cluster involved in DNA restriction modification between cj1720 and cj1729c of NCTC11168 (supplementary fig. S1E, Supplementary Material online). FixL (CJJ5070_14950c) showed homology with DNA adenine/modification methylases in Campylobacter rectus (100% coverage, 62% amino acid identity), Riemerella anatipestifer (99% coverage, 61% identity), and Gallibacterium anatis (95% coverage, 57% identity). This gene cluster was also found to include a homolog of IceA1/NlaIII (hypothetical protein, CJJ5070_14940c) with high sequence similarities with, for example, Helicobacter hepaticus (98% coverage, 73% identity), R. anatipestifer (97% coverage, 73% identity), G. anatis (93% coverage, 67% identity), and Helicobacter pylori (94% coverage, 62% identity). The ORF predicted as ulcer associated adenine-specific DNA methyltransferase by RAST (CJJ5070_14910c) also showed high sequence similarities with H. hepaticus (99% coverage, 68% identity), R. anatipestifer (99% coverage, 58% identity), and Campylobacter upsaliensis (99% coverage, 66% identity) and was identified as an ortholog of the CATG-specific methyltransferase hpyIM of H. pylori. The same position and gene synteny were present for the whole gene cluster in all ST-677 CC isolates included in this study. Furthermore, we screened all available C. jejuni genomes (GenBank, EMBL, Campylobacter PubMLST/BIGSdb) for this cluster of genes and found it only in isolates of ST-22 (ST-22 CC), ST-403 (ST-403 CC), and ST-1080 in addition to the ST-677 CC.
Another unique orphan DNA methyltransferase for C. jejuni was identified in the ST-677 CC isolates (CJJ5070_08940), homologs of which were found to be only present in C. coli and C. upsaliensis isolates and no other C. jejuni strains except LMG9872 in the nr protein database.
Discussion
Overall, the ST-677 CC was identified as a highly clonal lineage with relatively small differences in the genomes of strains even in some cases isolated 25 years apart (LMG9872 vs. farm A). The epidemiologically unrelated clinical isolates and the farm isolates differed only from 9 to 41 SNPs in the wgMLST analysis suggesting a high level of genome stability among this lineage. However, the main differences at the genome level were located in the CJIE1- and CJIE2-like integrated elements. The CJIE2 was present in both STs isolated from chicken but in only one of the clinical isolates, raising the question of whether strains carrying this element have a competitive disadvantage in human infections relative to strains in which this element is absent. The ClonalFrame genealogy of the whole-genome sequences revealed an association of a large internal deletion in the more conserved CJIE1 with one of the clusters of ST-677 isolates, which could indicate that this integrated element was obtained from a common ancestor and has since been carried in this CC, in contrast to the CJIE2, which seems to be more recently acquired. Supporting this view is that the CJIE2 was more divergent, unevenly distributed among the isolates and located in different positions in different STs.
Campylobacter jejuni is generally considered to be naturally competent for DNA uptake (Wang and Taylor 1990), a mechanism contributing to genetic diversity. Nevertheless, nonnaturally transformable strains and relatively stable clonal lineages exist (Asakura et al. 2012). In a previous study, the majority of nonnaturally transformable strains of C. jejuni was shown to express DNase activity, encoded by cje0256 present in the CJIE1, whereas all naturally competent strains lacked this activity (Gaasbeek et al. 2009). In vitro we found very low levels of natural transformation (<10−9) among isolates of the ST-677 CC and between ST-677 CC and other CCs (ST-45 CC, ST-21 CC) (data not shown). In agreement with this finding, the ST-677 CC genomes contained a homolog of CJE0256 (100% amino acid identity). Two other DNA/RNA nonspecific endonucleases encoded by CJE0566 and CJE1441 (located on CJIE2 and CJIE4, respectively) of strain RM1221 have also been shown to result in decreased efficiency of natural transformation in C. jejuni (Gaasbeek et al. 2010). The majority (6/8) of the ST-677 isolates, including all farm A isolates, contained also the CJE0566 homolog (90% amino acid identity). Furthermore, all genomes were missing the competence family protein (cj1211) shown previously (Jeon et al. 2008) to be required for natural transformation and transfer of antibiotic resistance determinants in C. jejuni. These results are in line with the observed high clonality and susceptibility to antimicrobial agents (data not shown) in ST-677 CC.
Clusters of regularly interspaced short palindromic repeats and associated genes, the CRISPR-Cas, have been proposed to function as a prokaryotic immune system, conferring resistance to exogenous genetic elements such as plasmids and phages (Bondy-Denomy and Davidson 2014). We found that the CRISPR-Cas system of the ST-677 CC isolates was highly degenerated and likely nonfunctional. Previously, the ganglioside-like lipooligosaccharide (LOS) structures of GBS-associated C. jejuni strains were shown to confer efficient bacteriophage resistance and the presence of sialyltransferases correlated with the apparent degeneration of type II CRISPR-Cas (Louwen et al. 2013). However, our results do not support this finding, as the ST-677 CC has not only a degenerated CRISPR-Cas system but also a nonsialylated LOS class O biosynthetic locus. We suggest that the degradation of the CRISPR-Cas system and the type I restriction–modification systems (data not shown) could be associated with the presence of CJIEs, some of which are of phage or plasmid origin, and more specifically, the DNA/RNA endonucleases and methyltransferases encoded by the integrated elements. This remains, however, to be shown with a larger and more diverse strain collection. This view is supported by an earlier study demonstrating that C. jejuni strains with degenerated CRISPR loci often carry integrated elements (prophage) or plasmids not present in the strains with an active CRISPR-Cas system (Dugar et al. 2013).
The C. jejuni farm isolates showed SNPs in only one gene and homopolymeric tract length variation in a few genes associated with the capsule, LOS, and flagella. In addition, only one point mutation leading to a synonymous substitution in a hypothetical protein located in the CJIE1 was identified. Phase variation in genes encoding surface-associated molecules, mainly the capsule, flagella and LOS, has been shown to represent an important strategy for C. jejuni to promote host colonization, avoid bacteriophage infection, and cause increased virulence (Jerome et al. 2011; Holst Sorensen et al. 2012). Previously, phase variation has been shown to occur in contingency genes cj0045c and cj0170 in association with passage through the chicken or mouse gastrointestinal tract and to be associated with colonization and disease potential in a mouse model (Kim et al. 2012). We found that in the ST-677 CC isolates the homolog of cj0170 was absent (similar to strain 81116), and furthermore, the Cj0045 sequence showed only 84% identity (BLASTP) and was missing part of the sequence (7.5%, encoding for the C-terminus of the protein), including the poly-G tract.
The CPS of C. jejuni is highly variable and is considered to be the major determinant of the Penner serotyping scheme (Moran and Penner 1999; Karlyshev et al. 2000). In our previous study, we showed that the Penner serotype HS4 complex was associated with ST-677 CC and ST-48 CC (Kärenlampi et al. 2007). In this study, we showed that the ST-677 CC capsule biosynthetic locus resembles that of ATCC43432, which is the type strain of Penner serotype HS4. In addition, the capsule locus of a ST-48 CC isolate (strain 32488, ST-1460, ST-48 CC) was found to be nearly identical to the capsule locus of ST-677 CC. Earlier, a higher proportion of C. jejuni HS4 strains was reported among blood isolates relative to fecal isolates, suggesting that these strains were likely to be more serum-resistant and/or invasive (Skirrow et al. 1993). This is in line with our findings that the ST-677 CC was more common among patients requiring hospitalization and a longer stay in hospital (Kärenlampi et al. 2007) and a recent study reporting that ST-677 CC was associated with bacteremia in Finnish patients (Feodoroff et al. 2013). Moreover, the capsule biosynthesis locus contained a large insertion predicted to be involved in capsular polysaccharide biosynthesis and modification and showing high sequence similarities with CDSs from C. jejuni subsp. doylei (strain 269.97) just outside the cps cluster of this strain. This is an interesting finding since C. jejuni subsp. doylei has been the major cause of bacteremia in Australia (85.2% of bacteremia cases caused by Campylobacter spp. or related organisms) (Morey 1996) and also a more frequent finding in blood cultures (24%) compared with Campylobacter-positive stool cultures (9.7%) at Red Cross Children’s Hospital in Cape Town South Africa (Lastovica 1996). The detection of CPS-related genes located outside the cps cluster has previously been reported (Karlyshev et al. 2013), suggesting that CPS structural variability may be determined not only by the genes present in this cluster but also by the genes in other parts of the genome. It has also been suggested that genes outside the cps gene cluster may form a pool of mobile genes capable of integration into the cps region and inducing structural rearrangements (Karlyshev et al. 2013). How this modulates the capsule structure and host–pathogen interactions remains to be elucidated.
The capsule has been shown to be important for resisting the human complement, and LOS to be more important for protection against killing mediated by cationic antimicrobial peptides and proteins (Keo et al. 2011). We identified variation in the poly-G homopolymeric tract lengths of GDP-mannose 4,6-dehydratase (wcbK) gene in the capsule biosynthesis locus in C. jejuni ST-677 CC. Previously dmhA (synonymous to wcbK) has been shown to be phase variable in some strains of C. jejuni (Karlyshev et al. 2005). Furthermore, wcbK was demonstrated to be missing from Campylobacter fetus subsp. venerealis; however, it was also shown to be involved in lipopolysaccharide (LPS) biosynthesis in C. fetus subsp. fetus type B isolates and to correlate with serum susceptibility (Kienesberger et al. 2014). Furthermore, the knockout mutant of wcbK was shown to be serum-resistant (Kienesberger et al. 2014). It is tempting to hypothesize that this phase-variable gene is associated with the varying degrees of serum resistance observed between the ST-677 CC isolates identified previously (Feodoroff et al. 2013) through genotypic switching resulting from slipped strand mispairing. Furthermore, the wcbK mutant of C. fetus (ATCC27374) was earlier established to be more acid-sensitive than the wild type (Kienesberger et al. 2014). In addition to the capsule, a mosaic organization has also been reported for the LOS gene clusters in C. jejuni and gene transfer between the LOS and capsule locus to occur (Parker et al. 2008; Karlyshev et al. 2013). The phase-variable wcbK in ST-677 CC could have a major role in pathogenesis by conferring acid resistance during transmission of the bacterium through the stomach, helping evade the host immune defense (affecting recognition by the immune system), and conferring serum resistance during systemic infections. Our finding that the poly-G tract was missing in the majority of the C. jejuni alleles for this locus (and the lack of the locus in several genomes, e.g., NCTC11168) supports the idea that this gene could be associated with evolution and niche specialization, however, the function and particular selective pressures associated with this phase-variable gene during infection remain to be proven experimentally.
In all ST-677 CC isolates, the cdt operon was highly degenerated, with the cdtA and cdtB missing nearly completely and cdtC truncated. The cdt operon has previously been shown to be conserved among most C. jejuni subsp. jejuni isolates and missing or highly degraded in C. jejuni subsp. doylei (cdtA truncated, cdtB and cdtC missing) and a link has been proposed between the absence of cdt genes or missing CDT activity in C. jejuni subsp. doylei and bacteremia (Parker et al. 2007). Supporting this view, AbuOun et al. (2005) have reported the degeneration of the cdt locus in relation to CDT-negative C. jejuni strains isolated from blood samples of campylobacteriosis patients with underlying medical problems.
Campylobacter jejuni requires iron for successful colonization of the host; however, it is not known to produce any endogenous siderophores of its own. Iron acquisition relies on iron scavenging and outer-membrane receptors specific for particular iron sources (Miller et al. 2009). Similar to the highly invasive strain 81-176 (Hofreuter et al. 2006), the ST-677 CC was missing the ferric enterobactin uptake receptor (cfrA) as well as the adjacent tonB3 and the tonB1 transport system (exbB1/exbD1/tonB1). Instead, the ST-677 CC isolates and strain 81-176 encode for CfrB which has been shown to be a functional ferric enterobactin receptor in 81-176 (Xu et al. 2010). Cj1613c and ChuA have been shown to be required for hemin and hemoglobin utilization by C. jejuni (Ridley et al. 2006). Interestingly chuA (cjj5070_13760, annotated as hemin uptake system outer-membrane receptor by RAST) in ST-677 CC showed sequence divergence in similar regions as 81-176 relative to NCTC11168. The variable regions have previously been identified as the exposed loops between membrane-embedded β sheets, and the observed diversity has been speculated to affect the interaction of ChuA with the siderophore or to result from the selective pressure by the host immune system (Hofreuter et al. 2006). Whether differences in the affinities of the different homologs of ChuA exist in binding to hemin remains to be studied.
Interestingly, we identified a conserved hypothetical protein belonging to the Pfam family Peptidase_S6 and an associated autotransporter beta domain containing protein CJJ5070_17030/CJJ5070_17040, which encode a novel putative type Vb secretion system (T5bSS, alternatively known as TPS system) in C. jejuni ST-677 CC. The Peptidase_S6 family consists of T5SS autotransporter proteins associated with many important bacterial virulence factors, for example, the immunoglobulin A1 protease proteins capable of cleaving immunoglobulin IgA found in pathogenic bacteria such as Neisseria gonorrhoeae (Pohlner et al. 1987) and Neisseria meningitidis; EspP, an extracellular (plasmid-encoded) serine protease of Escherichia coli O157:H7 capable of cleaving pepsin A and human coagulation factor V, which may contribute to the mucosal hemorrhage observed in hemorrhagic colitis (Brunder et al. 1997); and the hemoglobin-binding protease Hbp autotransporter of E. coli EB1 shown to interact with hemoglobin, degrade it, and subsequently bind the released heme (Otto et al. 1998). In T5bSS, the effector protein and translocator domain are produced as separate polypeptides (Henderson et al. 2004). Our group previously reported a similar serine protease in C. coli 73699; however, in that strain both domains resided in the same ORF (Skarp-de Haan et al. 2014), similar to type Va secretion system proteins (Henderson et al. 2004). Whether a similar kind of function as that seen in, for example, E. coli is conferred by CJJ5070_17030 in C. jejuni and/or C. coli and whether it could be associated with the more frequent appearance of the ST-677 CC among blood culture isolates remain unknown.
Interestingly, we also identified two further homologous syntenic gene regions, including two filamentous hemagglutinin domain proteins and two putative associated transport proteins (another T5bSS) conserved among the ST-677 CC while degraded in the majority of C. jejuni isolates. In Bordetella pertussis, the FHA has been shown to play a critical role by acting as a major virulence attachment factor containing at least three distinct domains that exhibit specific affinities for different ligands or receptors; one specific for macrophages, one specific for interactions with cilia, and one with heparin-binding activity that may be important for the interaction of B. pertussis with epithelial cells or extracellular matrices (Locht et al. 1993). FHA has also been shown to function as an immunomodulator, affecting host specificity in Bordetella (Inatsuka et al. 2005). In Japan, C. jejuni ST-4526 host cell binding was demonstrated to be increased for FHA-positive strains compared with FHA-truncated strains (Asakura et al. 2012). Furthermore, the N-terminus region was apparently required for the increased binding affinity to heparan sulfate (Asakura et al. 2012), a linear polysaccharide chain covalently O-linked to serine residues within the core proteins (so-called heparan sulfate proteoglycans), which are produced by almost all mammalian cells and occur in close proximity to the cell surface. The role of the FHAs in the adhesion, virulence and immune evasion of the ST-677 CC remains to be studied.
Of special interest was the discovery of the IceA1/NlaIII gene cluster (including two putative DNA methyltrasferases and an endonuclease) showing homology between C. jejuni isolates of the ST-677 CC, Helicobacter spp., Neisseria lactamica, R. anatipestifer, and G. anatis. Riemerella anatipestifer is a Gram-negative bacterial pathogen causing septicemic disease primarily in ducks, geese, turkeys, and waterfowl (Segers et al. 1993). Gallibacterium anatis, previously known as Pasteurella anatis, is associated with pathological lesions, including salpingitis with or without peritonitis, septicemia, pericarditis, hepatitis, respiratory tract lesions, and enteritis in chickens (Christensen et al. 2003). Gallibacterium anatis has been shown to be responsible for decreased egg production and increased mortality in commercial laying hens. Recently, the first case of bacteremia related to G. anatis in an immunocompromised patient was described (Aubin et al. 2013). This is the first study to report a homolog of IceA1/NlaIII endonuclease and the type II methyltransferase (ortholog of hpyIM) in C. jejuni (CJJ5070_14940c and CJJ5070_14910c, respectively). Further studies using the PubMLST database showed that this gene cluster was limited to only ST-22 and ST-403 of C. jejuni in addition to the ST-677 CC. In H. pylori, the iceA1 has been suggested to affect virulence in vivo through transcriptional regulation of the associated hpyIM expression levels, which may result in specific variations in DNA methylation patterns, leading to alteration in the expression of genes involved in virulence or pathogenesis (Donahue et al. 2000). Mutation of the hpyIM methylase in H. pylori altered the expression of the stress-responsive dnaK operon and decreased survival (associated with increased RNA degradation) in stationary phase cultures and cells adherent to AGS gastric epithelial cells in vitro, suggesting a role for this gene in transcriptional regulation (Donahue et al. 2002). Previously, at least one putative DNA methyltransferase in C. jejuni (cj1461, homolog present also in the ST-677 CC isolates) has been demonstrated to have a role in regulating virulence characteristics in C. jejuni (Kim et al. 2008). A mutant (Δcj1461) of strain 81-176 showed reduced motility and was 7-fold more adherent but 50-fold less invasive in INT-407 human epithelial cells compared with the wild type (Kim et al. 2008). The importance and potential role of these additional DNA methyltransferases and endonuclease in gene regulation, stress response, and virulence in Campylobacter spp. remain to be clarified.
To conclude, maintenance of the genome in the C. jejuni ST-677 CC lineage seems to be highly controlled, reflected in the high genetic similarity of unrelated genomes and the minor differences (one synonymous SNP and poly-G/C tract length differences in contingency genes) in epidemiologically related isolates. The main differences between the epidemiologically unrelated isolates were located in the CJIEs. Novel interesting gene candidates, including two putative T5bSS proteins and an RM/methyltransferase gene cluster, potentially linked to the association of this CC with bacteremia were identified. Furthermore, some contingency genes present in NCTC11168 were either found to be divergent and lacking the poly-G tract or were missing completely from the ST-677 CC. However, we identified the phase-variable gene, GDP-mannose 4,6-dehydratase (EC 4.2.1.47) (wcbK, CAMP1649), which may be a key component in serum resistance and niche adaptation in this lineage. Further studies are needed to confirm the association of these features with blood-borne infections and to characterize their roles in host–pathogen interactions in more detail.
Supplementary Material
Supplementary figure S1 and tables S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Joana Revez for advice and support, and Urszula Hirvi for excellent technical assistance. This work was supported by an Academy of Finland grant on behalf of CoE-MiFoSa (No. 11411405) and Ministry of Agriculture and Forestry grant (No. MMM776/312/2012). This publication made use of the Campylobacter Multi Locus Sequence Typing website (http://pubmlst.org/campylobacter/) developed by Keith Jolley and sited at the University of Oxford (Jolley & Maiden 2010, BMC Bioinformatics, 11:595). The development of this site has been funded by the Wellcome Trust.
Literature Cited
- Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38:W7–W13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AbuOun M, et al. Cytolethal distending toxin (CDT)-negative Campylobacter jejuni strains and anti-CDT neutralizing antibodies are induced during human infection but not during colonization in chickens. Infect Immun. 2005;73:3053–3062. doi: 10.1128/IAI.73.5.3053-3062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Asakura H, et al. Molecular evidence for the thriving of Campylobacter jejuni ST-4526 in Japan. PLoS One. 2012;7:e48394. doi: 10.1371/journal.pone.0048394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin GG, Haloun A, Treilhaud M, Reynaud A, Corvec S. Gallibacterium anatis bacteremia in a human. J Clin Microbiol. 2013;51:3897–3899. doi: 10.1128/JCM.01638-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Lenski RE. Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harb Symp Quant Biol. 2009;74:119–129. doi: 10.1101/sqb.2009.74.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, et al. GenBank. Nucleic Acids Res. 2013;41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ. Epidemiologic and clinical features of Campylobacter jejuni infections. J Infect Dis. 1997 doi: 10.1086/513780. 176(Suppl 2):S103–S105. [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy J, Davidson AR. To acquire or resist: the complex biological effects of CRISPR-Cas systems. Trends Microbiol. 2014;22:218–225. doi: 10.1016/j.tim.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Bonfield JK, Whitwham A. Gap5—editing the billion fragment sequence assembly. Bioinformatics. 2010;26:1699–1703. doi: 10.1093/bioinformatics/btq268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol Microbiol. 1997;24:767–778. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- Carver TJ, et al. ACT: the Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Christensen H, Bisgaard M, Bojesen AM, Mutters R, Olsen JE. Genetic relationships among avian isolates classified as Pasteurella haemolytica, “Actinobacillus salpingitidis” or Pasteurella anatis with proposal of Gallibacterium anatis gen. nov., comb. nov. and description of additional genomospecies within Gallibacterium gen. nov. Int J Syst Evol Microbiol. 2003;53:275–287. doi: 10.1099/ijs.0.02330-0. [DOI] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan CP, Kivistö R, Hakkinen M, Rautelin H, Hänninen M-L. Decreasing trend of overlapping multilocus sequence types between human and chicken Campylobacter jejuni isolates over a decade in Finland. Appl Environ Microbiol. 2010;76:5228–5236. doi: 10.1128/AEM.00581-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan CPA, Kivistö RI, Rautelin H, Hänninen M-L. Chapter 18: How molecular typing has changed our understanding on sources and transmission routes of campylobacteriosis in Finland. In: Sheppard SK, editor. Campylobacter ecology and evolution. Norfolk (United Kingdom): Caister Academic Press; 2014. pp. 241–252. [Google Scholar]
- Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Lawson D, Darling A, Falush D. Inference of homologous recombination in bacteria using whole-genome sequences. Genetics. 2010;186:1435–1449. doi: 10.1534/genetics.110.120121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue JP, et al. Analysis of iceA1 transcription in Helicobacter pylori. Helicobacter. 2000;5:1–12. doi: 10.1046/j.1523-5378.2000.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue JP, Israel DA, Torres VJ, Necheva AS, Miller GG. Inactivation of a Helicobacter pylori DNA methyltransferase alters dnaK operon expression following host-cell adherence. FEMS Microbiol Lett. 2002;208:295–301. doi: 10.1111/j.1574-6968.2002.tb11097.x. [DOI] [PubMed] [Google Scholar]
- Dugar G, et al. High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet. 2013;9:e1003495. doi: 10.1371/journal.pgen.1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority), ECDC (European Centre for Disease Prevention and Control) The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2012. EFSA J. 2014;12(2):3547. [Google Scholar]
- EFSA Panel on Biological Hazards (BIOHAZ) Scientific opinion on Campylobacter in broiler meat production: control options and performance objectives and/or targets at different stages of the food chain. EFSA J. 2011;9(4):2105. [Google Scholar]
- Feodoroff B, et al. Clonal distribution and virulence of Campylobacter jejuni isolates in blood. Emerg Infect Dis. 2013;19:1653–1655. doi: 10.3201/eid1910.121537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts DE, et al. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaasbeek EJ, et al. A DNase encoded by integrated element CJIE1 inhibits natural transformation of Campylobacter jejuni. J Bacteriol. 2009;191:2296–2306. doi: 10.1128/JB.01430-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaasbeek EJ, et al. Nucleases encoded by the integrated elements CJIE2 and CJIE4 inhibit natural transformation of Campylobacter jejuni. J Bacteriol. 2010;192:936–941. doi: 10.1128/JB.00867-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griekspoor P, Engvall EO, Olsen B, Waldenstrom J. Multilocus sequence typing of Campylobacter jejuni from broilers. Vet Microbiol. 2010;140:180–185. doi: 10.1016/j.vetmic.2009.07.022. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala’Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev. 2004;68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreuter D, et al. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect Immun. 2006;74:4694–4707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst Sorensen MC, et al. Phase variable expression of capsular polysaccharide modifications allows Campylobacter jejuni to avoid bacteriophage infection in chickens. Front Cell Infect Microbiol. 2012;2:11. doi: 10.3389/fcimb.2012.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Larget B, Alfaro ME. Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. Mol Biol Evol. 2004;21:1123–1133. doi: 10.1093/molbev/msh123. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Inatsuka CS, Julio SM, Cotter PA. Bordetella filamentous hemagglutinin plays a critical role in immunomodulation, suggesting a mechanism for host specificity. Proc Natl Acad Sci U S A. 2005;102:18578–18583. doi: 10.1073/pnas.0507910102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon B, Muraoka W, Sahin O, Zhang Q. Role of Cj1211 in natural transformation and transfer of antibiotic resistance determinants in Campylobacter jejuni. Antimicrob Agents Chemother. 2008;52:2699–2708. doi: 10.1128/AAC.01607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerome JP, et al. Standing genetic variation in contingency loci drives the rapid adaptation of Campylobacter jejuni to a novel host. PLoS One. 2011;6:e16399. doi: 10.1371/journal.pone.0016399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley KA, Maiden MC. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärenlampi R, Rautelin H, Schönberg-Norio D, Paulin L, Hänninen M-L. Longitudinal study of Finnish Campylobacter jejuni and C. coli isolates from humans, using multilocus sequence typing, including comparison with epidemiological data and isolates from poultry and cattle. Appl Environ Microbiol. 2007;73:148–155. doi: 10.1128/AEM.01488-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlyshev AV, et al. Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses. Mol Microbiol. 2005;55:90–103. doi: 10.1111/j.1365-2958.2004.04374.x. [DOI] [PubMed] [Google Scholar]
- Karlyshev AV, Linton D, Gregson NA, Lastovica AJ, Wren BW. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol Microbiol. 2000;35:529–541. doi: 10.1046/j.1365-2958.2000.01717.x. [DOI] [PubMed] [Google Scholar]
- Karlyshev AV, Quail MA, Parkhill J, Wren BW. Unusual features in organisation of capsular polysaccharide-related genes of C. jejuni strain X. Gene. 2013;522:37–45. doi: 10.1016/j.gene.2013.03.087. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keo T, Collins J, Kunwar P, Blaser MJ, Iovine NM. Campylobacter capsule and lipooligosaccharide confer resistance to serum and cationic antimicrobials. Virulence. 2011;2:30–40. doi: 10.4161/viru.2.1.14752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienesberger S, et al. Comparative genome analysis of Campylobacter fetus subspecies revealed horizontally acquired genetic elements important for virulence and niche specificity. PLoS One. 2014;9:e85491. doi: 10.1371/journal.pone.0085491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, et al. Role of the Campylobacter jejuni Cj1461 DNA methyltransferase in regulating virulence characteristics. J Bacteriol. 2008;190:6524–6529. doi: 10.1128/JB.00765-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, et al. Passage of Campylobacter jejuni through the chicken reservoir or mice promotes phase variation in contingency genes Cj0045 and Cj0170 that strongly associates with colonization and disease in a mouse model. Microbiology. 2012;158:1304–1316. doi: 10.1099/mic.0.057158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittl S, Heckel G, Korczak BM, Kuhnert P. Source attribution of human Campylobacter isolates by MLST and fla-typing and association of genotypes with quinolone resistance. PLoS One. 2013;8:e81796. doi: 10.1371/journal.pone.0081796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuusi M, Nakari U-M. Campylobacter. In: Jaakola S, Lyytikäinen O, Rimhanen-Finne R, Salmenlinna S, Vuopio J, Roivainen M, Löflund J-E, Kuusi M, Ruutu P, editors. Infectious diseases in Finland 2011. National Institute for Health and Welfare (THL) Report 38/2012. Juvenes Print. Tampere (Finland): Tampere University Press; 2012. 14 pp. [Google Scholar]
- Kwan PS, et al. Molecular epidemiology of Campylobacter jejuni populations in dairy cattle, wildlife, and the environment in a farmland area. Appl Environ Microbiol. 2008;74:5130–5138. doi: 10.1128/AEM.02198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastovica AJ. Campylobacter/Helicobacter bacteremia in Cape Town, South Africa, 1977-1995. In: Newell DG, Ketley JM, Feldman RA, editors. Campylobacters, Helicobacters, and related organisms. New York: Plenum Press; 1996. pp. 475–479. [Google Scholar]
- Locht C, Bertin P, Menozzi FD, Renauld G. The filamentous haemagglutinin, a multifaceted adhesion produced by virulent Bordetella spp. Mol Microbiol. 1993;9:653–660. doi: 10.1111/j.1365-2958.1993.tb01725.x. [DOI] [PubMed] [Google Scholar]
- Louwen R, et al. A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barre syndrome. Eur J Clin Microbiol Infect Dis. 2013;32:207–226. doi: 10.1007/s10096-012-1733-4. [DOI] [PubMed] [Google Scholar]
- Miller CE, Williams PH, Ketley JM. Pumping iron: mechanisms for iron uptake by Campylobacter. Microbiology. 2009;155:3157–3165. doi: 10.1099/mic.0.032425-0. [DOI] [PubMed] [Google Scholar]
- Moran AP, Penner JL. Serotyping of Campylobacter jejuni based on heat-stable antigens: relevance, molecular basis and implications in pathogenesis. J Appl Microbiol. 1999;86:361–377. doi: 10.1046/j.1365-2672.1999.00713.x. [DOI] [PubMed] [Google Scholar]
- Morey F. Five Years of Campylobacter bacteraemia in Central Australia. In: Newell DG, Ketley JM, Feldman RA, editors. Campylobacters, Helicobacters, and related organisms. New York: Plenum Press; 1996. pp. 491–494. [Google Scholar]
- Norrby R, McCloskey RV, Zackrisson G, Falsen E. Meningitis caused by Campylobacter fetus ssp jejuni. Br Med J. 1980;280:1164. doi: 10.1136/bmj.280.6224.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto BR, van Dooren SJ, Nuijens JH, Luirink J, Oudega B. Characterization of a hemoglobin protease secreted by the pathogenic Escherichia coli strain EB1. J Exp Med. 1998;188:1091–1103. doi: 10.1084/jem.188.6.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CT, Gilbert M, Yuki N, Endtz HP, Mandrell RE. Characterization of lipooligosaccharide-biosynthetic loci of Campylobacter jejuni reveals new lipooligosaccharide classes: evidence of mosaic organizations. J Bacteriol. 2008;190:5681–5689. doi: 10.1128/JB.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CT, Miller WG, Horn ST, Lastovica AJ. Common genomic features of Campylobacter jejuni subsp. doylei strains distinguish them from C. jejuni subsp. jejuni. BMC Microbiol. 2007;7:50. doi: 10.1186/1471-2180-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlner J, Halter R, Beyreuther K, Meyer TF. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature. 1987;325:458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- Quevillon E, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley KA, Rock JD, Li Y, Ketley JM. Heme utilization in Campylobacter jejuni. J Bacteriol. 2006;188:7862–7875. doi: 10.1128/JB.00994-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Segers P, et al. Riemerella anatipestifer gen. nov., comb. nov., the causative agent of septicemia anserum exsudativa, and its phylogenetic affiliation within the Flavobacterium-Cytophaga rRNA homology group. Int J Syst Bacteriol. 1993;43:768–776. doi: 10.1099/00207713-43-4-768. [DOI] [PubMed] [Google Scholar]
- Simpson JT, et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarp-de Haan CP, et al. Comparative genomics of unintrogressed Campylobacter coli clades 2 and 3. BMC Genomics. 2014;15:129. doi: 10.1186/1471-2164-15-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirrow MB, Jones DM, Sutcliffe E, Benjamin J. Campylobacter bacteraemia in England and Wales, 1981-91. Epidemiol Infect. 1993;110:567–573. doi: 10.1017/s0950268800050986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeds L, Kunstner A. ConDeTri—a content dependent read trimmer for Illumina data. PLoS One. 2011;6:e26314. doi: 10.1371/journal.pone.0026314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- Wang Y, Taylor DE. Natural transformation in Campylobacter species. J Bacteriol. 1990;172:949–955. doi: 10.1128/jb.172.2.949-955.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Zeng X, Haigh RD, Ketley JM, Lin J. Identification and characterization of a new ferric enterobactin receptor, CfrB, in Campylobacter. J Bacteriol. 2010;192:4425–4435. doi: 10.1128/JB.00478-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.