Abstract
The non-recombining Y chromosome is expected to degenerate over evolutionary time, however, gene gain is a common feature of Y chromosomes of mammals and Drosophila. Here, we report that a large palindrome containing interchromosomal segmental duplications is located in the vicinity of the first amplicon detected in the Y chromosome of D. melanogaster. The recent appearance of such amplicons suggests that duplications to the Y chromosome, followed by the amplification of the segmental duplications, are a mechanism for the continuing evolution of Drosophila Y chromosomes.
Keywords: Y chromosome, palindrome, pericentromeric region, segmental duplications, D. melanogaster
Almost a century ago, Muller (1914) first suggested that the Y chromosome is a highly degenerate X chromosome. Since then, several models have tried to explain how the heteromorphic sex chromosomes have evolved from a pair of autosomes (Ohno 1967; Charlesworth 1978; Rice 1994; Graves 1995). The prevailing theory holds that after the acquisition of a sex-determining gene by one autosome, suppression of recombination leads to its degeneration by the accumulation of deleterious mutations. Although the relative importance of the mechanisms governing this evolutionary transition is still an open issue, the presence of X-degenerated genes on the primate Y chromosome (Skaletsky et al. 2003; Hughes et al. 2010) and Z-degenerated sequences on the avian W chromosome (Fridolfsson et al. 1998) provide empirical evidence for this hypothesis. The three major evolutionary processes that account for the degradation of Y-linked genes are Muller's ratchet, genetic hitchhiking, and background selection (Charlesworth B and Charlesworth D 2000; Bachtrog 2004). Furthermore, the relative contributions of these evolutionary forces over the course of Y chromosome degeneration have recently been formally addressed (Bachtrog 2008).
In Drosophila melanogaster, the lack of a male-determining gene on the Y chromosome and the lack of sequence homology, other than the ribosomal DNA, between the X and Y chromosomes have given rise to the hypothesis that the Drosophila Y chromosome evolved from a supernumerary B chromosome (Hackstein et al. 1996; Carvalho 2002). The few D. melanogaster Y-linked genes so far identified have male-related functions and all originated from duplication of autosomal genes (Kennison 1981; Gepner and Hays 1993; Carvalho 2002; Koerich et al. 2008). However, the acquisition of autosomal genes is not restricted to noncanonical Y chromosomes; in fact, the deleted in azoospermia gene cluster on the human Y chromosome was the first clear example of an autosomal fertility gene that moved onto a Y chromosome (Saxena et al. 1996).
The sequencing of the human (Skaletsky et al. 2003), chimpanzee (Hughes et al. 2010), and mouse (Alföldi 2008) Y chromosomes led to the discovery of ampliconic regions (made of large repeat units) containing multicopy genes required for male fertility. In primates, these amplicons are principally arrayed as palindromes, and their sequence identity is maintained by frequent gene conversion between repeated regions (Rozen et al. 2003; Hughes et al. 2010). Thus, although these Y chromosomes do not recombine with the X chromosomes, their ampliconic regions are proposed to conserve gene functions over evolutionary time by means of gene conversion (Charlesworth 2003; Rozen et al. 2003). Recently, theoretical and computer simulation studies have shown that low rates of gene conversion can oppose the degeneration of Y-linked genes (Connallon and Clark 2010; Marais et al. 2010).
Assessing whether amplicons and large palindromic regions are a common feature of Y chromosome evolution requires further studies in other nonmammalian species. In this context, it has been shown in D. melanogaster that the Y-linked Su(Ste) repeats, which affect male fertility, derive from a recent duplication and amplification of the autosomal SSL gene (Kalmykova et al. 1997; Gvozdev et al. 2005). Likewise, Russell and Kaiser (1993) found that a Y chromosome pericentromeric region also derives from a recent segmental duplication of the Mst77F locus and subsequent amplification. Recently, Carvalho et al. have shown that this ampliconic region contains at least 18 members of the Mst77Y gene family, 8 of which are potentially functional (Krsticevic et al. 2010). However, previous analyses of the assembled genome sequence have not found any large palindromes on the Y chromosome or any other chromosome (Carlson JW, Celniker SE, personal communication).
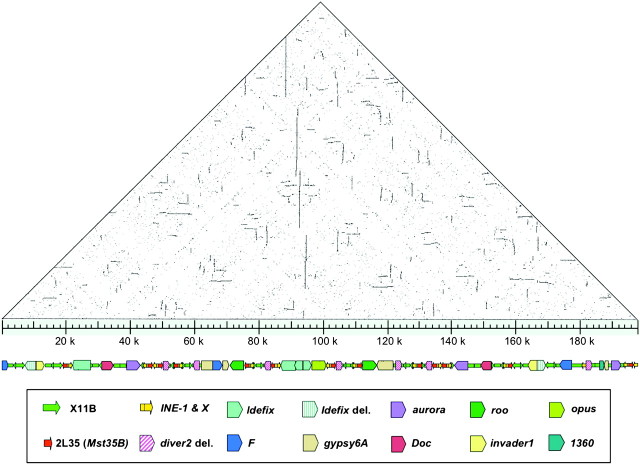
To investigate the presence of additional pericentromeric amplicons on the D. melanogaster Y chromosome and their potential palindromic organization, we isolated several pericentromeric bacterial artificial chromosomes (BACs) using heterochromatin P-element insertions (Yan et al. 2002), given that the Mst77Y amplicon had previously impeded extension of the Y centromeric region by chromosome walking (Abad et al. 2004). Using the flanking sequence of B783.2, a P-element inserted at the h17–18 region, four BACs were detected by an “in silico” search (supplementary fig. S1A, Supplementary Material online). DNA restriction analysis of the biggest ones, BACR36F19 and BACR07N15, showed that they derived from the same region, and fluorescence in situ hybridization mapping on mitotic chromosomes confirmed their pericentromeric origin (supplementary fig. S1B–C, Supplementary Material online). To investigate the chromosomal organization of this region, the clone BACR07N15 (∼200 kb) was sequenced (GenBank: CU457433) and compared with the D. melanogaster genome (Release 5.1). Sequence analysis showed that the region contained in this BAC is not part of the publicly available Y chromosome sequences. In addition, dotplot analysis revealed the presence of a large palindrome (fig. 1).
FIG. 1.
Schematic representation of the BACR07N15 sequence. The sequence organization of the large palindrome is represented by the horizontal diagram at the bottom. Sequence similarity within BACR07N15 is depicted above as a triangular dot plot (made with the program “dotter”) in which the BACR07N15 sequence is compared with itself. Direct repeats appear as horizontal lines and inverted repeats as vertical lines.
We reconstructed the history of this region and found that this large palindrome arose by a series of distinct molecular processes (fig. 2): 1) transposition to the Y of two interchromosomal duplications, one from the cytological band 35B (chromosome arm 2L) and the other from 11B (chromosome X), 2) intrachromosomal amplification of a fragment containing these segmental duplications, 3) small inversions and deletions through the entire region, 4) insertion of multiple transposable element (TE) sequences, and 5) a large tandem duplication resulting from a single recombination event within two Idefix long terminal repeats, followed by a large inversion resulting from intrachromatid pairing of two Idefix elements, double-strand breaks inside these elements and nonhomologous end-joining between the broken elements. This reconstruction is supported by the detailed annotation of TEs in BACR07N15. The majority of these TEs are from the same family and in the same relative inverted positions in both arms of the palindrome (fig. 1), and thus, we assume their insertion predates the origin of the palindrome since the alternative model that these TE insertions arose in one arm and were copied to the other arm by gene conversion is less parsimonious. Under this model, insertions that are unique to only one or the other arm of the palindrome are assumed to have inserted after the palindrome formation.
FIG. 2.
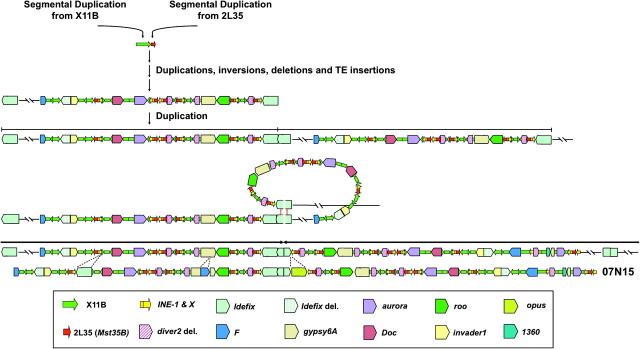
Proposed model for palindrome formation. The evolutionary processes hypothesized to be involved in the generation of the palindrome are a large tandem duplication followed by a large inversion. In the middle of the palindrome, there is an inverted duplication of an Idefix element with a small portion of sequence missing at the breakage-fusion junction. Because a double-strand break repaired with concomitant loss of sequence information is characteristic of nonhomologous end-joining, this is the mechanism proposed for the origin of the palindrome. The 12 TE families and the 2 segmental duplications are indicated.
To provide an estimate of the sequence divergence between arms of the palindrome, we compared the 72 kb of unambiguously alignable DNA from each arm present in BACR07N15. This analysis provided an overall divergence between arms of 0.0036 sub/site using Kimura's 2-parameter method. This arm-to-arm nucleotide sequence divergence is greater than in human Y palindromes (<0.06%, Rozen et al. 2003) but similar to the 0.5% arm-to-arm divergence observed for some chimpanzee Y palindromes (Hughes et al. 2010). The arms of the BACR07N15 palindrome show high sequence similarity either because of recent origin and/or ongoing gene conversion. Assuming that TE insertions present in both arms predate the palindrome formation, the age of TE insertions that are present in either one or both arms of the palindrome provides an independent method to place lower and upper bounds on the age of the palindrome (supplementary results and fig. S2, Supplementary Material online). This analysis, together with analysis of the CG12717/CG41423 gene family (see below), favors the interpretation of a recent origin for the palindrome as the primary reason why arms of the palindrome exhibit high sequence similarity. If this interpretation is correct and assuming a molecular clock of 0.011 subs/Ma (Tamura et al. 2004), we obtain an age estimate of ∼0.33 Ma for the origin of the palindrome. This result is consistent with the palindrome being melanogaster specific because the divergence of D. melanogaster from its sister species D. simulans occurred ∼5.5 Ma (Tamura et al. 2004). We note, however, that despite providing evidence for a recent origin, our data cannot conclusively rule out the effects of subsequent gene conversion between the palindrome arms.
We next addressed the extent of the palindrome by further physical mapping using probes derived from BACR07N15. Sequence comparison of the duplications and their ancestral loci suggests that the segmental duplication 2L35B derived from a genomic fragment that spanned at least the first intron and the second exon of Mst35Ba (one of the two protamine genes in tandem) (supplementary fig. S3A, Supplementary Material online), whereas the segmental duplication X11B derived from a fragment encompassing the whole CG12717 gene (encoding a SUMO protease) and parts of the neighbor genes ade5 and CG3812 (supplementary fig. S3B, Supplementary Material online). A search for other CG12717-related gene models in the D. melanogaster genome revealed the presence of CG41423, which is located in the unmapped whole-genome shotgun scaffold AABU01001566. Because the X11B derived CG12717 fragments in BACR07N15 are highly similar to CG41423 (see supplementary fig. S3C, Supplementary Material online), we screened the BAC library RPCI-98 with a probe from the Mst35Y intron (supplementary methods, Supplementary Material online) to see if the AABU01001566 scaffold belongs to another region of the palindrome identified in BACR07N15. Restriction analyses of the seven strongly hybridizing BACs allowed us to build a contig of about 500 kb with a palindromic region of at least 350 kb (supplementary fig. S4A, Supplementary Material online). By means of polymerase chain reaction amplification, an entire CG41423 gene was isolated from each of the BACs that partially overlap BACR07N15 and extend in both directions (supplementary fig. S4B, Supplementary Material online). Moreover, an “in silico” search detected two BACs from library DM1 (DME1-001M04 and DME1-007E02) that extend the palindrome to the Mst77Y amplicon (Krsticevic et al. 2010). As the average insert size of this library is 100 kb, the palindrome identified here could be located at less than 100 kb of the Mst77Y amplicon.
Finally, we addressed the potential functionality of gene sequences in the BACR07N15 palindrome by molecular evolutionary analysis. BlastN searches in the genome of sequenced species of the melanogaster subgroup have failed to identify any CG41423-like gene, other than the one from the X chromosome. This fact and the phylogenetic position of CG41423 (supplementary fig. S5, Supplementary Material online) strongly suggests that the initial duplication from the X to the Y chromosome occurred after the split of the D. melanogaster and D. simulans lineages, supporting the inference of recent origin in D. melanogaster for the palindrome itself. CG41423 has evolved under purifying selection (dN/dS = 0.3837) suggesting recent functional constraint. Although CG41423 transcripts have been detected by the modENCODE project, no cDNA has yet been found in expressed sequence tag databases. The potential SUMO protease encoded by CG41423 could be implicated in spermiogenesis-related sumoylation events.
Evolutionary analysis of the decayed Mst35Y genes shows, as happens in primate protamine genes (Retief and Dixon 1993), that their exons are more variable than their introns. Although the Mst35Y genes in BACR07N15 appear nonfunctional, it remains possible that the missing portion of the palindrome, or a flanking region might contain a functional Mst35Y, as has been shown for Mst77Y (Krsticevic et al. 2010), which would act in redundancy to Mst35Ba and Mst35Bb. This interpretation is consistent with the fact that the Mst35Y genes in BACR07N15 show evidence of functional constraint (dN/dS = 0.0114). The existence of a functional protamine gene on the Y chromosome would also explain why D. melanogaster males homozygous for a deletion of both protamine genes, Mst35Ba and Mst35Bb, are fertile (Rathke et al. 2010). Lastly, we note that the two segmental duplications found at the pericentromeric region of the D. melanogaster Y (Krsticevic et al. 2010; this work) show functional as well as structural (see above) coherence because Mst35B and Mst77F encoded for a protamine and a sperm-specific linker histone-like protein, respectively (Raja and Renkawitz-Pohl 2005).
We emphasize that the palindromic structure identified here in the Drosophila Y chromosome differs from its mammalian counterparts in two essential aspects. First, our data provide no direct evidence for gene conversion between arms of the palindrome. Although the lack of meiotic crossing in D. melanogaster males (Morgan 1912) would seem to preclude the possibility of gene conversion between palindromic regions on the Y chromosome, mitotic recombination can occur in D. melanogaster males and lead to gene conversion (Preston and Engels 1996). Additionally, there is molecular evidence of intrachromosomal gene conversion among copies of the Y-linked Su(Ste) locus in D. melanogaster (Balakireva et al. 1992; McKee and Satter 1996) and duplicates of the Y-linked kl2 gene in D. simulans complex (Kopp et al. 2006). Second, whereas the human Y palindromes contain functional genes essential for male fertility, the selective advantage for gene conversion to preserve function on the Drosophila Y palindrome is likely to be small because it has not been shown to contain essential genes. Nevertheless, the discovery of palindromic structures on the Drosophila Y chromosome that contain genes provides the first step toward establishing the existence of functional palindromes in nonmammalian species.
Supplementary Material
Supplementary results, methods, references, and figures S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We would like to thank Keith Maggert for providing the flanking sequence of the P-element insertion B783.2 and Patrizio Dimitri for confirming the chromosomal location of BACR07N15. We are also grateful to Jane Rogers for her support. This work was supported by the Ministerio de Ciencia e Innovación (BFU2008-02947-C02-01/BMC to A.V.); an institutional grant from the Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa”; the Natural Environment Research Council (NERC NE/G000158/1 to C.M.B.); and The Wellcome Trust. The GenBank accession number for the sequence reported in this paper is CU457433.
References
- Abad JP, de Pablos B, Agudo M, Molina I, Giovinnazo G, Martín-Gallardo A, Villasante A. Genomic and cytological analysis of the Y chromosome of Drosophila melanogaster: telomere-derived sequences at internal regions. Chromosoma. 2004;113:295–304. doi: 10.1007/s00412-004-0318-0. [DOI] [PubMed] [Google Scholar]
- Alföldi JE. Sequence of the mouse Y chromosome [dissertation] Cambridge, Massachusetts: Massachusetts Institute of Technology; 2008. [Google Scholar]
- Bachtrog D. Evidence that positive selection drives Y-chromosome degeneration in Drosophila miranda. Nat Genet. 2004;36:518–522. doi: 10.1038/ng1347. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics. 2008;179:1513–1525. doi: 10.1534/genetics.107.084012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakireva MD, Shevelev YY, Nurminsky DI, Livak KJ, Gvozdev VA. Structural organization and diversification of Y-linked sequences comprising Su(Ste) gene in Drosophila melanogaster. Nucleic Acids Res. 1992;20:3731–3736. doi: 10.1093/nar/20.14.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AB. Origin and evolution of the Drosophila Y chromosome. Curr Opin Genet Dev. 2002;12:664–668. doi: 10.1016/s0959-437x(02)00356-8. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Model for evolution of Y chromosomes and dosage compensation. Proc Natl Acad Sci U S A. 1978;75:5618–5622. doi: 10.1073/pnas.75.11.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. The organization and evolution of the human Y chromosome. Genome Biol. 2003;4:226. doi: 10.1186/gb-2003-4-9-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, Clark AG. Gene duplication, gene conversion and the evolution of the Y chromosome. Genetics. 2010;186:277–286. doi: 10.1534/genetics.110.116756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson AK, Cheng H, Copeland NG, Jenkins NA, Liu HC, Raudsepp T, Woodage T, Chowdhary B, Halverson J, Ellegren H. Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc Natl Acad Sci U S A. 1998;95:8147–8152. doi: 10.1073/pnas.95.14.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepner J, Hays TS. A fertility region on the Y chromosome of Drosophila melanogaster encodes a dynein microtubule motor. Proc Natl Acad Sci U S A. 1993;90:11132–11136. doi: 10.1073/pnas.90.23.11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JAM. The origin and function of the mammalian Y chromosome and Y-borne genes—an evolving understanding. Bioessays. 1995;17:311–320. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- Gvozdev VA, Kogan GL, Usakin LA. The Y chromosome as a target for acquired and amplified genetic material in evolution. Bioessays. 2005;27:1256–1262. doi: 10.1002/bies.20321. [DOI] [PubMed] [Google Scholar]
- Hackstein JHP, Hochstenbach R, Hauschteck-Jugen E, Beukeboom LW. Is the Y chromosome of Drosophila an evolved supernumerary chromosome? BioEssays. 1996;18:317–323. doi: 10.1002/bies.950180410. [DOI] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Pyntikova T, et al. (14 co-authors) Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature. 2010;463:536–539. doi: 10.1038/nature08700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova AI, Shevelyov YY, Dobritsa AA, Gvozdev VA. Acquisition and amplification of a testis-expressed autosomal gene, SSL, by the Drosophila Y chromosome. Proc Natl Acad Sci U S A. 1997;94:6297–6302. doi: 10.1073/pnas.94.12.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison JA. The genetic and cytological organization of the Y chromosome of Drosophila melanogaster. Genetics. 1981;98:529–548. doi: 10.1093/genetics/98.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerich LB, Wang X, Clark AG, Carvalho AB. Low conservation of gene content in the Drosophila Y chromosome. Nature. 2008;456:949–951. doi: 10.1038/nature07463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A, Frank AK, Barmina O. Interspecific divergence, intrachromosomal recombination, and phylogenetic utility of Y-chromosomal genes in Drosophila. Mol Phylogenet Evol. 2006;38:731–741. doi: 10.1016/j.ympev.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Krsticevic FJ, Santos HL, Januário S, Schrago CG, Carvalho AB. Functional copies of the Mst77F gene on the Y chromosome of Drosophila melanogaster. Genetics. 2010;184:295–307. doi: 10.1534/genetics.109.107516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais GA, Campos PR, Gordo I. Can intra-Y gene conversion oppose the degeneration of the human Y chromosome? A simulation study. Genome Biol Evol. 2010;2:347–57. doi: 10.1093/gbe/evq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee BD, Satter MT. Structure of the Y chromosomal Su(Ste) locus in Drosophila melanogaster and evidence for localized recombination among repeats. Genetics. 1996;142:149–161. doi: 10.1093/genetics/142.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan TH. Complete linkage in the second chromosome of the male of Drosophila. Science. 1912;36:719–720. [Google Scholar]
- Muller HJ. A gene for the fourth chromosome of Drosophila. J Exp Zool. 1914;17:325–336. [Google Scholar]
- Ohno S. Sex chromosomes and sex linked genes. New York: Springer-Verlag; 1967. [Google Scholar]
- Preston CR, Engels WR. P-element-induced male recombination and gene conversion in Drosophila. Genetics. 1996;144:1611–1622. doi: 10.1093/genetics/144.4.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja SJ, Renkawitz-Pohl R. Replacement by Drosophila melanogaster protamines and Mst77F of histones during chromatin condensation in late spermatids and role of sesame in the removal of these proteins from the male pronucleus. Mol Cell Biol. 2005;25:6165–6177. doi: 10.1128/MCB.25.14.6165-6177.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathke C, Barckmann B, Burkhard S, Raja SJ, Roote J, Renkawitz-Pohl R. Distinct functions of Mst77F and protamines in nuclear shaping and chromatin condensation during Drosophila spermiogenesis. Eur J Cell Biol. 2010;89:326–338. doi: 10.1016/j.ejcb.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Retief JD, Dixon GH. Evolution of pro-protamine P2 genes in primates. Eur J Biochem. 1993;214:609–615. doi: 10.1111/j.1432-1033.1993.tb17960.x. [DOI] [PubMed] [Google Scholar]
- Rice WR. Degeneration of a nonrecombining chromosome. Science. 1994;263:230–232. doi: 10.1126/science.8284674. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Page DC. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- Russell SRH, Kaiser K. Drosophila melanogaster male germ line-specific transcripts with autosomal and Y-linked genes. Genetics. 1993;134:293–308. doi: 10.1093/genetics/134.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, Brown LG, Hawkins T, et al. (11 co-authors) The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat Genet. 1996;14:292–299. doi: 10.1038/ng1196-292. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, et al. (40 co-authors) The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- Yan CM, Dobie KW, Le HD, Konev AY, Karpen GH. Efficient recovery of centric heterochromatin P-element insertions in Drosophila melanogaster. Genetics. 2002;161:217–229. doi: 10.1093/genetics/161.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]