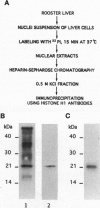
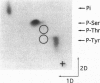
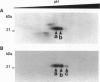
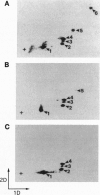
Abstract
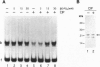
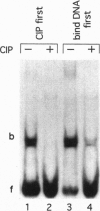
We have previously shown that estradiol treatment of roosters resulted in a rapid loss of binding activity of the repressor MDBP-2-H1 (a member of the histone H1 family) to methylated DNA that was not due to a decrease in MDBP-2-H1 concentration. Here we demonstrate that MDBP-2-H1 from rooster liver nuclear extracts is a phosphoprotein. Phosphoamino acid analysis reveals that the phosphorylation occurs exclusively on serine residues. Two-dimensional gel electrophoresis and tryptic phosphopeptide analysis show that MDBP-2-H1 is phosphorylated at several sites. Treatment of roosters with estradiol triggers a dephosphorylation of at least two sites in the protein. Phosphatase treatment of purified rooster MDBP-2-H1 combined with gel mobility shift assay indicates that phosphorylation of MDBP-2-H1 is essential for the binding to methylated DNA and that the dephosphorylation can occur on the protein bound to methylated DNA causing its release from DNA. Thus, these results suggest that in vivo modification of the phosphorylation status of MDBP-2-H1 caused by estradiol treatment may be a key step for the down regulation of its binding to methylated DNA.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ball D. J., Gross D. S., Garrard W. T. 5-methylcytosine is localized in nucleosomes that contain histone H1. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5490–5494. doi: 10.1073/pnas.80.18.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992 Jul 10;70(1):5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Boyle W. J., van der Geer P., Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- Boyle W. J., van der Geer P., Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M., Inglis R. J., Matthews H. R. Control of cell division by very lysine rich histone (F1) phosphorylation. Nature. 1974 Feb 1;247(5439):257–261. doi: 10.1038/247257a0. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M. Reversible histone modifications and the chromosome cell cycle. Bioessays. 1992 Jan;14(1):9–16. doi: 10.1002/bies.950140103. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Green G. R., Lee H. J., Poccia D. L. Phosphorylation weakens DNA binding by peptides containing multiple "SPKK" sequences. J Biol Chem. 1993 May 25;268(15):11247–11255. [PubMed] [Google Scholar]
- Grunstein M. Nucleosomes: regulators of transcription. Trends Genet. 1990 Dec;6(12):395–400. doi: 10.1016/0168-9525(90)90299-l. [DOI] [PubMed] [Google Scholar]
- Hill C. S., Packman L. C., Thomas J. O. Phosphorylation at clustered -Ser-Pro-X-Lys/Arg- motifs in sperm-specific histones H1 and H2B. EMBO J. 1990 Mar;9(3):805–813. doi: 10.1002/j.1460-2075.1990.tb08177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost J. P., Hofsteenge J. The repressor MDBP-2 is a member of the histone H1 family that binds preferentially in vitro and in vivo to methylated nonspecific DNA sequences. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9499–9503. doi: 10.1073/pnas.89.20.9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost J. P., Saluz H. P., Pawlak A. Estradiol down regulates the binding activity of an avian vitellogenin gene repressor (MDBP-2) and triggers a gradual demethylation of the mCpG pair of its DNA binding site. Nucleic Acids Res. 1991 Oct 25;19(20):5771–5775. doi: 10.1093/nar/19.20.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost J. P., Seldran M. Association of transcriptionally active vitellogenin II gene with the nuclear matrix of chicken liver. EMBO J. 1984 Sep;3(9):2005–2008. doi: 10.1002/j.1460-2075.1984.tb02082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I., Lieman-Hurwitz J., Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986 Feb 28;44(4):535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Langan T. A. Characterization of highly phosphorylated subcomponents of rat thymus H1 histone. J Biol Chem. 1982 Dec 25;257(24):14835–14846. [PubMed] [Google Scholar]
- Langan T. A. Methods for the assessment of site-specific histone phosphorylation. Methods Cell Biol. 1978;19:127–142. doi: 10.1016/s0091-679x(08)60018-7. [DOI] [PubMed] [Google Scholar]
- Laybourn P. J., Kadonaga J. T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991 Oct 11;254(5029):238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- Lewis J. D., Meehan R. R., Henzel W. J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992 Jun 12;69(6):905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Lewis J. D., Meehan R. R., Henzel W. J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992 Jun 12;69(6):905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Meehan R. R., Lewis J. D., McKay S., Kleiner E. L., Bird A. P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989 Aug 11;58(3):499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. Z., Goodman H. M., O'Farrell P. H. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977 Dec;12(4):1133–1141. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. Z., Goodman H. M., O'Farrell P. H. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977 Dec;12(4):1133–1141. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- Pawlak A., Bryans M., Jost J. P. An avian 40 KDa nucleoprotein binds preferentially to a promoter sequence containing one single pair of methylated CpG. Nucleic Acids Res. 1991 Mar 11;19(5):1029–1034. doi: 10.1093/nar/19.5.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak A., Bryans M., Jost J. P. An avian 40 KDa nucleoprotein binds preferentially to a promoter sequence containing one single pair of methylated CpG. Nucleic Acids Res. 1991 Mar 11;19(5):1029–1034. doi: 10.1093/nar/19.5.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991 Sep;55(3):451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S. Y., Allis C. D. Chromatin condensation: does histone H1 dephosphorylation play a role? Trends Biochem Sci. 1992 Mar;17(3):93–98. doi: 10.1016/0968-0004(92)90243-3. [DOI] [PubMed] [Google Scholar]
- Saluz H. P., Feavers I. M., Jiricny J., Jost J. P. Genomic sequencing and in vivo footprinting of an expression-specific DNase I-hypersensitive site of avian vitellogenin II promoter reveal a demethylation of a mCpG and a change in specific interactions of proteins with DNA. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6697–6700. doi: 10.1073/pnas.85.18.6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccaro M., Pawlak A., Jost J. P. Positive and negative regulatory elements of chicken vitellogenin II gene characterized by in vitro transcription competition assays in a homologous system. Proc Natl Acad Sci U S A. 1990 Apr;87(8):3047–3051. doi: 10.1073/pnas.87.8.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P. Transcription: in tune with the histones. Cell. 1994 Apr 8;77(1):13–16. doi: 10.1016/0092-8674(94)90229-1. [DOI] [PubMed] [Google Scholar]
- Zhang X. Y., Asiedu C. K., Supakar P. C., Khan R., Ehrlich K. C., Ehrlich M. Binding sites in mammalian genes and viral gene regulatory regions recognized by methylated DNA-binding protein. Nucleic Acids Res. 1990 Nov 11;18(21):6253–6260. doi: 10.1093/nar/18.21.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J., Yaneva J. Histone H1-DNA interactions and their relation to chromatin structure and function. DNA Cell Biol. 1991 May;10(4):239–248. doi: 10.1089/dna.1991.10.239. [DOI] [PubMed] [Google Scholar]