Abstract
Background/Aims
Tropomyosin-related kinase B receptor (TrkB)-mediated signaling is vital for neuronal differentiation, survival, plasticity, and cognition. In this study, the focus was placed on TrkB-Shc, a neuron-specific transcript, to determine if microRNAs (miRNAs) play a role in TrkB-Shc regulation.
Methods
A combination of bioinformatics and molecular gene expression analysis techniques was used to assess the effect of miR-409-3p and miR-216b on TrkB-Shc expression.
Results
miR-409-3p and miR-216b were found to regulate the TrkB-Shc 3′UTR through the identified putative binding sites. When the effect of the miRNAs on TrkB was assessed using SHSY5Y neuronal cells, differential effects were observed between mRNA and protein expression.
Conclusion
This study highlights the importance of miRNA-mediated regulation in TrkB signaling.
Key Words: Alzheimer's disease, TrkB, Gene expression, Luciferase, Neurotrophin
Introduction
Tropomyosin-related kinase B receptor (TrkB)-mediated signaling is vital for neuronal differentiation, survival, plasticity, and cognition of a variety of neurons throughout the brain [1,2,3,4]. Following transcription, the TrkB pre-mRNA is alternatively spliced to generate three alternative transcripts, TrkB-TK+, TrkB-TK-, and TrkB-Shc [5]. All three alternative transcripts code for receptors possessing a ligand-binding domain, but each varies in length at the 3′ end. There is evidence to suggest that the expression pattern of TrkB in the brain differs depending on the isoform. For instance, TrkB-TK-is expressed in both neurons and glia [6,7,8], whereas TrkB-Shc expression is neuron specific [5]. Surprisingly, while the existence of TrkB-Shc has been known for some time, much less is known about its regulation and function in both normal and diseased states. Recent evidence has demonstrated that TrkB transcripts may also be substantially regulated by microRNAs (miRNAs).
miRNAs are small noncoding RNAs that function as gene expression modulators in regulating cell growth and differentiation [9,10]. Interestingly, the brain was identified as the organ expressing the highest variety of miRNAs, suggesting an important role for miRNAs in the development of the nervous system [11,12]. Guidi et al. [13] reported isoform-specific regulation of the TrkB receptor in the neuronal cell-line SHSY5Y. The study identified and demonstrated that miR-151-3p could repress TrkB-TK+ mRNA levels and that miR-128, −485-3p, −765, and −768-5p could repress TrkB-TK-mRNA levels [13]. In a recent study, TrkB-TK-expression was also found to be regulated by specific miRNAs [14]. Maussion et al. [14] found increased levels of miR-185* in the frontal cortex of suicide completers and identified five putative miR-185*-binding sites in the TrkB-TK-3′UTR. These findings provide clear evidence that TrkB alternative transcripts are susceptible to miRNA-mediated regulation, and importantly, they also show that these processes can occur in the brain as well as in brain-related disorders. To date, no study has yet assessed whether miRNAs are capable of regulating TrkB-Shc transcripts. Previously, expression of TrkB-Shc transcripts was shown to be regulated by the RNA splice regulatory protein Srp20 during pre-mRNA processing [15]. Moreover, it was found that changes in TrkB-Shc expression through Srp20 were associated with Alzheimer's disease (AD).
AD is the most prevalent form of dementia characterized by the formation of neuritic amyloid plaque deposits and neurofibrillary tangles, progressive loss in brain volume, and cognitive deficits [16,17]. In a previous study, a number of miRNAs in mouse primary hippocampal neurons were found to be dysregulated following amyloid beta treatment [18]. Considering that TrkB signaling has been found to be affected early in AD, this study aimed to determine whether TrkB-Shc can be regulated by miRNAs and whether expression changes identified in the miRNAs can modulate TrkB expression. Considering that TrkB-Shc is a neuron-specific isoform, determining which miRNAs are capable of regulating its expression may shed much needed light on how TrkB signaling is regulated, and consequently, how it can be disrupted in disease conditions.
Materials and Methods
Cell Culture
Cell-lines were obtained from the American Type Culture Collection and grown at 37°C in a 5% CO2 atmosphere. HEK293 cells were cultured in Dulbecco's modified Eagle's medium, while SHSY5Y cells were cultured in RPMI medium. All culture media were supplemented with 10% (v/v) FBS and glutamax (2 mM; Gibco-BRL). For experiments, SHSY5Y cells were differentiated by addition of all-trans retinoic acid (10 μM) to the culture medium and incubated for 9 days. No cell toxicity was observed in any of the treatment conditions at the concentrations employed.
Plasmid Constructs
The plasmid pGL3-SV40-TrkB-Shc-3′UTR was constructed as previously described [19] by amplifying the entire 6,080 bp 3′UTR of the human TrkB-Shc transcript (accession No. NM_001018066) from 9 days differentiated SHSY5Y cDNA. Primers were TrkB-ShcF1: TAATAATTTGGTATTTGGAGGCTC; TrkB-ShcF2: TTTGGGTCTAGATAATAATTTGGTATTTG, and TrkB-ShcR2: CCCGGGTCTAGATATATGACAGTC. The miRNA-binding sequences of each putative miR-409-3p- or miR-216b-binding site contained in the TrkB-Shc 3′UTR were cloned into pGL3-SV40 (Promega) as previously described [19,20].
Luciferase Reporter Assay
Luciferase reporter assays were carried out as described previously [19]. On day 1, cells were seeded in 48-well plates (density: 3 × 104 HEK293). On day 2, cells were cotransfected with either pGL3-SV40 (empty vector), pGL3-SV40-TrkB-Shc-3′UTR, pGL3-SV40-miR-409-3p, or pGL3-SV40-miR-216b (125 ng/well) miR-409-3p, miR-216b, or miRNA control (5 pmol/well; Sigma-Aldrich; category No. HMC0002), and phRL-TK renilla (12.5 ng/well; Promega) for 24 h using Lipofectamine 2000 (0.5 μl/well). The phRL-TK renilla internal control plasmid was cotransfected for normalization of transfection efficiency. On day 3, cells were washed and resuspended in 50 µl of 1× passive lysis buffer. Luciferase assays were performed using the Dual Luciferase Assay Reporter System according to the manufacturer's instructions in a FlourStar OPTIMA (BMG). Results were normalized to the renilla control and expressed as relative luciferase activity with respect to the empty vector control-miRNA transfected condition, which was set to one. All assays were conducted with no less than three independent experiments and with replicate cultures.
RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR
SHSY5Y cells were seeded in 12-well plates (density: 1 × 106), differentiated for 9 days and transfected with miRNAs (miR-409-3p, miR-216b, miRNA control) for 24 h using Lipofectamine 2000. Total RNA extraction, cDNA synthesis, and quantitative real-rime PCR (qPCR) were conducted as previously described but with minor modifications [21]. Reverse transcriptase-PCR was performed according to the manufacturer's protocol for iScript™ Select cDNA Synthesis Kit (BioRad). qPCR was performed using Sso Fast Evagreen Supermix (BioRad) on a LightCycler 480 with a 96-well format (Roche). PCR cycling conditions and primer pairs used for gene expression analyses are detailed in the study by Wong et al. [21]. Control reactions with no template were included, which produced no signal. Melt curve analysis and agarose gel electrophoresis were performed to confirm the production of a single product. Changes in gene expression levels were determined by normalizing mRNA levels of the gene of interest to the mRNA level of the housekeeping gene β-actin using the 2-ΔΔCT method [22]. β-Actin did not vary with miRNA transfections.
Western Blotting
Total cell protein extraction and Western blotting were performed as previously described [21]. The primary antibody used was TrkB antibody (1:3,000 dilution; Cell Signaling Technology; category No. 4,603). The secondary antibody used was mouse peroxidase-conjugated affinity purified secondary antibody (Dako). Membranes were incubated with enhanced chemiluminescence reagent (Millipore) and visualized by autoradiography. Protein loading was assessed by stripping and reprobing for β-actin (1:10,000 dilution; Sigma Aldrich; category No. A2066). Bands were quantitated by densitometry using Image J (version 1.37v; National Institutes of Health, USA). The brightness/contrast of images has been adjusted using Adobe Photoshop CS (version 8).
Statistical Analysis
Cell culture experiments are from three replicate cultures representative of 3-6 independent experiments. Values are presented as mean ± standard error of the mean (SEM). Outliers were determined and removed from subsequent analyses using the Grubb's test or if data points were greater or less than two standard deviations from the mean. Statistical analyses were conducted using Statistica 7 (StatSoft Inc., 2000; STATISTICA for Windows). Two-tailed unpaired t tests were used to assess significance between two groups. A p value <0.05 (two-tailed) was considered statistically significant.
Results
miR-409-3p and miR-216b as Putative Regulators of the TrkB-Shc 3′UTR
A number of miRNAs in mouse primary hippocampal neurons were previously found to be dysregulated following amyloid beta treatment [18]. In particular, miR-409-3p was found to be altered in adult mice (2 months old), and the changes were maintained in aged mice (13 months old). In light of previous findings suggesting that TrkB signaling may be altered in the disease process [21,23,24], miR-409-3p was assessed to determine if it could target and potentially regulate TrkB alternative transcripts.
To ascertain if the TrkB transcripts may be regulated by miR-409-3p, an in silico screen of the 3′UTR of the three major TrkB alternative transcripts was conducted using microRNA.org, a database with a comprehensive resource of miRNA target predictions and expression profiles (www.microrna.org). Interestingly, the TrkB-Shc 3′UTR was the only TrkB 3′UTR found to contain two putative miR-409-3p-binding sites (table 1). However, miR-409-3p was not one of the miRNAs identified by the database algorithm to have a high likelihood of targeting the TrkB-Shc 3′UTR. Instead, miR-216b was found to have the highest probability of targeting the TrkB-Shc 3′UTR with six putative miRNA-binding sites (table 1).
Table 1.
Identification of putative miRNA-binding sites in the TrkB-Shc 3′UTR
 |
The mirSVR downregulation scores are interpreted as an empirical probability of downregulation [25]. In addition, the composite approach of miRanda-generated alignments is utilized in conjunction with the mirSVR scores to enable conservative predictions of noncanonical binding sites without increasing the incidence of false-positive predictions. The database algorithm utilized in this study included all target site predictions, which had either a 6-mer or better seed site, or a mirSVR score ≤-0.1. Of the two potential binding sites identified for miR-409-3p, only one had a mirSVR score ≤-0.1 (miR-409-3p-BS2) (table 1). Similarly, three of the six potential miRNA-binding sites identified for miR-216b also had mirSVR scores ≤-0.1 (miR-216b-BS3, miR-216b-BS4, miR-216b-BS5). After considering the regulatory implication of miR-409-3p and miR-216b on TrkB-Shc expression, the regulatory effect of the two miRNAs on TrkB-Shc expression was assessed.
Targeted miRNA Regulation in vitro
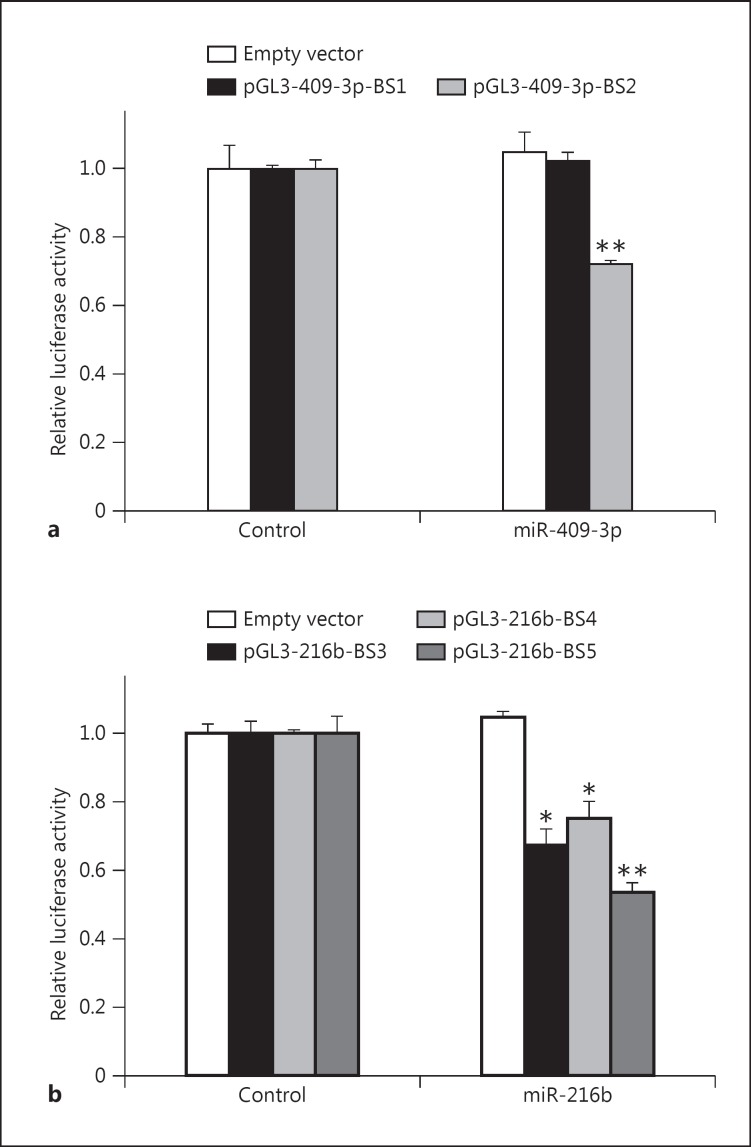
Considering that the putative binding sites in the TrkB-Shc 3′UTR with mirSVR scores ≤-0.1 have a greater probability of being targeted by their specific miRNAs (table 1), it was next determined whether the miRNAs identified with mirSVR scores ≤-0.1 could regulate target mRNA levels in a biological setting using HEK293 cells. The putative TrkB-Shc 3′UTR miR-409-3p- and miR-216b-binding sites identified from microrna.org (table 1) with mirSVR scores ≤-0.1 were cloned into pGL3-SV40 luciferase reporter constructs. Then, these constructs were cotransfected into HEK293 cells either with the luciferase reporter construct containing the putative binding site of interest or an empty vector in conjunction with miR-409-3p or miR-216b, or control miRNA. Consistent with the mirSVR scores, miR-409-3p-BS2 showed a significant decrease in luciferase reporter activity following miR-409-3p transfection (t = 10.6, d.f. = 4, p = 0.0004) (fig. 1a). Similarly, miR-216b-BS3, miR-216b-BS4, and miR-216b-BS5 showed significant decreases in luciferase reporter activity following miR-216b transfection (t ≥ 6.52, d.f. = 4, p < 0.003) (fig. 1b).
Fig. 1.
Assessment of putative miR-409-3p- and miR-216b-binding sites. HEK293 cells were cotransfected with pGL3-SV40 (empty vector; white column) or a pGL3-SV40 construct containing the putative miR-409-3p- (a) and miR-216b-binding sites (b) identified in table 1 in combination with miR-409-3p, miR-216b, or control miRNA. Data are normalized to the respective empty vector controls and expressed as relative luciferase activity. Values are presented as mean ± SEM and are representative of n = 3 independent experiments from three replicate cultures. * p < 0.005 (pGL3-216b-BS3: p = 0.002; pGL3-216b-BS3: p = 0.003); ** p < 0.001 (pGL3-409-3p-BS2: p = 0.0004; pGL3-216b-BS5: p = 0.0008).
As an experimental control, a miR-409-3p-binding site with a mirSVR score of −0.0022 (miR-409-3p-BS1) was also cloned. miR-409-3p-BS1 showed no significant change in luciferase reporter activity (t = −0.90, d.f. = 4, p = 0.42). Moreover, no significant changes were observed in the empty vector control transfections (miR-409-3p: t = −0.56, d.f. = 4, p = 0.61; miR-216b: t = −1.42, d.f. = 4, p = 0.23) (fig. 1a, b).
Putative miRNA Regulation of the TrkB-Shc 3′UTR
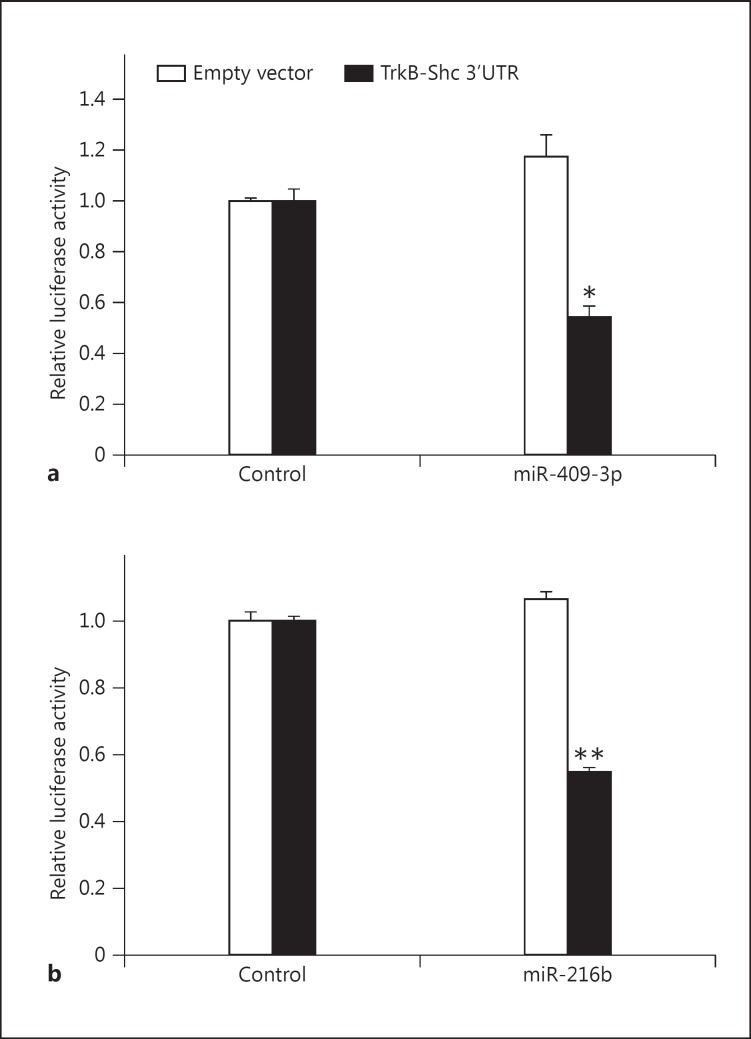
It was next determined whether miR-409-3p and miR-216b could regulate TrkB-Shc transcript expression through its 3′UTR. The TrkB-Shc 3′UTR was cloned into the 3′UTR of the pGL3-SV40 luciferase reporter vector. Then, HEK293 cells were cotransfected with either this luciferase reporter construct or an empty vector with either miR-409-3p, miR-216b, or control miRNA. Highly significant decreases in luciferase reporter activity were observed following miR-409-3p and miR-216b transfection (miR-409-3p: t = 7.10, d.f. = 4, p = 0.002; miR-216b: t = 23.3, d.f. = 4, p = 0.00002) (fig. 2a, b). No significant changes were observed in the empty vector control transfections (miR-409-3p: t = −2.07, d.f. = 4, p = 0.0.11; miR-216b: t = −1.87, d.f. = 4, p = 0.14) (fig. 2a, b). Consistent with observations using only the putative mi409-3p- and miR-216b-binding sites, it was found that both miR-409-3p and miR-216b could decrease luciferase reporter activity in cells transfected with constructs containing the endogenous TrkB-Shc 3′UTR.
Fig. 2.
Regulation of the TrkB-Shc 3′UTR by miR-409-3p and miR-216b. HEK293 cells were cotransfected with pGL3-SV40 (empty vector; white column) or a pGL3-SV40 construct containing the TrkB-Shc 3′UTR (black column) in combination with a miR-409-3p or control miRNA and b miR-216b or control miRNA. Data are normalized to the respective empty vector controls and expressed as relative luciferase activity. Values are presented as mean ± SEM and are representative of n = 3 independent experiments from three replicate cultures. * p = 0.002; ** p = 0.00002.
Effect of miR-409-3p and miR-216b on TrkB Expression
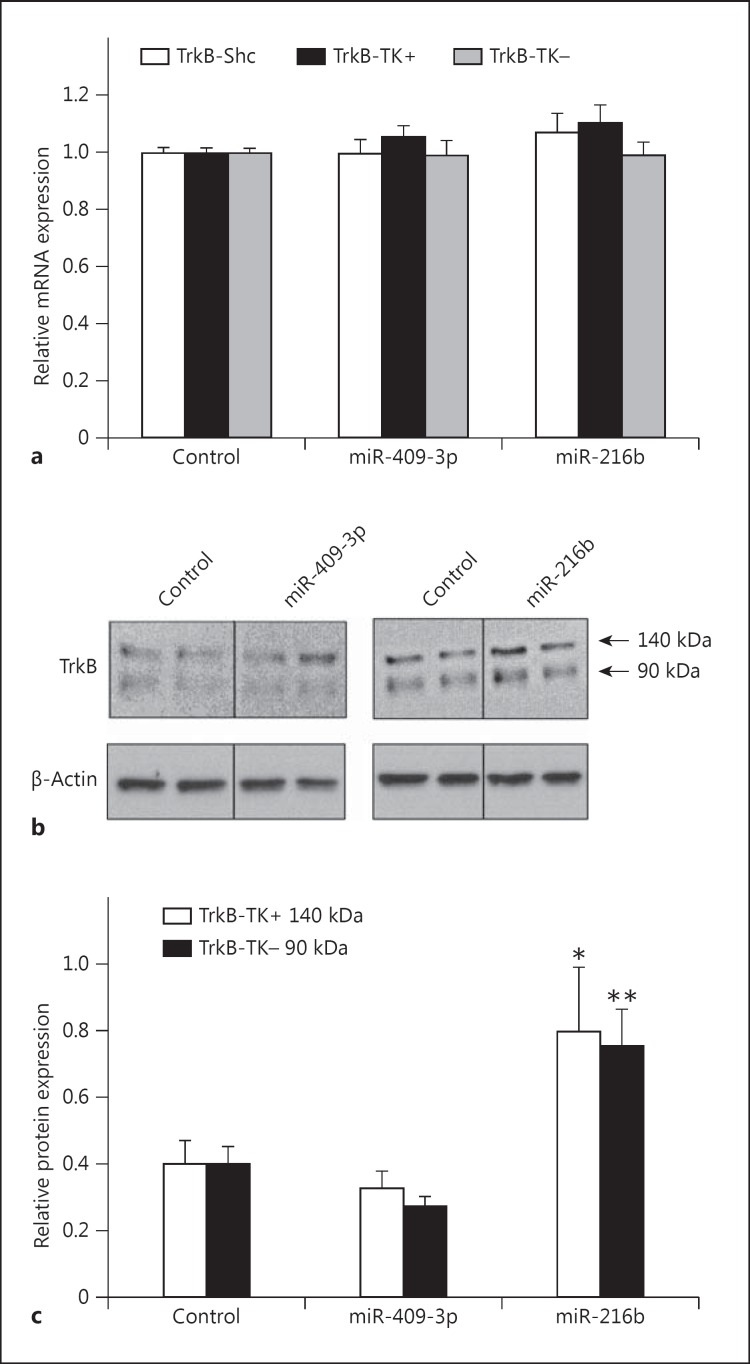
TrkB-Shc is a neuron-expressed TrkB isoform [5]. Thus, to determine whether miR-409-3p and miR-216b can regulate endogenous TrkB-Shc mRNA and protein expression in a neuronal setting, changes in TrkB-Shc expression were assessed using the neuronal cell-line SHSY5Y. SHSY5Y cells were transiently transfected with miR-409-3p and miR-216b for 48 h, and total RNA and protein were extracted to assess gene expression changes. Interestingly, no significant change in TrkB-Shc transcript levels was found in SHSY5Y cells following miR-409-3p and miR-216b transfections (miR-409-3p: t = 0.08, d.f. = 14, p = 0.94; miR-216b: t = −0.96, d.f. = 15, p = 0.35) (fig. 3a). Moreover, no significant changes were observed for the TrkB-TK+ or TrkB-TK-transcripts (miR-409-3p: TrkB-TK+: t = −1.42, d.f. = 15, p = 0.18; TrkB-TK-: t = 0.18, d.f. = 15, p = 0.86; miR-216b: TrkB-TK+: t = −1.65, d.f. = 16, p = 0.12; TrkB-TK-: t = 0.20, d.f. = 15, p = 0.84).
Fig. 3.
Effect of miR-409-3p and miR-216b on TrkB expression. a SHSY5Y cells were transfected with miR-409-3p, miR-216b, or control miRNA. Expression of TrkB-Shc (white column), TrkB-TK+ (black column), and TrkB-TK-(grey column) mRNA expression was measured by qPCR. Data are expressed as TrkB mRNA normalized to β-actin mRNA and presented as mean ± SEM. Values are representative of n = 3 independent experiments from three replicate cultures. b Representative Western blots of SHSY5Y cells probed with a TrkB antibody, which can detect the predicted 140-kDa protein (white column) representing the full length TrkB proteins including the TrkB-TK+ isoform as well as the 90-kDa protein (black column) representing the truncated TrkB proteins including the TrkB-TK- and TrkB-Shc isoforms. β-Actin was probed as a protein loading control. c Bands were quantified by densitometry. TrkB protein expression was normalized to β-actin protein and presented as mean ± SEM. * p = 0.03; ** p = 0.004.
However, when TrkB protein levels were assessed, significant changes were observed. Previously, it was shown by Western blotting that TrkB immunoreactive bands migrate at ∼140 kDa (corresponding to full-length TrkB) and ∼90 kDa (corresponding to truncated TrkB) (fig. 3b) [21,26]. Both TrkB-TK- and TrkB-Shc isoforms are similar in size and both migrate at ∼90 kDa.
Following miR-216b transfection, significant increases were observed in the 140- and 90-kDa bands corresponding to the full-length and truncated TrkB protein isoforms, respectively (miR-216b: 140 kDa: t = −2.36, d.f. = 15, p = 0.03; 90 kDa: t = −3.36, d.f. = 16, p = 0.004) (fig. 3c). In contrast, levels of the 140- and 90-kDa TrkB proteins were not significantly changed following miR-409-3p transfection (miR-409-3p: 140 kDa: t = 0.72, d.f. = 15, p = 0.48; 90 kDa: t = 1.66, d.f. = 16, p = 0.12). No significant changes in β-actin protein levels were observed following miRNA transfections.
Discussion
The mirSVR score was found to be a good indicator of miRNA binding. Experimentally, the putative miRNAs with scores ≤-0.1 were able to elicit a downregulatory effect on gene expression levels, whereas those with mirSVR scores ≥-0.1 did not alter gene expression levels. Importantly, the present findings substantiated this using miR-409-3p-BS1, a putative miRNA of interest with a mirSVR score >-0.1.
miRNAs and TrkB Protein Expression
Whilst reductions in luciferase reporter activity were observed for constructs regulated by the TrkB-Shc 3′UTR, decreases in the 90-kDa TrkB protein would have been expected. Instead, increased expression levels of both the 140- and 90-kDa TrkB protein isoforms were observed. This may have occurred as selective reduction in TrkB-Shc transcripts may lead to increased translation of TrkB-TK+ and TrkB-TK-transcripts, thus leading to increased protein levels. At present, there is no specific antibody available to detect the TrkB-Shc protein. Moreover, TrkB-Shc is predicted to have the same molecular weight as the truncated TrkB-TK-protein and other truncated TrkB proteins lacking the kinase domain [5]. Thus, any decrease in the TrkB-Shc protein is likely to be masked by increased expression of other truncated TrkB proteins. The observed increase in the 140-kDa TrkB protein supports this rationale. Furthermore, the reductions quantified in luciferase reporter levels detected for TrkB-Shc 3′UTR-regulated luciferase constructs support the likelihood that TrkB-Shc protein levels can be regulated by miR-216b. It is notable that changes in TrkB protein levels were likely to be specific for TrkB as no significant changes were detected in β-actin protein levels in the experimental conditions tested.
Differential Effect of miRNAs on TrkB Protein Expression
Changes in TrkB protein levels were unlikely to have occurred as a result of mRNA degradation. Gene expression regulation by miRNAs can either be due to the inhibition of translation of existing mRNA and/or due to mRNA degradation [27,28]. In this study, discordant regulation was observed between TrkB-Shc transcripts and TrkB proteins in miR-216b transfections. These findings are consistent with early studies in Caenorhabditis elegans, which showed that miRNAs may also repress gene expression by decreasing target protein levels, but without affecting the stability of the mRNA [29]. Moreover, the present findings are also consistent with a recent study by To et al. [28], which showed that miR-519c was capable of downregulating ABCG2 expression through both translation repression and mRNA degradation in HEK293 cells. However, they found that repression of ABCG2 gene expression by miR-519c was more pronounced through translation inhibition compared to increases in mRNA degradation [28]. Interestingly, while modulation of luciferase activity was observed in miR-409-p and miR-216b transfections using constructs cloned in the absence or presence of the TrkB-Shc 3′UTR, in contrast to miR-216b, no changes in TrkB protein levels were observed in transfection experiments using miR-409-3p. In early studies using C. elegans, small noncoding RNAs such as lin-4 and let-7 were shown to repress gene expression through imperfect complementary matches to sequences within the 3′UTRs of their target mRNAs [29]. To et al. [28] found that the functional distinction through which miR-519c operated could be predicted through the degree of base pairing between the miRNA and the miRNA-binding site in the 3′UTR. Additionally, they also found that it was the suboptimal degree of base pairing between miR-519c and the human ABCG2 3′UTR, which was critical for the observed changes [28]. When the miR-519c-binding site was replaced with a sequence complementary to miR-519c in the same ABCG2 expression vector, ABCG2 was repressed predominantly by mRNA degradation [28]. In this current study, suboptimal pairing was observed between the three putative miR-216b-binding sites in the TrkB-Shc 3′UTR and miR-216b, consistent with the mechanism proposed by To et al. In contrast, miR-409-3p showed partial but optimal base pairing with the TrkB-Shc 3′UTR.
It was previously found that spacing of the seed sites within the 3′UTR may play a significant role in the function of miRNAs, as sequences adjacent to the miRNA recognition elements (MREs) and/or the secondary structure of the 3′UTR of the target mRNA may affect the direct binding of miRNAs to the target sequence and/or affect regulatory protein binding, such as miRNPs, so that they cannot efficiently ‘unwind’ structured RNA areas; thus, indirectly preventing miRNAs from binding to sites that are embedded in such structured areas [29,30]. Additionally, multiple MREs for the same or different miRNAs within the identical 3′UTR can function cooperatively to enhance repression [29,30]. In this current study, six putative miR-216b-binding sites were identified in the TrkB-Shc 3′UTR of which three showed regulatory activity. In contrast, two miR-409-3p-binding sites were identified in the TrkB-Shc 3′UTR of which only one demonstrated regulatory activity. Thus, the different regulatory activity observed between miR-409-3p and miR-216b on TrkB protein expression may be due to the individual or combined effects of suboptimal base-pairing and multiple MREs.
While previous findings have shown that treatment of primary hippocampal neurons of mice with amyloid beta is associated with dramatic dysregulation of multiple miRNAs (including miR-409-3p) [18], the fact that miR-409-3p had no significant impact on TrkB protein expression in a neuronal setting may suggest that the effects of this miRNA on TrkB are subtle or may require interaction with other miRNAs or cellular processes. This may be plausible, since transfection experiments using miR-409-3p were able to reduce luciferase reporter activity when regulated by the TrkB-Shc 3′UTR. In contrast, miR-216 had a pronounced effect on TrkB protein expression. This current study provides experimental evidence to confirm that miR-216b can regulate TrkB-Shc expression and suggests that dysregulation in its expression may play a role in disease processes.
Conclusion
The miRNAs identified in this study highlights the importance of miRNA-mediated regulation on TrkB expression. On a general level, it reaffirms the relevance of miRNAs in the regulation of alternative transcript expression in biological cell systems.
Disclosure Statement
There are no conflicts of interest and financial disclosures.
Acknowledgements
J.W. was supported by a National Health and Medical Research Council of Australia (NHMRC) Postdoctoral Training Fellowship (No. 568884) and grants from the Illawarra Health and Medical Research Institute (University of Wollongong), the Centre for Medical Bioscience (University of Wollongong), and the Alzheimer's Australia Dementia Research Foundation (AADRF). Dr. Carol Dobson-Stone provided assistance in optimizing the luciferase assay and Dr. John B.J. Kwok contributed to the editing of the manuscript.
References
- 1.Barbacid M. The Trk family of neurotrophin receptors. J Neurobiol. 1994;25:1386–1403. doi: 10.1002/neu.480251107. [DOI] [PubMed] [Google Scholar]
- 2.Minichiello L, Korte M, Wolfer D, et al. Essential role for TrkB receptors in hippocampus-mediated learning. Neuron. 1999;24:401–414. doi: 10.1016/s0896-6273(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 3.Lu Y, Christian K, Lu B. BDNF: a key regulator for protein synthesis-dependent LTP and long-term memory? Neurobiol Learn Mem. 2008;89:312–323. doi: 10.1016/j.nlm.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marty S, Carroll P, Cellerino A, et al. Brain-derived neurotrophic factor promotes the differentiation of various hippocampal nonpyramidal neurons, including Cajal-Retzius cells, in organotypic slice cultures. J Neurosci. 1996;16:675–687. doi: 10.1523/JNEUROSCI.16-02-00675.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoilov P, Castren E, Stamm S. Analysis of the human TrkB gene genomic organization reveals novel TrkB isoforms, unusual gene length, and splicing mechanism. Biochem Biophys Res Commun. 2002;290:1054–1065. doi: 10.1006/bbrc.2001.6301. [DOI] [PubMed] [Google Scholar]
- 6.Ohira K, Shimizu K, Yamashita A, et al. Differential expression of the truncated TrkB receptor, T1, in the primary motor and prefrontal cortices of the adult macaque monkey. Neurosci Lett. 2005;385:105–109. doi: 10.1016/j.neulet.2005.05.033. [DOI] [PubMed] [Google Scholar]
- 7.Ohira K, Kumanogoh H, Sahara Y, et al. A truncated tropomyosin-related kinase B receptor, T1, regulates glial cell morphology via Rho GDP dissociation inhibitor 1. J Neurosci. 2005;25:1343–1353. doi: 10.1523/JNEUROSCI.4436-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohira K, Hayashi M. Expression of TrkB subtypes in the adult monkey cerebellar cortex. J Chem Neuroanat. 2003;25:175–183. doi: 10.1016/s0891-0618(02)00096-0. [DOI] [PubMed] [Google Scholar]
- 9.Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 10.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 11.Miska EA, Alvarez-Saavedra E, Townsend M, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smirnova L, Grafe A, Seiler A, et al. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 13.Guidi M, Muinos-Gimeno M, Kagerbauer B, et al. Overexpression of miR-128 specifically inhibits the truncated isoform of NTRK3 and upregulates BCL2 in SH-SY5Y neuroblastoma cells. BMC Mol Biol. 2010;11:95. doi: 10.1186/1471-2199-11-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maussion G, Yang J, Yerko V, et al. Regulation of a truncated form of tropomyosin-related kinase B (TrkB) by Hsa-miR-185* in frontal cortex of suicide completers. PLoS One. 2012;7:e39301. doi: 10.1371/journal.pone.0039301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong J, Garner B, Halliday GM, et al. Srp20 regulates TrkB pre-mRNA splicing to generate TrkB-Shc transcripts with implications for Alzheimer's disease. J Neurochem. 2012;123:159–171. doi: 10.1111/j.1471-4159.2012.07873.x. [DOI] [PubMed] [Google Scholar]
- 16.Hoozemans JJ, Chafekar SM, Baas F, et al. Always around, never the same: pathways of amyloid beta induced neurodegeneration throughout the pathogenic cascade of Alzheimer's disease. Curr Med Chem. 2006;13:2599–2605. doi: 10.2174/092986706778201585. [DOI] [PubMed] [Google Scholar]
- 17.Shankar GM, Walsh DM. Alzheimer's disease: synaptic dysfunction and Abeta. Mol Neurodegener. 2009;4:48. doi: 10.1186/1750-1326-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schonrock N, Ke YD, Humphreys D, et al. Neuronal microRNA deregulation in response to Alzheimer's disease amyloid-beta. PLoS One. 2010;5:e11070. doi: 10.1371/journal.pone.0011070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong J, Duncan C, Beveridge NJ, et al. Expression of NPAS3 in the human cortex and evidence of its posttranscriptional regulation by miR-17 during development, with implications for schizophrenia. Schizophr Bull. 2012. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 20.Beveridge NJ, Tooney PA, Carroll AP, et al. Down-regulation of miR-17 family expression in response to retinoic acid induced neuronal differentiation. Cell Signal. 2009;21:1837–1845. doi: 10.1016/j.cellsig.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 21.Wong J, Higgins MJ, Halliday G, et al. Amyloid beta selectively modulates neuronal TrkB alternative transcript expression with implications for Alzheimer's disease. Neuroscience. 2012;210:363–374. doi: 10.1016/j.neuroscience.2012.02.037. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Allen SJ, Wilcock GK, Dawbarn D. Profound and selective loss of catalytic TrkB immunoreactivity in Alzheimer's disease. Biochem Biophys Res Commun. 1999;264:648–651. doi: 10.1006/bbrc.1999.1561. [DOI] [PubMed] [Google Scholar]
- 24.Wong J, Garner B. Evidence that truncated TrkB isoform, TrkB-Shc can regulate phosphorylated TrkB protein levels. Biochem Biophys Res Commun. 2012;420:331–335. doi: 10.1016/j.bbrc.2012.02.159. [DOI] [PubMed] [Google Scholar]
- 25.Betel D, Koppal A, Agius P, et al. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong J, Rothmond DA, Webster MJ, et al. Increases in two truncated TrkB isoforms in the prefrontal cortex of people with schizophrenia. Schizophr Bull. 2013;39:130–140. doi: 10.1093/schbul/sbr070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valencia-Sanchez MA, Liu J, Hannon GJ, et al. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 28.To KK, Zhan Z, Litman T, et al. Regulation of ABCG2 expression at the 3′ untranslated region of its mRNA through modulation of transcript stability and protein translation by a putative microRNA in the S1 colon cancer cell-line. Mol Cell Biol. 2008;28:5147–5161. doi: 10.1128/MCB.00331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Felekkis K, Touvana E, Stefanou C, et al. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14:236–240. [PMC free article] [PubMed] [Google Scholar]
- 30.Lal A, Navarro F, Maher CA, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to ‘seedless’ 3′UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]