Abstract
Viruses are thought to spread across susceptible cells through an iterative process of infection, replication, and release, so that the rate of spread is limited by replication kinetics. Here, we show that vaccinia virus spreads across one cell every 75 minutes, fourfold faster than its replication cycle would permit. To explain this phenomenon, we found that newly infected cells express two surface proteins that mark cells as infected and, via exploitation of cellular machinery, induce the repulsion of superinfecting virions away toward uninfected cells. Mechanistically, early expression of proteins A33 and A36 was critical for virion repulsion and rapid spread, and cells expressing these proteins repelled exogenous virions rapidly. Additional spreading mechanisms may exist for other viruses that also spread faster than predicted by replication kinetics.
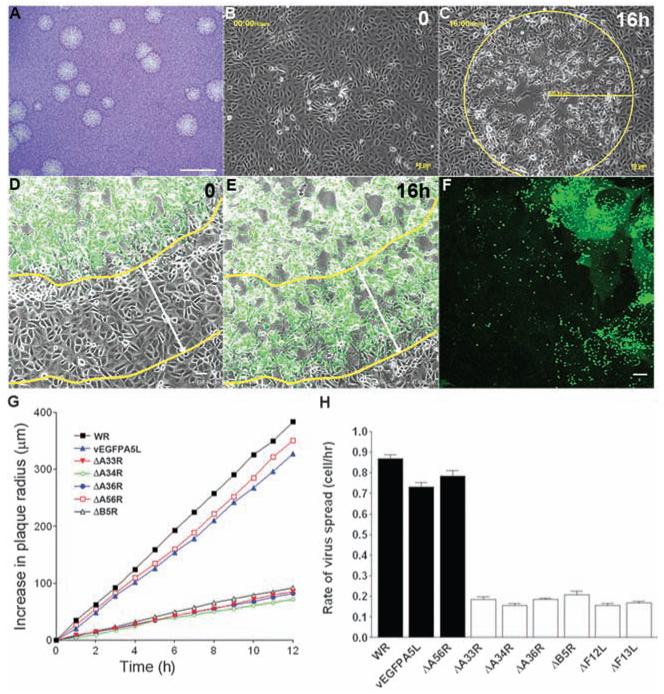
Mechanisms enhancing the cell-to-cell spread of intracellular pathogens are important for virulence and are targets for development of antimicrobial therapeutics. Vaccinia virus (VACV) is a poxvirus and is the live vaccine used to eradicate smallpox (1). VACV replication is unusual in that it produces both single- and double-enveloped virions (2, 3). The single-enveloped virions, called intracellular mature virus (IMV), remain intracellular until cell lysis and spread slowly from cell to cell. In contrast, the double-enveloped virions, called cell-associated enveloped virus (CEV) and extracellular enveloped virus (EEV), are released rapidly and mediate efficient cell-to-cell spread and long-range dissemination (3, 4). VACV spreading mechanisms include virus-induced cell motility (5) and the formation of actin projections (6-8) that propel VACV particles toward other cells late during infection (9). However, we wondered whether either mechanism could explain how VACV Western Reserve (WR) spreads rapidly to form a plaque of diameter 2.90 ± 0.07 mm (SEM, nine experiments, n = 11 to 12 plaques) in 3 days (Fig. 1A). The distance between nuclei of adjacent BSC-1 cells was 37.26 ± 1.02 μm (SEM, n = 25 single cells and the 5 to 8 cells in contact with it), so that VACV was spreading across each cell in <2 hours. To study this further, live video microscopy was used to measure the spread of VACV-induced cytopathic effect after infection with VACV WR (Fig. 1, B and C) or VACV expressing enhanced green fluorescent protein (EGFP) fused to core protein A5 that is expressed late during infection (vEGFPA5L) (Fig. 1, D to F, and movies S1 to S5) (10, 11). A linear increase in plaque size with time was observed (Fig. 1G), and the mean rate of spread was 32.36 ± 0.74 μm/hour (SEM, n = 9 plaques) for VACV WR. Knowing the distance between nuclei of adjacent cells, this indicated that VACV crossed one cell every 1.2 hours. This rate of spread is inconsistent with VACV replication kinetics, in which new virions are formed only 5 to 6 hours after infection (12), or virus-induced cell motility, in which cells start to move 5 to 6 hours after infection (5). These observations demonstrated that another mechanism to accelerate spread must exist. Mutants defective in actin tail formation (ΔA36R, ΔA33R, ΔA34R, ΔB5R, ΔF13L, and ΔF12L) (3) spread much more slowly (Fig. 1, G and H), infecting only one cell every 5 to 6 hours, which is consistent with replication kinetics. This reaffirmed the importance of actin tails for VACV spread but did not explain the rapid dissemination because actin tails are produced only late during infection after new virions are formed.
Fig. 1.
VACV spreads more rapidly than predicted. (A) VACV plaques 3 days after infection in BSC-1 cells. Scale bar, 5 mm. (B and C) Live cell imaging recording plaque formation at 0 (B) and 16 (C) hours later. (D and E) Live cell imaging of vEGFPA5L-infected cells confirmed the correlation between cytopathic effect (cpe) and virus infection. Yellow lines indicate the boundary between infected and uninfected cells and white arrows indicate the distance this has moved over 16 hours. (F) Confocal image showing the spread of EGFP-tagged virus particles (single green dots) far from the center of infection. (G) Increase in plaque radius formed by VACV WR and mutants with time; n = 6 to 11 plaques. (H) Diagram showing the rate of spread (BSC-1 cell per hour) with indicated viruses. White bars indicate viruses with a defect in actin tail formation. Error bars are SEM, with n = 6 to 11 plaques. Scale bar, (A) 5 mm, (B) to (E) 50 μm, and (F), 10 μm.
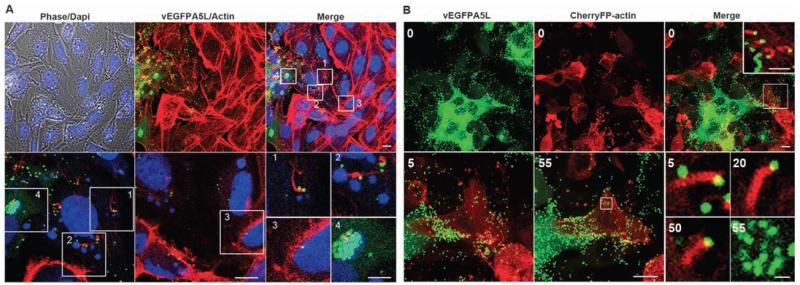
Inspection of plaques formed by vEGFPA5L revealed EGFP-positive virions (green dots) several cells away from EGFP-positive cells, where virions are formed, showing that VACV particles spread rapidly to distal cells (Fig. 1F). To investigate this phenomenon, actin was stained with phalloidin, and confocal optical sections revealed virus-tipped actin tails on cells producing new virions but also on distal cells lacking virus factories and so not producing virions (no EGFP expression) (Fig. 2A). This result was reproduced in different cell lines (BSC-1, RK13, ECV, and CEF) and using different VACV strains (WR and Lister) (figs. S1 and S2). To be certain that actin tails on cells that lack virus factories were not derived from distant virus-producing cells, a lawn of cells was formed in which some cells expressed cherry fluorescent protein fused to actin (cherry-actin) and thus produced red actin tails after virus infection (Fig. 2B and movies S6 and S7). After infection of such monolayers at low multiplicity, cells containing green factories adjacent to a red cell (cherry-actin positive) were studied. This revealed green virions on tips of red actin tails originating from a red cell that lacked any green factory and therefore new virions. Careful examination of z stacks of these cells confirmed no virus factory was present. Therefore, the virions and actin tail originated from different cells. Further examination by means of time-lapse microscopy revealed virus-tipped red actin tails on a cell 5, 20, and 50 min before green factory formation (55 min) (Fig. 2B), confirming that actin tails appeared before virion production. Furthermore, virions on a red actin tail were observed recontacting the same red cell and inducing another actin tail (movie S7). Thus, virions can be repelled repeatedly, thereby accelerating spread until an uninfected cell is found.
Fig. 2.
Cells form actin tails before production of new virions. (A) Confocal images showing the edge of vEGFPA5L plaque (green) on BSC-1 cells stained for actin (red) or DNA (blue). Bottom panel shows zoomed areas (white squares 1 to 4). Actin tails are on cells with nascent factories (cytoplasmic blue) but that are not producing any virus particles (green) (squares 1 and 2), and on a cell with no virus factory (square 3), whereas square 4 shows a productive virus factory (green). Scale bars, top row, 10 μm; bottom row, 10 μm; and insets 1 to 4, 5 μm. (B) Actin tails (red) present at the surface of a cell expressing cherryFP-actin but with no green virus factory [time (t) = 0, white square and zoomed inset]. Bottom panels show zoomed images of this cell with actin tails detected 5, 20, and 50 min later, before the appearance of virus factories at 55 min as indicated by the white square. Scale bars, top row, 10 μm; top right inset, 5 μm; bottom row, 10 μm; and bottom right inset, 1 μm.
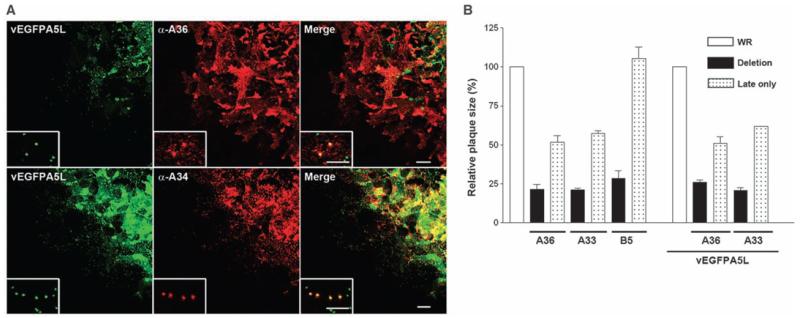
To understand this mechanism, we searched for the required VACV proteins. Proteins K2 and A56 inhibit entry of IMV by binding the membrane fusion complex on the IMV surface (13-15), but this is masked on EEV or CEV, and mutants lacking these genes form normal-sized plaques (16, 17). Better candidates would be early, cell-surface proteins that are needed for actin tail formation. Although proteins A33, A34, A36, B5, F12, and F13 are needed for efficient actin tail formation, only surface proteins B5 (18), A33 (19), and A36 (20) are expressed early (and late). Immunostaining of plaques formed by vEGFPA5L demonstrated that A33 and A36 are expressed early during infection at the periphery of plaques before expression of late proteins (A34, EGFP-A5, and B5) (Fig. 3A and fig. S3). A34 and B5 were nevertheless detectable on virions spreading toward noninfected cells, as expected. Thus, A33 and A36 seemed candidates for early induction of actin tails.
Fig. 3.
Early expression of A33 and A36 is important for VACV spread. (A) Images of edge of plaque showing A36, but not A34, is expressed early during infection. A36 was detected in cells where no late protein A5 (green) was present, whereas A34 was expressed late during infection in cells that also express A5. (Insets) Zoomed images of virions (single green dots) relative to A36 and A34 distribution. (B) Graph showing the size of plaques formed by recombinant viruses in which A33R, A36R, or B5R are under a late promoter only (4b) or deleted (Δ) as compared with parental viruses WR or vEGFPA5L. Error bars are SEM mean values from three experiments with n = 11 to 12 plaques. Scale bars, 20 μm; insets, 5 μm.
The importance of early expression of A33 and A36 was investigated by generating recombinant viruses in which A33R, A36R, or B5R genes were driven only by a late promoter (21). Infection of cells by these viruses (v4b-A33, v4b-A36, and v4b-B5) confirmed expression of these proteins only late during infection (fig. S4) and showed that plaques formed by v4b-A33 and v4b-A36 were much smaller than wild type and closer to those formed by deletion mutants lacking either gene (Fig. 3B). In contrast, v4b-B5 formed plaques similar to wild type. Thus, early expression of A33 and A36, but not B5, is critical for efficient VACV spread. Similar results were obtained with viruses in which 4b-A33R and 4b-A36R were inserted into vEGFP-A5L, allowing direct visualization of spreading virions via EGFP. Virions released from cells infected by these viruses (vEGFPA5L/4b-A33 and vEGFPA5L/4b-A36) spread poorly as compared with vEGFPA5L and induced actin tails only on cells with a virus factory (fig. S5).
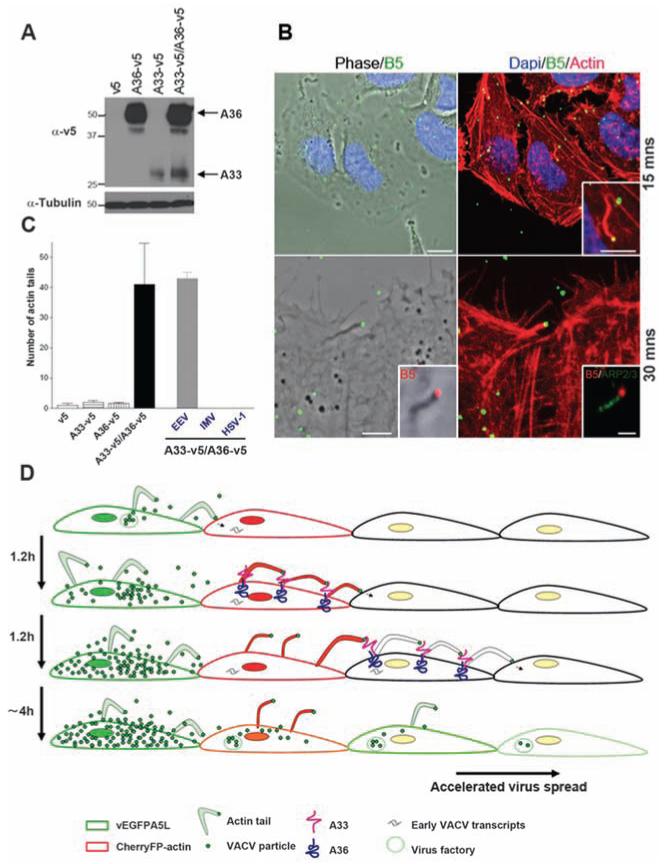
Next, we investigated whether A33 and A36 are sufficient to induce actin tails upon contact with an EEV particle. Both proteins are expressed on the plasma membrane of VACV-infected cells (22-24) and interact with each other (25-27). Lentivirus vectors expressing A33 or A36 fused to a C-terminal V5 tag were used to generate HeLa cells expressing cell surface A33-v5, A36-v5, or A36-v5 and A33-v5 (Fig. 4A and fig. S6). These cells were incubated with EEV particles and then stained with phalloidin and a monoclonal antibody to B5. Actin tails were detected on cells expressing A33-v5 and A36-v5 within 15 to 30 min (Fig. 4, B and C). Similar results were obtained with haemagglutinin (HA)–tagged A33. No actin tails were detected if only one protein was expressed or if the cells were incubated with IMVor with GFP-tagged herpes simplex virus 1 (HSV-1). Thus, A33 and A36 are both necessary and sufficient to induce actin tails after binding EEV particles. Currently, we are investigating the effect of ectopic expression of A33 and A36 on VACV spread by measuring plaque size using additional cell lines that form clearer plaques.
Fig. 4.
Expression of A33 and A36 is sufficient for actin tail formation. (A) Immunoblot showing A33 and A36 expression. (B) Actin tails present 15 and 30 min after spinoculation of EEV particles onto HeLa cells expressing A33 and A36 proteins. Staining for Arp2/3 shows actin polymerisation machinery. Scale bars, top row, 10 μm; inset, 5 μm; bottom row, 5 μm; inset, 1 μm. (C) Graph showing the mean number of actin tails detected per coverslip in the different cell lines. Actin tails were not formed by IMV or HSV-1. Error bars are SEM; n = 3 experiments. (D) Model showing how VACV spreads rapidly. The first infected cell expresses EGFP-A5 late during infection and releases green virions, which infect an adjacent cell expressing cherry actin (red). Early after infection, A33 and A36 are expressed at the cell surface and mark the cell as infected. Upon contact with new CEV/EEV particles, the A33/A36 complex induces the formation of red actin tails, which repel these virions toward uninfected cells. Superinfecting virions may be repelled from multiple infected cells before an uninfected cell is found that can be infected.
Here, we demonstrate that VACV has evolved a mechanism (Fig. 4D) by which infected cells repel superinfecting CEV/EEV particles on actin tails toward neighboring cells. Two outcomes are then possible: (i) If the neighboring cell is un-infected, the virion enters and starts a new cycle of replication; (ii) alternatively, if the cell is already infected then superinfection is blocked, and a new actin tail is formed, propelling the virus further away until it reaches uninfected cells. This mechanism accelerates virus spread and explains how VACV can cross one cell every 1.2 hours as determined by means of live cell imaging. Early expression of proteins A33 and A36 is required, and viruses expressing either protein only late during infection form small plaques. These plaques are closer in size to those formed by the deletion mutants lacking either gene than to wild type, indicating that the formation of actin tails upon superinfection is more important for virus spread than the production of actin tails on cells releasing new virions. All mutations that cause VACV strains to spread poorly and form small plaques also cause dramatic attenuation in vivo, showing the biological importance of rapid spread for VACV virulence (18, 20, 28, 29).
Plaque assays were first described more than 50 years ago (30), and many animal viruses form plaques of size comparable with VACV. Some of these viruses, for instance HSV-1 (31), have replication kinetics similar to VACV, suggesting that other viruses also spread faster than predicted by their replication kinetics. The mechanisms underlying cell-to-cell spread of many viruses remain poorly understood, and the elucidation of such mechanisms could lead to the discovery of novel therapeutics.
Supplementary Material
Acknowledgments
We thank Professor Rick E. Randall, University of St. Andrews for the lentivirus vectors. GLS is a Wellcome Trust Principal Research Fellow. This work was supported by the UK Medical Research Council.
Footnotes
www.sciencemag.org/cgi/content/full/science.1183173/DC1
Materials and Methods
Figs. S1 to S6
References Movies S1 to S7
References and Notes
- 1.Moss B. In: Fields Virology. Knipe DM, editor. Vol. 2. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 2905–2946. [Google Scholar]
- 2.Condit RC, Moussatche N, Traktman P. Adv. Virus Res. 2006;66:31. doi: 10.1016/S0065-3527(06)66002-8. [DOI] [PubMed] [Google Scholar]
- 3.Smith GL, Vanderplasschen A, Law M. J. Gen. Virol. 2002;83:2915. doi: 10.1099/0022-1317-83-12-2915. [DOI] [PubMed] [Google Scholar]
- 4.Roberts KL, Smith GL. Trends Microbiol. 2008;16:472. doi: 10.1016/j.tim.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Sanderson CM, Way M, Smith GL. J. Virol. 1998;72:1235. doi: 10.1128/jvi.72.2.1235-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blasco R, Cole NB, Moss B. J. Virol. 1991;65:4598. doi: 10.1128/jvi.65.9.4598-4608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiller G, Weber K, Schneider L, Parajsz C, Jungwirth C. Virology. 1979;98:142. doi: 10.1016/0042-6822(79)90533-6. [DOI] [PubMed] [Google Scholar]
- 8.Stokes GV. J. Virol. 1976;18:636. doi: 10.1128/jvi.18.2.636-643.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cudmore S, Cossart P, Griffiths G, Way M. Nature. 1995;378:636. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 10.Carter GC, et al. J. Gen. Virol. 2003;84:2443. doi: 10.1099/vir.0.19271-0. [DOI] [PubMed] [Google Scholar]
- 11.Materials and methods are available as supporting material on Science online.
- 12.Payne LG, Kristenson K. J. Virol. 1979;32:614. doi: 10.1128/jvi.32.2.614-622.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner PC, Moyer RW. Virology. 2008;380:226. doi: 10.1016/j.virol.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagenaar TR, Moss B. J. Virol. 2007;81:6286. doi: 10.1128/JVI.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagenaar TR, Moss B. J. Virol. 2009;83:1546. doi: 10.1128/JVI.01684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Law KM, Smith GL. J. Gen. Virol. 1992;73:549. doi: 10.1099/0022-1317-73-3-549. [DOI] [PubMed] [Google Scholar]
- 17.Law M, Hollinshead R, Smith GL. J. Gen. Virol. 2002;83:209. doi: 10.1099/0022-1317-83-1-209. [DOI] [PubMed] [Google Scholar]
- 18.Engelstad M, Smith GL. Virology. 1993;194:627. doi: 10.1006/viro.1993.1302. [DOI] [PubMed] [Google Scholar]
- 19.Roper RL, Wolffe EJ, Weisberg A, Moss B. J. Virol. 1998;72:4192. doi: 10.1128/jvi.72.5.4192-4204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkinson JE, Smith GL. Virology. 1994;204:376. doi: 10.1006/viro.1994.1542. [DOI] [PubMed] [Google Scholar]
- 21.Rosel J, Moss B. J. Virol. 1985;56:830. doi: 10.1128/jvi.56.3.830-838.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorenzo MM, Galindo I, Griffiths G, Blasco R. J. Virol. 2000;74:10535. doi: 10.1128/jvi.74.22.10535-10550.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roper RL, Payne LG, Moss B. J. Virol. 1996;70:3753. doi: 10.1128/jvi.70.6.3753-3762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Eijl H, Hollinshead M, Smith GL. Virology. 2000;271:26. doi: 10.1006/viro.2000.0260. [DOI] [PubMed] [Google Scholar]
- 25.Perdiguero B, Blasco R. J. Virol. 2006;80:8763. doi: 10.1128/JVI.00598-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Röttger S, Frischknecht F, Reckmann I, Smith GL, Way M. J. Virol. 1999;73:2863. doi: 10.1128/jvi.73.4.2863-2875.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolffe EJ, Weisberg AS, Moss B. J. Virol. 2001;75:303. doi: 10.1128/JVI.75.1.303-310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McIntosh AA, Smith GL. J. Virol. 1996;70:272. doi: 10.1128/jvi.70.1.272-281.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang WH, Wilcock D, Smith GL. J. Virol. 2000;74:11654. doi: 10.1128/jvi.74.24.11654-11662.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dulbecco R. Proc. Natl. Acad. Sci. U.S.A. 1952;38:747. doi: 10.1073/pnas.38.8.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elliott G, O’Hare P. J. Virol. 1999;73:4110. doi: 10.1128/jvi.73.5.4110-4119.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.