Abstract
Soybean is sensitive to flooding stress and exhibits reduced growth under flooding conditions. To better understand the flooding-responsive mechanisms of soybean, the effect of exogenous calcium on flooding-stressed soybeans was analyzed using proteomic technique. An increase in exogenous calcium levels enhanced soybean root elongation and suppressed the cell death of root tip under flooding stress. Proteins were extracted from the roots of 4-day-old soybean seedlings exposed to flooding stress without or with calcium for 2 days and analyzed using gel-free proteomic technique. Proteins involved in protein degradation/synthesis/posttranslational modification, hormone/cell wall metabolisms, and DNA synthesis were decreased by flooding stress; however, their reductions were recovered by calcium treatment. Development, lipid metabolism, and signaling-related proteins were increased in soybean roots when calcium was supplied under flooding stress. Fermentation and glycolysis-related proteins were increased in response to flooding; however, these proteins were not affected by calcium supplementation. Furthermore, urease and copper chaperone proteins exhibited similar profiles in 4-day-old untreated soybeans and 4-day-old soybeans exposed to flooding for 2 days in the presence of calcium. These results suggest that calcium might affect the cell wall/hormone metabolisms, protein degradation/synthesis, and DNA synthesis in soybean roots under flooding stress.
Keywords: soybean, flooding, calcium, proteomics, root
Introduction
Flooding events caused by heavy rainfall have increased globally over the past six decades as a consequence of climate change (Bailey-Serres et al., 2012). Flooding severely affects the productivity of farmland, because most agriculturally important crops are intolerant to flooding stress (Setter and Waters, 2003). One of the main effects of flooding is a marked reduction in oxygen availability due to the slower diffusion of gas in water as compared to air (Dat et al., 2004). Thus, when plants are exposed to flooding conditions, the roots initially suffer from oxygen deficiency (Sauter, 2013), leading to the inhibition of root respiration and a marked decrease in the energy status of root cells (Ashraf, 2012). In response to flooding, plants activate an alternative fermentation metabolic pathway to produce ATP and regenerate NAD+ (Gibbs and Greenway, 2003), as evidenced by the drastic increase in alcohol dehydrogenase activity in soybean under flooding stress (Komatsu et al., 2011). Although the flooding-induced changes in energy metabolism are important processes in adaptation to flooding conditions, metabolism induced by flooding and adaption to flooding in soybean remain unclear.
Calcium is an essential plant nutrient that determines the structure of cell wall and membranes, and plays a role in regulation of growth and development (Hepler, 2005). Calcium also has functions in protecting the integrity of cell membranes, reducing membrane permeability, and preventing ion leakage caused by biotic and abiotic stresses (Lin et al., 2008). Exogenous calcium alleviated the suppression of plant growth and kept plant to maintain and modulate cellular function by relieving gene repression under salt (Henriksson and Nordin Henriksson, 2005), anoxic (Aurisano et al., 1995), and chilling stresses (Gao et al., 2004). Calcium served the regulated optimal amounts of antioxidative enzymes and antioxidants to the antioxidative systems in leaves of sweet potato under waterlogging stress (Lin et al., 2008). Furthermore, improved tolerance to short-term hypoxia by calcium-mediated reduction of polyamine degradation, elevation of nitrate uptake, and accelerated synthesis of heat-stable proteins and polyamines was reported in muskmelon roots (Gao et al., 2011). Calcium was also shown to be effective in soybean for reducing Phytophthora stem rot disease, which is caused by Phytophthora soja, in flooded soil (Sugimoto et al., 2005). These observations indicate that study of the roles of calcium in protecting plants against environmental stresses, including flooding, will aids in the understanding of stress tolerance mechanisms in soybean.
Soybean is an important crop worldwide as it serves an abundant source of both protein and oil for animal and human consumption (Hartman et al., 2011). However, soybean is particularly sensitive to flooding (Sullivan et al., 2001), which markedly reduces the growth and productivity of plants (Githiri et al., 2006). Studies on the flooding-responsive mechanisms in soybean using proteomics-based approaches have revealed that the levels of several proteins involved in signal transduction, glucose degradation/sucrose accumulation, alcohol fermentation, and cell wall loosening were changed under flooding conditions (Komatsu et al., 2012a). Specifically, proteins involved in energy production were increased, whereas proteins involved in protein folding and cell structure maintenance were decreased in response to flooding stress in soybean (Nanjo et al., 2012). In addition, these proteomic studies identified a number of calcium-related proteins that may play important roles in flooding stress-responsive mechanisms in soybean.
To further understand calcium-related signaling pathway under flooding condition, Komatsu et al. (2013a) investigated calcium-related proteins in soybean cotyledon under flooding conditions using a proteomic technique and suggested that calcium might play a role in flooding-induced signal transduction through heat shock protein 70. Annexin, a calcium-dependent membrane-binding protein, was identified in soybean under flooding stress with abscisic acid supplementation (Komatsu et al., 2013b). The levels of calreticulin, a calcium-binding protein with chaperone functions (Menegazzi et al., 1993), were also lower in soybean under flooding stress (Komatsu et al., 2009), and a number of annexin proteins, calcium-transporting ATPase 4, calnexin, luminal-binding protein, and calcium ion-binding protein have been identified in flooded soybean root using gel-based and gel-free proteomic techniques (Komatsu et al., 2012b). These changes have effect on the calcium signaling under flooding stress and appear to be contributed flooding-responsive mechanism. Various calcium-related proteins, including annexin, calreticulin, calcium-binding EF-hand family protein, and calcium-dependent lipid-binding family protein, are changed in soybean under flooding stress (Oh et al., 2014a). Taken together, these results indicate that calcium has functions as a key signaling regulator in response to flooding by controlling calcium-related proteins in soybean. However, calcium-induced flooding response metabolism and response in soybean remain to be determined.
Because of the importance of calcium-regulatory mechanisms in plants to adjust to adverse abiotic stresses, including flooding, several studies have examined the involvement of calcium in stress-response mechanisms. Although calcium has been shown to ameliorate stress-induced damage in other crop species (He et al., 2012), calcium-mediated flooding-responsive mechanisms are poorly understood in soybean. In the present study, a gel-free proteomic technique was used to investigate the effect of calcium on soybean under flooding stress.
Materials and methods
Plant material and treatment
Soybean (Glycine max L. cultivar Enrei) seeds were sterilized with 1% sodium hypochlorite solution, rinsed in water, and sown in a plastic case (180 × 140 × 45 mm) containing 500 mL quartz sand wetted with 125 mL water. Soybeans were grown in a growth chamber under white fluorescent light (160 μmol m−2s−1, 16 h light period/day) at 25°C and 70% relative humidity. Two-day-old soybeans were transferred to a glass tube (38 × 130 mm) containing 120 mL tap water supplemented without or with 1, 5, 10, and 50 mM CaCl2 for flooding stress treatment, and further grown at 25°C under dark conditions. For physiological experiments, 4, 6, and 8-day-old soybeans treated with flooding for 2, 4, and 6 days, respectively, were collected and the length and weight of roots, including the hypocotyl, were measured. At the time of collection, soybeans were also stained with Evans blue dye, and the amount of dye extracted from stained root tips was measured spectroscopically as described below. For proteomic analysis, roots were collected from 4-day-old soybeans flooded without or with 50 mM CaCl2 for 2 days. For quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis, roots, hypocotyls, and cotyledons were collected from 2-, 3-, and 4-day-old soybean flooded without or with 50 mM CaCl2 for 0, 1, and 2 days, respectively. For all experiments, non-treated equivalent soybeans were collected as controls, and three independent biological replicates were performed for each experiment (Supplemental Figure 1).
Evans blue staining for assay of cell death
Root tip cell death was evaluated by Evans blue staining, as described by Baker and Mock (1994) and Delisle et al. (2001). Briefly, soybeans were stained in a 0.25% aqueous solution of Evans blue for 15 min at room temperature. The stained samples were washed with water and immediately photographed. For quantitative assessment of staining, the terminal 5 mm of stained root tips was excised and immersed in 200 μL N,N-dimethylformamide for 24 h at 4°C. After the incubation, the absorbance of Evans blue released from the root tips was measured spectroscopically at 600 nm.
Protein extraction
A portion (0.5 g) of fresh roots was ground to a powder in liquid nitrogen with a mortar and pestle. The powder was added to an acetone solution containing 10% trichloroacetic acid and 0.07% 2-mercaptoethanol, and the resulting mixture was vortexed and then sonicated for 10 min. The suspension was incubated for 1 h at −20°C with vortexing every 15 min, and was then centrifuged at 9000× g for 20 min at 4°C. The resulting supernatant was discarded and the obtained pellet was washed twice with 0.07% 2-mercaptoethanol in acetone. The pellet was dried using a Speed-Vac concentrator (Savant Instruments, Hicksville, NY, USA) and resuspended in lysis buffer containing 7 M urea, 2 M thiourea, 5% CHAPS, and 2 mM tributylphosphine by vortexing for 1 h at 25°C. The suspension was centrifuged at 20,000× g for 20 min at 25°C, and the supernatant was collected as protein extract. Protein concentrations were determined using the Bradford method (Bradford, 1976) with bovine serum albumin as the standard.
Protein purification and digestion for mass spectrometry analysis
Extracted proteins (100 μg) were purified with methanol and chloroform to remove any detergent from the sample solutions, as previously described (Nanjo et al., 2012). Briefly, 400 μL methanol was added to 100 μL samples and then mixed. A total of 100 μL chloroform and 300 μL water were added to resulting mixture, mixed, and centrifuged at 20,000× g for 10 min to achieve phase separation. The upper phase was discarded, 300 μL methanol was added to lower phase, and the samples were further centrifuged at 20,000× g for 10 min. The supernatants were discarded, and the obtained pellets were dried. Dried samples were reduced with 50 mM dithiothreitol for 30 min at 56°C and the alkylated with 50 mM idoacetoamide for 30 min at 37°C in the dark. Alkylated proteins were digested with trypsin and lysyl endopeptidase at 1:100 enzyme/protein concentrations at 37°C for 16 h. The resulting tryptic peptides were acidified with formic acid, desalted with a C18-pipette tip (SPE C-TIP, Nikkyo Technos, Tokyo, Japan), and then analyzed by nano-liquid chromatography (LC)-tandem mass spectrometry (MS).
Nanoliquid chromatography-mass spectrometry analysis
Using an Ultimate 3000 Nano LC system (Dionex, Germering, Germany), peptides in 0.1% formic acid were loaded onto a C18 PepMap trap column (300 μm ID × 5 mm, Dionex). The peptides were eluted from the trap column with a linear acetonitrile gradient (8–30% in 120 min) in 0.1% formic acid at a flow rate of 200 nL/min, and then loaded onto a C18 NANO HPLC NTTC-360/75-3 capillary tip column (75 μm ID × 120 mm, Nikkyo Technos) using a spray voltage of 1.5 kV. A nanospray LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) was operated in data-dependent acquisition mode with the installed Xcalibur software (version 2.0.7, Thermo Fisher Scientific). Full-scan mass spectra were acquired in the Orbitrap MS over 400–1500 m/z with a resolution of 30,000. A lock mass function was used for high mass accuracy (Olsen et al., 2005). The 10 most intense precursor ions were selected for collision-induced fragmentation in the linear ion trap at a normalized collision energy of 35%. Dynamic exclusion was employed within 90 s to prevent the repetitive selection of peptides (Zhang et al., 2009).
Protein identification
Proteins were identified by Mascot searches (version 2.4.1, Matrix Science, London, UK) of a soybean peptide database (55,787 sequences) constructed from the soybean genome database (Phytozome version 9.1, http://www.phytozome.net/soybean) (Schmutz et al., 2010). The acquired raw data files were processed using Proteome Discover software (version 1.4, Thermo Fisher Scientific). The parameters used in the Mascot searches were as follows: carbamidomethylation of cysteine was set as a fixed modification and oxidation of methionine was set as a variable modification. Trypsin was specified as the proteolytic enzyme and one missed cleavage was allowed. Peptide mass tolerance was set at 5 ppm, fragment mass tolerance was set at 0.5 Da, and peptide charge was set at +2, +3, and +4. An automatic decoy database search was performed as part of the analysis. Mascot results were filtered with the Percolator function to improve the accuracy and sensitivity of peptide identification. False discovery rates for peptide identification in all searches were less than 1.0%. The Mascot results were imported for SIEVE analysis (version 2.1, Thermo Fisher Scientific), which was performed as described in the following section.
Data analysis of differential abundant proteins acquired using mass spectrometry
For differential analysis of the relative abundance of peptides and proteins between the control and treatment groups, the commercial label-free quantification package SIEVE was used. All SIEVE data analysis was acquired from 3 biological replicates of MS results. The chromatographic peaks detected by MS were aligned, and the peptide peaks were detected as a frame for all parent ions scanned by MS/MS using a frame time width of 5 min and frame m/z width of 10 ppm. Chromatographic peak areas within frames of each sample were compared, and the ratios between two sample groups for each frame were determined. The frames detected in the MS/MS scans were matched with the imported Mascot results. The ratios of peptides between samples were determined from the variance-weighted average of the ratios in frames that matched to the peptides with MS/MS spectrum. The ratios of peptides were further integrated with Ingenuity Pathways Analysis which is a widely-adopted application for 3 biological replications of MS into a ratio of protein to determine the ratio of the corresponding protein. In the differential analysis, total ion current was used for normalization. For the identification of differentially changed proteins, the minimum requirements for the identification of a protein were two matched peptides, and the protein quantities required a greater than two-fold difference with t-test significance (P < 0.05) between the flooding-treated and control samples.
Functional categorization analysis
Proteins were categorized based on function using MapMan bin codes (Usadel et al., 2005).
RNA extraction and quantitative reverse transcription-polymerase chain reaction analysis
A portion (100 mg) of soybean roots, hypocotyls, and cotyledons was ground into powder with a sterilized pestle and mortar in liquid nitrogen. Total RNA was extracted from the powdered tissue using an RNeasy Plant Mini kit (Qiagen, Valencia, CA, USA) and was then reverse-transcribed to cDNA using iScript Reverse Transcription Supermix (Bio-Rad, Hercules, CA, USA) according to the manufacturer's instructions. qRT-PCR was performed using the cDNA as template and SsoAdvanced SYBR Green Supermix (Bio-Rad) on a MyiQ Single-Color Real-Time PCR Detection system (Bio-Rad). The qRT-PCR was performed under the following conditions: 95°C for 30 s, followed by 45 cycles of 95°C for 10 s and 60°C for 30 s. Relative mRNA levels were calculated through normalization using 18S rRNA (X02623.1) abundance. Primer sequences were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/input.htm) (Rozen and Skaletsky, 2000) and are listed in Supplemental Table 1.
Statistical analysis
The statistical significance of the results was evaluated with one-way ANOVA followed by Duncan's multiple comparisons test, unless otherwise stated. All calculations were performed using SPSS software (version 22.0). A p-value of <0.05 was considered to be statistically significant.
Results and discussion
Effects of calcium on growth of soybean under flooding stress
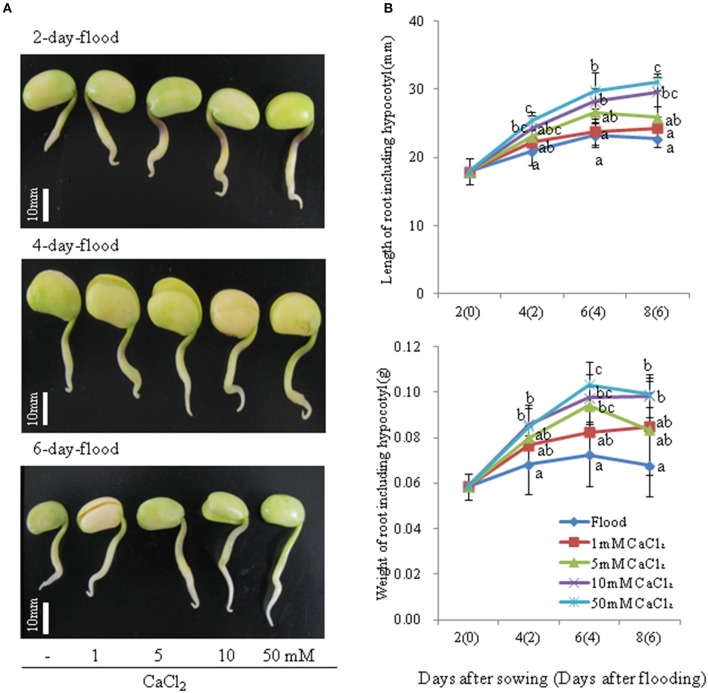
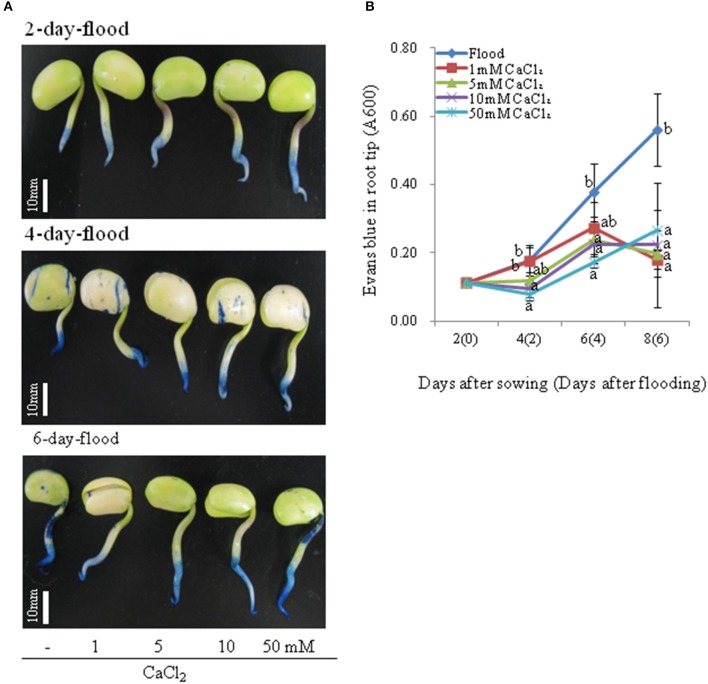
To investigate the effects of calcium on morphological changes induced by flooding in soybean and evaluate the corresponding degree of root tip damage, 2-day-old soybeans were flooded for 2, 4, and 6 days without or with 1, 5, 10, and 50 mM CaCl2. The total length of root including hypocotyl under flooding with various concentrations of CaCl2 was clearly longer than that under flooding without CaCl2 (Figure 1A). Although no marked differences in root length were detected between flooded soybean in the presence or absence of 1 mM CaCl2, the length after treatment with 10 and 50 mM CaCl2 was significantly longer than that under flooding without CaCl2 (Figure 1B). Consistent with this finding, the root weight of flooding-treated soybeans was also higher for plants exposed to CaCl2 during the treatment period (Figure 1B). The length and weight of root including hypocotyl were measured after each treatment in biological triplicates to assess reproducibility (Supplemental Figures 2A–C). The length and weight of root were gradually increased in 2-, 4-, and 6-day flooded soybean with 10 and 50 mM CaCl2. In particularly, 2-day-flooded soybean was most affected by CaCl2 (Figure 1). The flooding-treated soybeans were also stained with Evans blue dye to evaluate cell death (Figure 2). The degree of staining in root tips was dependent on the CaCl2 concentration and treatment period (Figure 2A). Cell death in the root tip was severely induced by flooding without CaCl2 compared to that in the presence of 50 mM CaCl2 (Figure 2B). The Evans blue staining was also performed in biological triplicate experiments to assess reproducibility (Supplemental Figures 3A–C). Even though cell death in the root tip was significantly suppressed by flooding with CaCl2 in 4 and 6 days, the degree of staining in root tips was most affected by CaCl2 in 2-day-flooded soybean (Figure 2).
Figure 1.
Effect of calcium on growth of soybean under flooding stress. Two-day-old soybeans were flooded without (dark blue) or with 1 (red), 5 (light green), 10 (purple), and 50 mM CaCl2 (light blue) for 2, 4, and 6 days. (A) Photographs show soybean seedlings after 2, 4, and 6 days of flooding. Bars indicate 10 mm. (B) Length and weight of roots, including the hypocotyl, were measured at the indicated time points. Data are means ± SE from three independent biological replications. Means with the same letter are not significantly different according to ANOVA (P < 0.05).
Figure 2.
Evaluation of cell death in flooding-stressed soybean root tips treated with calcium. Two-day-old soybeans were flooded without (dark blue) or with 1 (red), 5 (light green), 10 (purple), and 50 mM CaCl2 (light blue) for 2, 4, and 6 days. The roots were stained with 0.25% Evans blue dye, which was then extracted and measured spectroscopically at 600 nm. (A) Photographs show soybeans after 2, 4, and 6 days of flooding. Bars indicate 10 mm. (B) Absorbance of Evans blue in root tips at the indicated time points. Data are means ± SE from three independent biological replications. Means with the same letter are not significantly different according to ANOVA (P < 0.05).
Nanjo et al. (2013) reported that the amount of Evans blue uptake in the root tip region under 3 days flooding, which is evidence of cell death (Delisle et al., 2001), affected by the volume of floodwater and suggested that the loss of root tips in flooded soybean seedlings is due to flood-induced cell death. Consistent with this speculation, Evans blue uptake was markedly induced in soybean seedlings by 4 days flooding (Komatsu et al., 2013c). In the present study, the application of 50 mM CaCl2 clearly promoted root elongation and suppressed root tip cell death in soybeans during 6 days flooding stress, in particularly, in 2 days flooding treatment with CaCl2. Taken together, these findings indicate that flooding-induced root tip damage might be suppressed by exogenous calcium treatment. Based on the present results, for subsequent proteomic and qRT-PCR analyses, 50 mM CaCl2 was selected for the treatment in soybean under flooding.
Protein profiles in flooding-stressed soybean root treated with calcium
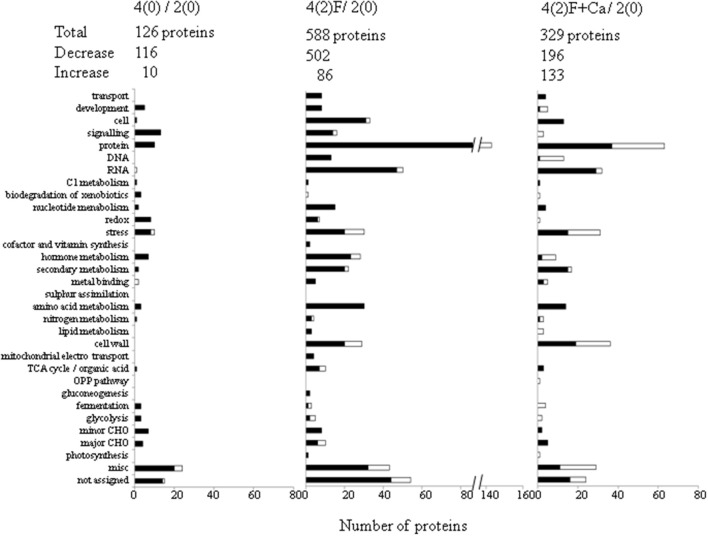
To investigate the effect of calcium on protein profiles in soybean roots under flooding stress, a gel-free proteomic technique was used. Because 2-day-old soybean was considered as starting point of each treatment, 2-day-old soybean was used for comparison. Proteins were extracted from the roots of 4-day-old soybeans treated with flooded in absence or presence of 50 mM CaCl2 for 2 days and analyzed by nanoLC-MS/MS. Based on 3 biological replicated SIEVE analysis (Supplemental Figure 4), a total of 126 differentially changed proteins were identified in 4-day-old untreated soybeans (Figure 3, Supplemental Table 2), 588 such proteins were identified in 2-day-flooded soybeans (Figure 3, Supplemental Table 3), and 329 such proteins were identified in 2-day-flooded soybeans treated with calcium (Figure 3, Supplemental Table 4) compared to 2-day-old soybeans.
Figure 3.
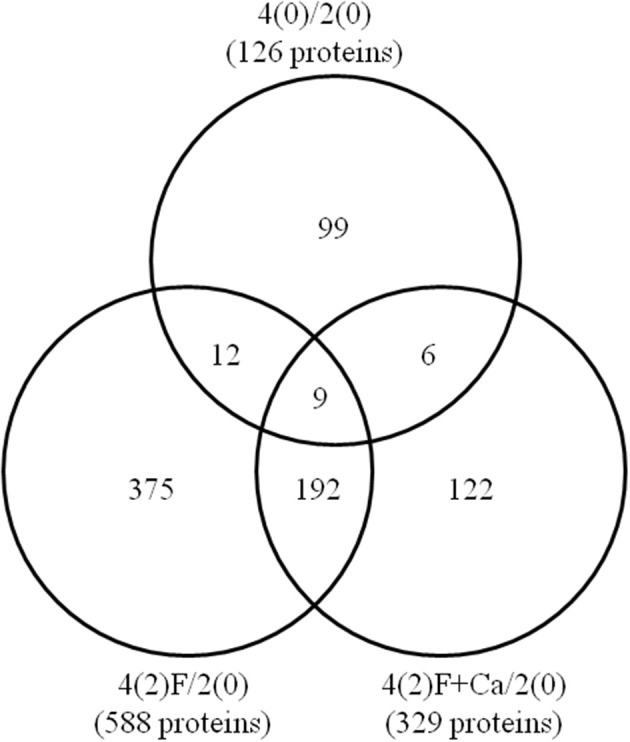
Venn diagram of differentially changed proteins between control and flooded soybeans treated without or with calcium. Two-day-old soybeans were flooded in the presence and absence of 50 mM CaCl2 for 2 days, and proteins extracted from roots were then analyzed by gel-free proteomics. The Venn diagram shows the number of differentially changed proteins in roots of 4-day-old soybeans without treatment [4(0)/2(0)] or with 2-day-flooding with [4(2)F+Ca/2(0)] and without CaCl2 [4(2)F/2(0)]. The overlapping regions denote common proteins among untreated and 2-day-flooded soybeans in the absence or presence of calcium. The numbers represent the number of identified proteins. The identified proteins are listed in Supplementary Tables 2–4.
To determine whether the altered proteins were different in the absence or presence of exogenous calcium, the root proteins identified under each condition were compared (Figure 3). A total of 99, 375, and 122 differentially changed proteins were unique to 4-day-old soybeans, 2-day-flooded soybeans, and 2-day-flooded soybeans treated with calcium, respectively. In addition, 9 proteins were commonly changed among the three conditions; 12 proteins were common between 4-day-old soybeans and 2-day-flooded soybeans; 192 proteins were common between 2-day-flooded soybeans treated without and with calcium; and only 6 proteins were common between 4-day-old soybeans and 2-day-flooded soybeans treated with calcium. Among the 122 proteins that were specifically changed in response to calcium treatment (Table 1), the predominant functional categories were cell wall (15%), protein degradation/synthesis (13%), DNA (11%), and stress (10%)-related proteins. Notably, the identified proteins included xyloglucan endotransglucosylase/hydrolase (XTH) (11/18), which was categorized within cell-wall proteins, and several ribosomal proteins (9/16), which were grouped within the protein degradation/synthesis functional category.
Table 1.
Differentially changed proteins in soybean in response to calcium supplementation under flooding.
| Protein ID | Description | M.P. | Ratio | P-value | Function | |
|---|---|---|---|---|---|---|
| DECREASED | ||||||
| 1 | Glyma09g15620.2 | Cellulose synthase family protein | 4 | 0.13 | 0.0165 | Cell wall |
| 2 | Glyma15g43040.1 | Cellulose synthase family protein | 4 | 0.13 | 0.0165 | Cell wall |
| 3 | Glyma02g46450.3 | Microtubule associated proteins 651 | 2 | 0.17 | 0.0009 | Cell |
| 4 | Glyma14g02180.1 | Microtubule associated proteins 651 | 2 | 0.17 | 0.0009 | Cell |
| 5 | Glyma14g02200.2 | Microtubule associated proteins 651 | 2 | 0.17 | 0.0009 | Cell |
| 6 | Glyma19g27036.1 | Heat shock protein 81-2 | 3 | 0.19 | 0.0079 | Stress |
| 7 | Glyma03g25621.1 | RNA binding (RRM/RBD/RNP motifs) family protein | 2 | 0.22 | 0.0223 | RNA |
| 8 | Glyma07g13211.1 | RNA binding (RRM/RBD/RNP motifs) family protein | 2 | 0.22 | 0.0223 | RNA |
| 9 | Glyma12g31620.1 | DNAJ homolog 2 | 5 | 0.25 | 0.0392 | Stress |
| 10 | Glyma03g37240.4 | RNA binding (RRM/RBD/RNP motifs) family protein | 2 | 0.27 | 0.0185 | RNA |
| 11 | Glyma03g37280.1 | NAD(P) binding Rossmann fold superfamily protein | 2 | 0.29 | 0.0163 | Cell wall |
| 12 | Glyma16g04950.1 | Xyloglucan endotransglucosylase/hydrolase 5 | 10 | 0.30 | 0.0000 | Cell wall |
| 13 | Glyma19g28220.1 | Xyloglucan endotransglucosylase/hydrolase 5 | 9 | 0.32 | 0.0000 | Cell wall |
| 14 | Glyma04g01244.1 | Thymidylate synthase 1 | 2 | 0.33 | 0.0086 | C1 |
| 15 | Glyma10g33350.2 | ARABIDOPSIS THALIANA PEROXYGENASE 2 | 2 | 0.33 | 0.0302 | Development |
| 16 | Glyma02g02170.1 | NAD(P) binding Rossmann fold superfamily protein | 3 | 0.33 | 0.0291 | Cell wall |
| 17 | Glyma19g39870.1 | NAD(P) binding Rossmann fold superfamily protein | 3 | 0.33 | 0.0291 | Cell wall |
| 18 | Glyma17g03030.1 | NAD(P) binding Rossmann fold superfamily protein | 2 | 0.34 | 0.0297 | Cell wall |
| 19 | Glyma14g11760.1 | Early nodulin like protein 15 | 2 | 0.34 | 0.0012 | Misc |
| 20 | Glyma17g34040.1 | Early nodulin like protein 15 | 2 | 0.34 | 0.0012 | Misc |
| 21 | Glyma19g36580.1 | Pyridoxal-dependent decarboxylase family protein | 5 | 0.34 | 0.0009 | Amino acid metabolism |
| 22 | Glyma18g52860.1 | O Glycosyl hydrolases family 17 protein | 4 | 0.34 | 0.0065 | Misc |
| 23 | Glyma19g39850.1 | RNA binding (RRM/RBD/RNP motifs) family protein | 3 | 0.35 | 0.0047 | RNA |
| 24 | Glyma03g02240.1 | SIN3 associated polypeptide P18 | 2 | 0.36 | 0.0171 | RNA |
| 25 | Glyma12g03570.1 | Subtilisin like serine protease 2 | 6 | 0.38 | 0.0143 | Protein |
| 26 | Glyma02g06400.1 | Succinate dehydrogenase 11 | 3 | 0.39 | 0.0034 | TCA/org transformation |
| 27 | Glyma15g03761.1 | Leucine rich repeat (LRR) family protein | 2 | 0.39 | 0.0326 | Not assigned |
| 28 | Glyma02g07610.1 | Xyloglucan endotransglucosylase/hydrolase 5 | 5 | 0.39 | 0.0000 | Cell wall |
| 29 | Glyma06g10180.1 | P450 reductase 1 | 2 | 0.39 | 0.0483 | Misc |
| 30 | Glyma06g01280.2 | Thymidylate synthase 2 | 2 | 0.39 | 0.0075 | Nucleotide metabolism |
| 31 | Glyma18g53470.1 | Rad23 UV excision repair protein family | 3 | 0.40 | 0.0296 | DNA |
| 32 | Glyma02g14450.1 | Chalcone and stilbene synthase family protein | 5 | 0.41 | 0.0004 | Secondary metabolism |
| 33 | Glyma11g07250.1 | Succinate dehydrogenase 11 | 6 | 0.43 | 0.0004 | TCA/org transformation |
| 34 | Glyma01g38200.1 | Succinate dehydrogenase 11 | 5 | 0.43 | 0.0008 | TCA/org transformation |
| 35 | Glyma06g04100.2 | RNA binding protein 47C | 2 | 0.43 | 0.0190 | RNA |
| 36 | Glyma07g12190.1 | Hexokinase 1 | 4 | 0.44 | 0.0313 | Major CHO metabolism |
| 37 | Glyma17g37270.2 | Beta galactosidase 5 | 4 | 0.46 | 0.0023 | Misc |
| 38 | Glyma14g07700.1 | Beta galactosidase 5 | 2 | 0.47 | 0.0090 | Misc |
| 39 | Glyma08g03730.1 | Hexokinase 1 | 8 | 0.48 | 0.0181 | Major CHO metabolism |
| 40 | Glyma11g33880.1 | DegP protease 7 | 5 | 0.48 | 0.0027 | Protein |
| 41 | Glyma18g04400.1 | DegP protease 7 | 5 | 0.48 | 0.0027 | Protein |
| 42 | Glyma01g22880.1 | Chalcone and stilbene synthase family protein | 6 | 0.49 | 0.0011 | Secondary metabolism |
| 43 | Glyma05g28610.1 | Chalcone and stilbene synthase family protein | 6 | 0.49 | 0.0011 | Secondary metabolism |
| 44 | Glyma08g11530.1 | Chalcone and stilbene synthase family protein | 6 | 0.49 | 0.0011 | Secondary metabolism |
| 45 | Glyma08g11620.1 | Chalcone and stilbene synthase family protein | 6 | 0.49 | 0.0011 | Secondary metabolism |
| 46 | Glyma09g08780.1 | Chalcone and stilbene synthase family protein | 6 | 0.49 | 0.0011 | Secondary metabolism |
| 47 | Glyma11g01350.1 | Chalcone and stilbene synthase family protein | 6 | 0.49 | 0.0011 | Secondary metabolism |
| 48 | Glyma08g11610.1 | Chalcone and stilbene synthase family protein | 5 | 0.49 | 0.0016 | Secondary metabolism |
| 49 | Glyma02g35640.1 | CTC interacting domain 11 | 2 | 0.49 | 0.0052 | RNA |
| INCREASED | ||||||
| 50 | Glyma19g31590.1 | Beta-1,3-glucanase 1 | 4 | 2.05 | 0.0403 | Misc |
| 51 | Glyma09g00711.1 | Glycine cleavage T protein family | 3 | 2.06 | 0.0390 | Photosynthesis |
| 52 | Glyma11g14950.1 | Heat shock protein 70 | 25 | 2.22 | 0.0017 | Stress |
| 53 | Glyma08g36700.1 | ARF-GAP domain 8 | 6 | 2.26 | 0.0012 | Protein |
| 54 | Glyma17g14950.1 | Lactate/malate dehydrogenase family protein | 4 | 2.28 | 0.0009 | Fermentation |
| 55 | Glyma03g02760.1 | N.D.* | 6 | 2.29 | 0.0000 | Not assigned |
| 56 | Glyma15g07850.3 | Aluminum induced protein with YGL and LRDR motifs | 2 | 2.30 | 0.0347 | Hormone metabolism |
| 57 | Glyma18g52650.2 | Heat shock cognate protein 70-1 | 22 | 2.31 | 0.0000 | Stress |
| 58 | Glyma18g52610.1 | Heat shock cognate protein 70-1 | 25 | 2.31 | 0.0046 | Stress |
| 59 | Glyma13g19331.1 | Heat shock cognate protein 70-1 | 26 | 2.32 | 0.0002 | Stress |
| 60 | Glyma02g41890.3 | Type one serine/threonine protein phosphatase 4 | 3 | 2.33 | 0.0039 | Protein |
| 61 | Glyma14g07080.3 | Type one serine/threonine protein phosphatase 4 | 3 | 2.33 | 0.0039 | Protein |
| 62 | Glyma01g41920.2 | Lactate/malate dehydrogenase family protein | 3 | 2.33 | 0.0019 | Fermentation |
| 63 | Glyma13g40890.1 | Histone H2A 12 | 2 | 2.33 | 0.0002 | DNA |
| 64 | Glyma13g40900.1 | Histone H2A 12 | 2 | 2.33 | 0.0002 | DNA |
| 65 | Glyma13g40940.1 | Histone H2A 12 | 2 | 2.33 | 0.0002 | DNA |
| 66 | Glyma15g04530.1 | Histone H2A 12 | 2 | 2.33 | 0.0002 | DNA |
| 67 | Glyma15g04540.1 | Histone H2A 12 | 2 | 2.33 | 0.0002 | DNA |
| 68 | Glyma03g34830.1 | Enolase | 19 | 2.46 | 0.0016 | Glycolysis |
| 69 | Glyma02g04800.1 | Calcium dependent phosphotriesterase superfamily protein | 2 | 2.58 | 0.0371 | Secondary metabolism |
| 70 | Glyma16g22650.1 | Calcium dependent phosphotriesterase superfamily protein | 2 | 2.58 | 0.0371 | Secondary metabolism |
| 71 | Glyma17g29320.1 | Peroxidase family protein | 2 | 2.63 | 0.0199 | Misc |
| 72 | Glyma09g06350.1 | Peroxidase superfamily protein | 5 | 2.65 | 0.0034 | Misc |
| 73 | Glyma19g42760.1 | Gamma histone variant H2AX | 3 | 2.72 | 0.0000 | DNA |
| 74 | Glyma20g28460.1 | Cupin family protein | 2 | 2.72 | 0.0006 | Development |
| 75 | Glyma20g28640.1 | Cupin family protein | 2 | 2.72 | 0.0006 | Development |
| 76 | Glyma04g38590.1 | Beta galactosidase 10 | 6 | 2.77 | 0.0011 | Misc |
| 77 | Glyma06g16420.2 | Beta galactosidase 10 | 5 | 2.78 | 0.0020 | Misc |
| 78 | Glyma02g00850.3 | Type one serine/threonine protein phosphatase 4 | 2 | 2.79 | 0.0012 | Protein |
| 79 | Glyma05g33410.2 | Aldolase-type TIM barrel family protein | 2 | 2.84 | 0.0077 | OPP |
| 80 | Glyma19g34780.1 | RmlC like cupins superfamily protein | 3 | 2.91 | 0.0287 | Developmen |
| 81 | Glyma19g32690.1 | Ribosomal protein S10p/S20e family protein | 4 | 2.95 | 0.0335 | Protein |
| 82 | Glyma03g30440.1 | Histone superfamily protein | 2 | 2.99 | 0.0000 | DNA |
| 83 | Glyma12g34360.1 | Histone H2A 10 | 2 | 2.99 | 0.0000 | DNA |
| 84 | Glyma12g34370.2 | Histone H2A 2 | 2 | 2.99 | 0.0000 | DNA |
| 85 | Glyma13g36180.1 | Histone H2A 10 | 2 | 2.99 | 0.0000 | DNA |
| 86 | Glyma13g36190.1 | Histone H2A 10 | 2 | 2.99 | 0.0000 | DNA |
| 87 | Glyma19g33360.1 | Gamma histone variant H2AX | 2 | 2.99 | 0.0000 | DNA |
| 88 | Glyma17g03360.1 | N.D.* | 7 | 3.03 | 0.0000 | Not assigned |
| 89 | Glyma02g47210.1 | HEAT SHOCK PROTEIN 81.4 | 16 | 3.11 | 0.0008 | Stress |
| 90 | Glyma03g32380.2 | Ribosomal protein L14p/L23e family protein | 6 | 3.14 | 0.0025 | Protein |
| 91 | Glyma13g18830.1 | Ribosomal protein L14p/L23e family protein | 6 | 3.14 | 0.0025 | Protein |
| 92 | Glyma14g01530.1 | HEAT SHOCK PROTEIN 81.4 | 17 | 3.25 | 0.0010 | Stress |
| 93 | Glyma14g04840.1 | Ribosomal protein S10p/S20e family protein | 3 | 3.34 | 0.0309 | Protein |
| 94 | Glyma19g32680.1 | Ribosomal protein S10p/S20e family protein | 3 | 3.34 | 0.0309 | Protein |
| 95 | Glyma08g44590.1 | HEAT SHOCK PROTEIN 81.4 | 16 | 3.71 | 0.0014 | Stress |
| 96 | Glyma03g28870.1 | Beta-1,3-glucanase 1 | 3 | 3.87 | 0.0415 | Misc |
| 97 | Glyma13g01140.1 | Xyloglucan endotransglucosylase/hydrolase family protein | 6 | 3.94 | 0.0005 | Cell wall |
| 98 | Glyma18g08220.1 | HEAT SHOCK PROTEIN 81.4 | 16 | 4.02 | 0.0001 | Stress |
| 99 | Glyma08g11300.2 | Xyloglucan endotransglucosylase/hydrolase 16 | 3 | 4.05 | 0.0003 | Cell wall |
| 100 | Glyma11g36730.2 | Xyloglucan endotransglucosylase/hydrolase 16 | 3 | 4.05 | 0.0003 | Cell wall |
| 101 | Glyma18g00630.2 | Xyloglucan endotransglucosylase/hydrolase 16 | 3 | 4.05 | 0.0003 | Cell wall |
| 102 | Glyma15g17620.1 | Peroxidase superfamily protein | 5 | 4.14 | 0.0012 | Misc |
| 103 | Glyma13g01120.1 | Xyloglucan endotransglycosylase 6 | 3 | 4.14 | 0.0027 | Cell wall |
| 104 | Glyma17g07240.1 | Xyloglucan endotransglycosylase 6 | 3 | 4.14 | 0.0027 | Cell wall |
| 105 | Glyma13g01150.1 | Xyloglucan endotransglucosylase/hydrolase family protein | 3 | 4.18 | 0.0045 | Cell wall |
| 106 | Glyma17g07270.1 | Xyloglucan endotransglycosylase 6 | 3 | 4.18 | 0.0045 | Cell wall |
| 107 | Glyma19g31580.1 | beta-1,3-glucanase 1 | 2 | 4.82 | 0.0205 | Misc |
| 108 | Glyma13g34540.1 | D mannose binding lectin protein with Apple like carbohydrate binding domain | 3 | 5.24 | 0.0291 | Misc |
| 109 | Glyma03g32030.1 | RmlC like cupins superfamily protein | 5 | 5.35 | 0.0160 | Development |
| 110 | Glyma07g02720.1 | Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein | 7 | 6.05 | 0.0034 | Protein |
| 111 | Glyma08g23260.3 | Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein | 7 | 6.06 | 0.0033 | Protein |
| 112 | Glyma13g44690.1 | Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein | 7 | 6.06 | 0.0033 | Protein |
| 113 | Glyma15g00610.1 | Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein | 7 | 6.06 | 0.0033 | Protein |
| 114 | Glyma13g32300.1 | Quinone reductase family protein | 6 | 6.14 | 0.0133 | Lipid metabolism |
| 115 | Glyma15g07040.1 | Quinone reductase family protein | 6 | 6.14 | 0.0133 | Lipid metabolism |
| 116 | Glyma08g45510.1 | Kunitz family trypsin and protease inhibitor protein | 3 | 6.65 | 0.0161 | Stress |
| 117 | Glyma04g39930.1 | Manganese superoxide dismutase 1 | 4 | 7.27 | 0.0490 | Redox |
| 118 | Glyma09g30370.1 | Glutamine synthase clone R1 | 8 | 7.86 | 0.0038 | N-metabolism |
| 119 | Glyma03g22260.1 | Auxin responsive family protein | 2 | 11.51 | 0.0081 | Hormone metabolism |
| 120 | Glyma08g06570.1 | Flavodoxin like quinone reductase 1 | 2 | 14.54 | 0.0044 | Lipid metabolism |
| 121 | Glyma04g37140.1 | SNF1 related protein kinase regulatory subunit gamma 1 | 2 | 14.59 | 0.0493 | Cell wall |
| 122 | Glyma09g29300.1 | Kunitz family trypsin and protease inhibitor protein | 7 | 22.58 | 0.0124 | Stress |
Protein ID, according to the Phytozome database; M.P., number of matched peptide; Ratio, relative abundance of protein; Function, functional classification by MapMan bin code;
N.D., No description in Phytozome database; protein, protein synthesis/targeting/degradation/post-translational modification; DNA, DNA synthesis; RNA, RNA processing/binding; C1, one carbon; TCA, tricarboxylic acid; OPP, oxidative pentose phosphate; CHO, carbohydrates; misc, miscellaneous.
In salt-stressed soybean, exogenous calcium treatment restored root growth by maintaining pectin levels and increasing the calcium concentration in the cell wall, suggesting that calcium plays a role in maintaining cell wall composition to protect from salt toxicity (An et al., 2014). XTH, which acts as a cell wall-loosening enzyme (van Sandt et al., 2007), functions in cell-wall elongation and reconstruction through rearranging the bonds between xyloglucan chains (Fry et al., 1992; Nishitani and Tominaga, 1992) and also involves in cell-wall metabolism during flooding-induced aerenchyma development. In Capsicum annuum, increased tolerance to drought stress by overexpression of CaXTH3 resulted from the change of cell-wall extensibility of guard cells mediated by the cell-wall remodeling activity of CaXTH3 (Choi et al., 2011), corresponding with increased abundance of XTH in present study. In mungbean, expression of VrXTH1, which is an auxin-inducible gene isolated from mungbean, was closely related to plant growth and modulated by the cytosolic calcium concentration (Yun et al., 2007). Furthermore, changes in calcium ion levels influenced the molecular size of xyloglucans by modifying the expression of VaXTHS4 in azuki bean (Soga et al., 2007). Taken together, these results suggest that the regulation of cell wall-related proteins, such as XTH, by exogenous calcium might promote the elongation of soybean roots under flooding stress. In this study, except for XTH, other cell wall-related proteins such as glycosyl hydrolase, which was decreased in flood-stressed soybean, was exhibited a similar tendency in previous study (Oh et al., 2014a).
A number of ribosomal proteins play roles in cell metabolism/division, and plant growth and fitness (Whittle and Krochko, 2009). Ribosomal proteins are generally decreased in plants in response to abiotic stresses (Rogalski et al., 2008; Falcone Ferreyra et al., 2010), leading to retarded growth and productivity (Kawaguchi et al., 2004). Ribosomal L19 protein, which has 60S ribosomal subunit, is a calcium-calmodulin interacting protein implicated in translational processes (Sonnemann et al., 1991). Mutation of the RPS6A gene, which encodes a component of the small ribosomal 40S subunit (Zhao et al., 2003), led to higher intracellular calcium concentrations in response to exogenous calcium (Zhao et al., 2013). Flooding stress also caused a decrease in the abundance of ribosomal proteins in soybean, indicating that ribosomal proteins resulted in alterations of protein synthesis (Oh et al., 2014b). In present study, ribosomal proteins were increased in soybean roots exposed to exogenous calcium under flooding stress, suggesting that ribosomal proteins in association with calcium-dependent proteins and/or molecules might enhance protein synthesis in response to flooding stress.
Functional categorization of proteins in calcium-treated soybean root under flooding
To determine the biological processes of proteins that were altered in flooding-stressed soybean roots by calcium treatment, the identified proteins were functionally characterized using MapMan bin codes (Figure 4). Under flooding stress, the number of proteins related to hormone metabolism, DNA synthesis, cell wall, and protein degradation/synthesis/posttranslational modification was decreased; however, the exposure of plants to calcium under flooding conditions increased the number of these proteins. Similarly, the number of proteins related to development, signaling, and lipid metabolism was increased when exogenous calcium was added to the roots of flooding-stressed plants. In addition, the number of proteins related to fermentation and glycolysis was increased under flooding stress; however, calcium supplementation had little effect on the number of differentially changed proteins (Figure 4). Proteins within the hormone category that were changed in response to flooding in absence or presence of calcium included lipoxygenase and auxin-responsive family protein.
Figure 4.
Functional categorization of flooding-responsive proteins in soybean roots treated with calcium. Two-day-old soybeans were flooded without or with 50 mM CaCl2 for 2 days. Untreated soybeans were used as control. Proteins extracted from roots were analyzed using gel-free proteomics. The following comparisons were made: 4(0)/2(0), 2-day-old soybeans and 4-day-old soybeans; 4(2)F/2(0), 2-day-old soybeans and 2-day-flooded soybeans without calcium; and 4(2)F+Ca/2(0), 2-day-old soybeans and 2-day-flooded soybeans treated with calcium. Identified proteins were categorized using MapMan bin codes: protein, protein synthesis/targeting/degradation/post-translational modification; DNA, DNA synthesis; RNA, RNA processing/binding; C1, one carbon; TCA, tricarboxylic acid; OPP, oxidative pentose phosphate; CHO, carbohydrates; misc, miscellaneous. The number of differentially changed proteins is indicated on the x-axis of the graph. Black and white bars indicate decreased and increased proteins, respectively.
Under flooding stress, most lipoxygenases were decreased in soybean roots and were not induced by calcium supplementation. Lipoxygenases, which are a group of non-heme iron-containing dioxygenases (Brash, 1999), are involved in increasing stress resistance and boosting defense reactions in both Arabidopsis and soybean (Melan et al., 1994; Park et al., 1994). Komatsu et al. (2010) reported that two lipoxygenases were decreased in soybean in response to flooding stress and suggested that these enzymes affected cell wall metabolism due to suppression of lignification. The present findings indicate that lipoxygenase may regulate hormone pathways for stress tolerance and involve in defense reaction in soybean roots under flooding stress.
Auxin-responsive family protein was increased under flooding conditions in soybean roots treated with calcium compared to non-treated roots. In Arabidopsis, the gene encoding auxin-responsive family protein was expressed during the early stages of lateral root formation (Laskowski et al., 2006), and the corresponding protein interacted with other redox partners within the plasma membrane to form a redox link between the cytoplasm and the apoplast (Preger et al., 2009). Auxin-responsive family protein was also shown to be glycosylated in Arabidopsis and involved in the formation of a glycosylphosphatidylinositol anchor to the external side of the plasma membrane (Borner et al., 2003). As calcium ions have an important role in polar auxin transport and gravitropic responses (Toyota et al., 2008), together, these results indicate that calcium supplementation may control hormone metabolic pathways by regulating activation of these proteins in soybean under flooding stress.
In the present study, DNA synthesis-related proteins, include histone, were decreased by flooding stress, but were increased by the treatment of soybean roots with calcium. Histones, which are small and basic proteins associated with DNA to form chromatin (Thuleau et al., 2012), undergo posttranslational modification, such as acetylation/deacetylation. Epigenetic mechanisms, including histone modifications, play a decisive role in regulating plant responses to abiotic stresses (Luo et al., 2012). Servet et al. (2010) reported that histone acetyltransferase AtGCN5/HAG1 in Arabidopsis was essential to regulate gene expression during development processes and in response to environmental stresses. In Nicotiana tabacum, type-2 histone deacetylases act as negative regulators of programmed cell death induced by the defense elicitor cryptogein (Bourque et al., 2011). The results of an Evans blue assay indicated that flooding-induced root tip cell death of soybean was suppressed by exogenous calcium. It has also been reported that histone modifications were activated by a transient influx of calcium in response to heat (Mach, 2012). Based on these results, calcium-induced stabilization of DNA synthesis via histone modification may be a critical plant defense response to flooding stress in soybean.
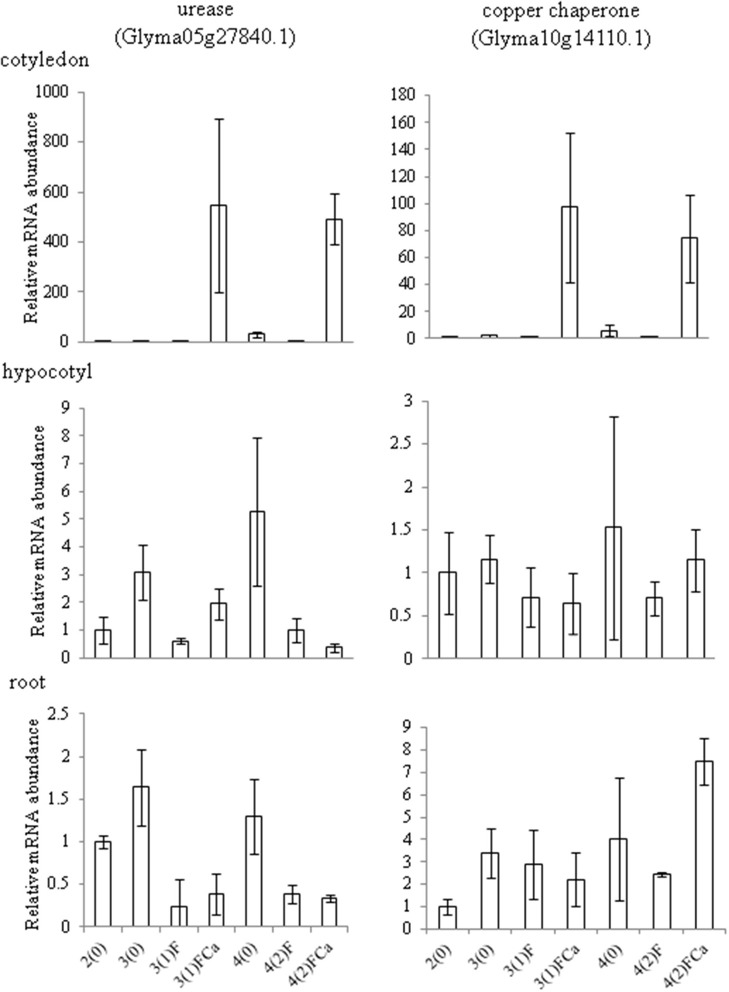
Effects of calcium on mRNA expression level of key proteins in different organs under flooding stress
Six differentially changed proteins were common between 4-day-old soybeans and 2-day-flooded soybeans treated with calcium (Figure 3). The abundance of these proteins was calculated among 4-day-old soybeans, 2-day-flooded soybeans, and 2-day-flooded soybeans treated with 50 mM CaCl2 (Supplemental Figure 5). The analysis of protein abundance indicated that urease (Glyma05g27840.1) and two copper chaperones (Glyma10g14110.1 and Glyma02g19380.1) exhibited the same profiles in 4-day-old soybeans and 2-day-flooded soybeans treated with 50 mM CaCl2. To determine whether the changes in protein abundance were regulated at the transcriptional level, the mRNA expression levels of these proteins were analyzed in root, hypocotyl, and cotyledon under flooding for 0, 1, and 2 days (Figure 5). Total RNAs extracted from roots, hypocotyls, and cotyledons of soybeans were analyzed using qRT-PCR. In roots, the mRNA level of urease was down-regulated by 1 and 2 days flooding; however, the level was not changed by calcium supplementation. In hypocotyls, the mRNA level of urease was down-regulated by 1 day of flooding and significantly up-regulated by 1 day flooding with calcium; however, there was no significant between 2 days flooding without and with calcium. The mRNA expression level of copper chaperone was similar to their protein level in the root and hypocotyl under 2 days flooding with calcium. In roots and hypocotyls, the level of copper chaperone was down-regulated by 1 day flooding without and with calcium; whereas the level was significantly up-regulated by 2 days flooding with calcium. In cotyledons, the mRNA levels of urease and copper chaperone were significantly up-regulated by 1 and 2 day flooding when calcium was exogenously added (Figure 5).
Figure 5.
Effects of calcium on the mRNA expression levels of urease and copper chaperone proteins in different organs of soybean under flooding stress. Two-day-old soybeans were flooded without or with 50 mM CaCl2 for 1 and 2 days. Untreated soybeans were used as a control. RNAs extracted from roots, hypocotyls, and cotyledons of the soybeans were analyzed by qRT-PCR with specific primers for urease and copper chaperone (Supplemental Table 1). Relative mRNA abundance was normalized against that of 18S rRNA. Data are shown as means ± SD from three independent biological replicates. Means with the same letter are not significantly different according to ANOVA (P < 0.05).
Urease is a nickel-dependent metalloenzyme that catalyzes the hydrolysis of urea to form ammonia and carbon dioxide (Sirko and Brodzik, 2000). Plants typically have abundant urease (Yata et al., 2014), particularly in seedlings, in which it plays a pivotal role in nitrogen metabolism (Sirko and Brodzik, 2000). It has been reported that plant ureases are associated with plant defense mechanisms against insect predation (Follmer et al., 2004). In soybean, a urease was identified in developing embryos and was designated embryo-specific urease, which was later found to be encoded by the Eu1 gene (Meyer-Bothling and Polacco, 1987; Torisky et al., 1994; Polacco et al., 2011). The suppression of embryo-specific urease in soybean led to increased susceptibility to fungal infection, demonstrating that urease plays a role in plant defense (Wiebke-strohm et al., 2012). Medeiros-Silva et al. (2014) reported that a urease deficiency in soybean altered the physiology of root nodules and adversely affected nitrogen fixation. Consistent with our present findings, Jack bean urease activity was affected by intra- and extracellular calcium concentrations (Staniscuaski et al., 2009). These results, together with the present findings, suggest that flooding stress decreases the urease activity in soybean, and that exogenously applied calcium appears to increase urease activity under flooding stress.
Copper chaperone has a copper-binding motif and plays a role in the homeostatic regulation of copper within plant cells (Mira et al., 2001). In Arabidopsis, copper chaperone was up-regulated during leaf senescence, suggesting that it facilitates the transport of certain metal ions in leaves to other growing parts of the plant (Himelblau et al., 1998; Mira et al., 2001). In tomato, the copper chaperone gene, which encodes a copper chaperone for copper/zinc superoxide dismutase, is involved in defense mechanisms against oxidative stress (Company and Carmen, 2003). In pea, the copper/zinc superoxide dismutase gene was down-regulated when calcium deficiency was induced by heavy metal stress with high-concentration of cadmium (Rodriquez-Serrano et al., 2009). Copper chaperone plays a role in oxidant-responsive posttranslational regulation of superoxide dismutase activity in yeast (Brown et al., 2004). In poplar, copper chaperone specifically responds to certain metals and oxidative damage caused by abiotic stresses (Lee et al., 2005). In the present study, the copper chaperone gene was up-regulated in soybean roots by exogenous calcium under flooding stress, suggesting that this chaperone protein is one of the factors regulating flooding stress responses in a calcium-dependent manner in soybean root.
Concluding remarks
In the present study, proteins affected by calcium in flooded soybean were analyzed to better understand calcium-mediated flooding stress mechanisms in this agriculturally important crop. These main findings of this study are as follows: (i) cell wall, protein degradation/synthesis, hormone metabolism, and DNA synthesis-related proteins were decreased under flooding stress, but were increased by the addition of calcium; (ii) development, lipid metabolism, and signaling-related proteins were increased by calcium addition; (iii) fermentation- and glycolysis-related proteins were increased under flooding stress and were not affected by calcium addition; and (iv) urease and copper chaperone had similar abundances in untreated soybeans and flooded soybeans treated with calcium. Taken together, these results suggest that calcium might affect the cell wall/hormone metabolisms, protein degradation/synthesis, and DNA synthesis in soybean roots under flooding stress.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Drs. Keito Nishizawa and Makoto Tougou of the National Institute of Crop Science for their help with support and discussion.
Glossary
Abbreviations
- LC
liquid chromatography
- MS
mass spectrometry
- qRT-PCR
quantitative reverse transcription-polymerase chain reaction
- XTH
xyloglucan endotransglucosylase/hydrolase.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00559/abstract
Experimental design for this study. (A) For physiological analysis, 2-day-old soybeans were flooded without or with 1, 5, 10, and 50 mM CaCl2 for 2, 4, and 6 days. (B) For proteomic analysis, 2-day-old soybeans were flooded without or with 50 mM CaCl2 for 2 days. Two-day-old and 4-day-old soybeans without flooding were used as controls. (C) For transcriptional analysis, 2-day-old soybeans were flooded without or with 50 mM CaCl2 for 1 and 2 days. Two-day-old, 3-day-old, and 4-day-old soybeans without flooding were used as controls. Three independent experiments were performed as biological replicates.
Effect of calcium on the growth of soybean under flooding stress in three independent experiments (A–C). Two-day-old soybeans were flooded without (dark blue) or with 1 (red), 5 (light green), 10 (purple), and 50 mM CaCl2 (light blue) for 2, 4, and 6 days. Photographs show soybean seedlings after 2, 4, and 6 days of flooding. Bars indicate 10 mm. The length and weight of roots, including the hypocotyl, were measured at the indicated time points. Data are means ± SE from three independent biological replications (A–C). Means with the same letter are not significantly different according to ANOVA (P < 0.05).
Evaluation of cell death in flooding-stressed soybean roots treated with calcium in three independent experiments (A–C). Two-day-old soybeans were flooded without (dark blue) or with 1 (red), 5 (light green), 10 (purple), and 50 mM CaCl2 (light blue) for 2, 4, and 6 days. The root tips were stained with 0.25% Evans blue dye, which was then extracted and measured spectroscopically at 600 nm at the indicated time points. Photographs show soybean seedlings after 2, 4, and 6 days of flooding. Bars indicate 10 mm. Data are means ± SE from three independent biological replications (A–C). Means with the same letter are not significantly different according to ANOVA (P < 0.05).
Principle component analysis of the peptide profile for identified proteins. The data sets of soybean proteins from 2-day-old (blue circle), 4-day-old (red square), 4-day-old with 2-day-flood (yellow triangle), and 4-day-old with 2-day-flood with calcium (purple pentagon) were plotted using principle component analysis (PCA). PCA was fully integrated into SIEVE software. PCA loading plot shows the spectral regions responsible for separation of groups in the corresponding score plot.
Abundance of six differentially changed proteins that were common between untreated and flooding-stressed soybean exposed to calcium. Two-day-old soybeans were flooded without or with 50 mM CaCl2 for 2 days. Proteins extracted from roots were purified, digested, and analyzed using nanoLC MS/MS. The protein abundance was determined by differential analysis using SIEVE software. Six differentially changed proteins were commonly identified in 4-day-old soybeans and 2-day-flooded soybeans treated with calcium.
References
- An P., Li X., Zheng Y., Eneji E., Inanaga S. (2014). Calcium effects on root cell wall composition and ion contents in two soybean cultivars under salinity stress. Can. J. Plant Sci. 94, 733–740. 10.4141/cjps2013-291 [DOI] [Google Scholar]
- Ashraf M. A. (2012). Waterlogging stress in plants: a review. Afr. J. Agric. Res. 7, 1976–1981. 10.5897/AJARX11.084 [DOI] [Google Scholar]
- Aurisano N., Bertani A., Reggiani R. (1995). Involvement of calcium and calmodulin in protein and amino acid metabolism in rice roots under anoxia. Plant Cell Physiol. 36, 1525–1529 [Google Scholar]
- Bailey-Serres J., Fukao T., Gibbs D. J., Holdsworth M. J., Lee S. C., Licausi F., et al. (2012). Making sense of low oxygen sensing. Trends Plant Sci. 17, 129–138. 10.1016/j.tplants.2011.12.004 [DOI] [PubMed] [Google Scholar]
- Baker C. J., Mock N. M. (1994). An improved method for monitoring cell death in cell suspension and leaf disc assays using evans blue. Plant Cell Tissue Organ Cult. 39, 7–12. 10.1007/BF00037585 [DOI] [Google Scholar]
- Borner G. H., Lilley K. S., Stevens T. J., Dupree P. (2003). Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 132, 568–577. 10.1104/pp.103.021170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque S., Dutartre A., Hammoudi V., Blanc S., Dahan J., Jeandroz S., et al. (2011). Type-2 histone deacetylases as new regulators of elicitor-induced cell death in plants. New Phytol. 192, 127–139. 10.1111/j.1469-8137.2011.03788.x [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Brash A. R. (1999). Lipoxygenases: occurrence, functions, catalysis, and acquisition of substrate. J. Biol. Chem. 274, 23679–23682. 10.1074/jbc.274.34.23679 [DOI] [PubMed] [Google Scholar]
- Brown N. M., Torrese A. S., Doan P. E., O'Halloran T. V. (2004). Oxygen and the copper chaperone CCS regulate posttranslation activation of Cu, Zn superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 101, 5518–5523. 10.1073/pnas.0401175101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. Y., Seo Y. S., Kim S. J., Kim W. T., Shin J. S. (2011). Constitutive expression of CaXTH3, a hot pepper xyloglucan endotransglucosylase/hydrolase, enhanced tolerance to salt and drought stresses without phenotypic defects in tomato plants. Plant Cell Rep. 30, 867–877. 10.1007/s00299-010-0989-3 [DOI] [PubMed] [Google Scholar]
- Company P., Carmen G. B. (2003). Identification of a copper chaperone from tomato fruits infected with Botrytis cinerea by differential display. Biochem. Biophys. Res. Commun. 304, 825–830. 10.1016/S0006-291X(03)00680-6 [DOI] [PubMed] [Google Scholar]
- Dat J. F., Capelli N., Folzer H., Bourgeade P., Badot P. M. (2004). Sensing and signaling during plant flooding. Plant Physiol. Biochem. 42, 273–282. 10.1016/j.plaphy.2004.02.003 [DOI] [PubMed] [Google Scholar]
- Delisle G., Champoux M., Houde M. (2001). Characterization of oxalate oxidase and cell death in Al-sensitive and tolerant wheat roots. Plant Cell Physiol. 42, 324–333. 10.1093/pcp/pce041 [DOI] [PubMed] [Google Scholar]
- Falcone Ferreyra M. L., Pezza A., Biarc J., Burlingame A. L., Casati P. (2010). Plant L10 ribosomal proteins have different roles during development and translation under ultraviolet-B stress. Plant Physiol. 153, 1878–1894. 10.1104/pp.110.157057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follmer C., Real-Guerra R., Wasserman G. E., Olivera-Severo D., Carlini C. R. (2004). Jackbean, soybean and Bacillus pasteurii ureases: biological effects unrelated to ureolytic activity. Eur. J. Biochem. 271, 1357–1363. 10.1111/j.1432-1033.2004.04046.x [DOI] [PubMed] [Google Scholar]
- Fry S. C., Smith R. C., Renwick K. F., Martin D. J., Hodge S. K., Matthews K. J. (1992). Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem. J. 282, 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Chen C., Han L., Lin H. (2004). Calcium influence on chilling resistance of grafting eggplant seedlings. J. Plant Nutr. 27, 1327–1339. 10.1081/PLN-200025833 [DOI] [Google Scholar]
- Gao H., Jia Y., Guo S., Lv G., Wang T., Juan L. (2011). Exogenous calcium affects nitrogen metabolism in root-zone hypoxia-stressed muskmelon roots and enhances short-term hypoxia tolerance. J. Plant Physiol. 168, 1217–1225. 10.1016/j.jplph.2011.01.022 [DOI] [PubMed] [Google Scholar]
- Gibbs J., Greenway H. (2003). Review: mechanisms of anoxia tolerance in plants. I. Growth, survival and anaerobic catabolism. Funct. Plant Biol. 30, 1–47. 10.1071/PP98095 [DOI] [PubMed] [Google Scholar]
- Githiri S. M., Watanabe S., Harada K., Takahashi R. (2006). QTL analysis of flooding tolerance in soybean at an early vegetative growth stage. Plant Breed. 125, 613–618. 10.1111/j.1439-0523.2006.01291.x [DOI] [Google Scholar]
- Hartman G. L., West E. D., Herman T. K. (2011). Crops that feed the world 2. Soybean-worldwide production, use, and constraints caused by pathogens and pests. Food Sec. 3, 5–17. 10.1007/s12571-010-0108-x [DOI] [Google Scholar]
- He L., Lu X., Tian J., Yang Y., Li B., Li J., et al. (2012). Proteomic analysis of the effects of exogenous calcium on hypoxic-responsive proteins in cucumber roots. Proteome Sci. 10:42. 10.1186/1477-5956-10-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson E., Nordin Henriksson K. (2005). Salt-stress signalling and the role of calcium in the regulation of the Arabidopsis ATHB7 gene. Plant Cell Environ. 28, 202–210. 10.1111/j.1365-3040.2004.01263.x [DOI] [Google Scholar]
- Hepler P. K. (2005). Calcium: a central regulator of plant growth and development. Plant Cell 17, 2142–2155. 10.1105/tpc.105.032508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himelblau E., Mira H., Lin S. J., Culotta V. C., Penarrubia L., Amasino R. M. (1998). Identification of a functional homolog of the yeast copper homeostasis gene ATX1 from Arabidopsis. Plant Physiol. 117, 1227–1234. 10.1104/pp.117.4.1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R., Girke T., Bray E. A., Bailey-Serres J. (2004). Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J. 38, 823–839. 10.1111/j.1365-313X.2004.02090.x [DOI] [PubMed] [Google Scholar]
- Komatsu S., Han C., Nanjo Y., Altaf-Un-Nahar M., Wang K., He D., et al. (2013b). Label-free quantitative proteomic analysis of abscisic acid effect in early-stage soybean under flooding. J. Proteome Res. 12, 4769–4784. 10.1021/pr4001898 [DOI] [PubMed] [Google Scholar]
- Komatsu S., Hiraga S., Yanagawa Y. (2012a). Proteomics techniques for the development of flood tolerant crops. J. Proteome Res. 11, 68–78. 10.1021/pr2008863 [DOI] [PubMed] [Google Scholar]
- Komatsu S., Kobayashi Y., Nishizawa K., Nanjo Y., Furukawa K. (2010). Comparative proteomics analysis of differentially expressed proteins in soybean cell wall during flooding stress. Amino Acids 9, 1435–1449. 10.1007/s00726-010-0608-1 [DOI] [PubMed] [Google Scholar]
- Komatsu S., Kuji R., Nanjo Y., Hiraga S., Furukawa K. (2012b). Comprehensive analysis of endoplasmic reticulum-enriched fraction in root tips of soybean under flooding stress using proteomics techniques. J. Proteomics 77, 531–560. 10.1016/j.jprot.2012.09.032 [DOI] [PubMed] [Google Scholar]
- Komatsu S., Makino T., Yasue H. (2013a). Proteomic and biochemical analyses of the cotyledon and root of flooding-stressed soybean plants. PLoS ONE 8:e65301. 10.1371/journal.pone.0065301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu S., Nanjo Y., Nishimura M. (2013c). Proteomic analysis of the flooding tolerance mechanism in mutant soybean. J. Proteomics 79, 231–250. 10.1016/j.jprot.2012.12.023 [DOI] [PubMed] [Google Scholar]
- Komatsu S., Thibaut D., Hiraga S., Kato M., Chiba M., Hashiguchi A., et al. (2011). Characterization of a novel flooding stress-responsive alcohol dehydrogenase expressed in soybean roots. Plant Mol. Biol. 77, 309–322. 10.1007/s11103-011-9812-y [DOI] [PubMed] [Google Scholar]
- Komatsu S., Yamamoto R., Nanjo Y., Mikami Y., Yunokawa H., Sakata K. (2009). A comprehensive analysis of the soybean genes and proteins expressed under flooding stress using transcriptome and proteome techniques. J. Proteome Res. 8, 4766–4778. 10.1021/pr900460x [DOI] [PubMed] [Google Scholar]
- Laskowski M., Biller S., Stanley K., Kajstura T., Prusty R. (2006). Expression profiling of auxin-treated Arabidopsis roots: toward a molecular analysis of lateral root emergence. Plant Cell Physiol. 47, 788–792. 10.1093/pcp/pcj043 [DOI] [PubMed] [Google Scholar]
- Lee H., Lee J. S., Bae E. K., Choi Y. I., Noh E. W. (2005). Differential expression of a poplar copper chaperone gene in response to various abiotic stresses. Tree Physiol. 25, 395–401. 10.1093/treephys/25.4.395 [DOI] [PubMed] [Google Scholar]
- Lin K. H., Chiou Y. K., Hwang S. Y., Chen L. F. O., Lo H. F. (2008). Calcium chloride enhances the antioxidative system of sweet potato (Ipomoea batatas) under flooding stress. Ann. Appl. Biol. 152, 157–168. 10.1111/j.1744-7348.2007.00211.x [DOI] [Google Scholar]
- Luo M., Liu X., Singh P., Cui Y., Zimmerli L., Wu K. (2012). Chromatin modifications and remodeling in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 129–136. 10.1016/j.bbagrm.2011.06.008 [DOI] [PubMed] [Google Scholar]
- Mach J. (2012). Calcium channels and acquired thermotolerance: here comes the sun and it's all right. Plant Cell 24, 3167. 10.1105/tpc.112.240810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros-Silva M., Franck W. L., Borba M. P., Pizzato S. B., Strodtman K. N., Emerich D. W., et al. (2014). Soybean ureases, but not that of Bradyrhizobium japonicum, are involved in the process of soybean root nodulation. J. Agric. Food Chem. 62, 3517–3524. 10.1021/jf5000612 [DOI] [PubMed] [Google Scholar]
- Melan M. A., Enriquez A. L. D., Peterman T. K. (1994). The LOX1 gene of Arabidopsis is temporally and spatially regulated in germinating seedlings. Plant Physiol. 105, 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegazzi P., Guzzo F., Baldan B., Mariani P., Treves S. (1993). Purification of calreticulin-like protein(s) from spinach leaves. Biochem. Biophys. Res. Commun. 190, 1130–1135. 10.1006/bbrc.1993.1167 [DOI] [PubMed] [Google Scholar]
- Meyer-Bothling L. C., Polacco J. C. (1987). Mutational analysis of the embryo-specific urease locus of soybean. Mol. Gen. Genet. 209, 439–444. 10.1007/BF00331147 [DOI] [PubMed] [Google Scholar]
- Mira H., Martinez-Garcia F., Penarrubia L. (2001). Evidence for the plant-specific intercellular transport of the Arabidopsis copper chaperone CCH. Plant J. 25, 521–528. 10.1046/j.1365-313x.2001.00985.x [DOI] [PubMed] [Google Scholar]
- Nanjo Y., Nakamura T., Komatsu S. (2013). Identification of indicator proteins associated with flooding injury in soybean seedlings using label-free quantitative proteomics. J. Proteome Res. 12, 4758–4798. 10.1021/pr4002349 [DOI] [PubMed] [Google Scholar]
- Nanjo Y., Skultety L., Uvackova L., Klubicova K., Hajduch M., Komatsu S. (2012). Mass spectrometry-based analysis of proteomic changes in the root tips of flooded soybean seedlings. J. Proteome Res. 11, 372–385. 10.1021/pr200701y [DOI] [PubMed] [Google Scholar]
- Nishitani K., Tominaga R. (1992). Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J. Biol. Chem. 267, 21058–21064. [PubMed] [Google Scholar]
- Oh M. W., Nanjo Y., Komatsu S. (2014a). Analysis of soybean root proteins affected by gibberellic acid treatment under flooding stress. Protein Pept. Lett. 9, 911–947 10.2174/0929866521666140403122602 [DOI] [PubMed] [Google Scholar]
- Oh M. W., Nanjo Y., Komatsu S. (2014b). Identification of nuclear proteins in soybean under flooding stress using proteomic technique. Protein Pept. Lett. 21, 458–467. 10.2174/09298665113206660120 [DOI] [PubMed] [Google Scholar]
- Olsen J. V., de Godoy L. M. F., Li G., Macek B., Mortensen P., Pesch R., et al. (2005). Parts per million mass accuracy on an orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021. 10.1074/mcp.T500030-MCP200 [DOI] [PubMed] [Google Scholar]
- Park T. K., Holland M. A., Laskey J. G., Polacco J. C. (1994). Germination-associated lipoxygenase transcripts persist in maturing soybean plants and are induced by jasmonate. Plant Sci. 96, 109–117. 10.1016/0168-9452(94)90227-5 [DOI] [Google Scholar]
- Polacco J. C., Hyten D. L., Medeiros-Silva M., Sleper D. A., Bilyeu K. D. (2011). Mutational analysis of the major soybean UreF paralogue involved in urease activation. J. Exp. Bot. 62, 3599–3608. 10.1093/jxb/err054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preger V., Tango N., Marchand C., Lemaire S. D., Carbonera D., di Valentin M., et al. (2009). Auxin-responsive genes AIR12 code for a new family of plasma membrane b-type cytochromes specific to flowering plants. Plant Physiol. 150, 606–620. 10.1104/pp.109.139170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriquez-Serrano M., Romero-Puertas M. C., Pazmino D. M., Testillano P. S., Risuerio M. C., Del Rio L. A., et al. (2009). Cellular response of pea plants to cadmium toxicity: cross talk between reactive oxygen species, nitric oxide, and calcium. Plant Physiol. 150, 229–243. 10.1104/pp.108.131524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski M., Schottler M. A., Thiele W., Schulze W. X., Bock R. (2008). Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell 20, 2221–2237. 10.1105/tpc.108.060392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. J. (2000). Primer3 on the WWW for general users and for biologist programmers, in Bioinformatics Methods and Protocols, eds Krawetz S., Misener S. (New York, NY: Humana Press; ), 365–386 [DOI] [PubMed] [Google Scholar]
- Sauter M. (2013). Root responses to flooding. Curr. Opin. Plant Biol. 16, 282–286. 10.1016/j.pbi.2013.03.013 [DOI] [PubMed] [Google Scholar]
- Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., Nelson W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Servet C., Conde de Silva N., Zhou D. X. (2010). Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis. Mol. Plant 3, 670–677. 10.1093/mp/ssq018 [DOI] [PubMed] [Google Scholar]
- Setter T. L., Waters I. (2003). Review of prospects for germplasm improvement for waterlogging tolerance in wheat, barley and oats. Plant Soil 253, 1–34. 10.1023/A:1024573305997 [DOI] [Google Scholar]
- Sirko A., Brodzik R. (2000). Plant ureases: roles and regulation. Acta Biochim. Pol. 47, 1189–1195. [PubMed] [Google Scholar]
- Soga K., Wakabayashi K., Kamisaka S., Hoson T. (2007). Effects of hypergravity on expression of XTH genes in azuki bean epicotyls. Physiol. Plant. 131, 332–340. 10.1111/j.1399-3054.2007.00949.x [DOI] [PubMed] [Google Scholar]
- Sonnemann J., Bauerle A., Winckler T., Mutzel R. (1991). A ribosomal calmodulin-binding protein from Dictyostelium. J. Biol. Chem. 266, 23091–23096. [PubMed] [Google Scholar]
- Staniscuaski F., Brugge V. T., Carlini C. R., Orchard I. (2009). In vitro effect of Canavalia ensiformis urease and the derived peptide jaburetox-2Ec on Rhodnius prolixus Malpighian tubules. J. Insect Physiol. 55, 255–263. 10.1016/j.jinsphys.2008.12.002 [DOI] [PubMed] [Google Scholar]
- Sugimoto T., Aino M., Sugimoto M., Watanabe K. (2005). Reduction of Phytophthora stem rot disease on soybeans by the application of CaCl2 and Ca(NO3)2. J. Phytophathol. 153, 536–543. 10.1111/j.1439-0434.2005.01016.x [DOI] [Google Scholar]
- Sullivan M., van Toai T. T., Fausey N., Beuerlein J., Parkinson R., Soboyejo A. (2001). Evaluating on-farm flooding impacts on soybean. Crop Sci. 41, 93–100. 10.2135/cropsci2001.41193x [DOI] [Google Scholar]
- Thuleau P., Briere C., Mazar C. (2012). Recent advances in plant cell nuclear signaling. Mol. Plant 5, 968–970. 10.1093/mp/sss083 [DOI] [PubMed] [Google Scholar]
- Torisky R. S., Griffin J. D., Yenofsky R. L., Polacco J. C. (1994). A single gene (Eu4) encodes the tissue-ubiquitous urease of soybean. Mol. Gen. Genet. 242, 404–414. 10.1007/BF00281790 [DOI] [PubMed] [Google Scholar]
- Toyota M., Furuichi T., Tatsumi H., Sokabe M. (2008). Critical consideration on the relationshop between auxin transport and calcium transients in gravity perception of Arabidopsis seedlings. Plant Signal. Behav. 3, 521–524. 10.4161/psb.3.8.6339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B., Nagel A., Thimm O., Redestig H., Blaesing O. E., Palacios-Rojas N., et al. (2005). Extension of the visualization tool MapMan to allow statistical analysis of arrays; display of corresponding genes; and comparison with known responses. Plant Physiol. 138, 1195–1204. 10.1104/pp.105.060459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sandt V. S. T., Suslov D., Verbelen J. P., Vissenberg K. (2007). Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann. Bot. 100, 1467–1473. 10.1093/aob/mcm248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle C. A., Krochko J. E. (2009). Transcript profiling provides evidence of functional divergence and expression networks among ribosomal protein gene paralogs in Brassica napus. Plant Cell 21, 2203–2219. 10.1105/tpc.109.068411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebke-strohm B., Pasquali G., Margis-Pinheiro M., Bencke M., Bucker-Neto L., Becker-Ritt A. B., et al. (2012). Ubiquitous urease affects soybean susceptibility to fungi. Plant Mol. Biol. 79, 75–87. 10.1007/s11103-012-9894-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yata V. K., Thapa A., Mattaparthi V. S. (2014). Structural insight into the binding interactions of modeled structure of Arabidopsis thaliana urease with urea: an in silico study. J. Biomol. Struct. Dyn. 15, 1–7. 10.1080/07391102.2014.915765 [DOI] [PubMed] [Google Scholar]
- Yun H. S., Kwon C., Kim T. W., Joo S. H., Cho M. H., Kang B. G., et al. (2007). Regulation of VrXTH1 expression in mungbean. J. Plant Biol. 50, 65–69. 10.1007/BF03030602 [DOI] [Google Scholar]
- Zhang Y., Wen Z., Washburn M. P., Florens L. (2009). Effect of dynamic exclusion duration on spectral count based quantitative proteomics. Anal. Chem. 81, 6317–6326. 10.1021/ac9004887 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Du J., Zhao G., Jiang L. (2013). Activation of calcineurin is mainly responsible for the calcium sensitivity of gene deletion mutations in the genome of budding yeast. Genomics 101, 49–56. 10.1016/j.ygeno.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Sohn J. H., Warner J. R. (2003). Autoregulation in the biosynthesis of ribosomes. Mol. Cell. Biol. 23, 699–707. 10.1128/MCB.23.2.699-707.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental design for this study. (A) For physiological analysis, 2-day-old soybeans were flooded without or with 1, 5, 10, and 50 mM CaCl2 for 2, 4, and 6 days. (B) For proteomic analysis, 2-day-old soybeans were flooded without or with 50 mM CaCl2 for 2 days. Two-day-old and 4-day-old soybeans without flooding were used as controls. (C) For transcriptional analysis, 2-day-old soybeans were flooded without or with 50 mM CaCl2 for 1 and 2 days. Two-day-old, 3-day-old, and 4-day-old soybeans without flooding were used as controls. Three independent experiments were performed as biological replicates.
Effect of calcium on the growth of soybean under flooding stress in three independent experiments (A–C). Two-day-old soybeans were flooded without (dark blue) or with 1 (red), 5 (light green), 10 (purple), and 50 mM CaCl2 (light blue) for 2, 4, and 6 days. Photographs show soybean seedlings after 2, 4, and 6 days of flooding. Bars indicate 10 mm. The length and weight of roots, including the hypocotyl, were measured at the indicated time points. Data are means ± SE from three independent biological replications (A–C). Means with the same letter are not significantly different according to ANOVA (P < 0.05).
Evaluation of cell death in flooding-stressed soybean roots treated with calcium in three independent experiments (A–C). Two-day-old soybeans were flooded without (dark blue) or with 1 (red), 5 (light green), 10 (purple), and 50 mM CaCl2 (light blue) for 2, 4, and 6 days. The root tips were stained with 0.25% Evans blue dye, which was then extracted and measured spectroscopically at 600 nm at the indicated time points. Photographs show soybean seedlings after 2, 4, and 6 days of flooding. Bars indicate 10 mm. Data are means ± SE from three independent biological replications (A–C). Means with the same letter are not significantly different according to ANOVA (P < 0.05).
Principle component analysis of the peptide profile for identified proteins. The data sets of soybean proteins from 2-day-old (blue circle), 4-day-old (red square), 4-day-old with 2-day-flood (yellow triangle), and 4-day-old with 2-day-flood with calcium (purple pentagon) were plotted using principle component analysis (PCA). PCA was fully integrated into SIEVE software. PCA loading plot shows the spectral regions responsible for separation of groups in the corresponding score plot.
Abundance of six differentially changed proteins that were common between untreated and flooding-stressed soybean exposed to calcium. Two-day-old soybeans were flooded without or with 50 mM CaCl2 for 2 days. Proteins extracted from roots were purified, digested, and analyzed using nanoLC MS/MS. The protein abundance was determined by differential analysis using SIEVE software. Six differentially changed proteins were commonly identified in 4-day-old soybeans and 2-day-flooded soybeans treated with calcium.