Abstract
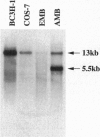
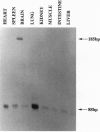
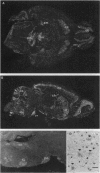
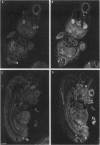
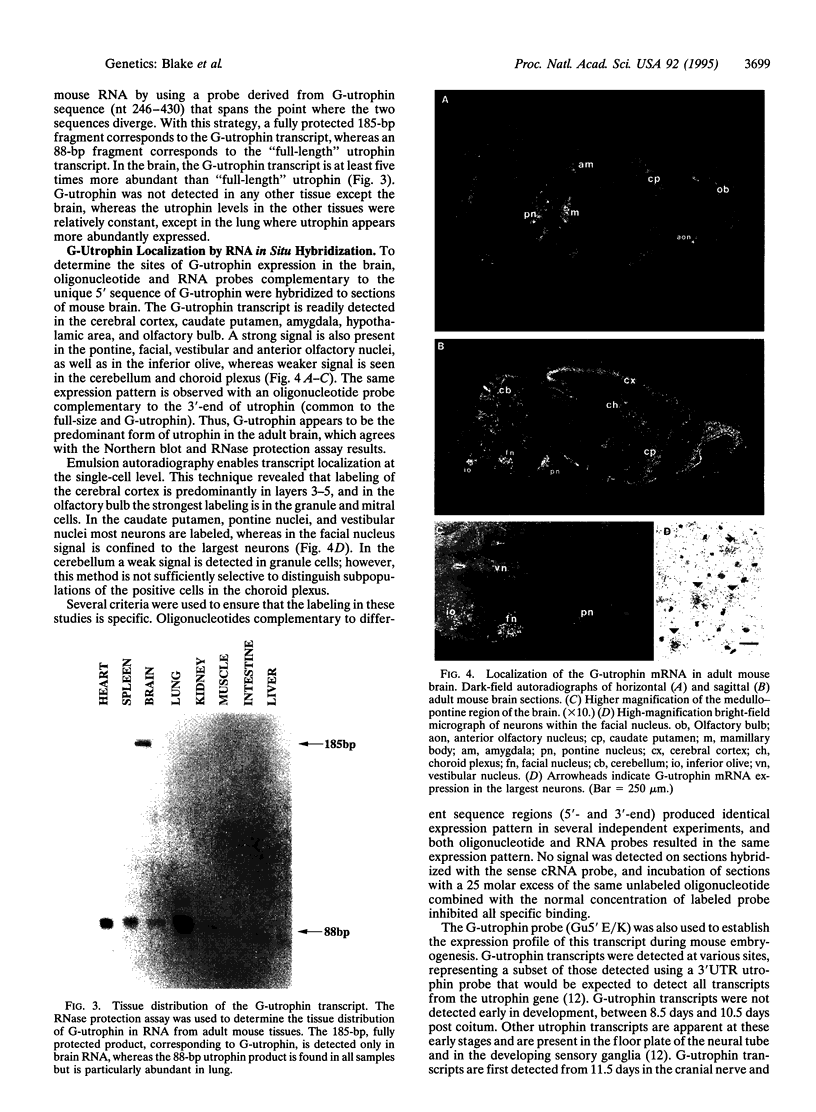
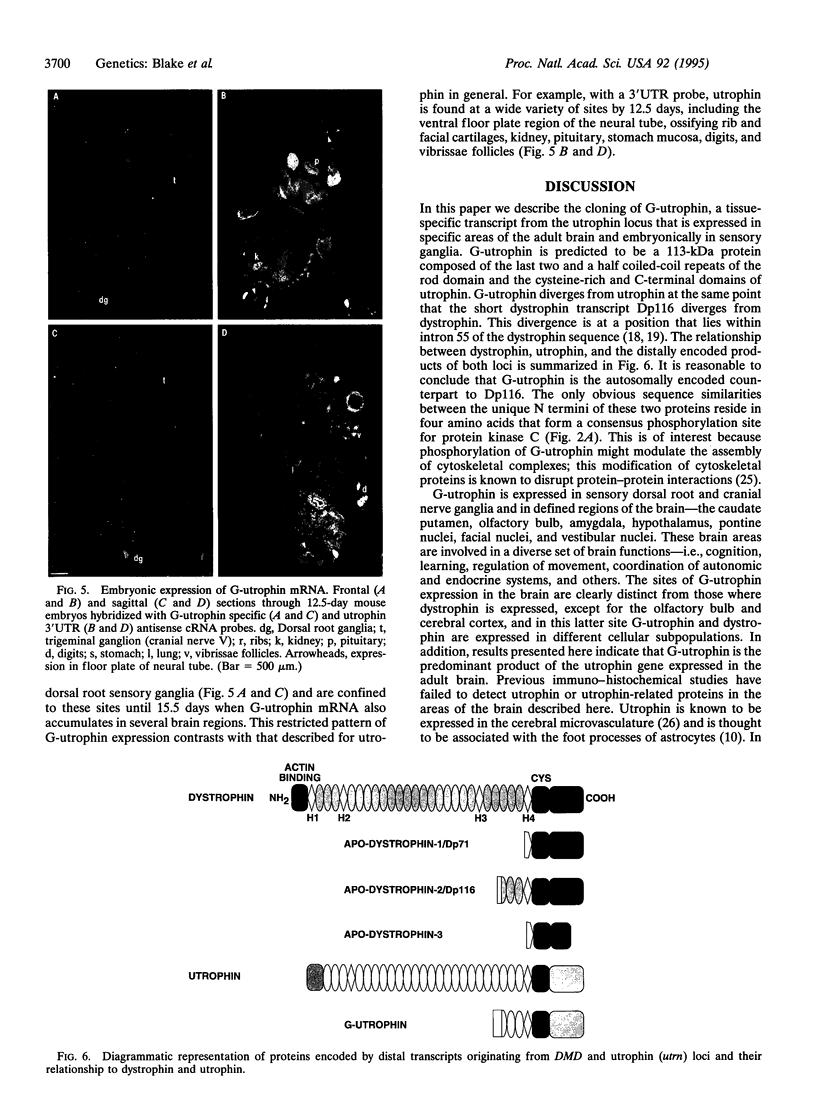
The utrophin gene is closely related to the dystrophin gene in both sequence and genomic structure. The Duchenne muscular dystrophy (DMD) locus encodes three 14-kb dystrophin transcripts in addition to several smaller isoforms, one of which, Dp116, is specific to peripheral nerve. We describe here the corresponding 5.5-kb mRNA from the utrophin locus. This transcript, designated G-utrophin, is of particular interest because it is specifically expressed in the adult mouse brain and appears to be the predominant utrophin transcript in this tissue. G-utrophin is expressed in brain sites generally different from the regions expressing beta-dystroglycan. During mouse embryogenesis G-utrophin is also seen in the developing sensory ganglia. Our data confirm the close evolutionary relationships between the DMD and utrophin loci; however, the functions for the corresponding proteins probably differ.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahn A. H., Kunkel L. M. The structural and functional diversity of dystrophin. Nat Genet. 1993 Apr;3(4):283–291. doi: 10.1038/ng0493-283. [DOI] [PubMed] [Google Scholar]
- Bewick G. S., Nicholson L. V., Young C., O'Donnell E., Slater C. R. Different distributions of dystrophin and related proteins at nerve-muscle junctions. Neuroreport. 1992 Oct;3(10):857–860. doi: 10.1097/00001756-199210000-00009. [DOI] [PubMed] [Google Scholar]
- Blake D. J., Love D. R., Tinsley J., Morris G. E., Turley H., Gatter K., Dickson G., Edwards Y. H., Davies K. E. Characterization of a 4.8kb transcript from the Duchenne muscular dystrophy locus expressed in Schwannoma cells. Hum Mol Genet. 1992 May;1(2):103–109. doi: 10.1093/hmg/1.2.103. [DOI] [PubMed] [Google Scholar]
- Blake D. J., Tinsley J. M., Davies K. E. The emerging family of dystrophin-related proteins. Trends Cell Biol. 1994 Jan;4(1):19–23. doi: 10.1016/0962-8924(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Bowe M. A., Deyst K. A., Leszyk J. D., Fallon J. R. Identification and purification of an agrin receptor from Torpedo postsynaptic membranes: a heteromeric complex related to the dystroglycans. Neuron. 1994 May;12(5):1173–1180. doi: 10.1016/0896-6273(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Byers T. J., Lidov H. G., Kunkel L. M. An alternative dystrophin transcript specific to peripheral nerve. Nat Genet. 1993 May;4(1):77–81. doi: 10.1038/ng0593-77. [DOI] [PubMed] [Google Scholar]
- Campanelli J. T., Roberds S. L., Campbell K. P., Scheller R. H. A role for dystrophin-associated glycoproteins and utrophin in agrin-induced AChR clustering. Cell. 1994 Jun 3;77(5):663–674. doi: 10.1016/0092-8674(94)90051-5. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Gee S. H., Montanaro F., Lindenbaum M. H., Carbonetto S. Dystroglycan-alpha, a dystrophin-associated glycoprotein, is a functional agrin receptor. Cell. 1994 Jun 3;77(5):675–686. doi: 10.1016/0092-8674(94)90052-3. [DOI] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Górecki D. C., Derry J. M., Barnard E. A. Dystroglycan: brain localisation and chromosome mapping in the mouse. Hum Mol Genet. 1994 Sep;3(9):1589–1597. doi: 10.1093/hmg/3.9.1589. [DOI] [PubMed] [Google Scholar]
- Górecki D. C., Monaco A. P., Derry J. M., Walker A. P., Barnard E. A., Barnard P. J. Expression of four alternative dystrophin transcripts in brain regions regulated by different promoters. Hum Mol Genet. 1992 Oct;1(7):505–510. doi: 10.1093/hmg/1.7.505. [DOI] [PubMed] [Google Scholar]
- Hugnot J. P., Gilgenkrantz H., Vincent N., Chafey P., Morris G. E., Monaco A. P., Berwald-Netter Y., Koulakoff A., Kaplan J. C., Kahn A. Distal transcript of the dystrophin gene initiated from an alternative first exon and encoding a 75-kDa protein widely distributed in nonmuscle tissues. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7506–7510. doi: 10.1073/pnas.89.16.7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakura K., Tadano Y., Kawai M., Ishiura S., Nakamura R., Miyamoto K., Nagata N., Sugita H. Dystrophin-related protein is found in the central nervous system of mice at various developmental stages, especially at the postsynaptic membrane. J Neurosci Res. 1994 Apr 15;37(6):728–734. doi: 10.1002/jnr.490370607. [DOI] [PubMed] [Google Scholar]
- Khurana T. S., Hoffman E. P., Kunkel L. M. Identification of a chromosome 6-encoded dystrophin-related protein. J Biol Chem. 1990 Oct 5;265(28):16717–16720. [PubMed] [Google Scholar]
- Khurana T. S., Watkins S. C., Kunkel L. M. The subcellular distribution of chromosome 6-encoded dystrophin-related protein in the brain. J Cell Biol. 1992 Oct;119(2):357–366. doi: 10.1083/jcb.119.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991 Nov;115(4):887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramarcy N. R., Vidal A., Froehner S. C., Sealock R. Association of utrophin and multiple dystrophin short forms with the mammalian M(r) 58,000 dystrophin-associated protein (syntrophin). J Biol Chem. 1994 Jan 28;269(4):2870–2876. [PubMed] [Google Scholar]
- Lederfein D., Levy Z., Augier N., Mornet D., Morris G., Fuchs O., Yaffe D., Nudel U. A 71-kilodalton protein is a major product of the Duchenne muscular dystrophy gene in brain and other nonmuscle tissues. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5346–5350. doi: 10.1073/pnas.89.12.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love D. R., Morris G. E., Ellis J. M., Fairbrother U., Marsden R. F., Bloomfield J. F., Edwards Y. H., Slater C. P., Parry D. J., Davies K. E. Tissue distribution of the dystrophin-related gene product and expression in the mdx and dy mouse. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3243–3247. doi: 10.1073/pnas.88.8.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura K., Campbell K. P. Dystrophin-glycoprotein complex: its role in the molecular pathogenesis of muscular dystrophies. Muscle Nerve. 1994 Jan;17(1):2–15. doi: 10.1002/mus.880170103. [DOI] [PubMed] [Google Scholar]
- Matsumura K., Ervasti J. M., Ohlendieck K., Kahl S. D., Campbell K. P. Association of dystrophin-related protein with dystrophin-associated proteins in mdx mouse muscle. Nature. 1992 Dec 10;360(6404):588–591. doi: 10.1038/360588a0. [DOI] [PubMed] [Google Scholar]
- Matsumura K., Shasby D. M., Campbell K. P. Purification of dystrophin-related protein (utrophin) from lung and its identification in pulmonary artery endothelial cells. FEBS Lett. 1993 Jul 12;326(1-3):289–293. doi: 10.1016/0014-5793(93)81810-m. [DOI] [PubMed] [Google Scholar]
- Matsumura K., Yamada H., Shimizu T., Campbell K. P. Differential expression of dystrophin, utrophin and dystrophin-associated proteins in peripheral nerve. FEBS Lett. 1993 Nov 22;334(3):281–285. doi: 10.1016/0014-5793(93)80695-q. [DOI] [PubMed] [Google Scholar]
- Nguyen T. M., Le T. T., Blake D. J., Davies K. E., Morris G. E. Utrophin, the autosomal homologue of dystrophin, is widely-expressed and membrane-associated in cultured cell lines. FEBS Lett. 1992 Nov 16;313(1):19–22. doi: 10.1016/0014-5793(92)81174-k. [DOI] [PubMed] [Google Scholar]
- Nigg E. A. Cellular substrates of p34(cdc2) and its companion cyclin-dependent kinases. Trends Cell Biol. 1993 Sep;3(9):296–301. doi: 10.1016/0962-8924(93)90011-o. [DOI] [PubMed] [Google Scholar]
- Nishio H., Takeshima Y., Narita N., Yanagawa H., Suzuki Y., Ishikawa Y., Ishikawa Y., Minami R., Nakamura H., Matsuo M. Identification of a novel first exon in the human dystrophin gene and of a new promoter located more than 500 kb upstream of the nearest known promoter. J Clin Invest. 1994 Sep;94(3):1037–1042. doi: 10.1172/JCI117417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce M., Blake D. J., Tinsley J. M., Byth B. C., Campbell L., Monaco A. P., Davies K. E. The utrophin and dystrophin genes share similarities in genomic structure. Hum Mol Genet. 1993 Nov;2(11):1765–1772. doi: 10.1093/hmg/2.11.1765. [DOI] [PubMed] [Google Scholar]
- Schofield J. N., Blake D. J., Simmons C., Morris G. E., Tinsley J. M., Davies K. E., Edwards Y. H. Apo-dystrophin-1 and apo-dystrophin-2, products of the Duchenne muscular dystrophy locus: expression during mouse embryogenesis and in cultured cell lines. Hum Mol Genet. 1994 Aug;3(8):1309–1316. doi: 10.1093/hmg/3.8.1309. [DOI] [PubMed] [Google Scholar]
- Schofield J., Houzelstein D., Davies K., Buckingham M., Edwards Y. H. Expression of the dystrophin-related protein (utrophin) gene during mouse embryogenesis. Dev Dyn. 1993 Dec;198(4):254–264. doi: 10.1002/aja.1001980403. [DOI] [PubMed] [Google Scholar]
- Tinsley J. M., Blake D. J., Roche A., Fairbrother U., Riss J., Byth B. C., Knight A. E., Kendrick-Jones J., Suthers G. K., Love D. R. Primary structure of dystrophin-related protein. Nature. 1992 Dec 10;360(6404):591–593. doi: 10.1038/360591a0. [DOI] [PubMed] [Google Scholar]
- Woodgett J. R., Gould K. L., Hunter T. Substrate specificity of protein kinase C. Use of synthetic peptides corresponding to physiological sites as probes for substrate recognition requirements. Eur J Biochem. 1986 Nov 17;161(1):177–184. doi: 10.1111/j.1432-1033.1986.tb10139.x. [DOI] [PubMed] [Google Scholar]