Abstract
Autism spectrum disorders (ASD) are a group of neurodevelopmental conditions characterized by dysfunction in social interaction, communication and stereotypic behavior. Genetic and environmental factors have been implicated in the development of ASD, but the molecular mechanisms underlying their interaction are not clear. Epigenetic modifications have been suggested as molecular mechanism that can mediate the interaction between the environment and the genome to produce adaptive or maladaptive behaviors. Here, using the Illumina 450 K methylation array we have determined the existence of many dysregulated CpGs in two cortical regions, Brodmann area 10 (BA10) and Brodmann area 24 (BA24), of individuals who had ASD. In BA10 we found a very significant enrichment for genomic areas responsible for immune functions among the hypomethylated CpGs, whereas genes related to synaptic membrane were enriched among hypermethylated CpGs. By comparing our methylome data with previously published transcriptome data, and by performing real-time PCR on selected genes that were dysregulated in our study, we show that hypomethylated genes are often overexpressed, and that there is an inverse correlation between gene expression and DNA methylation within the individuals. Among these genes there were C1Q, C3, ITGB2 (C3R), TNF-α, IRF8 and SPI1, which have recently been implicated in synaptic pruning and microglial cell specification. Finally, we determined the epigenetic dysregulation of the gene HDAC4, and we confirm that the locus encompassing C11orf21/TSPAN32 has multiple hypomethylated CpGs in the autistic brain, as previously demonstrated. Our data suggest a possible role for epigenetic processes in the etiology of ASD.
Introduction
Autism spectrum disorders (ASD) are defined by pervasive abnormalities in social interaction and communication, and by the presence of highly restricted interests and/or stereotypical repetitive behaviors.1 It is considered to be a highly heritable condition characterized by marked genetic heterogeneity and variable phenotype, ranging from mild-to-severe symptomatology. The etiology of ASD is mainly ascribed to different genetic variants such as copy number variations, single-nucleotide polymorphisms and only recently, de novo mutations.2,3 Despite its strong genetic component, several lines of evidence suggest that environmental factors may have a major role in the development of ASD. First, there is notable discordance within monozygotic twin pairs for diagnosed ASD, and often considerable symptom severity differences within ASD-concordant monozygotic twin pairs.4 Second, in separate studies, estimates of heritability among twin and sibling indicated a moderate genetic and a significantly larger environmental component to the etiology.5 In addition, according to recent epidemiological reports, only 54% of the recent increased incidence of ASD could be explained by known factors, including improved diagnostic power (25%),6 greater awareness by parents and pediatrician (15%),7 increased parental age (10%)8 and spatial clustering (4%).7 Therefore, 46% of increased incidence has been left unexplained, and unknown environmental factors may be involved in the increased ASD incidence. Putative environmental factors that have been correlated to ASD occurrence include maternal hospitalization for viral infections during the first trimester of pregnancy9 and prenatal exposure to sodium valproate.10 Other factors, like drugs, pollutants and several xenobiotics are strongly suspected.11 Therefore the molecular changes that are involved in the development of ASD are likely to be regulated by mechanisms which are, in part, modulated by environmental cues. In this scenario epigenetic mechanisms, such as DNA methylation and histone modifications, might be the ‘bridge' between the environmental influences and the manifest phenotype through the regulation of gene expression.
Brain development is a complex plastic process that requires regulated expression of specific gene sets in a space-temporal coordinated way. This task is fulfilled by transcription factors and by epigenetic machinery. Epigenetic modifications of both DNA and histone proteins are emerging as fundamental mechanisms by which neurons adapt their transcriptional response to developmental and environmental cues.12 Dysregulation of these mechanisms, caused by a genetic or environmental insult, can result in cognitive deficit and other features typical of neurodevelopmental disorders.13 Of particular interest, genome-sequencing studies in human neurodevelopmental and psychiatric disorders have uncovered mutations in many chromatin regulators.13 For example, mutation in methyl CpG binding protein 2 (MECP2), a protein product that binds methylated CpGs, causes Rett Syndome, a disorder with considerable phenotypic overlap with ASD.14 Therefore, there is a direct link between the machinery connected to DNA methylation and autistic-like behavior. Although recent genome-wide analyses provide evidence for epigenetic alterations in blood, ectodermal cells and in postmortem ASD brain, the field lacks a systematic investigation showing a direct implication of DNA methylation and correlative gene expression dysregulation in the development of ASD.15, 16, 17, 18
The central aim of this study is to determine the genome-wide DNA methylation patterns in autism brain and the significance of DNA methylation dysregulation in developing the disorder. To this end, we analyzed two brain areas, the prefrontal cortex (Brodmann Area 10; BA10) and the anterior cingulate gyrus (Brodmann Area 24; BA24), which have been previously linked to ASD through resting-state functional magnetic resonance imaging.19,20
Materials and methods
Brain tissue samples
Brain tissue samples from 13 autism cases and 12 controls were obtained from the Autism Tissue Program. All of these samples were received from the Harvard Brain Bank except for two autistic brain samples obtained from the UK Brain Bank for Autism (University of Oxford). For each brain, tissue was obtained from prefrontal cortex (BA10) and anterior cingulate gyrus (BA24), when available. Overall we had four distinct groups: 12 autism cases and 12 controls for BA10, and 11 autism cases and 11 controls for BA24 (Supplementary Table 1). For all autism cases and controls, clinical information is available upon request from the Autism Tissue Program (http:www.autismtissueprogram.org).
RNA extraction and quantitative RT–PCR
Total RNA was extracted from approximately 50 mg of liquid nitrogen pulverized tissue using the miRNeasy kit and treated with RNase-Free DNase (Qiagen, Valencia, CA, USA). RNA concentration was assessed by a NanoDrop spectrophotometer and 1 μg of total RNA was reverse transcribed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Real-time PCR (RT–PCR) was performed on an ABI ViiA7 Real-Time PCR detection system in 10 μl volume containing FastStart Universal SYBR Green Master (Roche, Basel, Switzerland) and primers at a concentration of 0.250 μM each. All primers used were designed using Primer-BLAST and tested for the efficiency through a standard curve. For genes with more than one isoform, primers were designed to encompass them all. All primer sequences are listed in Supplementary Table 2. The strategy used for the normalization of quantitative RT–PCR data from human genes was geometric averaging of multiple internal control genes according to the Vandesompele et al.21 method. We used four housekeeping genes (GAPDH, HPRT1, POLR2a and SDHA) that represent an accurate control for mRNA expression analysis of postmortem brain samples.22 For each housekeeping gene, we measured the gene stability (M value) and ranked it using the geNorm algorithm.21
DNA extraction and microarray
Genomic DNA was extracted from approximately 25 mg of liquid nitrogen pulverized tissue using the QIAamp DNA Mini Kit (Qiagen). Genomic DNA concentration was assessed by Qubit 2.0 Fluorometer using the Qubit dsDNA Broad RangeAssay. DNA samples were submitted to The Genomics Core Facility of The Rappaport Family Institute for Research in the Medical Sciences (Haifa, IL, USA), and processed by the following procedure. For each individual, genomic DNA (500 ng) extracted from brain tissue was treated with sodium bisulfite using the EZ96-DNA methylation kit (Zymo Research, Irvine, CA, USA) following the manufacturer's standard protocol. The bisulfite conversion reaction was performed in duplicate for each sample to minimize potential bias caused by variable conversion efficiency, and pooled bisulfite-treated DNA was used for subsequent array analysis. Genome-wide DNA methylation was assessed by the Infinium HumanMethylation450 BeadChip (Illumina, San Diego, CA, USA), which interrogates the DNA methylation profile of more than 485 000 methylation sites per sample at single-nucleotide resolution. Arrays were scanned by HiScan 2000 (Illumina). Illumina GenomeStudio software (Illumina) was used to extract signal intensities for each probe and to perform initial control quality checks. Internal Illumina controls were used to assess the quality of staining, extension, hybridization, bisulfite conversion and specificity. Probes targeting CpG sites with a single-nucleotide polymorphism in C or G or inside the probe sequence, that have a frequency >5% in the human population, were removed from the analysis according to Chen et al.23 A complete list of all the probes omitted from the analysis is available in Supplementary Table 3. The data from the microarrays have been deposited into the NCBI GEO (GSE53924).
Methylation microarray data analysis
Raw methylation β values were normalized using subset quantile normalization pipeline to correct for Infinium I and II bias, color bias, background correction, single-nucleotide polymorphism variation, allosomal positions and bead noise filtering.24 All additional computational and statistical analyses were performed using JMP-Genomic V6 (SAS Software). Because of the extensive normalization, about 0.05% of all data points had a β value either above or below 0. Hence this data points were adjusted to either 1 or 0 since this deviation might cause significant shift in results that have no biological relevance. Despite the subset quantile normalization 5% of the variance was still due to batch differences between the four chips. Using JMP Genomics batch normalization this variation was corrected. In addition, due to the strong effect of gender on DNA methylation, gender normalization was performed. We considered CpGs to be differentially methylated if they had a false discovery rate <0.05 and a difference in methylation >5% between the groups. To determine the overall similarity between samples, we used R software (http://www.r-project.org/) to perform principal component analysis that allowed us to project β values into an explanatory principal components.
Gene ontology analysis
Enrichment analysis for biological processes and InterPro categories were conducted using the GREAT online software (http://great.stanford.edu/great/public/html/).25 For differentially methylated CpG sites in both BA10 and BA24, the background was set to the total Illumina 450 K array to avoid any bias due to the different number of probes associated with each gene, and GRCh37 (UCSC hg19, Feb/2009) was used as human genome reference sequence. Only CpG sites with methylation Δβ value >5% at a false discovery rate <0.05 were included in the analysis. For each area, two independent analyses were conducted on hypermethylated and hypomethylated CpG sites in autism compared with control group.
Statistical analyses
All statistical analyses on gene expression and pyrosequencing methylation data were performed with SPSS software package (version 20.0; SPSS, Chicago, IL, USA). Levene's test was performed to assess the homogeneity of variance between the groups, and unequal variance was assumed whenever the test resulted significant (P-value <0.05). We used two-tailed independent t-test for groups with equal variance of distribution. The Mann–Whitney U-test was used to compare groups with unequal variance of distribution. For both statistical analyses P-values <0.05 were considered significant. Z-test and hypergeometric test were performed using an online statistical tool (http://www.socscistatistics.com/tests/ztest/Default2.aspx) and R software (http://www.r-project.org/), respectively. Cluster analysis and Treeview were utilized to cluster and visualize the results of the microarray experiment. Samples were clustered on the basis of hierarchical uncentered clustering of the genes utilizing the centroid linkage clustering method. Both programs were written by Michael Eisen (http://www.eisenlab.org/eisen/?page_id=42).
Methylation pyrosequencing
Pyrosequencing technology was used to further validate differences in DNA methylation detected with Infinium HumanMethylation450 BeadChip. We tested 12 CpG sites related to four different gene loci (C11orf21/TSPAN32, CTSZ, C1qA, IRF8) that demonstrate a large significant methylation Δβ value in autism versus control groups in BA10. DNA samples were submitted to the School of Medicine and Dentistry Genome Centre Barts and the London (London, UK) and processed by the following procedure. Briefly, 500 ng DNA from each individual was treated with sodium bisulfite using the EZ96-DNA methylation kit according to the manufacturer's recommendation, and amplified by a bisulfite polymerase chain reaction. Quantitative DNA methylation analysis of each CpG was conducted using PSQ96 Pyrosequencer (Qiagen). Genomic coordinates and sequencing primers for each single CpG are listed in Supplementary Table 4.
Results
To determine the genome-wide DNA methylation signature in the autistic brain, we analyzed two cortical regions, the prefrontal cortex (BA10) and the anterior cingulate gyrus (BA24) from individuals with an ADI-R-confirmed diagnosis of autism and matched controls. DNA was extracted from 12 BA10 and 11 BA24 brain samples from both control and autism groups, for a total of 46 samples. Clinical data from these individuals are found in Supplementary Table 1 and more in-depth case reports are available online (http://www.autismtissueprogram.org). There were no significant differences in the age and postmortem intervals between autism and control brain samples. DNA was converted with sodium bisulfite and probed with the Illumina 450 K methylation array. To validate the reproducibility of the assay, we probed a sodium bisulfite converted sample on two different batches of microarrays, and two different sodium bisulfite conversions on two different microarrays from the same batch. In both cases, the samples had extremely high correlation (Pearson's correlation coefficient, R2>0.99) (Supplementary Figures 1a and b).
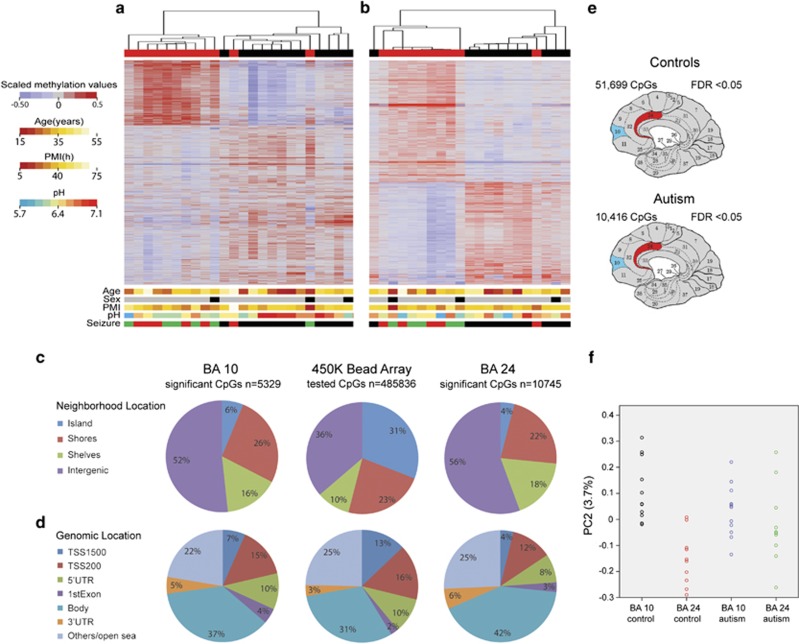
We performed filtering for high quality data which led to the exclusion of one sample, the BA24 region from individual AN04166, due to low signal-to-noise ratio in the sample. Following normalization procedures previously established for the Illumina 450 K methylation array (see Materials and Methods), we set cutoff parameters to detect differentially methylated CpG sites. We considered for further analyses only those CpGs that had a difference of at least 5% in DNA methylation after false discovery rate correction (false discovery rate <0.05). In addition, we removed probes that contained single-nucleotide polymorphisms that had a frequency >5% in human population, are cross-reactive or have previously been shown to display polymorphic binding (Supplementary Table 3).23 Applying these parameters, there were 5329 CpG sites differentially methylated between control and autism cohorts in BA10 (Figure 1a, Supplementary Table 5) and 10 745 in the BA24 (Figure 1b, Supplementary Table 6). Analysis of neighborhood location revealed that differentially methylated probes were more often found in areas of low CpG density, and rarely in CpG islands, compared with their representation on the microarray (Figure 1c). In addition, analysis of genomic location revealed a slight enrichment of differentially methylated probes in gene bodies and a significant decrease in transcription start site (TSS) here defined as TSS200 (Figure 1d).
Figure 1.
DNA methylation changes in autistic cerebral cortex regions. (a, b) Heat map of differentially methylated CpG sites between autism and control cohorts in prefrontal cortex (a) and cingulate gyrus (b). Scaled methylation values are color-coded according to the legend on the left. The dendrogram delineates the hierarchical clustering based on the >5% differentially methylated CpG sites after FDR (<0.05) correction. The top bar indicates the disease status: red, autism; black, control. The bottom bars evidence further variables for each sample: sex (gray, male; black, female), comorbidity of seizures (green, autism case with seizure disorder; red, autism case without seizure disorder; black, control), age, pH assessment and postmortem interval (PMI). The corresponding scale for the quantitative variables is shown on the left. (c, d) Pie charts depicting the neighborhood and genomic locations of CpGs represented in the microarray (middle) and those differentially methylated between controls and autism in BA10 (left) and BA24 (right). Only CpG sites >5% differentially methylated at an FDR <0.05 were plotted. (e) Diagram depicting the number of CpGs showing significant methylation differences between BA10 (blue) and BA24 (red) in control samples (top) and autism samples (bottom) at FDR <0.05 and difference of methylation >5%. (f) Scatter plot illustrating the second principle component (PC2; y axis) describing the divergence in methylation between two cortical areas, BA10 and BA24, in autism and control groups. BA, Brodmann area; FDR, false discovery rate.
Next, we analyzed the epigenetic distinction between the two cortical regions. We found that, in control brains, 51 699 CpG sites were differentially methylated between BA10 and BA24, demonstrating that the human brain differentiates into epigenetically distinct areas. In comparison, only 10 416 CpG sites were differentially methylated between the same two brain regions in the autistic brain (Figure 1e, Supplementary Tables 7 and 8), which is a highly significant deviation from the findings in the control brain (Z-test for difference in proportions, P<0.0001). To rule out the possibility that our findings were due to an issue of statistical threshold, we applied more stringent criteria. Remarkably, we found that 2615 and 31 532 CpG sites were differentially methylated between BA10 and BA24 in autistic and control respectively at a false discovery rate <0.01 (P<0.0001). To gain more statistical insight into our data, we performed a principal component analysis, an unsupervised learning method that is unaware of the identity of each sample.25 The β values from the microarray were projected into principal components that explain the source of variation in the data (Supplementary Table 9). Principal component number one accounts for 51% of the variation between the data and has an invariant loading profile among all the samples, which support the statistical accuracy of the microarray (Supplementary Figure 2a). Principal component number two, which explains 3.7% of the variance, distinguishes between the two brain regions in control subjects (Figure 1f). Interestingly, such differentiation does not occur in the autistic brain. This supports our finding that the autistic brain displays less region-specific identity, at the level of DNA methylation, as we reported above. Thus we may hypothesize a dysfunction in the developmental program that leads to less epigenetic distinction between cortical regions in the autistic compared with the control brain.
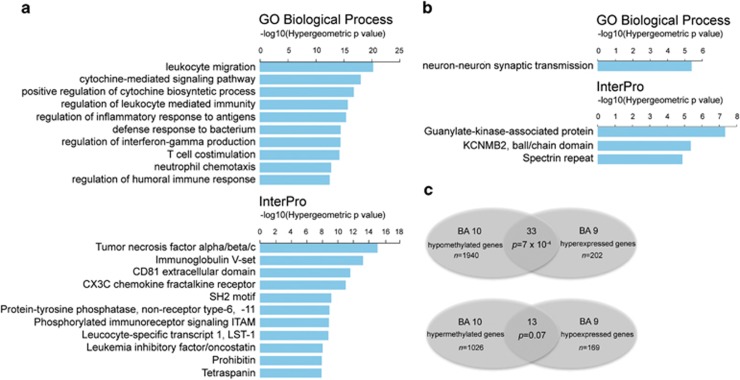
Our next aim was to determine which type of genomic regions, in respect to biological function, were differentially methylated between control and autistic brains. Gene Ontology analysis was carried out using the GREAT online tool (http://bejerano.stanford.edu/great/public/html/), which is designed for the study of cis-regulating factors, such as in epigenetic modifications.26 In BA10, CpGs that were hypomethylated in the autistic brain samples correspond with several categories related to immune response, including leukocyte migration, positive regulation of cytokine biosynthetic process and inflammatory response to antigens (Figure 2a). Hypermethylated regions in the autistic brain samples belong to the neuron–neuron synaptic transmission category, and include DLG2, DLGAP1 and DLGAP2 genes (Figure 2b). In contrast, GO analysis of the BA24 revealed a much more varied enrichment in several unrelated biological processes, with only a slight enrichment of some immune functions (Supplementary Figures 3a and b). Interestingly, in the BA10, there was a significantly greater number of CpGs that were hypomethylated with respect to those that were hypermethylated, whereas in BA24 we did not detect such difference (Supplementary Figure 4). Therefore, in the rest of this study, we concentrated on the BA10 region, both because of its well-established significance to autistic behavior and because of the very significant changes in immune system and synaptic categories, as revealed in the GREAT analysis.
Figure 2.
Gene Ontology analysis of differentially methylated CpGs in autistic prefrontal cortex. (a) Gene Ontology analysis for biological process and InterPro categories for CpG sites that were hypomethylated by >5% in the autism group at FDR <0.05. (b) Gene Ontology analysis for biological process and InterPro categories for CpG sites that were hypermethylated by >5% in the autism group at FDR <0.05. The analysis was performed using GREAT.26 (c) Venn diagram depicting the overlap between hypomethylated genes and hyperexpressed genes (top) and between hypermethylated genes and hypoexpressed genes (bottom), respectively from our methylation data (BA10) and Voineagu et al.27 transcriptome data (BA9). BA, Brodmann area; FDR, false discovery rate.
The data were further analyzed to determine if previously identified autism candidate genes were highly represented among regions of the genome that were differentially methylated in our samples. The Simons Foundation Autism Research Initiative (SFARI) database consists of a list of genes which have been linked to autism by genetic studies. SFARI genes were significantly underrepresented in the list of CpG sites that were hypomethylated in the autistic brain (3.49% representation of SFARI genes in hypomethylated regions, compared with 5.93% representation of SFARI genes in microarray; Z-test for difference in proportions, P<0.0001). This is not surprising, considering that immune system genes have not been highly implicated in the genetics of autism.27 In contrast, there was a minor hyperrepresentation of SFARI genes in the CpG sites that were hypermethylated in autism (7.46% representation of SFARI genes in hypermethylated genomic regions, P=0.0285).
Next we aimed to determine the relationship between gene expression and DNA methylation in the autistic brain. We compared our list of genes that displayed differentially methylated CpGs in BA10 of autistic subjects to a published list of genes from a genome-wide transcriptome study that investigated gene expression in a very close brain region, BA9.27 We found a very significant overlap between genes that were hypomethylated in our current study and those that were overexpressed in the transcriptome study, whereas there was only a tendency for an overlap between genes that were hypermethylated in our study and those that were underexpressed in the transcriptome study (Figure 2c, Supplementary Table 10). Interestingly, ‘immune response' was the GO category that was most significantly represented in both the transcriptome and our methylome analysis. It should be noted that our study has been conducted on BA10, while the expression data from Voineagu et al.27 referred to BA9 (a neighboring section of the prefrontal cortex), which may explain some of the differences in the results between the two studies. However, this data suggest that there is a strong relationship between hypomethylation and increased gene expression in the autistic brain.
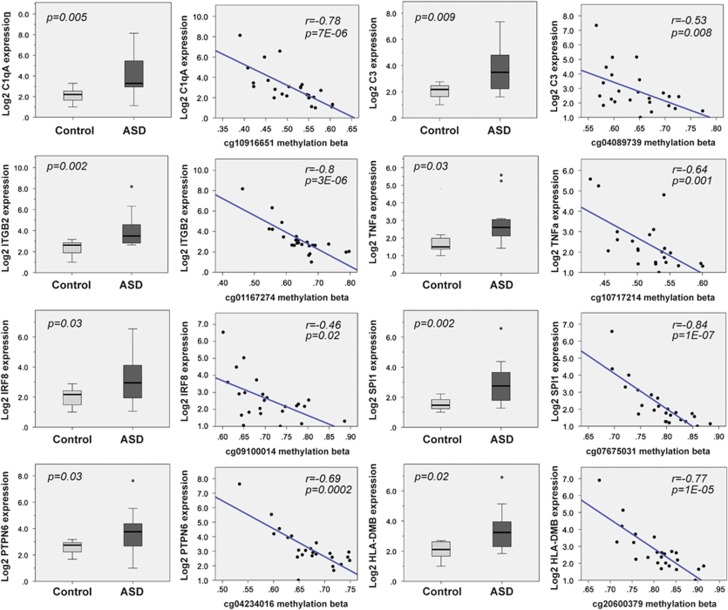
To further study the relationship between DNA methylation and gene expression, we performed RT–PCR analysis on a subset of genes that displayed dysregulated methylation at multiple CpG sites. We found that out of 17 genes that we tested, 12 displayed differential expression between control and autism groups (Figure 3, Supplementary Figure 5). Among these, C1qA, C3, ITGB2, TNF-α, IRF8, SPI1, PTPN6 and HLA-DMB were significantly overexpressed and showed highly significant inverse correlation between DNA methylation and gene expression (Figure 3, Supplementary Table 11). From the 17 genes we tested, 48 out of 93 CpG sites displayed a significant correlation between methylation levels and gene expression (Supplementary Table 11). However, of the 12 genes that displayed dysregulated gene expression in our RT–PCR analysis, the DNA methylation of 42 out of 61 CpGs was significantly correlated with gene expression. Therefore a subset of CpGs may have a significant influence on gene expression. Our expression data strongly indicates the presence of an altered immune response in the autistic brain that correlates well with epigenetic modulation of genomic regions relevant to immune function. To strengthen this hypothesis, we further explored the inflammatory status of the autistic brain by checking for expression of classic inflammatory markers. We found IL-1b and IBA-1, a marker of microglia activation, increased in autism brain (Supplementary Figure 6); this suggests the possibility of a neuroinflammatory process. To address the issue whether our methylation data may be affected by changes in cell-type heterogeneity in the autistic brain, we analyzed whether our differentially methylated CpGs are found in regions that are known to be differentially methylated between neurons and glia, according to a recent scientific report.28 We found that none of the genes that we have examined in this study are among those regions.
Figure 3.
Directional association between gene expression and methylation in prefrontal cortex. Gene expression analysis of eight immune system-related genes that displayed multiple hypomethylated CpGs in the autism group (C3, C1qA, ITGB2, TNF-α, IRF8, SPI1, PTPN6 and HLA-DMB). For each gene, we show the gene expression analysis (left) and Spearman's ρ correlation coefficient (right) between individuals' gene expression and DNA methylation levels. All of these genes are significantly overexpressed in the autism group and display significant inverse correlations between DNA methylation levels and gene expression levels.
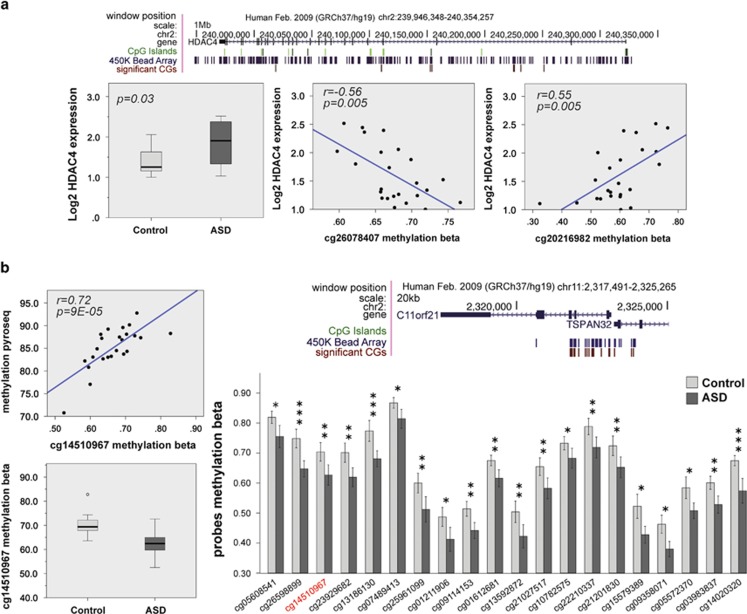
Among genes with several dysregulated CpGs in the autistic BA10, there was HDAC4. HDAC4 is also an interesting gene in the context of autism because it is known to downregulate the transcription of several synaptic genes,29 and genetic aberrations in this gene cause brachydactyly-mental retardation syndrome, including autistic features.30 Interestingly, HDAC4 did not show a specific methylation trend, with some CpGs being hypomethylated whereas others hypermethylated. We found that HDAC4 was significantly overexpressed in autistic subjects, and that methylation of some CpGs had a positive while others a negative correlation to its gene expression (Figure 4a, Supplementary Table 11). Therefore, there is a complex interaction between DNA methylation and gene expression of HDAC4 in the autistic brain.
Figure 4.
Dysregulated DNA methylation in the genes HDAC4 and TSPAN32. (a) Gene expression and directional association with β methylation values are depicted for HDAC4. The position of the significant CpGs in relation to the gene, CpG islands and the Illumina 450 K methylation array are shown on the top of the three box plots. Gene expression (left) and inverse (center) and direct (right) correlations between gene expression and CpG methylation levels are shown below. P-values for both expression and Spearman's ρ correlation coefficient were <0.05. (b) Differences in methylation between autistic and control cohorts are described for C11orf21/TSPAN32. The position of significant CpGs in relation to the gene, CpG islands and the Illumina 450 K methylation array are shown above. A bar chart provides a detailed description of each single CpG site for C11orf21/TSPAN32 for both autistic and control cohorts (below) with relative P-value indicated above each pair of bars (*P<0.05; **P<0.01; ***P<0.001). For one CpG site (highlighted in red) is shown a box plot describing the β methylation value in autism and control cohorts, and a Spearman's ρ correlation coefficient between the methylation β values (microarray) and the pyrosequencing methylation values. P-values for both methylation and Spearman's ρ correlation coefficient were <0.05.
In a very recent study performed by Ladd-Acosta et al.18 they determined significant DNA methylation differences at multiple CpGs related to C11orf21/TSPAN32, PRRT1 and SDHAP3 between control and autistic brains.18 In our data, we found only a few differentially methylated CpGs in the PRRT1 gene in both BA10 and BA24. However, there was a very large amount of CpGs in sites upstream the TSS of C11orf21/TSPAN32 that were hypomethylated specifically in the BA10 (Figure 4b). We validated seven of these CpG sites by pyrosequencing technology (Supplementary Figure 7, Supplementary Table 12), and we demonstrated a very significant correlation between β and pyrosequencing methylation values (Supplementary Table 13). Therefore, C11orf21/TSPAN32 has now been identified as differentially methylated by two independent studies. In addition, we validate the other three CpG sites associated to C1qA, IRF8 and CTSZ genes (Supplementary Figure 7; Supplementary Table 12), and also for these CpGs the correlation between β and pyrosequencing methylation values was very significant (Supplementary Table 13).
Discussion
The aim of this study was to interrogate the DNA methylation profile of two cortical brain areas, prefrontal cortex (BA10) and cingulate gyrus (BA24), in autism and control cohorts. We found that both brain regions have a profoundly distinctive epigenetic signature in the autistic brain with respect to their normal counterpart. Low-density CpG regions, such as intergenic regions, are highly represented among differentially methylated CpGs, whereas high-density CpG regions, such as CpG islands, are less abundant. In addition, we observed a significant underrepresentation of differentially methylated CpGs in the region just upstream the TSS, here defined as TSS200, and a slight enrichment in the gene bodies. This tendency was common to both brain areas suggesting a general trend rather than a technical artifact. The biological meaning of these findings is not well understood but some speculations can be made. First, regions with lower CpG density are considered more dynamic with respect to CpG islands that, on the contrary, are under stronger constraints.31 Second, a recent publication from Lister et al. that profiles the methylome of human and mouse prefrontal cortex throughout the development, demonstrated that half of the CpGs which are differentially methylated between fetal and adult prefrontal cortex are found within gene bodies, and there are very few in promoters and TSSs.32 This finding coincides with the enrichment of dysregulated CpGs in the gene body and depletion in promoter regions in our current study. Therefore, we may speculate that CpGs, which show dynamic methylation changes during development are even more liable to be effected by an early insult during this time window.
An earlier study performed by Voineagu et al.27 demonstrated that the typical differences in gene transcription between two cortical areas, temporal and prefrontal cortex, are attenuated in the autistic brain suggesting a lack of normal cortical differentiation into diverse regions. In addition, Stoner et al.33 in a recent explorative study found focal disruption of cortical laminar architecture in the cortexes of a majority of young children with autism, suggesting a probable dysregulation of layer formation and layer-specific neuronal differentiation at prenatal developmental stages. In light of these studies, we asked if DNA methylation might also be involved in the deficit of cortical region-specific differentiation. Both t-test analysis and principal component analysis performed between BA10 and BA24 in autistic and control groups revealed less divergence in the autistic brain pointing to an abnormal developmental patterning as potential pathophysiological driver in ASD. In neurons, widespread methylome reconfiguration occurs during development from fetal to young adult and is considered a critical process in defining the neuron molecular identity.33 As DNA methylation is involved in the programming of cellular differentiation,34 it is plausible that the dysregulated DNA methylation patterns may have a central role in the lack of cortical region-specific differentiation in the autistic brain.
Gene ontology analysis of differentially methylated CpG sites in BA10 highlights the ‘immune response' as the most enriched class, which strengthens the scientific evidence that a dysregulated immune system is one of the contributing factors in ASD.35 A comparison between our methylation data and the transcriptomic data from Voineagu et al.27 evidences a very significant overlap between genes characterized by DNA hypomethylation and increased expression level. We confirm the same trend checking the expression of 17 differentially methylated genes by RT–PCR. The finding that hypomethylation of many immune response-related genes is correlated with the increase in their expression suggests that epigenetic modulation may be particularly important for the changes in the immune response observed in individuals with ASD. This is particularly interesting in relation to the findings by Voineagu et al.27 that many neuronal genes that were underexpressed in the autistic brain, were found enriched in autism GWAS (genome-wide association studies), whereas immune response genes did not evidence a strong genetic etiology in autism. Therefore, the changes in immune response genes, proved by multiple studies, are likely to be induced through epigenetic mechanisms.
In addition, many of the immune response genes for which we checked DNA methylation and expression profiles, are involved both in microglial cell fate specification and in synaptic pruning during brain development. For example, genes like SPI1 (PU.1) and IRF8 are both indispensable for microgliogenesis36 and, as recently demonstrated for IRF8, they could also have a role in the adult brain.37 Of interest, the constitutive microglia marker, CX3CR1, although differentially methylated, did not show differential expression in the autistic brain in our data. Therefore, there appears to be an increased activation of microglia, confirmed also by increased expression of IBA-1, but not an increased number of microglial cells. A correct expression of immune mediators and a proper microglia phenotype are indispensable during human brain development.38,39 Microglial cells are the primary cells that come in contact with neurons during the first wave of embryonic synaptogenesis where they secrete trophic factors. During pre- and postnatal development, microglia actively engulf synaptic structures and exert a major role in synaptic pruning.40 Therefore its dysregulation, particularly during brain development, could impact negatively on the neural patterning and connectivity.
Of great interest, many of the same immune response-related genes that we found to be dysregulated in the autistic brain have also been recently identified as specific mediators of synaptic development. Crucial roles for the chemokine CX3CR1, for the members of the complement cascade C1q, C3 and their receptor ITGB2 (CR3), were demonstrated in pruning neuronal synapses during brain development.40, 41, 42, 43, 44 In fact, knockout mice for CX3CR1, C1q, C3 or ITGB2 display deficits in synaptic connectivity along with several behavioral impairments.40, 41, 42, 43, 44, 45, 46 In addition, TNF-α has been shown as potent effector of synaptic scaling in mature brain and seems to be involved in the regulation of synaptic plasticity,47 whereas CXCR3 is fundamental in removing synaptic structures after neuronal damage.48 Therefore, we suggest that many of the immune system changes that we detected in the autistic brain may be implicated in synaptic dysregulation, in addition to having a role in classic immune processes. Although there is ample evidence for nonimmunologic roles of these molecules during brain development, much less is known about their function in the adult brain. Further studies will be necessary to address this question.
One of the major challenges in understanding the biological consequences of our DNA methylation data is the fact that the human cortex consists of several cell types, including neurons and several glia lineages. We may assume that some of the immune response gene dysregulations are contributed by changes in the microglia, whereas dysregulation at the synaptic genes is attributed to changes in the neurons. However, there is ample evidence to consider that some of the immunological dysfunctions are representative of changes in the neurons themselves, and not only in immune cells. First, in the DNMT1/DNMT3a forebrain neuron knockout mice, immune system genes were found to be the most differentially methylated compared with wild-type mice.49 Therefore, in neurons, regions of the genome that are associated with immune function are particularly sensitive to dysregulation. Second, some of the immune response genes that were found to be dysregulated in the autistic brain are prone to expression in neurons during development, including C1q and C3.43 Third, a recent report has highlighted that, during brain development, more robust changes in DNA methylation occur in neurons rather than in glia, and that methylation in glia cells is considerably more stable.32 Considering all the above, we suggest that the immune-related epigenetic and gene expression differences in the autistic brain represent changes both in the neuronal and glia cell types.
It has been widely suggested that there is a linear causal relationship between environmental factors, epigenetic dysregulation and mental disorders. One of the environmental factors which have been implicated in human studies to give a predisposition to the development of ASD is a prenatal inflammatory event, such as maternal hospitalization during the first trimester due to viral infection.9 Similarly, stimulation of the immune response in mice during pregnancy leads to the development of autistic-like behaviors in the offspring.50 Therefore, this may lead to the hypothesis that early-life immunological insults may induce long-term changes in behavior through modulation of DNA methylation in genomic regions involved in the immune response, such as the hypomethylated regions we have seen in our data. Future research on this hypothesis should aim to understand the direct interaction and causal relationships between environmental insults, DNA methylation and neurodevelopment.
Acknowledgments
We thank Dr Nili Avidan from the Genomics Core Facility of The Rappaport Family Institute for Research in the Medical Sciences (Haifa, IL, USA) for the preliminary analysis of the Illumina 450K methylation array. In addition, we thank Professor Itzhak Haviv from the Bar Ilan University Faculty of Medicine (Safed, IL, USA) for his informative discussion which improved the quality of this work. We thank the Autism Tissue Program, the Harvard Brain Tissue Resource Center and Oxford University for the control and autism tissues used in this project. This research was supported by the ISRAEL SCIENCE FOUNDATION (grant No. 1047/12) and the National Institute of Psychobiology in Israel.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders: DSM-5TM (5th edn), 2013
- Stevens C, Lin C-F, Cook EH, Jr, Newsham I, Samocha KE, Zhao T, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt P, State MW. The conundrums of understanding genetic risks for autism spectrum disorders. Nat Neurosci. 2011;14:1499–1506. doi: 10.1038/nn.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald A, Hoekstra RA. Autism spectrum disorders and autistic traits: a decade of new twin studies. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:255–274. doi: 10.1002/ajmg.b.31159. [DOI] [PubMed] [Google Scholar]
- Torres A, Phillips J, Miller J, Cohen B, Lajonchere C, Fedele A, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry. 2011;68:1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M, Bearman P. Diagnostic change and the increased prevalence of autism. Int J Epidemiol. 2009;38:1224–1234. doi: 10.1093/ije/dyp261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearman PS, King MD. Socioeconomic status and the increased prevalence of autism in California. Am Sociol Rev. 2011;76:320–346. doi: 10.1177/0003122411399389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WSW, Masson G, Helgason H, Helgason A, Thorsteinsdottir U, Besenbacher S, et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atladóttir HO, Thorsen P, Østergaard L, Schendel DE, Lemcke S, Abdallah M, et al. Maternal infection requiring hospitalization during pregnancy and autism spectrum disorders. J Autism Dev Disord. 2010;40:1423–1430. doi: 10.1007/s10803-010-1006-y. [DOI] [PubMed] [Google Scholar]
- Christensen J, Grønborg TK, Sørensen MJ, Schendel D, Parner ET, Pedersen LH, et al. Prenatal valproate exposure and risk of autism spectrum disorders and childhood autism. JAMA. 2013;309:1696–1703. doi: 10.1001/jama.2013.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle JM. A genomic point-of-view on environmental factors influencing the human brain methylome. Epigenetics. 2011;6:862–869. doi: 10.4161/epi.6.7.16353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio A. Dynamic epigenetic regulation in neurons: enzymes, stimuli and signaling pathways. Nat Neurosci. 2010;13:1330–1337. doi: 10.1038/nn.2671. [DOI] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Wong CCY, Meaburn EL, Ronald A, Price TS, Jeffries AR, Schalkwyk LC, et al. Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Mol Psychiatry. 2013. pp. 1–9. [DOI] [PMC free article] [PubMed]
- Berko ER, Suzuki M, Beren F, Lemetre C, Alaimo CM, Calder RB, et al. Mosaic epigenetic dysregulation of ectodermal cells in autism spectrum disorder. PLoS Genet. 2014;10:e1004402. doi: 10.1371/journal.pgen.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulha HP, Lin CL, Guo Y, Lessard A, Akbarian S, Virgil D, et al. Epigenetic signatures of autism: trimethylated H3K4 landscapes in prefrontal neurons. Arch Gen Psychiatry. 2012;69:314–324. doi: 10.1001/archgenpsychiatry.2011.151. [DOI] [PubMed] [Google Scholar]
- Ladd-Acosta C, Hansen KD, Briem E, Fallin MD, Kaufmann WE, Feinberg AP. Common DNA methylation alterations in multiple brain regions in autism. Mol Psychiatry. 2014;19:862–871. doi: 10.1038/mp.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A, Yan C-G, Li Q, Denio E, Castellanos FX, Alaerts K, et al. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry. 2013. pp. 1–9. [DOI] [PMC free article] [PubMed]
- Di Martino A, Shehzad Z, Kelly C, Roy AK, Gee DG, Uddin LQ, et al. Relationship between cingulo-insular functional connectivity and autistic traits in neurotypical adults. Am J Psychiatry. 2009;166:891–899. doi: 10.1176/appi.ajp.2009.08121894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller S, Sarchiapone M, Zarrilli F, Videtic A, Ferraro A, Carli V, et al. Increased BDNF promoter methylation in the Wernicke area of suicide subjects. Arch Gen Psychiatry. 2010;67:258–267. doi: 10.1001/archgenpsychiatry.2010.9. [DOI] [PubMed] [Google Scholar]
- Chen Y, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tost J, Touleimat N. Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012;4:325–341. doi: 10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- Ringnér M. What is principal component analysis. Nat Biotechnol. 2008;26:303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guintivano J, Aryee MJ, Kaminsky ZA. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics. 2013;8:290–302. doi: 10.4161/epi.23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sando R, Gounko N, Pieraut S, Liao L, Yates J, Maximov A. HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell. 2012;151:821–834. doi: 10.1016/j.cell.2012.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SR, Aldred MA, Der Kaloustian VM, Halal F, Gowans G, McLeod DR, et al. Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. Am J Hum Genet. 2010;87:219–228. doi: 10.1016/j.ajhg.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino D, Loke YJ, Gordon L, Ollikainen M, Cruickshank MN, Saffery R, et al. Longitudinal, genome-scale analysis of DNA methylation in twins from birth to 18 months of age reveals rapid epigenetic change in early life and pair-specific effects of discordance. Genome Biol. 2013;14:R42. doi: 10.1186/gb-2013-14-5-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner R, Chow ML, Boyle MP, Sunkin SM, Mouton PR, Roy S, et al. Patches of disorganization in the neocortex of children with autism. N Engl J Med. 2014;370:1209–1219. doi: 10.1056/NEJMoa1307491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Depino AM. Peripheral and central inflammation in autism spectrum disorders. Mol Cell Neurosci. 2013;53:69–76. doi: 10.1016/j.mcn.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- Masuda T, Tsuda M, Yoshinaga R, Tozaki-Saitoh H, Ozato K, Tamura T, et al. IRF8 is a critical transcription factor for transforming microglia into a reactive phenotype. Cell Rep. 2012;1:334–340. doi: 10.1016/j.celrep.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn MW, Elmer BM, Garay PA, Liu X-B, Needleman LA, El-Sabeawy F, et al. MHCI negatively regulates synapse density during the establishment of cortical connections. Nat Neurosci. 2011;14:442–451. doi: 10.1038/nn.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M, Fujita Y, Tanaka T, Nakamura Y, Kikuta J, Ishii M, et al. Layer V cortical neurons require microglial support for survival during postnatal development. Nat. Neurosci. 2013;16:543–551. doi: 10.1038/nn.3358. [DOI] [PubMed] [Google Scholar]
- Kettenmann H, Kirchhoff F, Verkhratsky A. Microglia: new roles for the synaptic stripper. Neuron. 2013;77:10–18. doi: 10.1016/j.neuron.2012.12.023. [DOI] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- Zhan Y, Paolicelli RC, Sforazzini F, Weinhard L, Bolasco G, Pagani F, et al. Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nat Neurosci. 2014;17:400–406. doi: 10.1038/nn.3641. [DOI] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- Schafer DP, Kautzman AG, Greenberg ME, Koyama R, Ransohoff RM, Stevens B, et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Jin X, Parada I, Pesic A, Stevens B, Barres B, et al. Enhanced synaptic connectivity and epilepsy in C1q knockout mice. Proc Natl Acad Sci USA. 2010;107:7975–7980. doi: 10.1073/pnas.0913449107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Alcazar M, Daborg J, Stokowska A, Wasling P, Björefeldt A, Kalm M, et al. Altered cognitive performance and synaptic function in the hippocampus of mice lacking C3. Exp Neurol. 2014;253:154–164. doi: 10.1016/j.expneurol.2013.12.013. [DOI] [PubMed] [Google Scholar]
- Stellwagen D, Malenka RC. Synaptic scaling mediated by glial TNF-alpha. Nature. 2006;440:1054–1059. doi: 10.1038/nature04671. [DOI] [PubMed] [Google Scholar]
- Rappert A, Bechmann I, Pivneva T, Mahlo J, Biber K, Nolte C, et al. CXCR3-dependent microglial recruitment is essential for dendrite loss after brain lesion. J Neurosci. 2004;24:8500–8509. doi: 10.1523/JNEUROSCI.2451-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer U. Prenatal poly(I:C) exposure and other developmental immune activation models in rodent systems. Biol Psychiatry. 2013. pp. 1–9. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.