Abstract
Epigenetic processes such as DNA methylation have been implicated in the pathophysiology of neurodevelopmental disorders including schizophrenia and autism. Epigenetic changes can be induced by environmental exposures such as inflammation. Here we tested the hypothesis that prenatal inflammation, a recognized risk factor for schizophrenia and related neurodevelopmental conditions, alters DNA methylation in key brain regions linked to schizophrenia, namely the dopamine rich striatum and endocrine regulatory centre, the hypothalamus. DNA methylation across highly repetitive elements (long interspersed element 1 (LINE1) and intracisternal A-particles (IAPs)) were used to proxy global DNA methylation. We also investigated the Mecp2 gene because it regulates transcription of LINE1 and has a known association with neurodevelopmental disorders. Brain tissue was harvested from 6 week old offspring of mice exposed to the viral analog PolyI:C or saline on gestation day 9. We used Sequenom EpiTYPER assay to quantitatively analyze differences in DNA methylation at IAPs, LINE1 elements and the promoter region of Mecp2. In the hypothalamus, prenatal exposure to PolyI:C caused significant global DNA hypomethylation (t=2.44, P=0.019, PolyI:C mean 69.67%, saline mean 70.19%), especially in females, and significant hypomethylation of the promoter region of Mecp2, (t=3.32, P=0.002; PolyI:C mean 26.57%, saline mean 34.63%). IAP methylation was unaltered. DNA methylation in the striatum was not significantly altered. This study provides the first experimental evidence that exposure to inflammation during prenatal life is associated with epigenetic changes, including Mecp2 promoter hypomethylation. This suggests that environmental and genetic risk factors associated with neurodevelopmental disorders may act upon similar pathways. This is important because epigenetic changes are potentially modifiable and their investigation may open new avenues for treatment.
Introduction
Epigenetic processes are crucial for the regulation of genomic function and development.1 For example, DNA methylation is implicated in X-inactivation, genomic imprinting control and remote silencing of genomic regions.2 Tissue-specific epigenetic modifications through CpG methylation are thought to modulate brain development, circadian rhythm and disease status.3, 4, 5 Consistent with this, epigenetic anomalies have been linked to complex neurodevelopmental disorders including schizophrenia and autism spectrum disorders.6,7
In early brain development, environmental exposures such as inflammation,8 diet,9,10 toxins and contaminants11 disrupt developmental trajectories. For example, prenatal exposure to inflammation has been implicated in the etiology of schizophrenia12 autism13 and bipolar disorder.14 Direct evidence supporting a role for maternal immune activation (MIA) during prenatal life in neurodevelopmental conditions has come from rodent studies from our group and others.15, 16, 17 The consensus is that the MIA model mimics many features relevant to schizophrenia and autism in humans.18,19
Although not a genetic model, gene and protein expression differences have consistently been reported in MIA rodent models, including in the adult frontal cortex20 and the fetal whole brain.17,21,22 In an MIA model elicited by exposing pregnant rodents to the viral analog Polyinosinic:polycytidylic acid (PolyI:C), interleukin 6 (IL6)-induced inflammation has been shown to exert an epigenetic influence by regulating the methyltransferase gene.23 Consistent with this, IL6 is been shown to alter global DNA methylation in diseases such as oral cancer;24 however, it is unknown whether this occurs in the brain of offspring exposed to MIA.
One function of DNA methylation is to repress the transcription of repetitive retroviral elements in the genome including long interspersed element 1 (LINE1) or intracisternal A-particles (IAPs). IAPs are endogenous retroviral sequences, an important class of transposable elements that ‘jump' within the genome inducing genomic mutations and cell transformation. LINE1 elements are retrotransposons - a subclass of transposons that also modulate gene expression, especially in the developing brain.25 However, whether prenatal exposure to maternal inflammation alters methylation state of these repetitive elements has not been directly examined.
An important target for epigenetic modification in models of neurodevelopment is the Methyl CpG-binding protein2 (Mecp2).26 MECP2 is involved in the timely activation and repression of gene expression. It has an important role in control of methylation27 and has a regulatory role in neuronal transcription of LINE1.25 It is strongly associated with neurodevelopmental disorders such as Rett syndrome,28 autism,29 schizophrenia and neural tube defects.30,31 To date however, whether Mecp2 is epigenetically altered in the MIA model has not been investigated.
In this study we tested the hypothesis that prenatal exposure to MIA in the mouse results in global methylation differences in the brain and specifically alters DNA methylation in the promoter of Mecp2. We elected to study the striatum because of its established association with schizophrenia32 and autism,33 and the hypothalamus that has an important influence on the limbic system and is thought to have a critical role in the onset of prodromal features of schizophrenia during the pubertal/postpubertal period.34,35
Materials and methods
Animal model of neurodevelopmental disorders
Female and male C57BL/6N mice were bred and mated in the Laboratory Animal Unit, The University of Hong Kong. The holding space had a 12:12 h normal light–dark cycle (lights off at 1900 hours), and temperature and humidity-controlled (21±1 °C, 55±5%) animal vivarium. Pregnant females were not disturbed, except for weekly cage-cleaning. Animals were maintained under ad libitum food and water supplied by the Laboratory Animal Unit. All experiments were performed in accordance with the relevant institutional and national guidelines and regulations approved by the Committee on the Use of Live Animals in Teaching and Research at The University of Hong Kong, and every effort was made to minimize the number of animals used and their suffering.
The MIA model was prepared following the procedures previously reported in our laboratory and others.18,36 Potassium salt of PolyI:C was obtained from Sigma-Aldrich (Gillingham, UK) and dissolved in saline. A dose of 5 mg kg−1 in an injection volume of 5 ml kg−1, prepared on the day of injection, was administered to pregnant dams on gestation day 9 via the tail vein under gentle physical restraint. The resulting offsprings were weaned and sexed at postnatal day 21. The pups were weighed and littermates of the same sex were caged three to four per cage.
At 6 weeks, mice were killed by cervical dislocation, and their brains were removed quickly. The hypothalamus and striatum were collected in 1.5-ml tubes using careful dissection methods referring the Allen Mouse Brain Atlas37 and frozen in liquid nitrogen for storage. Ten to fifty milligrams of brain tissue were homogenized on ice in a new reaction tube with freshly made and autoclaved lysis buffer (10 mM Tris-HCl, 0.1 M EDTA, 0.5% sodium dodecyl sulfate) using a DNase-free, sterile plastic pestle. Samples were lysed by incubation with 300 ml of lysis buffer and 3.3 μl Proteinase K (18.5 mg ml−1) for 16 h in a water bath at 50 °C. Samples were mixed thoroughly to break up any unlysed tissue and 1.65 μl Proteinase K (18.5 mg ml−1) was added to the sample followed by 1-h incubation at 50 °C. Proteinase K was inactivated by incubating samples at 65 °C for 30 min.
Lysates were transferred to 2 ml eppendorf tubes and extracted with 300 ml of phenol–chloroform–isoamyl alcohol (25:24:1) buffered to pH 7.5 by inverting 20 times and centrifuging at 13 000 r.p.m. for 15 min. The aqueous layer was extracted twice with 300 ml of chloroform–isoamyl alcohol (24:1) in a new reaction tube, by inverting 20 times and centrifuging at 13 000 r.p.m. for 15 min. The aqueous layer was incubated with 75 μl of 10 M ammonium acetate (NH4Ac) and 600 ml ice-cold 100% ethanol in a new reaction tube at −20 °C for 1 h. The supernatant was removed after centrifugation at 13 000 r.p.m. for 15 min and the pellet was washed with 1 ml of 70% ethanol. DNA was pelleted by centrifuging at 13 000 r.p.m. for 10 min. The pellets were left to air dry for 1 h and resuspended in 100 μl of TE buffer. A NanoDrop ND-1000 was used to quantify the DNA samples, and the quality and size of samples were checked using a 1% agarose gel using gel electrophoresis.
Sodium bisulfite conversion
In all, 500 ng of genomic DNA from each sample was treated with sodium bisulfite in duplicate, using the EZ 96-DNA methylation kit (Zymo Research, Orange, CA, USA), following the manufacturer's standard protocol. The kit exploits the three-step reaction that takes place between cytosine and sodium bisulfite where unmethylated cytosine is converted to uracil. The DNA was resuspended in 30 μl of distilled water and stored at −20 °C until further use.
LINE1 and IAP assay
We measured highly repetitive element methylation, blinded to epidemiological exposure condition. Previously reported sequences of forward and reverse primers of LINE1 and IAP38,39 Sigma-Aldrich were used for analysis with the Sequenom EpiTYPER (San Diego, CA, USA), using universally methylated DNA as a methylated reference (EMD Millipore Corporation, Darmstadt, Germany), and an unmethylated DNA as negative control. The LINE1 and IAP element assays were designed to cover more than 600 000 genomic locations, which share a consensus sequence.40
Mecp2 promoter DNA methylation assay
Mecp2 promoter DNA methylation was quantified using the Sequenom EpiTYPER platform, and primers for the target region were designed using EpiDesigner (www.epidesigner.com, Sequenom). Six CpG sites were quantified in an amplicon spanning 282 bp in the promoter and the first exon (Please see Supplementary Table 1 for primer details). All EpiTYPER assays were tested using a standard curve with methylated and unmethylated DNA. Genomic DNA was isolated and 1 μg DNA was bisulfite-treated as described above using the EZ DNA methylation kit (Zymo Research).
Statistical analysis
Global DNA methylation was quantified in the two brain regions of interest (the hypothalamus and striatum) from 22 PolyI:C-exposed and 17 saline-exposed 6-week-old mice using the Sequenom EpiTYPER platform. The sample characteristics are shown in detail in Table 1. The samples were handled together to reduce experimental/handling bias at all stages of experiments. In all group comparisons each animal was treated as an individual subject. Both LINE1 and IAP assays measure a proxy of global DNA methylation, and Pearson correlation testing was used to identify any correlation between assays. A general linear model (GLM) was used to identify any main effect of group or sex or any interaction with unpaired t-tests to explore the data post hoc. Grubb's test using extreme studentized deviate method was used to detect outliers with an alpha cutoff >0.01. The criterion for statistical significance was set as P<0.05 and analyses were conducted using R (http://www.R-project.org/) and SPSS. The effect sizes reported are the absolute change (delta) in DNA methylation.
Table 1. Animal numbers.
| Group |
PolyI:C |
Saline |
|||
|---|---|---|---|---|---|
| Litter 6 weeks | M1 | M2 | M3 | M4 | M5 |
| Sex (male:female) | 3:3 | 3:6 | 2:5 | 4:5 | 3:5 |
| Total | 22 | 17 | |||
PolyI:C group (litter M1, M2 and M3) and saline group (M4 and M5) with the number of males and females in each litter. Animals used in the experiment were, as far as possible, kept free from any interventions that could further affect their epigenetic status.
Results
Altered global methylation in the adolescent brain following prenatal immune challenge
Quantitative DNA methylation was measured across LINE1 elements to give a proxy of global DNA methylation across ~600 000 repeats in the mouse genome.41,42
LINE1 methylation in the hypothalamus
There was a significant main effect of treatment (F (1, 36)=5.72, β=−0.516, P=0.022), with no effect of sex (F (1, 36)=1.27, β=0.248, P=0.266) on the mean LINE1 methylation, and 12.18% of variance was explained with the GLM used. This resulted from significant hypomethylation in the hypothalamus of the PolyI:C group (69.66%, s.d.=0.62) compared with the saline group (70.19%, s.d.=0.72; t=2.44, P=0.019, delta methylation=0.53; see Table 2), especially in females (t=3.04, P=0.005, PolyI:C group mean 69.45, s.d.=0.59, saline group mean 70.26, s.d.=0.71, delta methylation=0.81).
Table 2. PolyI:C and saline group comparison results of different assays.
| LINE1 | IAP | Mecp2 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| |
Mean |
s.d. |
P-value |
Mean |
s.d. |
P-value |
Mean |
s.d. |
P-value |
| Hypothalamus | |||||||||
| 0.696 | 0.006 | 0.265 | 0.067 | ||||||
| PolyI:C | F 0.694 | 0.005 | 0.935 | 0.014 | F 0.290 | 0.037 | |||
| 0.019* | 0.002** | ||||||||
| M 0.700 | 0.005 | M 0.148 | 0.042 | ||||||
| F 0.005** | 0.47 | F 0.000*** | |||||||
| 0.701 | 0.007 | 0.346 | 0.059 | ||||||
| M 0.885 | M 0.202 | ||||||||
| Saline | F 0.702 | 0.007 | 0.931 | 0.012 | F 0.362 | 0.046 | |||
| M 0.700 | 0.007 | M 0.266 | 0.068 | ||||||
| Striatum | |||||||||
| 0.707 | 0.011 | 0.930 | 0.050 | 0.295 | 0.179 | ||||
| PolyI:C | 0.29 | 0.85 | 0.37 | ||||||
| Saline | 0.704 | 0.010 | 0.936 | 0.066 | 0.345 | 0.121 | |||
Abbreviations: F, Female; IAP, intracisternal A particle; LINE1, long interspersed elements; M, Male; Mecp2, Methyl CpG Binding Protein2.
Mean methylation of all the markers for each assay. *P<0.05, **P<0.005 and ***P<0.0005.
The first CpG in the LINE1 element in the hypothalamus was differentially methylated in adolescent stage after PolyI:C administration in prenatal life, main effect of Group (F (1, 36)=5.77, β=−1.503, P=0.021). In all, 9.08% of the variance was explained by the GLM used. There was no significant main effect of sex (F (1, 36)=0.08, β=−0.183, P=0.775); however, there was a significant Group x Sex interaction (F (1, 35)=5.54, β=2.846, P=0.024) and the GLM explained 19.27% of variance. This was explained by significant hypomethylation in the PolyI:C group (t=2.42, delta methylation=1.49, P=0.02; PolyI:C group mean=27.68%, saline group=29.17%, s.d.=2.03). Again, differences were most pronounced in females (t=4.11, delta methylation=2.62, P<0.001).
LINE1 methylation in the striatum
There was no significant main effect of group, or sex, or any Group x Sex interaction on the mean LINE1 methylation in the striatum (PolyI:C group mean=70.79, s.d.=1.15 and saline group mean=70.41, s.d.=1.01).
IAP assay
IAP promoters were not differentially methylated in the hypothalamus or striatum in the PolyI:C and saline control groups, see Table 2. LINE1 and IAP assays for the mean global methylation were not correlated (P=0.298, r=−0.170).
Mecp2 promoter methylation
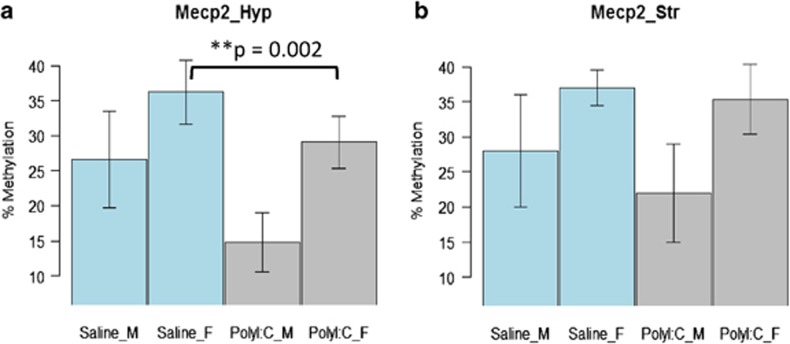
The mean methylation of Mecp2 in the polyI:C-exposed hypothalamus was lower than in saline groups (PolyI:C group=26.57%, s.d.=6.70; saline group=34.63%, s.d.=5.97). There was a significant effect of group and sex in the mean methylation of the Mecp2 promoter region in the hypothalamus: (F (1, 26)=24.08, β=−7.937, P<0.001) and (F (1, 26)=34.49, β=12.384, P<0.001), respectively. In total, 67.18% of variance in Mecp2 methylation was explained by the GLM used. There was no significant Group x Sex interaction. Post hoc tests confirmed Mecp2 promoter hypomethylation in the PolyIC group (t=3.39, delta methylation=8.05, P=0.003) and in females (t=4.35, delta methylation=7.14, P<0.001). Please see Figure 1 and Table 2.
Figure 1.
Hypomethylated Mecp2 promoter in the hypothalamus of PolyI:C-exposed offspring. y axis shows methylation in percentage with error bar as s.e.m., histograms represent saline- and polyI:C-treated male and female groups. (a) Mecp2 promoter methylation in the hypothalamus of polyI:C- and saline-exposed male and female groups. (**P-value is calculated from a stratified t-test between saline- and polyI:C-affected females). (b) Mecp2 promoter methylation in the striatum of polyI:C and saline-exposed male and female groups.
Individual CpG sites 2 and 4 were significantly differentially methylated in each group (F (1, 26)=13.12, β=−9.870, P=0.001; F (1, 26)=11.71, β=−6.244, P=0.002, respectively) and sex (F (1, 26)=10.84, β=11.294, P=0.003; F (1, 26)=13.08, β=8.604, P=0.001, respectively). The mean methylation for CpG site 2; PolyI:C group=20.35%, s.d.=8.71, saline group=30.33%, s.d.=7.24 and CpG site 4; PolyI:C group mean=30.58%, s.d.=6.21, saline group=36.91%, s.d.=5.19. There was no significant Group x Sex interaction. A total of 15.63% of variance in CpG 2 methylation and 45.22% of variance in CpG 4 methylation were explained by the GLM used.
Mean methylation of Mecp2 in polyI:C-exposed striatum was also lower than controls but this difference did not reach statistical significance (please see Figure 1 and Table 2).
Discussion
This is the first study to directly examine the epigenetic consequences of prenatal exposure to MIA in the adolescent rodent brain. We found significant DNA hypomethylation in the hypothalamus and a similar trend in the striatum of offspring exposed to MIA compared with saline-exposed controls. In addition, the Mecp2 gene promoter region was significantly hypomethylated in the hypothalamus of MIA-exposed group but not in the striatum. These effects were most pronounced in females.
LINE1 methylation
Approximately 50% of the mammalian genome comprises the retrotransposon-associated repeat sequences, with LINE1 elements and LTR elements (including IAP) accounting for 20%40 and 10%, respectively.43 LINEs are ~6-kb-long elements with an internal polymerase II promoter encoding two open reading frames and account for a great amount of somatic mosaicism.44 LINE1 elements transpose through RNA intermediates, assembling with their own encoded proteins through reverse transcription. LINE1 is highly active and can create indels, new splice sites which fine-tune gene expression.45 LINE1 transposons may also disrupt coding regions, and their epigenetic suppression by causing indels is vital during the early course of development.46 Inappropriate suppression caused by altered DNA methylation across these elements may lead to adverse consequences in the adult life.47
Consistent with this role in development, LINE1 has been reported to be differentially methylated and highly susceptible to retrotransposition in neurodevelopmental disorders.25 For example, hyperactive retrotransposition and higher copy number of LINE1 have been reported in schizophrenia and observed in the polyI:C MIA model.48 Our observation of LINE1 hypomethylation in the same MIA model would be expected to increase retrotransposition and may provide one possible path linking inflammation with the increased risk of schizophrenia.
Altered DNA methylation as a potential mediator of environmental risk for neurodevelopmental disorders fits with evidence from other systems. For example, prenatal exposure to the antiepilepsy drug valproate is associated with increased risk of autism spectrum disorders in the offspring,49 and this drug has been recognized to cause widespread epigenetic reprogramming especially DNA hypomethylation.50
IAP methylation
IAPs are autonomous retrotransposon elements with long terminal repeats and all the necessary transcriptional regulatory elements. These class-II endogenous retroviruses transpose through retroviral mechanisms in the cisternae of the endoplasmic reticulum.51 IAPs add a significant amount of genetic variabilities to the genome with differential tissue-specific expression and are capable of interfering gene function in the brain.52,53 The complexity and the size of IAP make it less common and less likely to occupy the vital regions in the genome. Thus, IAP assays target completely different elements than LINE1, with a lower representation over the genome,54 which may account for the sparsity of findings using this assay in our study, and the lack of evidence linking IAP alterations with neurodevelopmental disorders.
Mecp2 methylation
MECP2 has a well-established role in Rett's syndrome but has also been implicated, by our group and others, in idiopathic neurodevelopmental disorders such as autism and schizophrenia.55,56 MECP2 is an abundant neuronal protein that indirectly mediates gene transcription by recruiting transcription factor cAMP responsive element-binding protein 1, depending on the methylation status of the DNA.27 This action alters the expression of many genes in the hypothalamus57 and beyond, including LINE1. Aberrant promoter methylation of MECP2 itself is thought to contribute to a defective biological network and potentially to the etiology of neurodevelopmental disorders.58
Here we report a significant association between MIA and Mecp2 promoter hypomethylation. The polyI:C used in this MIA model is known to cause an increase in IL6.59 MECP2 in turn may suppress IL6 expression;60 therefore, we speculate that hypomethylation, leading to a higher level of MECP2, may be involved in silencing genes that are activated by the immune response. Changes in Mecp2 could theoretically influence the expression of other developmental genes previously reported to be altered in the MIA model, for example, brain-derived neurotrophic factor61 and Reelin, Glutamate decarboxylase 6762 and cAMP response element-binding protein;63 histone deacetylases and DNA (cytosine-5)-methyltransferase 1.26,64 A theoretical construct showing a functional protein association network potentially influenced by MECP2 is shown in Supplementary Information and Figure 1.65
More specifically, in terms of the dopamine system, closely linked to disorders such as schizophrenia, we speculate that removing a methylation ‘brake' on Mecp2 gene expression would promote or even overdevelop dopaminergic neurons. This is because the converse - low levels of MECP2 in dopamine neurons - results in fewer dendrites, less membrane surface area and even a compromised nigrostriatal pathway.66
PolyIC caused greater LINE1 hypomethylation in the hypothalamus of female offspring; however, although the Mecp2 promoter was more hypomethylated in females in both polyI:C- and saline-exposed groups, there was no significant Sex x Group interaction. Although neurodevelopmental disorders such as autism are generally more prevalent in males than females, this is not the case for schizophrenia,67 and Rett syndrome is predominantly diagnosed in females.68 Moreover, consistent with our results, genome-wide methylation studies of schizophrenia and other psychiatric conditions have reported global hypomethylation in females.69,70 This excess of epigenetic modification in the female sex may also be relevant to the suggestion that environmental exposures are relatively more important in females with neurodevelopmental disorders.71
Regional methylation changes
Hypomethylation of Mecp2 and LINE1 in an adolescent offspring exposed to MIA was most pronounced in the hypothalamus. Hypothalamus–pituitary–adrenal axis developmental changes, and especially hypothalamic dopamine systems, are strongly implicated in the onset of schizophrenia.35,72 Consistent with this evidence, MECP2 has been found to mediate dopaminergic precursor differentiation in the ventral midbrain especially in the developing hypothalamus via Dlk1.73 Similarly, aminergic neurotransmitter concentration is regulated by MECP2 through rate-limiting enzymes involved in synthesis and degradation, which may contribute to phenotypes relevant to schizophrenia and autism.74
The direction of Mecp2 methylation differences in the striatum of MIA exposed mice relative to controls was similar to those observed in the hypothalamus; however, group differences did not reach statistical significance. We also observed a main effect of sex - females were generally undermethylated relative to males in both PolyI:C and control groups in both regions of interest. Thus, whereas low power may have limited any group differences in methylation in the striatum, it is also possible that lower methylation in females of both groups may have obscured group differences in methylation in the striatum.
Advantages and limitations of the study
An organism's phenotype is produced by its genotype's interaction with the environment and thus epigenetic changes influence gene expression, DNA repair and recombination. Under the influence of these epigenetic mechanisms, genetically identical cells, tissues and organs show variation and become unique during the course of development.75 However, investigating epigenetic etiological mechanisms in the brain, and understanding the effect of epigenetic modifications resulting from the prenatal assaults in humans, is extremely challenging. This is partly because studies are indirect, relying upon peripheral blood, buccal swabs and other tissues.
Therefore, animal models can help to better control the environmental exposures and homogeneity of the sample. Moreover, the animal model allows DNA isolation from specific regions of the brain. This is important because patterns of DNA methylation are region- and tissue-specific,76,77 and peripheral tissue samples collected in humans are unlikely to reflect what is happening in the brain.78 However, although animal models allow us to sample brain, there are still technical constraints and it must be acknowledged that assays such as LINE145 and IAP provide only a proxy of global methylation.6
We also accept that the current study targeted restricted regions of the brain; therefore, we cannot say whether our findings are truly region-specific or more generalized. Even though carefully dissected out, DNA was isolated from tissue comprising a mix of cells with possibly different epigenetic profiles. Assays of single cell or minimal numbers of isolated cells could have improved cell type accuracy but also have restricted generalization. Furthermore, we used LINE1 and IAP assays only to obtain a proxy of global DNA methylation. These assays cannot reveal the individual genes affected, nor tell us whether the differences observed have functional implications. Future work will need to be conducted to ascertain the functionality of these methylation changes (especially the significant, but very small, group difference in LINE1 DNA methylation in the hypothalamus).
Finally, we acknowledge that alternative design would be to consider each litter as a single experimental unit with the assumption that the exposure of fetuses within one litter is identical. However, this assumption may not always hold as each fetus in the mouse uterus has been shown to have a distinct blood perfusion pattern;79 therefore, exposures may not necessarily be identical. On balance, we wished to optimize genetic similarity so that the focus would be on environmental exposure. We therefore elected to treat each an offspring as a ‘subject', and enter data from each littermate separately into the analysis.
Summary
This study examined epigenetic differences in the hypothalamus and striatum of the adolescent brain following exposure to MIA in prenatal life, a risk factor for schizophrenia and related neurodevelopmental conditions. We observed differences in both global methylation and Mecp2 promoter methylation that may have relevance to neurodevelopmental disorders. In particular, hypomethylation of the Mecp2 promoter may have widespread repercussions on gene expression and deserves further exploration as a possible biomarker for environmental modifications linked to prenatal assault. Further exploration of epigenetic mechanisms operating in neurodevelopmental conditions is warranted because these gene modifications are potentially modifiable and this approach may open new avenues for treatment.
Acknowledgments
We thank members of the Psychiatric Epigenetic group for Infrastructural support for aspects of the study at Social, Genetic and Developmental Psychiatry, Department of Forensic and Neurodevelopmental Sciences, Kings College London and Statistical Genetics group, Department of Psychiatry, The University of Hong Kong (HKU). PB is supported by a Post Graduate Scholarship, HKU and the Graduate Research Exchange Scheme, Faculty of Medicine, HKU. The experimental work was supported by Hong Kong Universities General Research Fund award GRF_HKU 774710M. We also acknowledge ZEE Foundation, Hong Kong for their financial support. GM is a member of the EU-AIMS consortium.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Waddington CH. The epigenotype. Int J Epidemiol. 2012;41:10–13. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Pidsley R, Dempster EL, Mill J. Brain weight in males is correlated with DNA methylation at IGF2. Mol Psychiatr. 2010;15:880–881. doi: 10.1038/mp.2009.138. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Grimaldi B, Sahar S, Hirayama J, Sassone-Corsi P. Signaling to the circadian clock: plasticity by chromatin remodeling. Curr Opin Cell Biol. 2007;19:230–237. doi: 10.1016/j.ceb.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Pidsley R, Mill J. Epigenetic studies of psychosis: current findings, methodological approaches, and implications for postmortem research. Biol Psychiatry. 2011;69:146–156. doi: 10.1016/j.biopsych.2010.03.029. [DOI] [PubMed] [Google Scholar]
- Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, Kane F, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20:4786–4796. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CC, Meaburn EL, Ronald A, Price TS, Jeffries AR, Schalkwyk LC, et al. Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Mol Psychiatry. 2013;19:495–503. doi: 10.1038/mp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M, Thanan R, Ma N, Kawanishi S. Role of nitrative and oxidative DNA damage in inflammation-related carcinogenesis. J Biomed Biotechnol. 2012;2012:623019. doi: 10.1155/2012/623019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CC, Cropley JE, Cowley MJ, Preiss T, Martin DI, Suter CM. A sustained dietary change increases epigenetic variation in isogenic mice. PLoS Genet. 2011;7:e1001380. doi: 10.1371/journal.pgen.1001380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Salas P, Cox SE, Prentice AM, Hennig BJ, Moore SE. Maternal nutritional status, C(1) metabolism and offspring DNA methylation: a review of current evidence in human subjects. Proc Nutr Soc. 2012;71:154–165. doi: 10.1017/S0029665111003338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AS, Begg MD, Gravenstein S, Schaefer CA, Wyatt RJ, Bresnahan M, et al. Serologic evidence of prenatal influenza in the etiology of schizophrenia. Arch Gen Psychiatry. 2004;61:774–780. doi: 10.1001/archpsyc.61.8.774. [DOI] [PubMed] [Google Scholar]
- Brown AS, Sourander A, Hinkka-Yli-Salomaki S, McKeague IW, Sundvall J, Surcel HM. Elevated maternal C-reactive protein and autism in a national birth cohort. Mol Psychiatry. 2013;19:259–264. doi: 10.1038/mp.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parboosing R, Bao Y, Shen L, Schaefer CA, Brown AS. Gestational influenza and bipolar disorder in adult offspring. JAMA Psychiatry. 2013;70:677–685. doi: 10.1001/jamapsychiatry.2013.896. [DOI] [PubMed] [Google Scholar]
- McAlonan GM, Li Q, Cheung C. The timing and specificity of prenatal immune risk factors for autism modeled in the mouse and relevance to schizophrenia. Neurosignals. 2010;18:129–139. doi: 10.1159/000321080. [DOI] [PubMed] [Google Scholar]
- Meyer U, Feldon J, Fatemi SH. In-vivo rodent models for the experimental investigation of prenatal immune activation effects in neurodevelopmental brain disorders. Neurosci Biobehav Rev. 2009;33:1061–1079. doi: 10.1016/j.neubiorev.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Reutiman TJ, Folsom TD, Huang H, Oishi K, Mori S, et al. Maternal infection leads to abnormal gene regulation and brain atrophy in mouse offspring: implications for genesis of neurodevelopmental disorders. Schizophr Res. 2008;99:56–70. doi: 10.1016/j.schres.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Cheung C, Wei R, Hui ES, Feldon J, Meyer U, et al. Prenatal immune challenge is an environmental risk factor for brain and behavior change relevant to schizophrenia: evidence from MRI in a mouse model. PLoS ONE. 2009;4:e6354. doi: 10.1371/journal.pone.0006354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer U, Feldon J, Dammann O. Schizophrenia and autism: both shared and disorder-specific pathogenesis via perinatal inflammation. Pediatr Res. 2011;69:26r–33r. doi: 10.1203/PDR.0b013e318212c196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng MY, Lam S, Meyer U, Feldon J, Li Q, Wei R, et al. Frontal-subcortical protein expression following prenatal exposure to maternal inflammation. PLoS ONE. 2011;6:e16638. doi: 10.1371/journal.pone.0016638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett KA, Hsiao EY, Kalman S, Patterson PH, Mirnics K. Effects of maternal immune activation on gene expression patterns in the fetal brain. Transl Psychiatry. 2012;2:e98. doi: 10.1038/tp.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer U, Nyffeler M, Yee BK, Knuesel I, Feldon J. Adult brain and behavioral pathological markers of prenatal immune challenge during early/middle and late fetal development in mice. Brain Behav Immun. 2008;22:469–486. doi: 10.1016/j.bbi.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Hodge DR, Xiao WH, Clausen PA, Heidecker G, Szyf M, Farrar WL. Interleukin-6 regulation of the human DNA methyltransferase (HDNMT) gene in human erythroleukemia cells. J Biol Chem. 2001;276:39508–39511. doi: 10.1074/jbc.C100343200. [DOI] [PubMed] [Google Scholar]
- Gasche JA, Hoffmann J, Boland CR, Goel A. Interleukin-6 promotes tumorigenesis by altering DNA methylation in oral cancer cells. Int J Cancer. 2011;129:1053–1063. doi: 10.1002/ijc.25764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MCN, Coufal NG, Oefner R, Yeo G, Nakashima K, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Agis-Balboa RC, Simonini MV, Grayson DR, Costa E, Guidotti A. Reelin and glutamic acid decarboxylase67 promoter remodeling in an epigenetic methionine-induced mouse model of schizophrenia. Proc Natl Acad Sci USA. 2005;102:12578–12583. doi: 10.1073/pnas.0505394102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Swanberg SE, Nagarajan RP, Peddada S, Yasui DH, LaSalle JM. Reciprocal co-regulation of EGR2 and MECP2 is disrupted in Rett syndrome and autism. Hum Mol Genet. 2009;18:525–534. doi: 10.1093/hmg/ddn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang F, Guan J, Le J, Wu LH, Zou JZ, et al. Relation between hypomethylation of long interspersed nucleotide elements and risk of neural tube defects. Am J Clin Nutr. 2010;91:1359–1367. doi: 10.3945/ajcn.2009.28858. [DOI] [PubMed] [Google Scholar]
- Tate P, Skarnes W, Bird A. The methyl-CpG binding protein MeCP2 is essential for embryonic development in the mouse. Nat Genet. 1996;12:205–208. doi: 10.1038/ng0296-205. [DOI] [PubMed] [Google Scholar]
- Thompson JL, Urban N, Slifstein M, Xu X, Kegeles LS, Girgis RR, et al. Striatal dopamine release in schizophrenia comorbid with substance dependence. Mol Psychiatry. 2013;18:909–915. doi: 10.1038/mp.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langen M, Schnack HG, Nederveen H, Bos D, Lahuis BE, de Jonge MV, et al. Changes in the developmental trajectories of striatum in autism. Biol Psychiatry. 2009;66:327–333. doi: 10.1016/j.biopsych.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Goldstein JM, Seidman LJ, Makris N, Ahern T, O'Brien LM, Caviness VS, Jr., et al. Hypothalamic abnormalities in schizophrenia: sex effects and genetic vulnerability. Biol Psychiatry. 2007;61:935–945. doi: 10.1016/j.biopsych.2006.06.027. [DOI] [PubMed] [Google Scholar]
- Thompson JL, Pogue-Geile MF, Grace AA. Developmental pathology, dopamine, and stress: a model for the age of onset of schizophrenia symptoms. Schizophr Bull. 2004;30:875–900. doi: 10.1093/oxfordjournals.schbul.a007139. [DOI] [PubMed] [Google Scholar]
- Meyer U, Feldon J, Schedlowski M, Yee BK. Towards an immuno-precipitated neurodevelopmental animal model of schizophrenia. Neurosci Biobehav Rev. 2005;29:913–947. doi: 10.1016/j.neubiorev.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Munoz-Lopez M, Macia A, Garcia-Canadas M, Badge RM, Garcia-Perez JL. An epi [c] genetic battle: LINE-1 retrotransposons and intragenomic conflict in humans. Mob Genet Elements. 2011;1:122–127. doi: 10.4161/mge.1.2.16730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng SH, Cokus SJ, Andrews S, Pellegrini M, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–U1126. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Hardies SC, Wang LP, Zhou LX, Zhao YP, Casavant NC, Huang SJ. LINE-1 (L1) lineages in the mouse. Mol Biol Evol. 2000;17:616–628. doi: 10.1093/oxfordjournals.molbev.a026340. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- McCarthy EM, McDonald JF. Long terminal repeat retrotransposons of Mus musculus. Genome Biol. 2004;5:R14. doi: 10.1186/gb-2004-5-3-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Gene Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- Faulk C, Dolinoy DC. Timing is everything The when and how of environmentally induced changes in the epigenome of animals. Epigenetics. 2011;6:791–797. doi: 10.4161/epi.6.7.16209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Gage FH. Generation of neuronal variability and complexity. Nature. 2006;441:1087–1093. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- Bundo M, Toyoshima M, Okada Y, Akamatsu W, Ueda J, Nemoto-Miyauchi T, et al. Increased L1 retrotransposition in the neuronal genome in schizophrenia. Neuron. 2013;81:306–313. doi: 10.1016/j.neuron.2013.10.053. [DOI] [PubMed] [Google Scholar]
- Christensen J, Gronborg TK, Sorensen MJ, Schendel D, Parner ET, Pedersen LH, et al. Prenatal valproate exposure and risk of autism spectrum disorders and childhood autism. JAMA. 2013;309:1696–1703. doi: 10.1001/jama.2013.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Chen Y, Gavin DP, Grayson DR, Guidotti A. Valproate induces DNA demethylation in nuclear extracts from adult mouse brain. Epigenetics-Us. 2010;5:730–735. doi: 10.4161/epi.5.8.13053. [DOI] [PubMed] [Google Scholar]
- Lueders KK, Kuff EL. Intracisternal A-particle genes: identification in the genome of Mus musculus and comparison of multiple isolates from a mouse gene library. Proc Natl Acad Sci USA. 1980;77:3571–3575. doi: 10.1073/pnas.77.6.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubatz JW, Arcement B, Cutler RG. Gene-expression of an endogenous retrovirus-like element during murine development and aging. Mech Ageing Dev. 1991;57:71–85. doi: 10.1016/0047-6374(91)90025-u. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Jin XD, Yan J, Jiao F, Li XM, Roe BA, et al. An insertion of intracisternal A-particle retrotransposon in a novel member of the phosphoglycerate mutase family in the lew allele of mutant mice. Genes Genet Syst. 2009;84:327–334. doi: 10.1266/ggs.84.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C, Barks A, Dolinoy DC. Phylogenetic and DNA methylation analysis reveal novel regions of variable methylation in the mouse IAP class of transposons. BMC Genomics. 2013;14:48. doi: 10.1186/1471-2164-14-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong EH, So HC, Li M, Wang Q, Butler AW, Paul B, et al. Common variants on Xq28 conferring risk of schizophrenia in han Chinese. Schizophr Bull. 2013;40:777–786. doi: 10.1093/schbul/sbt104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle JM, Yasui DH. Evolving role of MeCP2 in Rett syndrome and autism. Epigenomics. 2009;1:119–130. doi: 10.2217/epi.09.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum Mol Genet. 2009;18:2431–2442. doi: 10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan RP, Patzel KA, Martin M, Yasui DH, Swanberg SE, Hertz-Picciotto I, et al. MECP2 promoter methylation and X chromosome inactivation in autism. Autism Res. 2008;1:169–178. doi: 10.1002/aur.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao EY, Patterson PH. Activation of the maternal immune system induces endocrine changes in the placenta via IL-6. Brain Behav Immun. 2011;25:604–615. doi: 10.1016/j.bbi.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandrea M, Donadelli M, Costanzo C, Scarpa A, Palmieri M. MeCP2/H3meK9 are involved in IL-6 gene silencing in pancreatic adenocarcinoma cell lines. Nucleic Acids Res. 2009;37:6681–6690. doi: 10.1093/nar/gkp723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZL, Hong EJ, Cohen S, Zhao WN, Ho HYH, Schmidt L, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhubi A, Chen Y, Dong E, Cook EH, Guidotti A, Grayson DR. Increased binding of MeCP2 to the GAD1 and RELN promoters may be mediated by an enrichment of 5-hmC in autism spectrum disorder (ASD) cerebellum. Transl Psychiatry. 2014;4:e349. doi: 10.1038/tp.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M, Chen Y, Costa E, Grayson DR. DNA methyltransferase inhibitors coordinately induce expression of the human reelin and glutamic acid decarboxylase 67 genes. Mol Pharmacol. 2007;71:644–653. doi: 10.1124/mol.106.030635. [DOI] [PubMed] [Google Scholar]
- Kimura H, Shiota K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J Biol Chem. 2003;278:4806–4812. doi: 10.1074/jbc.M209923200. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, et al. STRING 8-a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz SC, Ford CP, Neve KA, Williams JT. Loss of Mecp2 in substantia nigra dopamine neurons compromises the nigrostriatal pathway. J Neurosci. 2011;31:12629–12637. doi: 10.1523/JNEUROSCI.0684-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Chant D, Welham J, McGrath J. A systematic review of the prevalence of schizophrenia. PLoS Med. 2005;2:e141. doi: 10.1371/journal.pmed.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg B. Rett's syndrome: prevalence and impact on progressive severe mental retardation in girls. Acta Paediatr Scand. 1985;74:405–408. doi: 10.1111/j.1651-2227.1985.tb10993.x. [DOI] [PubMed] [Google Scholar]
- Frieling H, Gozner A, Romer KD, Lenz B, Bonsch D, Wilhelm J, et al. Global DNA hypomethylation and DNA hypermethylation of the alpha synuclein promoter in females with anorexia nervosa. Mol Psychiatr. 2007;12:229–230. doi: 10.1038/sj.mp.4001931. [DOI] [PubMed] [Google Scholar]
- Burghardt KJ, Pilsner JR, Bly MJ, Ellingrod VL. DNA methylation in schizophrenia subjects: gender and MTHFR 677C/T genotype differences. Epigenomics. 2012;4:261–268. doi: 10.2217/epi.12.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Heath AC, Bucholz KK, Dinwiddie SH, Madden PAF, Statham DJ, et al. Major depressive disorder in a community-based twin sample—Are there different genetic and environmental contributions for men and women. Arch Gen Psychiat. 1999;56:557–563. doi: 10.1001/archpsyc.56.6.557. [DOI] [PubMed] [Google Scholar]
- Walker E, Mittal V, Tessner K. Stress and the hypothalamic pituitary adrenal axis in the developmental course of schizophrenia. Annu Rev Clin Psychol. 2008;4:189–216. doi: 10.1146/annurev.clinpsy.4.022007.141248. [DOI] [PubMed] [Google Scholar]
- Bauer M, Szulc J, Meyer M, Jensen CH, Terki TA, Meixner A, et al. Delta-like 1 participates in the specification of ventral midbrain progenitor derived dopaminergic neurons. J Neurochem. 2008;104:1101–1115. doi: 10.1111/j.1471-4159.2007.05037.x. [DOI] [PubMed] [Google Scholar]
- Samaco RC, Mandel-Brehm C, Chao HT, Ward CS, Fyffe-Maricich SL, Ren J, et al. Loss of MeCP2 in aminergic neurons causes cell-autonomous defects in neurotransmitter synthesis and specific behavioral abnormalities. Proc Natl Acad Sci USA. 2009;106:21966–21971. doi: 10.1073/pnas.0912257106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Ziller MJ, Gu HC, Muller F, Donaghey J, Tsai LTY, Kohlbacher O, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegawa S, Hinkal G, Kim HS, Shen LL, Zhang L, Zhang JX, et al. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:R14. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni R, Raz T, Biton IE, Kalchenko V, Garbow JR, Neeman M. Unique in utero identification of fetuses in multifetal mouse pregnancies by placental bidirectional arterial spin labeling MRI. Magn Reson Med. 2012;68:560–570. doi: 10.1002/mrm.23246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.