Abstract
Focal adhesion is critical for cell survival. The focal adhesion kinase (FAK, or PTK2) is an important component of the human interactome and thus is a potential longevity-related protein. Here we studied the association between two PTK2 gene single-nucleotide polymorphisms (SNPs) (rs7843014, rs7460) and exceptional longevity (EL). In addition to gaining insight into their functionality by determining luciferase gene reporter activity, we studied the genotype/allele frequency of these two SNPs among three different cohorts: (1) Spanish centenarians (n=175, 100–111 years, 144 women) and healthy controls (n=355, 20–50 years, 284 women); (2) Italian centenarians (n=79, 100–104 years, 40 women) and controls (n=316, 29–50 years, 156 women); and (3) Japanese centenarians (n=742, 100–116 years, 623 women) and healthy controls (n=499, 23–59 years, 356 women). Both SNPs had functional significance, with the A allele up-regulating luciferase activity compared to the other allele (rs7460 T allele and rs7843014 C allele, respectively). The A allele of both SNPs was negatively associated with EL in the Spanish cohort (rs7460, odds ratio [OR] adjusted by sex=0.40, 95% confidence intervals [CI] 0.3, 0.6, p<0.001); rs7843014, OR=0.37, 95% CI 0.3, 0.5, p<0.001). The OR of being a centenarian if having the rs7460-TT genotype was 6.68 (95% CI 4.1, 10.8, p<0.001). The rs7843014 CC genotype was also positively associated with EL (OR=7.58, 95% CI 4.6, 12.3, p<0.001]. No association was, however, found for the Italian or Japanese cohorts. Thus, two genotypes of the FAK gene, rs7460 TT and rs7843014 CC, are possibly associated with lower gene expression and might favor the likelihood of reaching EL in the Spanish population. Further research is needed to unveil the mechanisms by which FAK expression could perhaps influence the rate of aging.
Introduction
Centenarians are the paradigm of human extreme longevity (EL) and healthy aging, because they have postponed, if not avoided, major age-related diseases, e.g., cancer, neurodegeneration, or cardiovascular disease (CVD),1 as well as the onset of disability.2,3 Thus, the search for the genes and gene products (or longevityrelated proteins) that might influence the likelihood of reaching EL is of medical interest, particularly in helping to identify potential targets of highly specific anti-aging drugs.4
Growing evidence indicates that biological systems operate as complex networks of protein–protein interactions,5 which also applies to proteins associated with longevity and major age-related diseases such as cancer, Alzheimer's, or CVD.6,7 Although the human interactome is characterized by ∼8000 proteins and many more (>23,500) protein–protein interactions, the knowledge that some of these proteins are especially important because they act as hubs, i.e., key switchers or connectors between different signaling pathways, might help focus the search for potential longevity-related genes.4 An important hub is that implicated in cell-matrix adhesion or focal adhesion.4 Focal adhesion is indeed critical for survival of cells, as well as for crucial events such as cell adhesion, migration, and growth rate, which are regulated by signals from the surrounding milieu, i.e., adjacent cells and extracellular matrix (ECM). Cells (and their cytoskeleton) are connected to the ECM by adhesion proteins or integrins, such as the intracellular signaling protein focal adhesion kinase (FAK, also known as PTK2).
The FAK is a 125-kDa non-receptor and non-membrane protein tyrosine kinase that plays a central role in several pathways, including cytoskeleton-associated pathways, integrin signaling, and the phosphatidylinositol-3 kinase (PI3K)/Akt survival pathway.4 Activated FAK is involved in the ability of the cell to respond and adapt to the changing microenvironment and is activated in tumor cells, generating signals leading to cancer growth and metastasis.8 In fact, FAK inhibitors show promise for cancer therapy.9 A recent preliminary report showed that two single-nucleotide polymorphisms (SNPs) in the PTK2 gene encoding FAK—the intronic rs7843014 A/C and the 3′-untranslated region (3′-UTR) rs7460 A/T SNPs—were associated with individual variations in a human health-related phenotype, muscle strength.10 To the best of our knowledge, this is the only gene association study that has specifically reported the potential influence of PTK2 gene variants on human phenotypes, despite the rationale for postulating this gene as a candidate to influence several health/disease traits, including possibly EL. To gain insight into their functionality, here we determined luciferase gene reporter activity in the two above-mentioned SNPs, rs7843014 and rs7460. We then compared allele/genotype frequencies of rs7843014 and rs7460 among centenarians (cases) and disease-free controls of the same ethnic and geographic (Spanish) origin as well as in two other replication cohorts from Italy and Japan.
Methods
Participants
Written informed consent was obtained from each participant. The study protocol was approved by the corresponding institutional ethics committees (European University of Madrid [Spanish cohort], University of Pavia [Italian cohort], and National Institute of Health and Nutrition, Medical Research Institute and Keio University [Japanese cohort]) and was in accordance with the Declaration of Helsinki for Human Research of 1974 (last modified in 2008).
Spanish cohort
Two groups of Spanish subjects (most born and living in the central area of Spain, Meseta Castellana) were investigated: (1) 175 cases (centenarians, age range 100–111 years, 144 women, 31 men); and (2) 355 healthy controls (20–50 years, 284 women, 213 men). All of the Spanish participants were of the same Caucasian (Spanish) descent for three or more generations and spoke Spanish as their mother language.
Centenarians were living mainly in nursing residencies of the Spanish central area, where they were recruited during years 2009–2012 after we had access to the age lists of the people living in the residencies. The participants' ages were ascertained by the dates of birth as stated on identity cards. Their DNA was obtained from saliva samples. This group included the oldest European individual (111 years) alive in June of 2012 (www.grg.org/Adams/E.HTM) and ∼7% of the cohort was aged ≥105 years. The most prevalent diseases were osteoarthritis (66%), hypertension (57%), dementia (51%), and CVD (29%). Only two centenarians were free of any diagnosed disease.
The control group was a convenient sample composed of students and staff from the European University (Madrid, Spain). All of them were free of any major disease and had no known family history of high longevity (90+ years), as reported in a questionnaire.
Italian replication cohort
Two groups of Italian subjects born and living in northern Italy were studied: (1) 79 cases (healthy centenarians, 100–104 years, 40 women, 39 men); and (2) 316 healthy controls (29–50 years, 156 women, 160 men). The participants' ages were defined by the dates of birth as stated in identity cards. All patients and controls were Caucasian whites ascertained to be of Italian descent. The criterion of Italian descent was met when all the parents and grandparents of an individual originated from Italy.
The Italian centenarians were ascertained mainly via general practitioners in the community. These centenarians represented a convenience sample that has been previously described.11 The history of past and current diseases was accurately collected, and the centenarians' medical documentation and the current drug therapy were checked. Accordingly, all of the Italian centenarians were free of major age-related diseases, i.e., severe cognitive impairment, clinically evident cancer, CVD, renal insufficiency, or severe physical impairment. Only, part of this group had decreased visual or auditory acuity. Thus, all of the Italian centenarians were in good health relative to their very advanced age.
Controls were in apparent good physical health, with exclusion criteria including presence of major CVD or cerebrovascular disease, cancer, dementia, chronic autoimmune or inflammatory disorders, renal or hepatic failure, and major psychiatric disorders.
Japanese replication cohort
Two groups of Japanese subjects of the same Asian (Japanese) descent were studied: (1) 742 cases (centenarians, 100–116 years, 623 women, 119 men); and (2) 499 healthy controls (23–59 years, 356 women, 143 men). The group of cases was gathered from two prospective cohorts: The Tokyo Centenarians Study (TCS) and the Semi-Supercentenarians Study in Japan (SSC-J). A detailed description of population-based recruitments for the TCS has been previously reported.12 The TCS cohort included 304 centenarians (65 men, 239 women) aged 100–108 years.
The SSC-J is a nationwide longitudinal survey consisting mainly of individuals aged 105 years or older, which started in 2002 (with n=135 SSC). After 2002, the recruitment strategy has relied on responses to local governments and nursing homes in the whole country and direct enquiries to our research team. Consequently, a total of 450 centenarians (58 men, 392 women) were enrolled in the SSC-J by the end of November, 2011. The phenotype and disease characteristics of the Japanese centenarians are described elsewhere,13 with a prevalence of hypertension, CVD, and dementia of 63.6%, 28.8%, and 59.4%, respectively.
Inclusion criteria for the control group, which was recruited by advertising during years 2008–2012, were being a man or woman aged <60 years and free of diagnosed stroke, CVD, and chronic renal failure (as reported in a questionnaire).
Functional analysis—the luciferase reporter gene
The fragment (see below), including the allele, was directly inserted into the pGL3-promter at the restriction recognition sites SacI at the 5′ end and XhoI at the 3′ end (see below, in bold) of the sequences obtained from the genomes of: (1) One individual homozygous for the rs7460 A allele and one homozygous for the rs7460 T allele (see below, underlined); (2) one individual homozygous for the rs7843014 A allele and one homozygous for the rs7843014 C allele (see below, underlined). Fragments were: rs7460 AA, GAGCTCTGTGGATATGTGAAGCATTGGGTCGGGAACTAGCTGTAGAACACAACTAAAAACTCATGTCTTTTTTCACAGAATAATGTGCCAGTTTTTTGTAGCAATGATATTTCTCTTGGAAGCAGAAATGCTTTGTACCAGAGCACCTCCAAACTGCATTGAGGAGAAGTTCCAGAACCATCCCCTTTTTCCATTTTTATATAATTTCTCGAG; rs7460 TT, GAGCTCTGTGGATATGTGAAGCATTGGGTCGGGAACTAGCTGTAGAACACAACTAAAAAC TCATGTCTTTTTTCACAGAATAATGTGCCAGTTTTTTGTAGCAATGTTATTTCTCTTGGAAGCAGAAATGCTTTGTACCAGAGCACCTCCAAACTGCATTGAGGAGAAGTTCCAGAACCATCCCCTTTTTCCATTTTTATATAATTTCTCGAG; rs7843014 AA, GAGCTCGCTCATGGGAACGTTCTCATGTATGGATCAGAGAGTGATGGGACCTAAACCCATTGATTAGTCTTAACATCACAAAGAACAACCAGATGTACATCACCTAAGAAGTATTCTTGTTGTGGGGAGGGGGGGAATCAAGCCCAAACTGGATCAAACTTCTTTAGGTTCTAACAAGCAGCTGATGGGAAATAAACGGGCCAGAAACTCGAG; and rs7843014 CC, GAGCTCGCTCATGGGAACGTTCTCATGTATGGATCAGAGAGTGATGGGACCTAAACCCATTGATTAGTCTTAACATCACAAAGAACAACCAGATGTACATCA CCTACGAAGTATTCTTGTTGTGGGGAGGGGGGGAATCAAGCCCAAACTGGATCAAACTTCTTT AGGTTCTAACAAGCAGCTGATGGGAAATAAACGGGCCAGAAACTCGAG.
Mice skeletal muscle C2C12 cell lines were used to represent muscle-specific expression. Cell cultures, transfections, and dual-luciferase reporter assays were performed as previously described.14 We used the pRL-SV40 vector as an internal control for variations in transfection efficiency. The pGL3-promoter vector without an insert was used as a negative control. The transfected cells were harvested after 48 hr and assayed for firefly luciferase activity and Renilla luciferase activity using the dual-luciferase reporter assay system (Promega Biotech, Beijing, China), as suggested by the manufacturer using a luminometer (Tecan GENios™ Pro, Männedorf, Switzerland). From each measurement, relative luciferase activity was calculated by dividing the firefly luciferase activity reading by the Renilla luciferase activity reading. Experiments were performed in triplicate. Relative luciferase activity values were expressed as the means±standard deviation (SD) of the three different experiments.
Genotype assessment
Spanish cohort
In cases and controls, genomic DNA was extracted from buccal cells at the European University (Madrid, Spain) according to standard phenol/chloroform procedures followed by alcohol precipitation. rs7843014 and rs7460 SNPs were genotyped using TaqMan® SNP Genotyping Assays (assay ID, C__11605645_10 and C____243385_10, respectively) and the StepOneTM Real-Time PCR System (Applied Biosystems, Foster City, CA). A total of 5 μL of genotyping mixture containing 2.5 μL of GTXpress™ Master Mix, 0.25 μL of SNP assay mix (20×), and 1.25 μL of distilled water was mixed with 1 μL of genomic DNA (10 ng·μL−1) in each reaction. One or two negative controls were included on each plate. TaqMan® assays for genotype calls were analyzed using StepOne™ Software v. 2.1 (Applied Biosystems, Foster City, CA).
Italian cohort
Genomic DNA was purified from peripheral blood samples using the QiaAmp DNA Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Genotyping was performed using the TaqMan® rs7460 and rs7843014 SNP genotyping assays (Applied Biosystems, Foster City, CA). For quality control, genotyping analyses were done blind with respect to the centenarian/young control status of the study participants, and a random 20% of the samples were repeated. All genotyping repeated for quality control did not differ from the initial genotyping. Two investigators independently reviewed all the results.
Japanese cohort
Total DNA was isolated from venous blood by use of QIAamp DNA Blood Maxi Kit (QIAGEN, Hilden, Germany). The rs7843014 and rs7460 SNPs were genotyped using TaqMan® SNP Genotyping Assays (assay ID, C__11605645_10 and C____243385_10, respectively) and the StepOnePlus™ Real-Time PCR System (Applied Biosystems, Foster City, CA). A total of 5 μL of genotyping mixture containing 2.5 μL of GTXpress™ Master Mix, 0.125 μL of assay mix (40×), and 1.375 μL of distilled water was mixed with 1μL of genomic DNA (10 ng·μL−1) in each reaction. One or two negative controls were included on each plate. TaqMan® assays for genotype calls were analyzed using StepOne™ Software v. 2.1 (Applied Biosystems, Foster City, CA).
Statistical analysis
One-way analysis of variance (ANOVA) was used to compare the relative luciferase activity between the different plasmids of each SNP. The PTK2 allele frequencies were calculated from genotypes using the gene-counting method. Hardy–Weinberg equilibrium (HWE) was tested in the control groups of each cohort with the chi-squared test. Genotype/allele frequencies were compared between cases and controls in each of the three cohorts using the Fisher exact test. We compared each genotype and allele with all other genotypes and alleles. The above-mentioned analyses were also conducted separately for each sex within each cohort. We also conducted binary logistic regression (odds ratios [OR] and 95% confidence intervals [CI], all adjusted by sex) to examine the likelihood of reaching EL if carrying a given allele/genotype. A p value ≤0.05 was considered statistically significant in all of the statistical analyses, which were performed using the PASW (v. 18.0 for WINDOWS, Chicago, IL), except for statistical power, which we calculated with the StatMate software, v. 2.0 (GraphPad, San Diego, CA).
Results
Functional analysis
The results of luciferase report analyses are presented in Fig. 1. Both SNPs had functional significance, as shown by differences in luciferase activity between the constructs of the two SNPs (all p≤0.001). The A allele up-regulated luciferase activity compared to the other allele, i.e., rs7460 T allele and rs7843014 C allele, respectively.
FIG. 1.
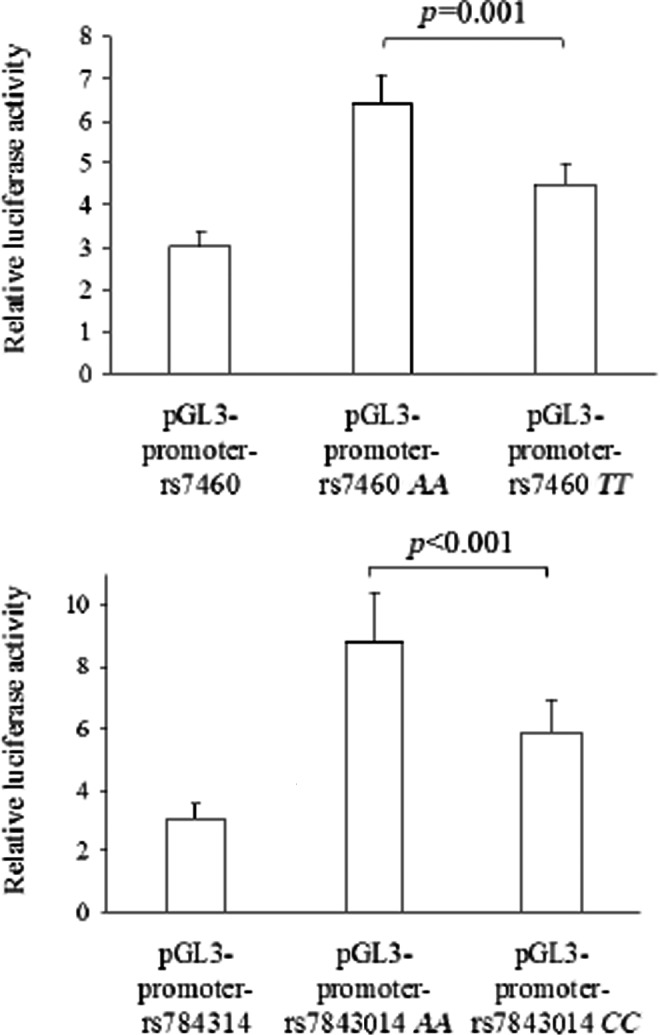
Comparison of relative luciferase activity (i.e., firefly luciferase activity divided by Renilla luciferase activity) between plasmids for the rs7460 (upper) and rs7843014 SNPs (lower). Values are means±standard deviation (SD) of three different experiments, each performed in triplicate. Significant differences (all p≤0.01) were found within each single-nucleotide polymorphism (SNP) for all comparisons between plasmids.
Spanish cohort
The failure rate of genotyping was 22.3% and 21.7% in cases and 7.3% and 3.9% in controls for rs7843014 and rs7460, respectively. The distribution of genotypes among controls was consistent with the HWE for the rs7460 SNP (p=0.29), but not for rs7843014 (p=0.008).
The results of genotype/allele frequency distributions (which remained essentially unchanged when both sexes were analysed together or separately) and binary logistic regression adjusted by sex are shown in Table 1. The A allele of both SNPs was negatively associated with EL (rs7460, OR=0.40, 95% CI 0.3, 0.6, p<0.01); rs7843014, OR=0.37, 95% CI 0.3, 0.5, p<0.01). The OR of being a Spanish centenarian if the subject had the rs7460 TT genotype was 6.68 (95% CI 4.1, 10.8, p<0.01). The rs7843014 CC genotype was also positively associated with EL (OR=7.58, 95% CI 4.6, 12.3, p<0.01).
Table 1.
Genotype/Allele Distributions of the PTK2 Gene and Results of Binary Logistic Regression Adjusted by Sex in the Spanish Cohort
| Cases | Controls | |||||||
|---|---|---|---|---|---|---|---|---|
| Model | rs7460 | n | % | n | % | Fisher exact test | OR (95% CI) | p value |
| Codom | T/T | 102 | 74 | 117 | 33 | 1 | ||
| A/T | 0 | 0 | 165 | 46 | <0.001 | NA (0.0–NA) | 0.99 | |
| A/A | 35 | 26 | 73 | 21 | 0.02 | 2.10 (1.2–3.6) | 0.007 | |
| Dom | T/T | 102 | 74 | 117 | 33 | 1 | ||
| A/T-A/A | 35 | 26 | 238 | 67 | <0.001 | 6.68 (4.1–10.8) | <0.001 | |
| Recess | T/T-A/T | 102 | 74 | 282 | 79 | 1 | ||
| A/A | 35 | 26 | 73 | 21 | 0.27 | 0.83 (0.5–1.4) | 0.46 | |
| Overdom | T/T-A/A | 137 | 100.0 | 190 | 54 | 1 | ||
| A/T | 0 | 0.0 | 165 | 46 | <0.001 | NA (0.0–NA) | <0.001 | |
| Log-add | — | — | — | 1.97 (1.5–2.6) | <0.001 | |||
| Allele | T | 204 | 74 | 399 | 56 | 1 | ||
| A | 70 | 26 | 311 | 44 | <0.001 | 0.40 (0.3–0.6) | <0.001 | |
| Cases | Controls | |||||||
|---|---|---|---|---|---|---|---|---|
| Model | rs7843014 | n | % | n | % | Fisher exact test | OR (95% CI) | p value |
| Codom | C/C | 94 | 69 | 92 | 26 | 1 | ||
| A/C | 0 | 0 | 152 | 43 | <0.001 | NA (0.0–NA) | <0.001 | |
| A/A | 43 | 31 | 111 | 31 | <0.001 | 3.22 (1.9–5.4) | <0.001 | |
| Dom | C/C | 94 | 69 | 92 | 26 | 1 | ||
| A/C-A/A | 43 | 31 | 263 | 74 | <0.001 | 7.58 (4.6–12.3) | <0.001 | |
| Recess | C/C-A/C | 94 | 69 | 244 | 69 | 1 | ||
| A/A | 43 | 31 | 111 | 31 | 1.00 | 1.09 (0.7–1.7) | 0.70 | |
| Overdom | C/C-A/A | 137 | 100 | 203 | 57 | 1 | ||
| A/C | 0 | 0 | 152 | 43 | <0.001 | NA (0.0–NA) | <0.001 | |
| Log-add | — | — | — | 2.04 (1.6–2.7) | <0.001 | |||
| Allele | C | 188 | 69 | 336 | 47 | <0.001 | 1 | |
| A | 86 | 31 | 374 | 53 | 0.37 (0.3–0.5) | <0.001 | ||
Bold denotes p<0.05.
Of note, the results of the Fisher test remained essentially unchanged when the two sexes were analyzed separately, except for the comparison of rs7843014 CC vs. AA genotypes in males, where statistical significance was not reached (p=0.08).
codom, codominant; dom, dominant; recess, recessive; overdo, overdominant; Log-add, additive; %, relative frequency; 95% CI, 95% confidence interval; NA, not applicable; OR, odds ratio.
On the basis of the observed prevalence of the A allele in the control group, the Spanish cohort's sample size had an 80% power to detect a relative likelihood of 1.4 (rs7460) and 1.3 (rs7843014) for being a centenarian between A allele carriers and non-carriers with a significance level (alpha) of 0.05 (two-tailed).
Italian cohort
The failure rate of genotyping was 0% in cases and controls for both rs7843014 and rs7460. Genotypes were in HWE among controls (rs7843014, p=0.08; rs7460, p=0.32). Table 2 shows the results of genotype/allele frequency distributions (which remained unchanged when both sexes were analyzed together or separately) and binary logistic regression adjusted by sex, where statistical significance was not reached.
Table 2.
Genotype/Allele Distributions of the PTK2 Gene and Results of Binary Logistic Regression Adjusted by Sex in the Italian Cohort
| Cases | Controls | |||||||
|---|---|---|---|---|---|---|---|---|
| Model | rs7460 | n | % | n | % | Fisher exact test | OR (95% CI) | p value |
| Codom | A/A | 22 | 28 | 98 | 31 | 1 | ||
| A/T | 42 | 53 | 148 | 47 | 0.47 | 0.77 (0.4–1.4) | 0.40 | |
| T/T | 15 | 19 | 70 | 22 | 1 | 1.03 (0.5–2.1) | 0.93 | |
| Dom | A/A | 22 | 28 | 98 | 31 | 1 | ||
| A/T-T/T | 57 | 72 | 218 | 69 | 0.68 | 0.83 (0.5–1.5) | 0.59 | |
| Recess | A/A-A/T | 64 | 81 | 246 | 78 | 1 | ||
| T/T | 15 | 19 | 70 | 22 | 0.65 | 1.19 (0.6–2.2) | 0.56 | |
| Overdom | A/A-T/T | 37 | 47 | 168 | 53 | 1 | ||
| A/T | 42 | 53 | 148 | 47 | 0.32 | 0.76 (0.5–1.3) | 0.32 | |
| Log-add | — | — | — | 0.74 (0.5–1.2) | 0.61 | |||
| Allele | T | 72 | 46 | 288 | 46 | 1 | ||
| A | 86 | 54 | 344 | 54 | 1 | 0.99 (0.7–1.5) | 0.78 | |
| Cases | Controls | |||||||
|---|---|---|---|---|---|---|---|---|
| Model | rs7843014 | n | % | n | % | Fisher exact test | OR (95% CI) | p value |
| Codom | C/C | 29 | 37 | 109 | 35 | 1 | ||
| A/C | 37 | 47 | 140 | 44 | 1 | 0.99 (0.6–1.7) | 0.96 | |
| A/A | 13 | 16 | 67 | 21 | 0.48 | 1.36 (0.6–2.8) | 0.41 | |
| Dom | C/C | 29 | 37 | 109 | 34 | 1 | ||
| A/C-A/A | 50 | 63 | 207 | 66 | 0.79 | 1.09 (0.6–1.8) | 0.72 | |
| Recess | C/C-A/C | 66 | 84 | 249 | 79 | 1 | ||
| A/A | 13 | 16 | 67 | 21 | 0.43 | 1.37 (0.7–2.6) | 0.32 | |
| Overdom | C/C-A/A | 42 | 53 | 176 | 56 | 1 | ||
| A/C | 37 | 47 | 140 | 44 | 0.71 | 0.91 (0.5–1.4) | 0.65 | |
| Log-add | — | — | — | 0.86 (0.6–1.4) | 0.58 | |||
| Allele | C | 95 | 60 | 358 | 57 | 1 | ||
| A | 63 | 40 | 274 | 43 | 0.47 | 1.12 (0.9–1.7) | 0.42 | |
Of note, the results of the Fisher test remained unchanged when the two sexes were analyzed separately (i.e., no significant p value was found).
codom, codominant; dom, dominant; recess, recessive; overdo, overdominant; Log-add, additive; %, relative frequency; 95% CI, 95% confidence interval; NA, not applicable; OR, odds ratio.
On the basis of the observed prevalence of the A allele in the control group, the Italian cohort's sample size had an 80% power to detect a relative likelihood of 1.4 (rs7460) and 1.3 (rs7843014) for being a centenarian between A allele carriers and non-carriers with a significance level (alpha) of 0.05 (two-tailed).
Japanese cohort
The failure rate of genotyping was 2.3% and 1.9% in cases and 0.6% and 0% in controls for rs7843014 and rs7460, respectively. The distribution of genotypes among controls was consistent with the HWE for the rs7460 SNP (p=0.18), but not for rs7843014 (p=0.008). Table 3 shows the results of genotype/allele frequency distributions (which remained unchanged when both sexes were analyzed together or separately) and binary logistic regression adjusted by sex, where statistical significance was not reached.
Table 3.
Genotype/Allele Distributions of the PTK2 Gene and Results of Binary Logistic Regression Adjusted by Sex in the Japanese Cohort
| Cases | Controls | |||||||
|---|---|---|---|---|---|---|---|---|
| Model | rs7460 | n | % | n | % | Fisher exact test | OR (95% CI) | p value |
| Codom | A/A | 300 | 41 | 214 | 43 | 1 | ||
| A/T | 329 | 44 | 215 | 43 | 0.49 | 0.91 (0.7–1.2) | 0.67 | |
| T/T | 113 | 15 | 70 | 14 | 0.43 | 0.88 (0.6–1.3) | 0.46 | |
| Dom | A/A | 300 | 40 | 214 | 43 | 1 | ||
| A/T-T/T | 442 | 50 | 285 | 57 | 0.41 | 0.90 (0.7–1.1) | 0.38 | |
| Recess | A/A-A/T | 629 | 85 | 429 | 86 | 1 | ||
| T/T | 113 | 15 | 70 | 14 | 0.57 | 0.92 (0.7–1.3) | 0.63 | |
| Overdom | A/A-T/T | 413 | 56 | 284 | 57 | 1 | ||
| A/T | 329 | 44 | 215 | 43 | 0.68 | 0.94 (0.8–1.2) | 0.60 | |
| Log-add | — | — | — | 0.93 (0.8–1.1) | 0.39 | |||
| Allele | T | 555 | 37 | 355 | 36 | |||
| A | 929 | 63 | 643 | 64 | 0.37 | 0.93 (0.8–1.1) | 0.37 | |
| Cases | Controls | |||||||
|---|---|---|---|---|---|---|---|---|
| Model | rs7843014 | n | % | n | % | Fisher exact test | OR (95% CI) | p value |
| Codom | C/C | 435 | 59 | 272 | 54 | 1 | ||
| A/C | 254 | 34 | 189 | 38 | <0.001 | 1.18 (0.9–1.5) | 0.38 | |
| A/A | 53 | 7 | 38 | 8 | <0.001 | 1.15 (0.7–1.8) | 0.55 | |
| Dom | C/C | 435 | 59 | 272 | 55 | 1 | ||
| A/C-A/A | 307 | 41 | 227 | 45 | <0.001 | 1.18 (0.9–1.5) | 0.17 | |
| Recess | C/C-A/C | 689 | 93 | 461 | 92 | 1 | ||
| A/A | 53 | 7 | 38 | 8 | <0.001 | 1.08 (0.7–1.7) | 0.75 | |
| Overdom | C/C-A/A | 488 | 66 | 310 | 62 | 1 | ||
| A/C | 254 | 34 | 189 | 38 | 0.001 | 1.16 (0.9–1.5) | 0.21 | |
| Log-add | — | — | — | 1.12 (0.9–1.3) | 0.22 | |||
| Allele | C | 1124 | 76 | 733 | 73 | |||
| A | 360 | 24 | 265 | 27 | 0.20 | 0.89 (0.7–1.1) | 0.21 | |
Bold denotes p<0.05.
Of note, the results of the Fisher test remained unchanged when both sexes were analyzed separately (i.e., no significant p value was found).
codom, codominant; dom, dominant; recess, recessive; overdo, overdominant; Log-add, additive; %, relative frequency; 95% CI, 95% confidence interval; NA, not applicable; OR, odds ratio.
On the basis of the observed prevalence of the A allele in the control group, the Japanese cohort's sample size had an 80% power to detect a relative likelihood of 1.1 (rs7460) and 1.5 (rs7843014) for being a centenarian between A allele carriers and non-carriers with a significance level (alpha) of 0.05 (two-tailed).
Discussion
Our main finding was that the two PTK2 genotypes that were possibly associated with lower gene expression, rs7460 TT and rs7843014 CC, might actually increase the likelihood of reaching EL in the Spanish population. However, the latter finding was not replicated in the two other independent cohorts, where we found no association between the PTK2 genotype and EL, suggesting that different environmental, epigenetic, or ethnic-related factors might influence the potential association between the PTK2 genotype and EL. The lack of replication in the Italian or Japanese cohort may also be explained, at least partly, by genetic heterogeneity among long-lived individuals due to cohort-specific differences in survival probability, as recently shown for the APOE and FOXO3A genes.15 Indeed, Nygaard et al. found a decrease in the allele frequency of these two gene variants in individuals from more recent birth cohorts, indicating that there are cohort differences in the effects of selection pressure on survival to the highest ages.15
Regarding the results of the functionality analyses (luciferase constructs), to the best of our knowledge this is the first attempt to gain insights into the potential functional consequences of these two PTK variants, which are not likely to influence the amino acid sequence of the protein product.10 Yet alterations in DNA sequence in the 3′-UTR of a gene, such as the PTK2 rs7460 A/T SNP, have the potential to alter the level, location, or timing of gene expression; and, intronic genomic variants, such as the PTK2 rs7843014 A/C SNP, can influence gene expression and thus phenotype by altering mRNA stability, alternative mRNA splicing, or the binding of transcription factors.16–19
We believe there is scientific rationale in postulating PTK2 as a potential longevity-related gene given the pivotal role that its product FAK plays in a potential hub of most tissue networks, as is focal adhesion. The latter is involved in functions that are necessary to the survival of cells, including adhesion, migration, and regulation of cell growth. The loss of cell growth regulation leads to diverse pathological states, including mainly the initiation and progression of the cascade of events leading to cancer.8 Focal adhesions have received considerable attention because they serve as a nexus between the contractile cytoskeleton and the ECM, with FAK mediating mechano-transduction signaling pathways between the cell and its environment.
Thus, FAK knockout mice exhibit an embryonic lethal phenotype with severe defects in mesoderm development.20 On the other hand, in many cancers there is a correlation between increased FAK expression and progression to a more malignant state, which is consistent with the importance of mechano-transduction pathways in tumor progression.21 Indeed, cancer progression requires several key events that are regulated by FAK-mediated signaling pathways, including the establishment of metastases at distant sites or the production of a rigid microenvironment encouraging cell proliferation. The possible role for FAK in the progression of tumors has, in fact, prompted the development of FAK inhibitors as potential cancer therapies, e.g., those being tested in models of hepatocellular carcinoma22 or metastatic prostate,23 breast,24 or bone tumors, 25 with some of these compounds now in clinical testing.9
The aging process is inevitably associated with functional decline in skeletal muscle tissue, i.e., sarcopenia, and acceleration of this process might increase mortality risk.26–28 As such, gene variations that are potentially associated with higher preservation of muscle function at late life, such as the K153R polymorphism in the myostatin (MSTN) gene,29 might also influence EL.30 The force generated by muscle fibers is transmitted laterally to the surrounding matrix of connective tissue31 via a special attachment system, i.e., protein complexes known as costameres,32 with the FAK playing a major role in costamere formation and turnover.33,34 Thus, a preliminary report by Erskine et al. in a small cohort of young men (n=51) suggested an association between the two SNPs studied here and individual variability in knee–extensor muscle strength, with the baseline specific muscle force being higher in 7843014 AA and rs7460 TT homozygotes compared with rs7843014 C allele and rs7460 A allele carriers, respectively.
With regard to this result, it is difficult to determine if higher or lower expression of the PTK2 gene might benefit muscle strength. Fibroblasts derived from FAK knockout mice form more robust adhesions compared to wild-type controls, suggesting that FAK is less important for focal adhesion assembly than it is for focal adhesion disassembly (turnover).20 Further studies have demonstrated that FAK−/− cells migrate more slowly and appear more contractile than wild-type cells, with both of these phenotypes being attributable to a lack of adhesion disassembly.35,36 Thus, the rs7460 TT and rs7843014 CC genotypes, which are possibly associated with lower gene expression as well as with EL in the Spanish cohort, might perhaps favor muscle fiber contraction and thus muscle force development, although much more research is obviously needed.
Besides the rationale for postulating the PTK2 gene as a candidate to influence EL or at least age-related diseases and the clear definition we used for the criterion of EL (with all cases being centenarians), a further strength from our study derives from the luciferase construct study we performed as well as from the fact that the analyses were conducted in three independent cohorts (with one of them, the Japanese cohort, including a very large sample of centenarians). Also, we studied both normal (Spanish/Japanese) and uniquely super healthy (Italian) centenarians.
There are, however, limitations on our design. First, we used convenience samples, thereby raising the risk of bias due to population stratification. Another limitation comes from the relatively high rate of genotyping failure in the Spanish cohort, particularly in cases indicating that DNA extraction from saliva in frail individuals such as centenarians (which we were ethically constrained to adopt) might result in small amounts of DNA available for analyses. The rs7843014 SNP did to meet HWE in Spanish and Japanese controls. With regard to this, failure to meet HWE is not necessarily a good proxy of genotyping errors,37,38 with ∼10% of all genotype–phenotype association studies showing in fact deviation from HWE.39 Nonetheless, violation of HWE might imply the use of a selected rather than a random sample. As such, our results should be corroborated with other cohorts whose genotype frequencies are in HWE. Finally, interpretation of gene association studies with centenarians may be flawed by differences in date of birth, e.g., centenarians and controls were born in the early 1900s and after 1930, respectively. Indeed, the interaction of genotype with environmental factors can be influenced by year of birth.40
On the other hand, it is now clear that epigenetic modifications might also affect longevity as well as inherited genetic variants because the former can affect gene expression without changing the DNA sequence.41 Epigenetic mechanisms may include DNA methylation, histone modifications, and altered expression of RNAs or small, non-coding RNAs that regulate gene expression profiles associated with longevity.42 Age-related hyper-methylation has been reported in promoter regions of genes involved in cell cycle regulation, tumor cell invasion, apoptosis, metabolism, cell signaling, or DNA repair, resulting in decreased mRNA levels.43–45 Although the possibility of the persistence of epigenetic marks through generations is a controversial issue,46 it seems that prenatal conditions such as maternal cigarette smoking during pregnancy or other factors, like diet, stress, prenatal nutrition, or prenatal drug exposure, may alter fetal growth and as well as gene expression through epigenetic mechanisms.47 Thus, epigenetic factors associated with trans-generational inheritance may be involved in longevity and should be also considered. Finally, our finding that there were essentially no sex-related differences in the possible effect of PTK2 gene variants on EL remains to be corroborated in other cohorts. Passarino et al.48 showed that genetic factors influence survival at advanced age in a sex-specific way, with genetic variability playing a stronger role in males than in females.
In summary, two genotypes of the FAK-coding gene (PTK2), rs7460 TT and rs7843014 CC, were possibly associated with lower gene expression (as suggested by the results of our luciferase construct) and were actually associated with higher likelihood of reaching EL in the Spanish population. The latter finding was, however, not corroborated in the two other replication cohorts, suggesting that different environment, epigenetic, ethnic, or specific cohort-related factors might influence the potential association between PTK2 genotype and EL. Further research is needed with larger cohorts of different geographical origins to unveil the mechanisms by which FAK could influence the rate of aging.
Acknowledgments
This study was partially supported by a grant from Fondo de Investigaciones Sanitarias (FIS, no. PI12/00914). This work was also supported by grants from the National Natural Science Foundation of China (grant code 31100853) and the China Institute of Sport Science (2013-13).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Salvioli S, Capri M, Santoro A, Raule N, Sevini F, Lukas S, Lanzarini C, Monti D, Passarino G, Rose G, De Benedictis G, Franceschi C. The impact of mitochondrial DNA on human lifespan: A view from studies on centenarians. Biotechnol J 2008;3:740–749 [DOI] [PubMed] [Google Scholar]
- 2.Christensen K, McGue M, Petersen I, Jeune B, Vaupel JW. Exceptional longevity does not result in excessive levels of disability. Proc Natl Acad Sci USA 2008;105:13274–13279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terry DF, Sebastiani P, Andersen SL, Perls TT. Disentangling the roles of disability and morbidity in survival to exceptional old age. Arch Intern Med 2008;168:277–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolfson M, Budovsky A, Tacutu R, Fraifeld V. The signaling hubs at the crossroad of longevity and age-related disease networks. Int J Biochem Cell Biol 2009;41:516–520 [DOI] [PubMed] [Google Scholar]
- 5.Barabasi AL, Oltvai ZN. Network biology: Understanding the cell's functional organization. Nat Rev Genet 2004;5:101–113 [DOI] [PubMed] [Google Scholar]
- 6.Budovsky A, Abramovich A, Cohen R, Chalifa-Caspi V, Fraifeld V. Longevity network: Construction and implications. Mech Ageing Dev 2007;128:117–124 [DOI] [PubMed] [Google Scholar]
- 7.Budovsky A, Tacutu R, Yanai H, Abramovich A, Wolfson M, Fraifeld V. Common gene signature of cancer and longevity. Mech Ageing Dev 2009;130:33–39 [DOI] [PubMed] [Google Scholar]
- 8.Tilghman RW, Parsons JT. Focal adhesion kinase as a regulator of cell tension in the progression of cancer. Semin Cancer Biol 2008;18:45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultze A, Fiedler W. Clinical importance and potential use of small molecule inhibitors of focal adhesion kinase. Anti-Cancer Agents Med Chem 2011;11:593–599 [DOI] [PubMed] [Google Scholar]
- 10.Erskine RM, Williams AG, Jones DA, Stewart CE, Degens H. Do PTK2 gene polymorphisms contribute to the interindividual variability in muscle strength and the response to resistance training? A preliminary report. J Appl Physiol 2012;112:1329–1334 [DOI] [PubMed] [Google Scholar]
- 11.Emanuele E, Fontana JM, Minoretti P, Geroldi D. Preliminary evidence of a genetic association between chromosome 9p21.3 and human longevity. Rejuvenation Res 2010;13:23–26 [DOI] [PubMed] [Google Scholar]
- 12.Gondo Y, Hirose N, Arai Y, Inagaki H, Masui Y, Yamamura K, Shimizu K, Takayama M, Ebihara Y, Nakazawa S, Kitagawa K. Functional status of centenarians in Tokyo, Japan: Developing better phenotypes of exceptional longevity. J Gerontol A Biol Sci Med Sci 2006;61:305–310 [DOI] [PubMed] [Google Scholar]
- 13.Takayama M, Hirose N, Arai Y, Gondo Y, Shimizu K, Ebihara Y, Yamamura K, Nakazawa S, Inagaki H, Masui Y, Kitagawa K. Morbidity of Tokyo-area centenarians and its relationship to functional status. J Gerontol A Biol Sci Med Sci 2007;62:774–782 [DOI] [PubMed] [Google Scholar]
- 14.He ZH, Hu Y, Li YC, Yvert T, Santiago C, Gómez-Gallego F, Ruiz JR, Lucia A. Are calcineurin genes associated with athletic status? A function, replication study. Med Sci Sports Exerc 2011;43:1433–1440 [DOI] [PubMed] [Google Scholar]
- 15.Nygaard M, Lindahl-Jacobsen R, Soerensen M, Mengel-From J, Andersen-Ranberg K, Jeune B, Vaupel JW, Tan Q, Christiansen L, Christensen K. Birth cohort differences in the prevalence of longevity-associated variants in APOE and FOXO3A in Danish long-lived individuals. Exp Gerontol 2014;57C:41–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knight JC. Regulatory polymorphisms underlying complex disease traits. J Mol Med (Berlin, Germany) 2005;83:97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mercado PA, Ayala YM, Romano M, Buratti E, Baralle FE. Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res 2005;33:6000–6010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasabe T, Furukawa A, Matsusita S, Higuchi S, Ishiura S. Association analysis of the dopamine receptor D2 (DRD2) SNP rs1076560 in alcoholic patients. Neurosci Lett 2007;412:139–142 [DOI] [PubMed] [Google Scholar]
- 19.Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet 2002;3:391–397 [DOI] [PubMed] [Google Scholar]
- 20.Ilić D, Furuta Y, Kanazawa S, Takeda N, Sobue K, Nakatsuji N, Nomura S, Fujimoto J, Okada M, Yamamoto T. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature 1995;377:539–544 [DOI] [PubMed] [Google Scholar]
- 21.Gabarra-Niecko V, Schaller MD, Dunty JM. FAK regulates biological processes important for the pathogenesis of cancer. Cancer Metastasis Rev 2003;22:359–374 [DOI] [PubMed] [Google Scholar]
- 22.Bagi CM, Christensen J, Cohen DP, Roberts WG, Wilkie D, Swanson T, Tuthill T, Andresen CJ. Sunitinib and PF-562,271 (FAK/Pyk2 inhibitor) effectively block growth and recovery of human hepatocellular carcinoma in a rat xenograft model. Cancer Biol Ther 2009;8:856–865 [DOI] [PubMed] [Google Scholar]
- 23.Sun H, Pisle S, Gardner ER, Figg WD. Bioluminescent imaging study: FAK inhibitor, PF-562,271, preclinical study in PC3M-luc-C6 local implant and metastasis xenograft models. Cancer Biol Ther 2010;10:38–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wendt MK, Smith JA, Schiemann WP. Transforming growth factor-beta-induced epithelial-mesenchymal transition facilitates epidermal growth factor-dependent breast cancer progression. Oncogene 2010;29:6485–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagi CM, Roberts GW, Andresen CJ. Dual focal adhesion kinase/Pyk2 inhibitor has positive effects on bone tumors: Implications for bone metastases. Cancer 2008;112:2313–2321 [DOI] [PubMed] [Google Scholar]
- 26.Metter EJ, Talbot LA, Schrager M, Conwit RA. Arm-cranking muscle power and arm isometric muscle strength are independent predictors of all-cause mortality in men. J Appl Physiol 2004;96:814–821 [DOI] [PubMed] [Google Scholar]
- 27.Ruiz JR, Sui X, Lobelo F, Morrow JR, Jr, Jackson AW, Sjöström M, Blair SN. Association between muscular strength and mortality in men: Prospective cohort study. Br Med J 2008;337:a439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metter EJ, Talbot LA, Schrager M, Conwit RA. Arm-cranking muscle power and arm isometric muscle strength are independent predictors of all-cause mortality in men. J Appl Physiol 2004;96:814–821 [DOI] [PubMed] [Google Scholar]
- 29.Garatachea N, Pinós T, Cámara Y, Rodríguez-Romo G, Emanuele E, Ricevuti G, Venturini L, Santos-Lozano A, Santiago-Dorrego C, Fiuza-Luces C, Yvert T, Andreu AL, Lucia A. Association of the K153R polymorphism in the myostatin gene and extreme longevity. Age 2013;35:2445–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garatachea N, Marin PJ, Lucia A. The ACE DD genotype and D-allele are associated with exceptional longevity: A meta-analysis. Ageing Res Rev 2013;12:1079–1087 [DOI] [PubMed] [Google Scholar]
- 31.Street SF. Lateral transmission of tension in frog myofibers: A myofibrillar network and transverse cytoskeletal connections are possible transmitters. J Cell Physiol 1983;114:346–364 [DOI] [PubMed] [Google Scholar]
- 32.Pardo JV, Siliciano JD, Craig SW. Vinculin is a component of an extensive network of myofibril-sarcolemma attachment regions in cardiac muscle fibers. J Cell Biol 1983;97:1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cary LA, Guan JL. Focal adhesion kinase in integrin-mediated signaling. Front Biosci 1999;4:D102–D113 [DOI] [PubMed] [Google Scholar]
- 34.Quach NL, Rando TA. Focal adhesion kinase is essential for costamerogenesis in cultured skeletal muscle cells. Dev Biol 2006;293:38–52 [DOI] [PubMed] [Google Scholar]
- 35.Chen BH, Tzen JT, Bresnick AR, Chen HC. Roles of Rho-associated kinase and myosin light chain kinase in morphological and migratory defects of focal adhesion kinase-null cells. J Biol Chem 2002;277:33857–33863 [DOI] [PubMed] [Google Scholar]
- 36.Ren XD, Kiosses WB, Sieg DJ, Otey CA, Schlaepfer DD, Schwartz MA. Focal adhesion kinase suppresses Rho activity to promote focal adhesion turnover. J Cell Sci 2000;113(Pt 20):3673–3678 [DOI] [PubMed] [Google Scholar]
- 37.Leal SM. Detection of genotyping errors and pseudo-SNPs via deviations from Hardy-Weinberg equilibrium. Genet Epidemiol 2005;29:204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou GY, Donner A. The merits of testing Hardy-Weinberg equilibrium in the analysis of unmatched case-control data: A cautionary note. Ann Hum Genet 2006;70(Pt 6):923–933 [DOI] [PubMed] [Google Scholar]
- 39.Trikalinos TA, Salanti G, Khoury MJ, Ioannidis JP. Impact of violations and deviations in Hardy-Weinberg equilibrium on postulated gene-disease associations. Am J Epidemiol 2006;163:300–309 [DOI] [PubMed] [Google Scholar]
- 40.Lewis SJ, Brunner EJ. Methodological problems in genetic association studies of longevity—the apolipoprotein E gene as an example. Int J Epidemiol 2004;33:962–970 [DOI] [PubMed] [Google Scholar]
- 41.Wolffe AP, Matzke MA. Epigenetics: Regulation through repression. Science 1999;286:481–486 [DOI] [PubMed] [Google Scholar]
- 42.Mango SE. Ageing: Generations of longevity. Nature 2011;479:302–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fraga MF, Esteller M. Epigenetics and aging: The targets and the marks. Trends Genet 2007;23:413–418 [DOI] [PubMed] [Google Scholar]
- 44.Bellizzi D, D'Aquila P, Montesanto A, Corsonello A, Mari V, Mazzei B, Lattanzio F, Passarino G. Global DNA methylation in old subjects is correlated with frailty. Age (Dordr) 2011;34:169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montesanto A, Dato S, Bellizzi D, Rose G, Passarino G. Epidemiological, genetic and epigenetic aspects of the research on healthy ageing and longevity. Immun Ageing 2012;9:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, Benayoun BA, Shi Y, Brunet A. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 2011;479:365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knopik VS, Maccani MA, Francazio S, McGeary JE. The epigenetics of maternal cigarette smoking during pregnancy and effects on child development. Dev Psychopathol 2012;24:1377–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Passarino G, Montesanto A, Dato S, Giordano S, Domma F, Mari V, Feraco E, De Benedictis G. Sex and age specificity of susceptibility genes modulating survival at old age. Hum Hered 2006;62:213–220 [DOI] [PubMed] [Google Scholar]


