Summary
Compromised systemic acquired resistance in eds1 mutant Arabidopsis plants is associated with a reduced ability of the mutant to accumulate azelaic acid and its precursor 9-oxo nonanoic acid.
Key words: Arabidopsis thaliana, azelaic acid, EDS1, lipid peroxidation, 9-oxo nonanoic acid, systemic acquired resistance.
Abstract
Systemic acquired resistance (SAR) is a form of inducible disease resistance that depends on salicylic acid and its upstream regulator ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1). Although local Arabidopsis thaliana defence responses activated by the Pseudomonas syringae effector protein AvrRpm1 are intact in eds1 mutant plants, SAR signal generation is abolished. Here, the SAR-specific phenotype of the eds1 mutant is utilized to identify metabolites that contribute to SAR. To this end, SAR bioassay-assisted fractionation of extracts from the wild type compared with eds1 mutant plants that conditionally express AvrRpm1 was performed. Using high-performance liquid chromatography followed by mass spectrometry, systemic immunity was associated with the accumulation of 60 metabolites, including the putative SAR signal azelaic acid (AzA) and its precursors 9-hydroperoxy octadecadienoic acid (9-HPOD) and 9-oxo nonanoic acid (ONA). Exogenous ONA induced SAR in systemic untreated leaves when applied at a 4-fold lower concentration than AzA. The data suggest that in planta oxidation of ONA to AzA might be partially responsible for this response and provide further evidence that AzA mobilizes Arabidopsis immunity in a concentration-dependent manner. The AzA fragmentation product pimelic acid did not induce SAR. The results link the C9 lipid peroxidation products ONA and AzA with systemic rather than local resistance and suggest that EDS1 directly or indirectly promotes the accumulation of ONA, AzA, or one or more of their common precursors possibly by activating one or more pathways that either result in the release of these compounds from galactolipids or promote lipid peroxidation.
Introduction
Plants protect themselves from pathogen invasion by innate immune mechanisms. In dicotyledonous plants, for example Arabidopsis thaliana, defence against biotrophic pathogens is dependent on the phytohormone salicylic acid (SA) and can be divided into local and systemic phases of immunity (Vlot et al., 2009; Spoel and Dong, 2012; Fu and Dong, 2013). Locally, plants respond to pathogen-associated molecular patterns (PAMPs) with PAMP-triggered immunity (PTI; Jones and Dangl, 2006). Alternatively, the recognition of pathogen effectors leads to effector-triggered immunity (ETI), which augments PTI (Tsuda et al., 2009; Tsuda and Katagiri, 2010). In contrast to PTI, ETI often results in hypersensitive response (HR)-associated death of the infected site and surrounding cells (Jones and Dangl, 2006; Maekawa et al., 2011). In ETI, pathogen effectors are recognized by plant nucleotide-binding leucine-rich repeat (NLR) receptors the majority of which possess N-terminal Toll-Interleukin1 Receptor-like (TIR) or coiled-coil (CC) domains, referred to as TNLs and CNLs, respectively (Maekawa et al., 2011; Bonardi and Dangl, 2012). PTI and ETI are associated with SA accumulation and a burst of reactive oxygen species (ROS; Jones and Dangl, 2006), and induce SA-dependent systemic acquired resistance (SAR) in systemic uninfected tissues (Cameron et al., 1994; Mishina and Zeier, 2007; Vlot et al., 2009; Liu et al., 2010; Fu and Dong, 2013; Breitenbach et al., 2014).
Long-distance acting metabolites reported to be associated with SAR include methyl salicylate (Park et al., 2007), the diterpenoid dihydroabietinal (Chaturvedi et al., 2012), the non-protein amino acid pipecolic acid (Navarova et al., 2012), the C9 dicarboxylic acid azelaic acid (AzA; Jung et al., 2009), and glycerol-3-phosphate (G3P; Chanda et al., 2011). In addition, the lipid transfer proteins AZELAIC ACID INDUCED 1 (AZI1; Jung et al., 2009; Yu et al., 2013), DEFECTIVE IN INDUCED RESISTANCE1 (DIR1), and DIR1-like (Maldonado et al., 2002; Champigny et al., 2013), as well as nitric oxide (NO) and ROS (Wang et al., 2014), have been implicated in long-distance SAR signalling. An increasing body of evidence suggests that some of these signals interact to coordinate SAR (Dempsey and Klessig, 2012; Shah and Zeier, 2013; Gao et al., 2014; Shah et al., 2014). DIR1 and AZI1, for example, physically interact and might act upstream of G3P accumulation, while G3P in turn appears to stabilize DIR1 and AZI1 transcripts and to act together with DIR1 to elicit SAR (Chaturvedi et al., 2008; Yu et al., 2013; Shah et al., 2014). AzA is thought to act upstream of the G3P–DIR1/AZI1 positive feedback loop (Yu et al., 2013), and NO and ROS were recently placed upstream of AzA in an SAR signalling pathway that appears to act in parallel with SA (Wang et al., 2014).
ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1), together with its sequence-related partners PHYTOALEXIN-DEFICIENT4 (PAD4) and SENESCENCE-ASSOCIATED GENE 101 (SAG101), is an important regulator of SA accumulation, as part of a feedback loop fortifying SA signalling (Falk et al., 1999; Feys et al., 2005; Vlot et al., 2009; Rietz et al., 2011). EDS1 contains a non-catalytic lipase-like domain with a classical α/β hydrolase-fold at its N-terminus and is essential for basal resistance to virulent pathogens as well as ETI mediated by TNL receptors and at least one CNL receptor (Aarts et al., 1998; Zhu et al., 2011; Wagner et al., 2013). EDS1 forms separate nucleocytoplasmic and nuclear heterodimers, respectively, with PAD4 and SAG101 (Feys et al., 2005; Wagner et al., 2013). EDS1 shuttles between the cytoplasm and nucleus via the nuclear pore machinery, and evidence suggests that both its nuclear and cytoplasmic pools contribute to defence (Garcia et al., 2010). Nuclear EDS1 accumulation is essential for TNL-mediated resistance and transcriptional activation of defence genes in ETI (Garcia, 2010). Moreover, EDS1 has been found in nuclear complexes with several TNL receptors as well as their recognized pathogen effectors, suggesting that EDS1 molecularly connects effector recognition to transcriptional defence reprogramming (Bhattacharjee et al., 2011; Heidrich et al., 2011; Zhu et al., 2011; Kim et al., 2012).
In resistance mediated by certain CNL receptors, EDS1 acts redundantly with SA (Bartsch et al., 2006; Venugopal et al., 2009; Roberts et al., 2013). SA-independent signalling roles of EDS1 have, for example, been associated with responses to the CNLs RPM1 (Bartsch et al., 2006) and HRT, recognizing the Turnip crinkle virus coat protein (Venugopal et al., 2009), and include a central role in the regulation of SAR (Truman et al., 2007; Rietz et al., 2011; Breitenbach et al., 2014). Both EDS1 and PAD4 are essential for SAR but not local ETI responses to the CNL receptors RPM1 and RPS2 (Aarts et al., 1998; Truman et al., 2007; Jing et al., 2011, Rietz et al., 2011). Recent analysis showed that EDS1 is necessary both for SAR signal generation in the locally infected tissue and for SAR signal perception in the systemic tissue in RPM1 resistance to Pseudomonas syringae expressing the effector AvrRpm1 (Breitenbach et al., 2014). Here, the SAR-specific phenotype of the eds1 mutant in response to AvrRpm1 was utilized to identify metabolites that are specifically associated with SAR (Fig. 1A). It is reported that the SAR defect of the eds1 mutant is in part due to a decreased ability to accumulate AzA and its precursors 9-hydroperoxy octadecadienoic acid (9-HPOD) and 9-oxo nonanoic acid (ONA). Application of exogenous ONA and AzA but not the AzA fragmentation product pimelic acid (PIM) induces systemic resistance in Arabidopsis. The data reinforce the close association between ONA, AzA, and SAR, and suggest that EDS1 influences the accumulation rate of immune-related lipid peroxidation precursors or products.
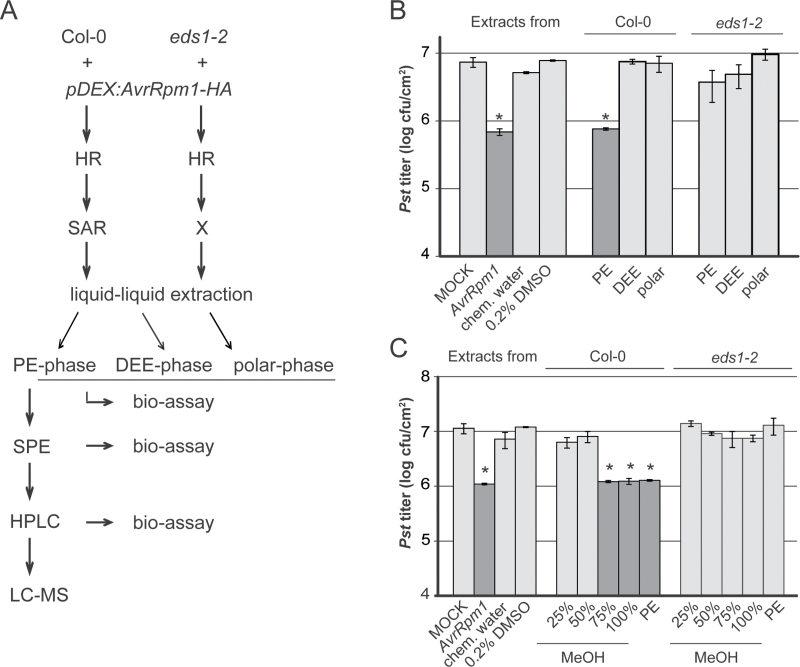
Fig. 1.
Extraction of SAR-related metabolites from pathogen-free SAR-induced plants. (A) Workflow. Metabolites were extracted with methanol (MeOH) from dexamethasone (DEX)-treated pDEX:AvrRpm1-HA Col-0 wild-type (wt) and pDEX:AvrRpm1-HA eds1-2 mutant plants. Metabolites were purified with a bioassay-assisted approach, including liquid–liquid extraction with petroleum ether (PE) and diethylether (DEE), solid-phase extraction (SPE), and high-performance liquid chromatography (HPLC) coupled with mass spectrometry (MS). (B) SAR bioassay after liquid–liquid extraction. Col-0 plants were locally treated with 10mM MgCl2 (MOCK), Pst/AvrRpm1 (AvrRpm1), chemical-treated water (chem. water), or 0.2% DMSO as controls or with metabolites from the PE, DEE, or polar phases (as indicated below the panel) derived from Col-0 or eds1-2 mutant plants (as indicated above the panel). Three days later, systemic leaves were infected with Pst and the resulting Pst titres are shown 4 d after infection (dpi). Plotted values are the average ±SD from two biologically independent experiments consisting of two replicates each. (C) SAR bioassay after SPE. Col-0 plants were locally treated with the same controls as in (B) or with metabolites from different SPE eluates as indicated below the panel derived from Col-0 or eds1-2 mutant plants as indicated above the panel. Three days later, systemic leaves were infected with Pst and the resulting Pst titres are shown at 4 dpi. Plotted values are the average ±SD of three replicates each. (B, C) Asterisks above the bars indicate statistically significant differences from the MOCK or 0.2% DMSO controls (*P<0.05, Student’s t-test). These experiments were repeated three times with similar results. HR, hypersensitive response; SAR, systemic acquired resistance.
Materials and methods
Plant material and growth conditions
All experiments were performed in A. thaliana ecotype Columbia-0 (Col-0). Mutants eds1-2, npr1-1, azi1-2, gly1-3, and sid2-1 as well as transgenic plants expressing haemagglutinin (HA)-tagged AvrRpm1 from a dexamethasone (DEX)-inducible transgene (pDEX:AvrRpm1-HA) in Col-0 and eds1-2 backgrounds were previously described (Cao et al., 1997; Wildermuth et al., 2001; Mackey et al., 2002; Kachroo et al., 2004; Bartsch et al., 2006; Jung et al., 2009; Breitenbach et al., 2014). Plants were grown on normal potting soil mixed with silica sand (ratio 5:1) in 10h light, 14h dark cycles at 70% relative humidity, 22 °C during the day at a light intensity of 100 μE m–2 s–1, and 18 °C during the night.
SAR bioassay
All infection experiments were performed in 4- to 5-week-old plants. Pseudomonas syringae pathovar tomato (Pst) and Pst/AvrRpm1 were maintained as described (Aarts et al., 1998). SAR was induced with Pst/AvrRpm1 and analysed with a secondary Pst infection as described (Breitenbach et al., 2014).
Metabolite isolation
Lawns of 3- to 4-week-old pDex:AvrRpm1-HA plants were sprayed with 30 μM DEX (Sigma Aldrich) dissolved in 0.01% Tween-20. Four to five hours later, 3g of above-ground tissue were harvested per sample and ground in liquid nitrogen. A 30ml aliquot of 100% methanol (MeOH; Merck) was added per sample, and samples were incubated for 1h in the dark while rotating at 28rpm at room temperature. Subsequently, samples were centrifuged at 2800 g at 4 °C for 10min and dried by evaporation. Pellets were dissolved in 10ml of MeOH:water (1:9 v/v) and extracted with an equal volume of petroleum ether (PE; Merck). The remaining material was extracted with an equal volume of diethyl ether (DEE, Merck). Both PE and DEE phases were dried by evaporation, and the dry matter was dissolved in 100 μl of dimethylsulphoxide (DMSO; Roth, Germany).
For subsequent solid-phase extraction (SPE), the PE phase in DMSO was diluted with 900 μl of MeOH:water (1:1 v/v). The sample was loaded onto a C18 cartridge (Agilent Technologies, 100mg bed mass, 1ml volume), which was consecutively washed with 5ml of 25, 50, 75, and 100% of MeOH followed by a wash with 5ml of PE. For further fractionation by high-performance liquid chromatography (HPLC), the 75% and 100% MeOH wash eluates and the final PE eluate were pooled, dried by evaporation, and the dry matter was dissolved in 600 μl of MeOH. Finally, the samples were centrifuged at the maximum speed (depending on the rotor) for 15min at 4 °C and the supernatant was used for HPLC.
Preparative RP18-HPLC-UV/ESI-MSn
Preparative HPLC was performed on a Jasco HPLC system (Jasco GmbH, Germany) consisting of two Jasco PU-2087 Plus pumps connected to a Jasco UV-2075 Plus variable wavelength detector set at 260nm, an Advantec CHF122SC fraction collector (Tokyo Seisakusho Kaisha Ltd, Japan), and an Agilent LC/MSD Trap XCT mass spectrometer. The (HP)LC column was a Synergi 4u Fusion-RP 80, 25 cm×21.5mm (Phenomenex). The HPLC solvents were 0.1% formic acid in water (A) and 0.1% formic acid in MeOH (B). For separation of compounds dissolved in 100% MeOH, a gradient was used from 100% A for 2min, then to 100% B in 28min, 20min at these conditions, returning to 100% A at a flow rate of 9.5ml min–1. The injection volume was 950 μl per HPLC run. Fractions (9.5ml) were collected at one fraction per minute. Data analysis was performed using the ChromPass Version 1.9.302.1124 software (Jasco GmbH, Germany).
Analytical LC-MS
A Bruker Daltonics esquire 3000plus ion trap mass spectrometer connected to an Agilent 1100 HPLC system equipped with a quaternary pump and a diode array detector was utilized. Components were separated with a Phenomenex Luna C-18 column (150mm long 2.0mm, particle size 5 μm) held at 28 °C. The injection volume was 5 μl. HPLC was performed with the following binary gradient system: solvent A, water with 0.1% formic acid; and solvent B, 100% MeOH with 0.1% formic acid. The gradient program was as follows: 0–30min, 100% A to 50% A/50% B; 30–35min, 50% A/50% B to 100% B, hold for 15min; 100% B to 100% A, in 5min, then hold for 10min. The flow rate was 0.2ml min−1. The full-scan mass spectra were measured in a mass-to-charge (m/z) scan range from 50 to 800 with a scan resolution of 13 000 m/z s–1 until the ICC target reached 20 000ms or 200ms, whichever was achieved first. The ionization parameters were as follows: the voltage of the capillary was 4000V and the end plate was set to –500V. The capillary exit was 121V and the Octopole RF amplitude 150 Vpp. The temperature of the dry gas (N2) was 330 °C at a flow of 9 litres min–1. Tandem mass spectrometry (MS) was carried out using helium as the collision gas (3.56×10−6 mbar) with 1V collision voltage. Auto-tandem MS was used to break down the most abundant [M-H]– or [M+HCOO]– ions of the different compounds. Metabolites were identified by their retention times, mass spectra, and product ion spectra compared with data of authentic reference materials. Data analysis was performed using DataAnalysis 3.1 (Bruker Daltonics).
12-Tesla FT-ICR-MS
Ultra-high resolution mass spectra were acquired using a Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer (Solarix, Bruker) with a 12 Tesla superconducting magnet (Magnex Scientific Varian Inc.). Samples dissolved in 70% MeOH were ionized by electrospray ionization (ESI, Apollo II; Bruker Daltonics) at a flow rate of 2 μl min–1. The temperature of the dry gas (N2) was 200 °C at a flow of 2 litres min–1. Mass spectra were recorded in a scan range of 128–1000 m/z with an ion accumulation time of 300ms. A total of 300 scans were accumulated for each MS acquisition. The FT-ICR-MS spectra were normalized by using the exact masses of known plant metabolites including C16 and C18 fatty acids with the Bruker Daltonics data analysis software. For linearization, absolute signal intensities were divided by the maximum amplitude of noise, yielding signal-to-noise (S/N) ratios.
Chemicals
AzA (Sigma Aldrich), ONA (Chrion AS, Norway), and PIM (Roth, Germany) were each dissolved in MeOH and kept at –80 ºC for a maximum period of 3 months.
Chemical SAR induction
The first two true leaves of 4- to 5-week-old plants were syringe-infiltrated with the appropriate concentration of a chemical compound or with fractions derived from plant extracts. Three days later, the next two ‘upper’ or systemic leaves were infiltrated with 105 cfu ml–1 of Pst. Resulting Pst titres were determined at 4 d post-infiltration (dpi) as described (Breitenbach et al., 2014). Primary treatments of plants with 0.1% MeOH, 0.2% DMSO, or chemical-treated water were included as negative controls. Chemical-treated water was generated by mixing equal volumes of PE, DEE, MeOH, and water, and evaporating the mixture to remove PE, DEE, and MeOH.
Cell death assay
Cell death was visualized by Trypan blue staining as described (Aarts et al., 1998) and observed under a light microscope (Olympus BX61).
RNA isolation and qRT–PCR
Total RNA was isolated using TRI-reagent (Sigma Aldrich) according to the manufacturer’s instructions. cDNA was generated using SuperscriptII reverse transcriptase (Invitrogen). Quantitative PCR (qPCR) was performed using the primers 5′CTACGCAGAACAACTAAGAGGCAAC3′ and 5′TTGGCACA TCCGAGTCTCACTG3′ for PATHOGENESIS-RELATED1 (PR1) and 5′GTACCTTGAAGCTTGCTAATCCTA3′ and 5′GTC AAAGGTGCAAAACCAAC3’ for TUBULIN (TUB) with the Sensimix SYBR low-rox kit (Bioline) on a 7500 real-time PCR system (Applied Biosystems). Transcript accumulation was analysed using relative quantification with the 7500 Fast System Software 1.3.1. Presented qPCR results are the average of three technical repetitions per sample ± the standard deviation.
Results
EDS1-dependent SAR is associated with apolar metabolites
It was previously shown that DEX treatment of pDEX:AvrRpm1-HA Col-0 wild type (wt) plants (Mackey et al., 2002) induces expression of AvrRpm1-HA in the treated leaves and EDS1-dependent SAR-like immunity in systemic AvrRpm1-HA-non-expressing leaves (Breitenbach et al., 2014; Fig. 1A). The leaves of pDEX:AvrRpm1-HA plants emitted SAR signals between 4h and 6h after DEX treatment (Breitenbach et al., 2014). Therefore, metabolite profiles in the above-ground tissue of DEX-treated pDEX:AvrRpm1-HA wt and eds1-2 mutant plants harvested at 4–5h after the DEX treatment were compared. First, metabolites were extracted in MeOH and separated into apolar and polar fractions by liquid–liquid extraction using PE followed by DEE (Fig. 1A). Metabolites in the PE and DEE phases were dried by evaporation and dissolved in DMSO. To allow in planta analysis of the SAR-inducing capacity of the metabolites, solutions were diluted with water to a final concentration of 0.2% DMSO. Additionally, PE or DEE remnants were removed from the remaining polar phase.
The SAR-inducing capacity of the different phases isolated from wt and eds1-2 mutant plants was tested after their infiltration into the first two true leaves of Col-0 wt recipient plants. As a positive control, plants were treated with Pst/AvrRpm1. As negative controls, plants were treated with 10mM MgCl2 (mock), 0.2% DMSO, or water treated with the chemicals used for liquid–liquid extraction. Three days later, SAR was measured by a challenge infection of the next two upper or systemic leaves of the treated plants with virulent Pst and quantification of the resulting Pst titres at 4 dpi. Primary treatment of plants with Pst/AvrRpm1 induced SAR, as indicated by reduced Pst titres in the systemic challenge-infected leaves compared with those in the mock-treated control plants (Fig. 1B). A similar degree of systemic resistance was observed in plants that were locally treated with the PE phase derived from DEX-treated pDEX:AvrRpm1-HA wt plants compared with the 0.2% DMSO- and chemical-treated water controls (Fig. 1B). In contrast, the DEE and polar phases from wt plants did not induce SAR. Similarly, the PE, DEE, or polar phases from eds1-2 mutant plants failed to induce SAR in wt plants (Fig. 1B). Thus, non-polar, PE-soluble SAR-inducing metabolites accumulated in extracts from DEX-treated pDEX:AvrRpm1-HA plants in an EDS1-dependent manner.
In the next purification step, metabolites contained in the PE phases from DEX-treated pDEX:AvrRpm1-HA wt and eds1-2 mutant plants were fractionated by SPE (Fig. 1A). C18 columns were loaded with the respective PE phases and consecutively washed with 25, 50, 75, and 100% MeOH followed by a final PE wash. Each wash eluate was dried by evaporation and dissolved in DMSO. Subsequently, the eluates were diluted with water to 0.2% DMSO and infiltrated into the first two true leaves of Col-0 plants. At 3 dpi, systemic leaves were challenged with Pst and the resulting Pst titres were determined at 4 dpi. Compounds derived from wt plants and eluting from C18 columns in 75% and 100% MeOH or in PE induced SAR (Fig. 1C). Compared with the respective negative control treatments, these eluates induced a similar reduction of Pst titres in the systemic challenge-infected tissue as the Pst/AvrRpm1-positive control treatment. In contrast, compounds eluting in 25% or 50% MeOH did not elicit SAR. Similarly, SPE eluates derived from eds1-2 mutant plants did not induce SAR (Fig. 1C). Together, these results confirmed the non-polar nature of EDS1-dependent SAR signalling components. In addition to a reduced capacity to accumulate apolar SAR-inducing compounds, the eds1-2 mutant also did not support systemic resistance in response to the SAR-inducing fractions derived from wt plant extracts (Supplementary Fig. S1 available at JXB online).
HPLC-assisted fractionation of SAR-inducing activities
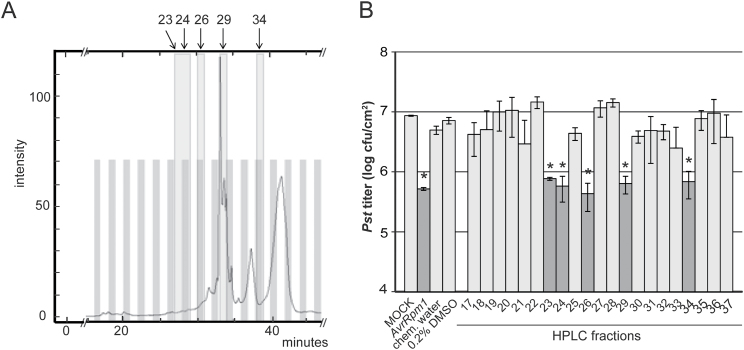
For HPLC, the 75% and 100% MeOH and final PE SPE eluates from 10–20 biologically independent extractions were pooled per plant genotype, dried by evaporation, and dissolved in MeOH. Thus, compounds derived from 30–60g of plant material per genotype were separated across a MeOH gradient in 10–20 consecutive preparative HPLC runs. During each run, 40 fractions were collected of 9–10ml each and the corresponding fractions of consecutive runs were pooled (Fig. 2A; Supplementary Fig. S2A at JXB online). Fractions 17–37 (corresponding to 75–100% MeOH) were dried by evaporation. For SAR assays, the solid matter in each fraction was dissolved in 200–300 μl of DMSO, diluted with water to 0.2% DMSO, and infiltrated into the first two true leaves of wt plants. SAR was then analysed as described above. In the experiment shown in Fig. 2B, primary treatments of plants with HPLC fractions 23, 24, 26, 29, and 34 derived from wt plants induced SAR, causing a reduction of Pst titres in systemic challenge-infected tissue to the same level as the positive control primary treatment with Pst/AvrRpm1. The corresponding HPLC fractions from eds1-2 mutant plants did not induce SAR (Supplementary Fig. S2B), providing further evidence that the HPLC-separable SAR-inducing activities derived from the wt plants are EDS1 dependent. Reciprocally, the SAR-inducing fractions derived from wt plants or the corresponding fractions derived from eds1-2 mutant plants did not induce SAR in eds1-2 mutant plants (Supplementary Fig. S2C), confirming that the eds1-2 mutant also does not respond to EDS1-dependent SAR signals derived from wt plants (Breitenbach et al., 2014).
Fig. 2.
HPLC-assisted separation of SAR-inducing metabolites. (A) UV absorption signal of an MeOH gradient HPLC chromatogram derived from DEX-treated pDEX:AvrRpm1-HA Col-0 plants. The signal intensity at 260nm (y-axis) is shown against the HPLC retention time in minutes (x-axis). One fraction was collected per minute and fractions 17–37 (analysed in B) are shown as alternating grey and white bars. SAR-inducing fractions are further highlighted in light grey and numbered above the panel. (B) SAR bioassay of HPLC fractions 17–37. Col-0 plants were locally treated with 10mM MgCl2 (MOCK), Pst/AvrRpm1 (AvrRpm1), chemical-treated water (chem. water), or 0.2% DMSO as controls or with HPLC fractions 17–37 derived from wt plants (A) in 0.2% DMSO. Three days later, systemic leaves were infected with Pst and the resulting Pst titres are shown 4 d after infection (dpi). Plotted values are the average ±SD of three replicates each. Asterisks above the bars indicate statistically significant differences from the MOCK or 0.2% DMSO controls (*P<0.05, Student’s t-test). This experiment was repeated three times with comparable results.
MS-assisted identification of SAR-related metabolites
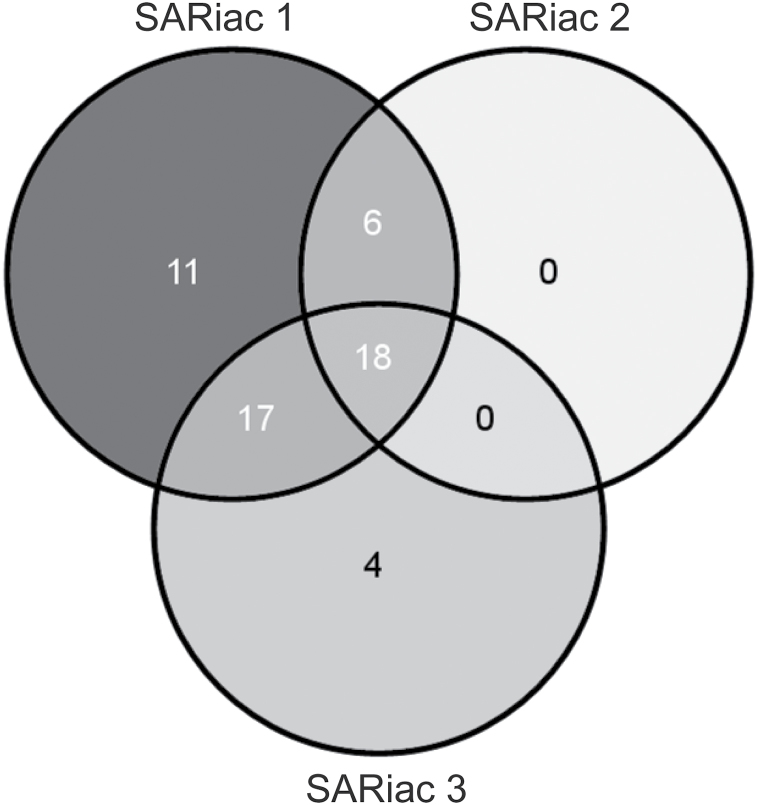
Because the SAR-inducing activity contained in HPLC fractions 23, 24, and 26 sometimes resolved in a single or two HPLC fractions, the SAR-inducing fractions from this range of the HPLC chromatogram were pooled and defined as ‘SAR-inducing activity 1’ (SARiac 1). Fractions 29 and 34 were analysed as SARiac 2 and 3, respectively. First, FT-ICR-MS was used to analyse negatively charged [M-H]– ions in SARiac 1–3 from wt plants compared with the corresponding HPLC fractions from the eds1-2 mutant. Mass spectra were acquired in the negative ionization mode focusing on organic compounds that bear hydroxyl or carboxyl groups and can be easily dissolved in MeOH. The generated mass spectra were normalized and the signal intensities converted to a linear S/N ratio scale (see the Materials and methods). Subsequently, masses were selected that accumulated in SARiac 1–3 in an EDS1-dependent manner if their S/N ratio was at least 5-fold higher in the fractions derived from wt plants compared with corresponding fractions from the eds1-2 mutant. The selected masses were queried against the KEGG, Knapsack, and Human Metabolome DataBase for annotation (Wishart et al., 2007; Afendi et al., 2012; Kanehisa et al., 2014). As a result, 56 annotated masses were found (Supplementary Table S1 at JXB online). Of these, 18 metabolites were associated with SARiac 1–3 (Fig. 3; Table 1). Notably, all of the EDS1-dependent metabolites identified in SARiac 2 were shared with SARiac 1. This result suggests that the separation of compounds by preparative HPLC was suboptimal. Nevertheless, 56 identified metabolites accumulated in SAR-inducing HPLC fractions from DEX-treated pDEX:AvrRpm1-HA plants in an EDS1-dependent manner and therefore are potentially associated with systemic immunity, possibly acting in concert and/or in a concentration-dependent manner.
Fig. 3.
Venn diagram of annotated metabolites accumulating in SAR-inducing activity (SARiac) 1–3 in an EDS1-dependent manner as detected by FT-ICR-MS (Oliveros, 2007). This experiment was repeated twice with similar results.
Table 1.
Putative SAR-related metabolites that are shared between SARiac 1, 2, and 3The identification number (ID) corresponds to numbering in Supplementary Table S1 at JXB online. This experiment was repeated twice with similar results.
| ID | Theoretical mass [M-H]– | Experimental mass [M-H]– | Annotated as | Chemical formula |
|---|---|---|---|---|
| 3 | 243.066285 | 243.066307 | 3,3′,4′5-Tetrahydroxystilbene | C14H12O4 |
| 7 | 269.04555 | 269.045579 | Sulphuretin | C15H10O5 |
| 17 | 315.087415 | 315.087402 | Cajanol | C17H16O6 |
| 20 | 405.11911 | 405.119227 | Astringin | C20H22O9 |
| 21 | 415.10346 | 415.103598 | Daidzin | C21H20O9 |
| 24 | 421.114025 | 421.114185 | Plicatic acid | C20H22O10 |
| 26 | 431.098375 | 431.098472 | Vitexin | C21H20O10 |
| 27 | 431.13476 | 431.134868 | 2-(2,4,5-Trimethoxyphenyl)-5,6,7,8-tetramethoxy-4H-1-benzopyran-4-one | C22H24O9 |
| 28 | 433.114025 | 433.114164 | Phlorizin chalcone | C21H22O10 |
| 29 | 435.09329 | 435.093389 | Irisxanthone | C20H20O11 |
| 32 | 445.114025 | 445.114141 | Biochanin A-β-d-glucoside | C22H22O10 |
| 36 | 449.10894 | 449.109062 | 2′,3,4,4′,6′-Peptahydroxychalcone 4′-O-glucoside | C21H22O11 |
| 38 | 461.10894 | 461.109081 | Isoscoparine | C22H22O11 |
| 43 | 477.103855 | 477.103989 | Isorhamnetin 3-O-β-d-glucopyranoside | C22H22O12 |
| 46 | 491.119505 | 491.119659 | Aurantio-obtusin β-d-glucoside | C23H24O12 |
| 49 | 519.18719 | 519.18737 | Brusatol | C26H32O11 |
| 54 | 563.140635 | 563.141039 | Apigenin 7-O-[β-d-apiosyl-(1→2)-β-D-glucoside] | C26H28O14 |
| 56 | 609.146115 | 609.146474 | Lucenin-2 | C27H30O16 |
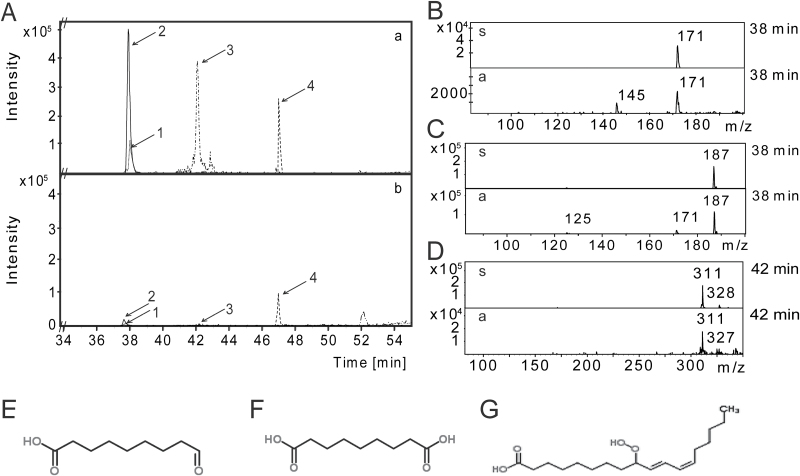
Because an SARiac-associated mass was detected by FT-ICR-MS that might correspond to the putative SAR signal AzA at a signal intensity close to the background noise, SARiac 1 was analysed further by liquid chromatography (LC) coupled with ion trap MS/MS (see the Materials and methods). Using this method, four masses were detected that were predominantly present in SARiac 1 from wt plants compared with corresponding fraction(s) from the eds1-2 mutant (Fig. 4A). By comparing the LC retention times, MS, and MS2 data with different standards, the peaks with pseudo-molecular ions at m/z 171, 187, and 311 were identified as ONA (Fig. 4B, E), AzA (Fig. 4C, F), and 9-HPOD (Fig. 4D, G), respectively. It was not possible to identify the fourth EDS1-dependent metabolite showing a pseudo-molecular ion at m/z 255. In contrast to the 56 metabolites identified by FT-ICR-MS, ONA, AzA, and 9-HPOD were found to be relatively unstable during storage of the samples. SARiac 1, for example, typically lost ONA and AzA and much of the 9-HPOD after 3 months of storage at –80 °C. Notably, this was associated with a loss of SAR-inducing activity. Initial evidence suggested that the SAR-inducing activity of SARiac 2 was similarly related to EDS1-dependent accumulation of ONA and AzA, but not 9-HPOD (Supplementary Fig. S3 at JXB online). Together, the data relate the SAR defect of eds1 mutant plants with reduced accumulation of ONA and AzA.
Fig. 4.
LC-MS analysis of SARiac 1 from DEX-treated pDEX:AvrRpm1-HA Col-0 plants and the corresponding fractions from DEX-treated pDEX:AvrRpm1-HA eds1-2 mutant plants. (A) Intensity peaks (y-axis) detected in the negative ionization mode and their LC retention time in minutes (x-axis) of masses that differentially accumulated in extracts from Col-0 (upper panel, a) and eds1-2 (lower panel, b) plants. (1) ONA, 9-oxo nonanoic acid; (2) AzA, azelaic acid; (3) 9-HPOD, 9-hydroperoxy octadecadienoic acid; and (4) an unknown compound. (B–D) LC-MS of metabolites 1–3 in fractions derived from wt plants (bottom half of each panel) compared with the respective ONA (B), AzA (C), and 9-HPOD (D) standards (upper half of each panel). Mass-to-charge (m/z) ratios are indicated above each peak and LC retention times in minutes to the right of each panel. (E–G) Chemical structures of ONA (E), AzA (F), and 9-HPOD (G) from www.chemspider.com (last accessed July 2014). This experiment was repeated twice with similar results.
Exogenous ONA induces SAR more efficiently than exogenous AzA
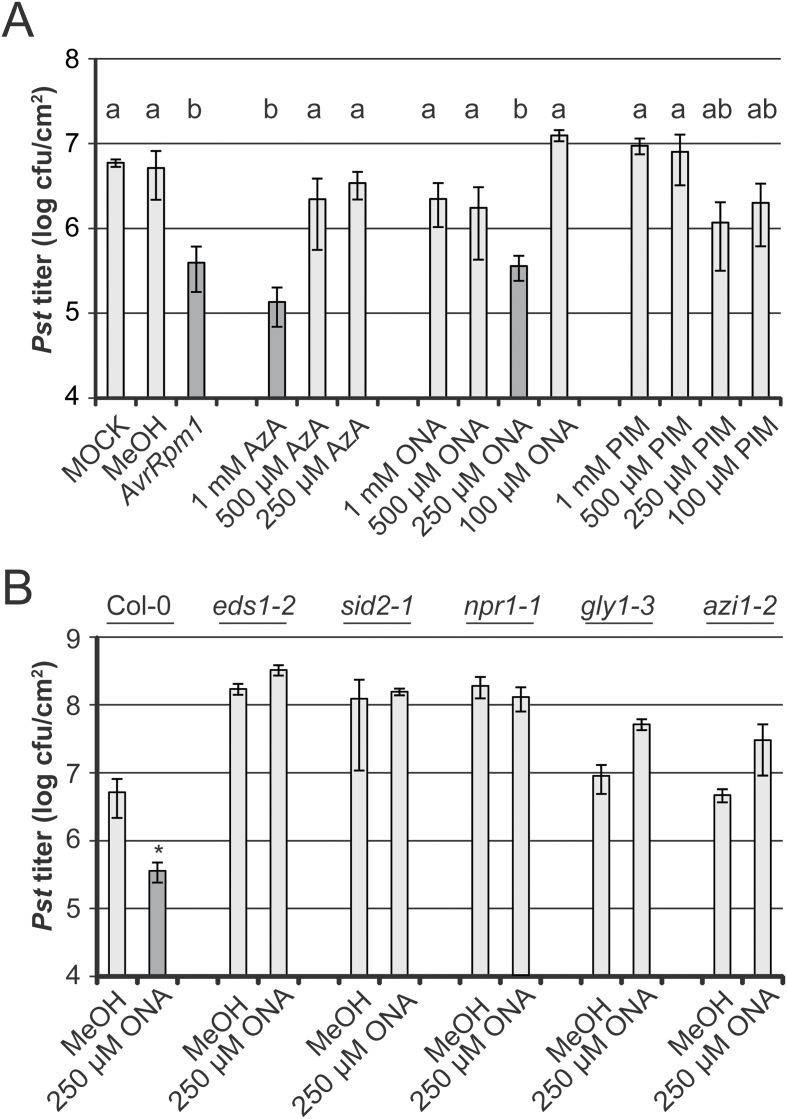
9-HPOD can be fragmented to yield ONA, and exogenous ONA is readily oxidized to AzA in Arabidopsis (Fig. 7; Zoeller et al., 2012; Farmer and Mueller, 2013; Yu et al., 2013; Wang et al., 2014). Additionally, it has been reported that exogenous AzA in Arabidopsis is converted within 24h into the C7 dicarboxylic acid PIM (Zoeller et al., 2012). Here, the SAR-inducing capacity of exogenously applied ONA, AzA, and PIM, was tested, but that of 9-HPOD could not be tested because a reasonable concentration of 9-HPOD in water could not be obtained for plant treatments. The first two true leaves of Col-0 plants were treated with different concentrations of ONA, AzA, or PIM. Alternatively, plants were treated with Pst/AvrRpm1 as a positive control or with 10mM MgCl2 or 0.1% MeOH as negative controls. SAR was analysed as above with a systemic Pst challenge infection. As previously observed (Jung et al., 2009; Chaturvedi et al., 2012), primary treatment of plants with 1mM AzA induced systemic resistance, causing a reduction in systemic Pst titres to a similar degree as the Pst/AvrRpm1 positive control primary treatment (Fig. 5A). Application of lower concentrations of AzA did not elicit SAR. Primary treatment of plants with 250 μM ONA induced systemic resistance to Pst, whereas the application of higher or lower concentrations of ONA did not (Fig. 5A). PIM did not trigger significant SAR when applied at the concentrations tested, although primary treatments of plants with 250 μM or 100 μM PIM induced an SAR trend that was not statistically different from the positive or negative controls (Fig. 5A). Taken together, the data show that application of ONA and AzA but not PIM induces SAR.
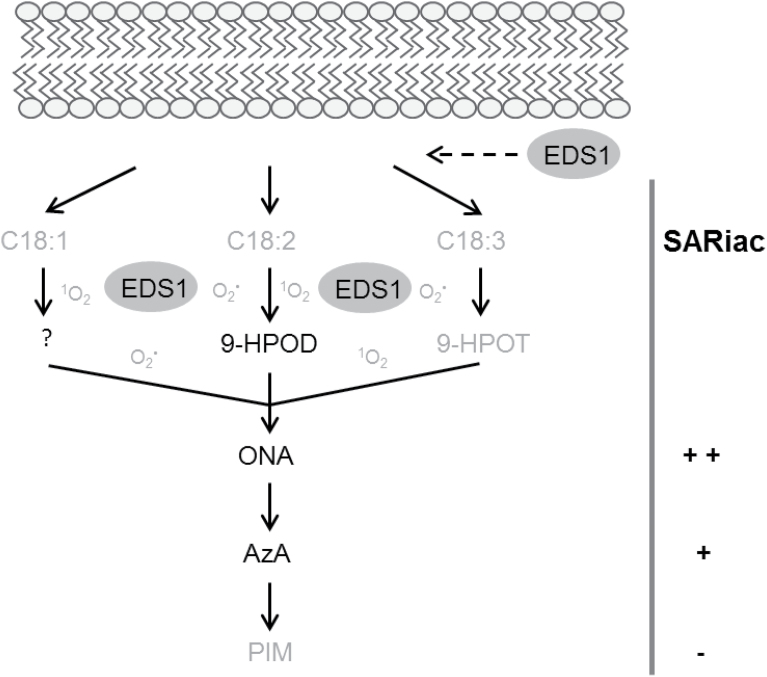
Fig. 7.
Working model. ONA and AzA are generated by peroxidation of C18 unsaturated fatty acids in an EDS1-regulated manner. Compounds identified here as differentially accumulating in extracts from AvrRpm1-HA-expressing wt and eds1 mutant plants are depicted in black; intermediates that were not identified in this study are depicted in grey. EDS1 may directly or indirectly affect the release of C18 unsaturated fatty acids or one or more of their downstream lipid peroxidation products from galactolipid bilayers (striped arrow below the lipid bilayer cartoon). Alternatively, EDS1 may directly or indirectly regulate auto-oxidation of ONA and AzA precursors by regulating ROS homeostasis. The SAR-inducing activity (SARiac) of exogenous ONA, AzA, and PIM is indicated to the right of the cartoon. 1O2, singlet oxygen, O2·–, superoxide radical
Fig. 5.
Induction of systemic resistance by ONA, AzA, and PIM application. (A) SAR bioassay in wt plants. Col-0 plants were locally treated with 10mM MgCl2 (MOCK), 0.1% MeOH, Pst/AvrRpm1 (AvrRpm1), or with different concentrations of AzA, ONA, or PIM as indicated below the panel. Three days later, systemic leaves were infected with Pst and the resulting Pst titres are shown at 4 dpi. Plotted values are the average ±SD of three replicates each. Results marked with different letters above the bars are statistically different (P<0.05, Student’s t-test). (B) ONA-induced SAR in different mutants. Col-0 plants or the mutants indicated above the panel were locally treated with 0.1% MeOH or 250 μM ONA as indicated below the panel. SAR was analysed as in (A). An asterisk above the bar indicates a statistically significant difference from the 0.1% MeOH control (*P<0.05, Student’s t-test). These experiments were repeated at least three times with similar results.
It is currently unclear why ONA did not trigger systemic resistance when applied at 1mM or 500 μM. Because SAR is often but not always associated with primary treatments that induce cell death (Cameron et al., 1994; Durrant and Dong, 2004; Mishina and Zeier, 2007; Liu et al., 2010), it was investigated whether the application of different concentrations of ONA or AzA induced different degrees of cell death by staining the treated leaves with Trypan blue (Supplementary Fig. S4 at JXB online). In contrast to the positive control treatment with Pst/AvrRpm1, which induced cell death, ONA and AzA treatments did not trigger more cell death than the negative control treatments with 10mM MgCl2 or 0.1% MeOH at any ONA and AzA concentration tested (Supplementary Fig. S4). Thus, SAR induced by ONA or AzA application does not appear to be associated with localized cell death, although it cannot be excluded that the accumulation of ONA or AzA during biologically induced SAR might be. Subsequently, the integrity of the commercial ONA used, which was kept in MeOH at –80 °C, was tested. After 3 months of storage, ~16% of ONA was oxidized to AzA, as determined by LC-MS (Supplementary Fig. S5A, B). Infiltration of this mixture into Col-0 leaves caused a rapid further oxidation of ONA within 4h post-infiltration (hpi), supporting previous findings using isotope-labelled ONA that ONA is readily converted to AzA in planta (Zoeller et al., 2012; Supplementary Fig. S5C). However, the possibility that exogenous ONA induced SAR independently of its in planta oxidation cannot be excluded, because ONA remained detectable and elevated compared with its basal level in leaf extracts until 72h after infiltration of leaves with 250 μM ONA (Supplementary Fig. S5C). Alternatively, exogenous ONA may be more membrane permeable than AzA and thus induce SAR via its oxidation, producing similar intracellular AzA accumulation when applied at ~250 μM (Supplementary Fig. S5A) compared with AzA applied at 1mM.
It was next investigated whether ONA contributes to SAR via a similar mechanism to AzA. As eds1-2, the SA biosynthesis mutant sid2-1 and the SA signalling mutant npr1-1 display enhanced susceptibility to Pst and are SAR defective (Fig. 5B; Cao et al., 1997; Wildermuth et al., 2001). Treatment of these and pad4 mutant plants with AzA did not enhance resistance against Pst, indicating that AzA acts upstream of SA (Jung et al., 2009). In the assays performed here, eds1-2, sid2-1, and npr1-1 mutant plants also failed to induce SAR in response to applications of 250 μM ONA (Fig. 5B). AzA-induced resistance was found to be dependent on G3P and AZI1 (Jung et al., 2009; Yu et al., 2013). For comparison, it was tested whether ONA application elicits systemic resistance in the gly1-3 mutant, which is compromised for G3P accumulation and SAR (Chanda et al., 2011), or in azi1-2 mutant plants. Both mutants displayed normal (wt-like) susceptibility to Pst, but did not support SAR in response to the application of 250 μM ONA (Fig. 5B). Taken together, these results suggest that the mechanisms leading to SAR downstream of ONA and AzA application are related since they are dependent on EDS1 and/or PAD4, SA, AZI1, and G3P.
Responses to AzA depend on the AzA concentration applied
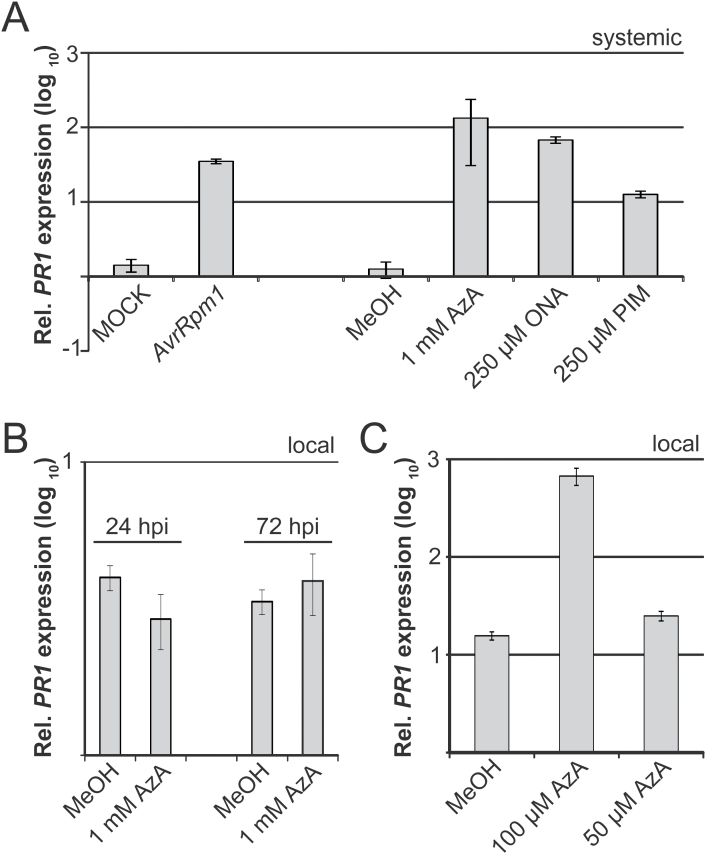
To investigate plant responses to ONA and AzA applications further, the transcript accumulation of the SAR marker gene PR1 was analysed in systemic untreated leaves at 3 d after a local treatment of the plants with 1mM AzA, 250 μM ONA, or 250 μM PIM, and this was compared with a positive control treatment with Pst/AvrRpm1 and negative control treatments with 10mM MgCl2 or 0.1% MeOH. Similar to the positive control treatment, ONA and AzA application induced PR1 transcript accumulation in systemic untreated leaves, whereas a local PIM application caused much lower systemic induction of PR1 transcripts (Fig. 6A). As previously shown (Jung et al., 2009), the same AzA treatment did not enhance local PR1 transcript accumulation in the treated tissue (Fig. 6B). Because up to 7% of exogenous AzA was reported to move systemically in Arabidopsis (Yu et al., 2013), the local response to applications of 50 μM and 100 μM AzA was investigated and it was observed that application of 100 μM but not 50 μM AzA locally induced PR1 transcript accumulation (Fig. 6C). These results indicate that responses to exogenous AzA depend on the AzA concentration applied and that exogenous AzA might induce SAR after travelling from the local treated site to the systemic site.
Fig. 6.
PR1 transcript accumulation in response to ONA, AzA, or PIM application (A) Systemic PR1 induction. Plants were locally treated with 10mM MgCl2 (MOCK), Pst/AvrRpm1 (AvrRpm1), 0.1% MeOH, 1mM AzA, 250 μM ONA, or 250 μM PIM. Three days later, PR1 transcript accumulation in systemic untreated leaves was analysed by qRT–PCR and normalized to that of the reference gene TUBULIN. The normalized expression is shown relative to that in leaf tissue from untreated Col-0 plants. (B, C) Local PR1 transcript accumulation in leaves treated with 0.1% MeOH or AzA at different concentrations as indicated below the panels. PR1 transcript accumulation was analysed as in (A) and samples were taken at the time points indicated in (B) or at 72 hpi (C). These experiments were repeated at least three times with similar results. Rel., relative; hpi, hours post-infiltration
Discussion
The C9 dicarboxylic acid AzA accumulates in infected leaves and petiole exudates of plants infected with P. syringae expressing the effector AvrRpt2 (Jung et al., 2009; Yu et al., 2013). Here, it is shown that AzA accumulates together with its immediate precursor ONA in extracts of AvrRpm1-HA-expressing plants in an EDS1-dependent manner (Fig. 4). Because SAR signal generation, but not local resistance in response to AvrRpm1, is compromised in eds1 mutant plants, these results associate ONA and AzA specifically with SAR rather than local resistance responses (Aarts et al., 1998; Truman et al., 2007; Rietz et al. 2011; Breitenbach et al., 2014). Along with ONA, AzA, and one of their precursors, 9-HPOD, 56 additional annotated metabolites were detected whose accumulation in extracts from AvrRpm1-expressing plants depended on EDS1 (Fig. 3; Supplementary Table S1 at JXB online). However, ONA and AzA appear to be important for the SAR-inducing activity of fractions from plant extracts because their loss during storage of samples correlated with a loss of SAR-inducing activity. Thus, although other EDS1-dependent metabolites may have supportive functions during SAR, the data suggest that the SAR defect of the eds1 mutant is in part caused by reduced accumulation of ONA and AzA.
Exogenous ONA induced SAR when applied at a 4-fold lower concentration compared with AzA (Fig. 5). The data reinforce previous findings that exogenous ONA is rapidly oxidized to AzA in Arabidopsis leaves (Zoeller et al., 2012). Nevertheless, ONA levels remained detectable and above basal levels for at least 72h after its application (Supplementary Fig. S5 at JXB online). Therefore, it is possible that ONA application induces SAR by actions that are independent of AzA. In contrast to AzA, ONA does not appear to accumulate in its free form in plants, but might, for example, remain esterified to galactolipids (Zoeller et al., 2012). In this context, the possibility cannot be excluded that C18 hydroperoxides such as 9-HPOD served as a substrate for fragmentation during the extraction procedure to yield ONA and AzA in AvrRpm1-HA-induced extracts (Figs 1, 2). In contrast, a small proportion of AzA accumulates in its free form in planta (Zoeller et al., 2012). Up to 7% of exogenous 14C-labelled AzA could be detected in systemic tissues away from the site of application, and most of the systemic [14C]AzA was detected in AzA derivatives (Yu et al., 2013; Gao et al., 2014). Gao et al. (2014) questioned the biological significance of AzA mobility in plants and proposed that AzA might enhance systemic resistance via a local function upstream of G3P in the primary infected tissues. In support of this idea, AzA application locally induces transcript accumulation of AZI1 (Jung et al., 2009; Yu et al., 2013), which is required for SAR signal emission from the primary infected leaves, but not for systemic SAR signal perception (Jung et al., 2009). Also, in preliminary experiments, no induction of AZI1 was detected in systemic untreated leaves of plants treated locally with ONA or AzA (Supplementary Fig. S6 at JXB online). A local signalling function of membrane-tethered ONA would fit well with a putative SAR-specific signalling event in the primary infected tissue that is independent of the systemic mobility of AzA. Similar to exogenous AzA, exogenous ONA also appears to depend on accumulation of the putative mobile SAR signal G3P to induce systemic resistance (Fig. 5; Yu et al., 2013). Thus, G3P might be the mobile compound transferring signalling from locally infected to systemic tissues in response to localized actions of ONA or AzA (Gao et al., 2014). Alternatively, exogenously applied ONA might act in SAR via its oxidation to AzA and could elicit SAR when applied at a lower concentration due to its enhanced membrane permeability compared with AzA. Whereas exogenous AzA might act locally, low levels of AzA moving systemically in the plant could suffice for eliciting systemic responses. This is supported by the local PR1 induction observed upon application of 100 μM AzA which was comparable with the level of systemic PR1 induced by a local application of 1mM AzA (Fig. 6).
It was previously proposed that AzA primes immunity by enhancing SA and PR1 transcript accumulation upon P. syringae challenge infection of AzA-treated Arabidopsis leaves (Jung et al., 2009). Priming could be detected from 6h until 18h (for SA) or 24h (for PR1 transcript accumulation) after the challenge infection of the AzA-treated leaves. A second independent study reported a very modest priming effect of AzA on SA and PR1 transcript accumulation detected at 6h or 12h after challenge infection of AzA-treated tissue (Yu et al., 2013). Notably, neither study reported an induction of PR1 transcript accumulation in the AzA-treated leaves before the challenge infection. Here, PR1 transcript accumulation was detected in systemic untreated leaves of plants locally treated with either ONA or AzA (Fig. 6). This induction was similar to that in systemic uninfected leaves of locally Pst/AvrRpm1-infected plants. Sometimes a further priming of PR1 transcript accumulation was detected at 6h after challenge infection of the systemic tissue, but priming was marginal and not always reproducible, and the data were therefore not included here. Application of the AzA fragmentation product PIM was considerably less effective compared with ONA and AzA applications (Figs 5, 6). PIM application moderately induced systemic PR1 expression, but a significant SAR response was not recorded. Although additional fragmentation products derived from C18 unsaturated fatty acids have been associated with systemic resistance (Vicente et al., 2012), the present data suggest that the lipid peroxidation products ONA and AzA promote SAR associated with the systemic accumulation of PR1 transcripts but not necessarily priming. Additionally, the sensitivity of immunity-related responses to the concentration of AzA applied might explain the inverse correlation between AzA levels and the extent of SAR discussed by Zoeller et al. (2012).
Transcriptomic and proteomic studies have investigated EDS1-dependent responses to Pst/AvrRpm1 or AvrRpm1-HA in order to delineate local and SAR-related events (Bartsch et al., 2006; Breitenbach et al., 2014). Until now, three genes identified in these studies have been related to SAR in planta. FLAVIN-DEPENDENT MONOOXYGENASE 1 (FMO1) is essential for SAR, acting upstream of SA in the systemic tissue (Mishina and Zeier, 2006). Locally, FMO1 affects resistance downstream of EDS1, in parallel with SA (Bartsch et al., 2006). APOPLASTIC, EDS1-DEPENDENT 1 (AED1) and LEGUME LECTIN-LIKE PROTEIN 1 (LLP1) act, negatively and positively, respectively, in SAR with limited effects if any on local resistance responses to different P. syringae strains (Armijo et al., 2013; Breitenbach et al., 2014). The data suggest that LLP1 promotes SAR by acting in parallel with SA (Breitenbach et al., 2014). EDS1 was also found to act redundantly with SA in resistance mediated by the CNL receptor HRT (Venugopal et al., 2009), and a related action might regulate the accumulation of ONA and AzA in response to AvrRpm1. Non-enzymatic peroxidation of C18 unsaturated fatty acids is believed to be the main source of AzA in planta (Fig. 7; Zoeller et al., 2012; Yu et al., 2013; Wang et al., 2014). The alternative enzymatic route downstream of 9-lipoxygenase (9-LOX) activity was excluded because a double mutant lacking both Arabidopsis 9-LOX enzymes accumulated normal AzA levels in response to Pst/AvrRpm1 (Zoeller et al., 2012). Recent evidence suggests that peroxidation of C18 unsaturated fatty acids is promoted by ROS downstream of NO (Wang et al., 2014). Because NO and ROS trigger systemic resistance via a pathway acting in parallel with SA and upstream of G3P and presumably AzA (Yu et al., 2013; Wang et al., 2014), it is conceivable that SA-independent ROS-driven accumulation of ONA and AzA is promoted by EDS1 in SAR.
An increasing body of evidence suggests that EDS1-mediated signalling affects ROS homeostasis, for example downstream of the non-canonical CNL protein ACTIVATED DISEASE RESISTANCE1 (ADR1; Roberts et al., 2013). ADR1 promotes SA accumulation in resistance mediated by TNL receptors and the CNL receptor RPS2 but not RPM1 (Bonardi et al., 2011). A function of EDS1 acting in parallel with SA regulates signalling downstream of ADR1 and this might be associated with the role of EDS1 in the run-away cell death (RCD) phenotype of the lesion simulating disease1 (lsd1) mutant (Roberts et al., 2013). RCD in lsd1 mutant plants can be initiated by various biotic and abiotic stresses and is thought to depend on EDS1 promoting H2O2 accumulation (Rustérucci et al., 2001; Mateo et al., 2004; Mühlenbock et al., 2008; Wituszynska et al., 2013). Similar to SAR, AvrRpm1-induced lsd1 RCD does not appear to be associated with localized EDS1-dependent HR-related responses (Rustérucci et al., 2001), suggesting that EDS1 functioning in ROS homeostasis might be associated with SAR. However, H2O2 is probably not a strong enough radical to support fragmentation of C18 unsaturated fatty acids, which is induced in vitro by singlet oxygen (1O2) and to a lesser extent by superoxide radicals (O2·–) but not by H2O2 (Mueller et al., 2006; Farmer and Mueller, 2013; Wang et al., 2014). Available evidence places EDS1 downstream of both 1O2 and O2·– (Ochsenbein et al., 2006; Straus et al., 2010). In the conditional flu mutant that hyperaccumulates 1O2 upon a dark-to-light shift, an EDS1-dependent pathway appears to ‘quench’ 1O2, contributing to recovery of the flu mutant from oxidative stress (Ochsenbein et al., 2006). Additionally, Straus et al. (2010) provided evidence that EDS1 responds to chloroplast-derived O2·– to coordinate SA- and H2O2-associated cell death and immune signalling. Although spatial separation of different ROS and a possible role of EDS1 upstream of 1O2 or O2·– at specific sites (Fig. 7; Straus et al., 2010) cannot be ruled out, a putative role for EDS1 promoting lipid peroxidation requires further investigation.
Alternatively, EDS1 might affect the release of ONA, AzA, or one or more of their common precursors from galactolipids (Fig. 7) because EDS1 and its partner PAD4 each have a conserved esterase catalytic triad embedded within an α/β-fold hydrolase topology (Falk et al., 1999; Wagner et al., 2013). However, mutation of the predicted catalytic residues of EDS1 and PAD4 did not compromise their functions in ETI or basal resistance responses, and no EDS1 hydrolase activity has so far been detected (Wagner et al., 2013). Taken together, it seems likely that EDS1 indirectly promotes ONA and AzA accumulation in SAR, for example by activating one or more signalling pathways. The nudix hydrolase NUDT7 is induced in Arabidopsis by Pst/AvrRpm1 downstream of EDS1 and possibly acts in parallel with SA to suppress immune-related cell death associated with ROS (Bartsch et al., 2006; Straus et al., 2010). However, the nature of EDS1-dependent, possibly SA-independent pathways that promote ONA and AzA accumulation, including a putative role for NUDT7 in SAR, require further investigation.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. SAR bioassays in eds1-2 mutant plants.
Figure S2. HPLC-assisted separation of SAR-inducing metabolites and their dependency on EDS1.
Figure S3. The SAR-inducing activity of SARiac 2 is associated with the accumulation of ONA and AzA.
Figure S4. Trypan blue staining of ONA- and AzA-treated leaves.
Figure S5. LC-MS of ONA after storage at –80 ºC and after infiltration into plants.
Figure S6. Systemic AZI1 expression in response to local ONA and AzA applications.
Table S1. Annotated metabolites identified by FT-ICR-MS in SARiac 1–3.
Acknowledgements
We thank Dr Werner Heller for helpful discussions, and Katrin Franz for technical support. This work was funded by the Deutsche Forschungsgemeinschaft (DFG) as part of SFB924 (grants to ACV and WS) and the Max Planck Society (to JEP). ACV thanks the EU and EMBO for the following fellowships to initiate this work in the lab of JEP: Marie Curie fellowship MEIF-CT-2006–040357 and EMBO fellowship ALTF 137–2006.
Glossary
Abbreviations:
- AzA
azelaic acid
- DEE
diethylether
- DEX
dexamethasone
- DMSO
dimethylsulphoxide
- EDS1
ENHANCED DISEASE SUSCEPTIBILITY1
- ETI
effector-triggered immunity
- FT-ICR
Fourier transform ion cyclotron resonance
- G3P
glycerol-3-phosphate
- HPLC
high-performance liquid chromatography
- 9-HPOD
9-hydroperoxy octadecadienoic acid
- HR
hypersensitive response
- MeOH
methanol
- MS
mass spectrometry
- ONA
9-oxo nonanoic acid
- PAMP
pathogen-associated molecular pattern
- PE
petroleum ether
- PIM
pimelic acid
- PTI
PAMP-triggered immunity
- PR1
PATHOGENESIS-RELATED1
- Pst
Pseudomonas syringae pathovar tomato
- ROS
reactive oxygen species
- SA
salicylic acid
- SAR
systemic acquired resistance
- SPE
solid-phase extraction.
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. 1998. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proceedings of the National Acadamy of Sciences, USA 95, 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afendi FM, Okada T, Yamazaki M, et al. 2012. KNApSAcK family databases: integrated metabolite–plant species databases for multifaceted plant research. Plant and Cell Physiology 53, e1. [DOI] [PubMed] [Google Scholar]
- Armijo G, Salinas P, Monteoliva MI, Seguel A, García C, Villarroel-Candia E, Song W, Van der Krol AR, Álvarez ME, Holuigue L. 2013. A salicylic acid-induced lectin-like protein plays a positive role in the effector-triggered immunity response of Arabidopsis thaliana to Pseudomonas syringae Avr-Rpm1. Molecular Plant-Microbe Interactions 26, 1395–1406 [DOI] [PubMed] [Google Scholar]
- BartschS M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE. 2006. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the nudix hydrolase NUDT7. The Plant Cell 18, 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Halane MK, Kim SH, Gassmann W. 2011. Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334, 1405–1408 [DOI] [PubMed] [Google Scholar]
- Bonardi V, Dangl JL. 2012. How complex are intracellular immune receptor signaling complexes? Frontiers in Plant Science 3, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL. 2011. Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proceedings of the National Academy of Sciences, USA 108, 16463–16468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitenbach HH, Wenig M, Wittek F, et al. 2014. Contrasting roles of apoplastic aspartyl protease APOPLASTIC, ENHANCED DISEASE SUSCEPTIBILITY1-DEPENDENT1 and LEGUME LECTIN-LIKE PROTEIN1 in Arabidopsis systemic acquired resistance. Plant Physiology 165, 791–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron RK, Dixon RA, Lamb CJ. 1994. Biologically induced systemic acquired resistance in Arabidopsis thaliana. The Plant Journal 5, 715–725 [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. 1997. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63 [DOI] [PubMed] [Google Scholar]
- Champigny MJ, Isaacs M, Carella P, Faubert J, Fobert PR, Cameron RK. 2013. Long distance movement of DIR1 and investigation of the role of DIR1-like during systemic acquired resistance in Arabidopsis. Frontiers in Plant Science 4, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda B, Xia Y, Mandal MK, et al. 2011. Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nature Genetics 43, 421–427 [DOI] [PubMed] [Google Scholar]
- Chaturvedi R, Krothapalli K, Makandar R, Nandi A, Sparks AA, Roth MR, Welti R, Shah J. 2008. Plastid omega 3-fatty acid desaturase-dependent accumulation of a systemic acquired resistance inducing activity in petiole exudates of Arabidopsis thaliana is independent of jasmonic acid. The Plant Journal 54, 106–117 [DOI] [PubMed] [Google Scholar]
- Chaturvedi R, Venables B, Petros RA, Nalam V, Li M, Wang X, Takemoto LJ, Shah J. 2012. An abietane diterpenoid is a potent activator of systemic acquired resistance. The Plant Journal 71, 161–172 [DOI] [PubMed] [Google Scholar]
- Dempsey DA, Klessig DF. 2012. SOS—too many signals for systemic acquired resistance? Trends in Plant Science 17, 538–545 [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X. 2004. Systemic acquired resistance. Annual Reviews of Phytopathology 42, 185–209 [DOI] [PubMed] [Google Scholar]
- Falk A, Feys BJ, Frost LN, Jones JDG, Daniels MJ, Parker JE. 1999. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proceedings of the National Academy of Sciences, USA 96, 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Mueller MJ. 2013. ROS-mediated lipid peroxidation and RES-activated signaling. Annual Review of Plant Biology 64, 429–450 [DOI] [PubMed] [Google Scholar]
- Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, Cabral A, Parker JE. 2005. Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. The Plant Cell 17, 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Dong X. 2013. Systemic acquired resistance: turning local infection into global defense. Annual Review of Plant Biology 64, 839–863 [DOI] [PubMed] [Google Scholar]
- Gao Q-M, Kachroo A, Kachroo P. 2014. Chemical inducers of systemic immunity in plants. Journal of Experimental Botany 65, 1849–1855 [DOI] [PubMed] [Google Scholar]
- Garcia AV, Blanvillain-Baufume S, Huibers RP, Wiermer M, Li G, Gobbato E, Rietz S, Parker JE. 2010. Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathogens 6, e1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich K, Wirthmueller L, Tasset C, Pouzet C, Deslandes L, Parker JE. 2011. Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science 334, 1401–1404 [DOI] [PubMed] [Google Scholar]
- Jing B, Xu S, Xu M, Li Y, Li S, Ding J, Zhang Y. 2011. Brush and spray: a high-throughput systemic acquired resistance assay suitable for large-scale genetic screening. Plant Physiology 157, 973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444, 323–329 [DOI] [PubMed] [Google Scholar]
- Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT. 2009. Priming in systemic plant immunity. Science 324, 89–91 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Venugopal SC, Lapchyk L, Falcone D, Hildebrand D, Kachroo P. 2004. Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proceedings of the National Academy of Sciences, USA 101, 5152–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Research 42, D199–D205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-H, Kunz H-H, Bhattacharjee S, et al. 2012. Natural variation in small molecule-induced TIR-NB-LRR signaling induces root growth arrest via EDS1- and PAD4-complexed R protein VICTR in Arabidopsis. The Plant Cell 24, 5177–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PP, Bhattacharjee S, Klessig DF, Moffett P. 2010. Systemic acquired resistance is induced by R gene-mediated responses independent of cell death. Molecular Plant Pathology 11, 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Holt BF, Wiig A, Dangl JL. 2002. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P. 2011. NLR functions in plant and animal immune systems: so far and yet so close. Nature Immunology 12, 817–826 [DOI] [PubMed] [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK. 2002. A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419, 399–403 [DOI] [PubMed] [Google Scholar]
- Mateo A, Mühlenbock P, Rustérucci C, Chang CC-C, Miszalski Z, Karpinska B, Parker JE, Mullineaux PM, Karpinski S. 2004. LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiology 136, 2818–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Zeier J. 2006. The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiology 141, 1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Zeier J. 2007. Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. The Plant Journal 50, 500–513 [DOI] [PubMed] [Google Scholar]
- Mueller MJ, Mène-Saffrané L, Grun C, Karg K, Farmer EE. 2006. Oxylipin analysis methods. The Plant Journal 45, 472–489 [DOI] [PubMed] [Google Scholar]
- Mühlenbock P, Szechynska-Hebda M, Plaszczyca M, Baudo M, Mateo A, Mullineaux PM, Parker JE, Karpinska B, Karpinski S. 2008. Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. The Plant Cell 20, 2339–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarova H, Bernsdorff F, Doring AC, Zeier J. 2012. Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. The Plant Cell 24, 5123–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenbein C, Przybyla D, Danon A, Landgraf F, Gobel C, Imboden A, Feussner I, Apel K. 2006. The role of EDS1 (enhanced disease susceptibility) during singlet oxygen-mediated stress responses of Arabidopsis. The Plant Journal 47, 445–456 [DOI] [PubMed] [Google Scholar]
- Oliveros JC. 2007. VENNY. An interactive tool for comparing lists with Venn Diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html
- Park SW, Kaimoyo E, Kumar D, Mosher S, Klessig DF. 2007. Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318, 113–116 [DOI] [PubMed] [Google Scholar]
- Rietz S, Stamm A, Malonek S, Wagner S, Becker D, Medina-Escobar N, Vlot AC, Feys BJ, Niefind K, Parker JE. 2011. Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytologist 191, 107–119 [DOI] [PubMed] [Google Scholar]
- Roberts M, Tang S, Stallmann A, Dangl JL, Bonardi V. 2013. Genetic requirements for signaling from an autoactive plant NB-LRR intracellular innate immune receptor. PLoS Genetics 9, e1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustérucci C, Aviv DH, Holt BF, III, Dangl JL, Parker JE. 2001. The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. The Plant Cell 13, 2211–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Chaturvedi R, Chowdhury Z, Venables B, Petros RA. 2014. Signaling by small metabolites in systemic acquired resistance. The Plant Journal (in press). [DOI] [PubMed] [Google Scholar]
- Shah J, Zeier J. 2013. Long-distance communication and signal amplification in systemic acquired resistance. Frontiers in Plant Science 4, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Dong X. 2012. How do plants achieve immunity? Defence without specialized immune cells. Nature Reviews Immunology 12, 89–100 [DOI] [PubMed] [Google Scholar]
- Straus MR, Rietz S, Ver Loren van Themaat E, Bartsch M, Parker JE. 2010. Salicylic acid antagonism of EDS1-driven cell death is important for immune and oxidative stress responses in Arabidopsis. The Plant Journal 62, 628–640 [DOI] [PubMed] [Google Scholar]
- Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M. 2007. Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proceedings of the National Acadamy of Sciences, USA 104, 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Katagiri F. 2010. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Current Opinion in Plant Biology 13, 459–465 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. 2009. Network properties of robust immunity in plants. PLoS Genetics 5, e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal SC, Jeong RD, Mandal MK, et al. 2009. Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene-mediated signaling. PLoS Genetics 5, e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente J, Cascon T, Vicedo B, Garcia-Agustin P, Hamberg M, Castresana C. 2012. Role of 9-lipoxygenase and alpha-dioxygenase oxylipin pathways as modulators of local and systemic defense. Molecular Plant 5, 914–928 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. 2009. Salicylic acid, a multifaceted hormone to combat disease. Annual Review of Phytopathology 47, 177–206 [DOI] [PubMed] [Google Scholar]
- Wagner S, Stuttmann J, Rietz S, Guerois R, Brunstein E, Bautor J, Niefind K, Parker JE. 2013. Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host and Microbe 14, 619–630 [DOI] [PubMed] [Google Scholar]
- Wang C, El-Shetehy M, Shine MB, Yu K, Navarre D, Wendehenne D, Kachroo A, Kachroo P. 2014. Free radicals mediate systemic acquired resistance. Cell Reports 7, 348–355 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. 2001. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565 [DOI] [PubMed] [Google Scholar]
- Wishart DS, Tzur D, Knox C, et al. 2007. HMDB: the human metabolome database. Nucleic Acids Research 35, D521–D526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wituszynska W, Slesak I, Vanderauwera S, et al. 2013. LESION SIMULATING DISEASE1, ENHANCED DISEASE SUSCEPTIBILITY1, and PHYTOALEXIN DEFICIENT4 conditionally regulate cellular signaling homeostasis, photosynthesis, water use efficiency, and seed yield in Arabidopsis. Plant Physiology 161, 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Soares JM, Mandal MK, Wang C, Chanda B, Gifford AN, Fowler JS, Navarre D, Kachroo A, Kachroo P. 2013. A feedback regulatory loop between G3P and lipid transfer proteins DIR1 and AZI1 mediates azelaic-acid-induced systemic immunity. Cell Reports 3, 1266–1278 [DOI] [PubMed] [Google Scholar]
- Zhu S, Jeong RD, Venugopal SC, Lapchyk L, Navarre D, Kachroo A, Kachroo P. 2011. SAG101 forms a ternary complex with EDS1 and PAD4 and is required for resistance signaling against turnip crinkle virus. PLoS Pathogens 7, e1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoeller M, Stingl N, Krischke M, Fekete A, Waller F, Berger S, Mueller MJ. 2012. Lipid profiling of the Arabidopsis hypersensitive response reveals specific lipid peroxidation and fragmentation processes: biogenesis of pimelic and azelaic acid. Plant Physiology 160, 365–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.