Summary
Differentially expressed miRNAs and ta-siRNAs between hybrid rice and its parents play important roles in hybrid vigour of super-hybrid rice by regulating their target genes controlling the auxin-mediated signalling pathway.
Key words: Auxin signalling pathway, differentially expressed gene, heterosis, QTL mapping, rice, small RNA, transcriptional profile.
Abstract
Heterosis is an important biological phenomenon; however, the role of small RNA (sRNA) in heterosis of hybrid rice remains poorly described. Here, we performed sRNA profiling of F1 super-hybrid rice LYP9 and its parents using high-throughput sequencing technology, and identified 355 distinct mature microRNAs and trans-acting small interfering RNAs, 69 of which were differentially expressed sRNAs (DES) between the hybrid and the mid-parental value. Among these, 34 DES were predicted to target 176 transcripts, of which 112 encoded 94 transcription factors. Further analysis showed that 67.6% of DES expression levels were negatively correlated with their target mRNAs either in flag leaves or panicles. The target genes of DES were significantly enriched in some important biological processes, including the auxin signalling pathway, in which existed a regulatory network mediated by DES and their targets, closely associated with plant growth and development. Overall, 20.8% of DES and their target genes were significantly enriched in quantitative trait loci of small intervals related to important rice agronomic traits including growth vigour, grain yield, and plant architecture, suggesting that the interaction between sRNAs and their targets contributes to the heterotic phenotypes of hybrid rice. Our findings revealed that sRNAs might play important roles in hybrid vigour of super-hybrid rice by regulating their target genes, especially in controlling the auxin signalling pathway. The above finding provides a novel insight into the molecular mechanism of heterosis.
Introduction
Heterosis, or hybrid vigour, is the superior performance of hybrids in biomass, resistance, and fertility compared with their parents (Shull, 1908; Bruce, 1910). Heterosis has been widely applied in the breeding of agricultural crops, and its underlying mechanism has been studied for over 100 years (Darwin, 1876). Hybrid rice is one of the most successful examples taking advantage of heterosis in crop breeding. Recent statistics showed that the planting area of hybrid rice accounts for more than 50% of the total rice area in China (Cheng et al., 2007), further indicating that hybrid rice plays an important role in international food security by increasing grain yield. Therefore, there is an urgent need to explore the molecular mechanisms associated with heterosis, to provide a foundation for further utilization of heterosis. Rice has a relatively small genome size, high-quality sequences, and has colinearity with other cereal species in the Poaceae family (Gale and Devos, 1998; International Rice Genome Sequencing, 2005). Importantly, there are also many databases and bioinformatic tools for rice, including rice genome annotation and gene expression, pathway, mutant library, and phenotype information data (Zhang et al., 2013). Thus, hybrid rice is an optimal model plant for genome-wide studies of heterosis.
The mechanism of heterosis has been studied extensively and there are three classic hypotheses to explain heterosis, with each explaining the genetic mechanism to some extent (Goff and Zhang, 2013): the dominance (Bruce, 1910; Jones, 1917), overdominance (Shull, 1908; East, 1936), and epistasis (Schnell and Cockerham, 1992; Yu et al., 1997) hypotheses. With the rapid development of genomic tools, the molecular mechanism of heterosis has been further investigated at the ‘omics’ level. Genome-wide comparative transcriptional profiling between hybrids and their parental lines has been studied in many plants, such as Arabidopsis (Wang et al., 2006), maize (Swanson-Wagner et al., 2006), rice (Huang et al., 2006b ; Wei et al., 2009; Song et al., 2010; Zhai et al., 2013), and Medicago sativa (Li et al., 2009b ). These studies have revealed that the differentially expressed genes (DGs) caused by the interaction of two different sets of parental genomes are potentially involved in controlling the superior performance of heterosis (Zhang et al., 2013). In addition, genomic dosage effects (Yao et al., 2013), allelic variations (Springer and Stupar, 2007; Schnable and Springer, 2013), and epigenetic modifications (He et al., 2013) also contribute to the molecular mechanism of heterosis.
Non-coding small RNAs (sRNAs) in plants, mainly including the microRNAs (miRNAs) (Bartel, 2004) and small interfering RNAs (siRNAs) (Hamilton and Baulcombe, 1999), play important roles in development, stress response (Mallory and Vaucheret, 2006), and heterosis (Baranwal et al., 2012; Ng et al., 2012; Chen, 2013) by inhibiting gene expression. Ha et al. (2009) first reported sRNAs involved in the molecular mechanism of heterosis in Arabidopsis interspecific hybrids and allopolyploids. Their results suggested that the expression variation of miRNAs, as well as a new class of endogenous siRNAs, trans-acting siRNAs (ta-siRNAs), derived from the TAS loci via the miRNA-dependent biogenesis pathway (Allen et al., 2005; Liu et al., 2007), led to changes in target gene expression and might result in the phenotypic variation in hybrids. He et al. (2010) and Chen et al. (2010) compared the expression profiles of sRNAs in seedlings between two rice subspecies (japonica cv. Nipponbare and indica cv. 93-11) and their reciprocal hybrids by high-throughput sequencing and microarray technology, respectively, and found differential expression of miRNAs between hybrids and their parents. The miRNA transcriptomes were also studied in cultivated and wild tomato seedlings and their hybrids (Shivaprasad et al., 2012), as well as in embryos of a maize hybrid and its parental inbred lines (Ding et al., 2012). However, the interaction between miRNAs and their potential target genes in heterosis of hybrid rice is poorly described.
Previously, we investigated the transcriptional and physiological metabolism changes between super-hybrid rice and its parents (Wei et al., 2009; Song et al., 2010) and found that DGs were significantly enriched in photosynthesis and carbon-fixation pathways, and most of the key genes in the carbon-fixation pathway were upregulated in F1 hybrid rice. DGs were mapped to the yield-related quantitative trait loci (QTL), and were involved in the circadian-rhythms and light-signalling regulatory network, suggesting a relationship between DGs and phenotypic changes in hybrid rice. In this study, to determine the roles of sRNAs and their interaction with their target genes in heterosis of hybrid rice, we performed transcriptomic analysis of sRNA and mRNA of flag leaves and panicles of the F1 super-hybrid rice LYP9 and its parents at the grain-filling stage, using next-generation sequencing technology.
Materials and methods
Plant materials
Super-hybrid rice Liang-You-Pei-Jiu (LYP9) and its parental lines, the sterile line Pei-Ai64s (PA64s) and the restorer line 93-11 (Lu and Zou, 2000; Yu et al., 2002), were planted in a rice field under the same environmental conditions. Each sample had at least three biological replications to minimize systematic errors, with around six to eight flag leaves and panicles per plant, and were harvested at the grain-filling stage. All the plant tissues were pooled in each sample, frozen in liquid nitrogen and stored at –80 °C for total RNA extraction.
sRNA library construction and sequencing
Total RNAs were extracted from the collected flag leaf and panicle samples at the grain-filling stage of the hybrid rice and its parents using TRIzol Reagent (Invitrogen) according to the manufacturer’s protocol. sRNAs of 18–30 nt were isolated from total RNA by polyacrylamide gel electrophoresis, and ligated to sequencing adapters for reverse transcription (RT)-PCR amplification to generate sRNA libraries as described previously (Hafner et al., 2008). sRNA libraries were subjected to Illumina deep sequencing at Beijing Genomics Institute (Shenzhen, China).
Sequencing data processing and analysis
After removing the low quality reads and reads <18 nt, and trimming adaptor sequences, clean sequencing reads from sRNA libraries were summarized for length distribution and common/specific sequences between samples, and aligned with the Oryza sativa L. ssp. indica cv. 93-11 genome (Yu et al., 2002) using the short oligonucleotide analysis package (SOAP) release 2.20 (Li et al., 2009a ).
Annotation and classification of sRNAs
sRNA clean reads were aligned with non-coding RNAs [ncRNAs, including rRNAs, tRNAs, small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs)] in NCBI GenBank release 189 (ftp://ftp.ncbi.nih.gov/genbank/) and Rfam release 10.1 (http://rfam.sanger.ac.uk/); rice miRNA precursors in miRBase release 19 (http://www.mirbase.org/); rice trans-acting siRNA 3 (TAS3) genes (Zhu et al., 2008), rice natural antisense transcripts (NATs) in PlantNATsDB release 1.3 (Chen et al., 2012), repeat sequences extracted from rice genome by RepeatMasker release 3.2.9 (http://www.repeatmasker.org/); and gene transcripts in Rice Genome Annotation release 6.1 (Ouyang et al., 2007), for ncRNA, miRNA, ta-siRNA, natural antisense transcript-generated siRNA (nat-siRNA), repeat-associated siRNA (ra-siRNA), and protein-coding gene annotation, respectively. sRNA annotation followed a priority rule for classification to avoid redundancy: ncRNA (in which GenBank > Rfam) > known miRNA > ta-siRNA > cis-nat-siRNA > trans-nat-siRNA > ra-siRNA > exon > intron.
Expression profiling of known miRNAs and ta-siRNAs
The distinct clean reads of sRNAs that perfectly matched the mature sequences of known rice miRNAs (miRBase release 19) and ta-siARFs (Zhu et al., 2008) were used to estimate the expression level of each rice miRNA and ta-siRNA. The initial counts of clean reads from different samples were normalized into reads per million mapped reads (RPM) for expression amounts. Differentially expressed sRNAs (DES) were analysed by comparing the expression amounts between hybrid and the mid-parental value (MPV), using the edgeR package (Robinson et al., 2010) version 3.5.27. The resultant P values were adjusted for false discovery rate (FDR) and only adjusted P values of ≤0.01 were considered statistically significant. The hierarchical clustering tree of DES in different libraries was generated by MultiExperiment View (Saeed et al., 2006) version 4.9 with the average linkage method.
RNA sequencing (RNA-Seq) analysis
The total RNAs were extracted as described above, with the same materials as used in the sRNA sequencing. The oligo(dT)-enriched mRNA was fragmented into short fragments of approximately 200 nt for cDNA synthesis and sequencing adapters ligation. The libraries were subjected to Illumina deep sequencing at Beijing Genomics Institute. After quality control, raw reads of RNA-Seq were filtered into clean reads and aligned with the gene models in the Rice Genome Annotation release 6.1 with SOAP release 2.20 (Li et al., 2009a ). The gene expression level was calculated by using reads per kb of transcriptome per million mapped reads (RPKM) method (Mortazavi et al., 2008), with unique position matched reads. DGs were defined by comparing the expression levels between hybrid and MPV, using the edgeR package (Robinson et al., 2010) version 3.5.27, with a significance threshold of FDR-adjusted P values of ≤0.01. Functional category analysis of DGs was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology-based annotation system (KOBAS) version 2.0 (Xie et al., 2011), using the NCBI Entrez gene ID of DGs as input.
Prediction and functional analysis of target genes
Target genes of DES were predicted from the gene transcripts in the Rice Genome Annotation release 6.1 (Ouyang et al., 2007) by the plant sRNA analysis toolbox psRobot (Wu et al., 2012) with the additional threshold of target site conservation and degradome data supported. Correlations between the expression level of DES and corresponding DG targets were analysed via the Pearson correlation coefficient method (Rodgers and Nicewander, 1988). Transcription factor (TF) annotations and classifications of DES targets were according to the PlantTFDB release 2.0 (Zhang et al., 2011). Gene Ontology (GO) enrichment analysis of DES targets was performed using the rice oligonucleotide array database (ROAD) web tools (Cao et al., 2012) with a significance threshold of P≤0.01 calculated by the hypergeometric statistical test method.
Real-time quantitative RT-PCR (qRT-PCR) analysis
Total RNA was extracted from flag leaves at the grain-filling stage of the LYP9 hybrid rice combination, and used as template for reverse transcription with miRNA-specific stem–loop RT primers (Chen et al., 2005) or gene-specific RT primers (Supplementary Table S12 at JXB online) to synthesize the first-strand cDNAs, which were used for SYBR Green (Invitrogen)-monitored qRT-PCR performed on an ABI PRISM 7900HT Fast Real-Time System (Applied Biosystems). The experiment was performed with three biological replicates, using U6 snRNA and ACT1 as internal references for qRT-PCR of DES and target genes, respectively. The relative expression values of DES or genes in different samples were calculated by the 2–ΔΔCT method (Livak and Schmittgen, 2001).
Regulatory network analysis
Pathway Studio software (Nikitin et al., 2003) version 9.0 (Ariadne Genomics, Elsevier) was applied to build the regulatory network of DES and their target genes with the NCBI Entrez gene ID (Maglott et al., 2011) as input, via global literature analysis by searching the direct interactions in the ResNet Plant Database version 4.0 release 2012H2.
Mapping the rice genes to QTL
Rice QTL data, including physical positions on the MSU rice genome release 6 and classification of trait category, were obtained from the Gramene database (Youens-Clark et al., 2011). Coordinates of rice MIR and TAS3 genes were acquired by alignment with the MSU rice genome release 6 (Ouyang et al., 2007) using the BLAST-Like Alignment Tool (BLAT) (Kent, 2002). Based on the genomic positions of both gene loci and QTL, rice genes were mapped to the QTL. QTL of small intervals of <100 genes were extracted and subjected to co-localization with genes in the rice chromosomes. P values in QTL trait enrichment analysis of DES and target genes in QTL of small intervals were calculated by the hypergeometric test of the GeneProf webtool (Halbritter et al., 2012), using total gene loci including MIR and TAS3 loci in rice genome as reference.
Accession numbers
The high-throughput sequence data reported in this paper has been deposited in GEO with accession number GSE51468.
Results
sRNA sequencing and data processing
Flag leaves and panicles at the grain-filling stage play important roles in rice yield. To dissect the role of sRNAs in hybrid vigour, we constructed sRNA sequencing libraries of flag leaves and panicles of the super-hybrid rice LYP9 combination at the grain-filling stage. The above hybrid rice combination includes F1 hybrid LYP9 and its parental lines including the male-sterile line PA64s and the restorer line 93-11. After sequencing, we obtained 63 356 633 high-quality 18–30 nt sRNA clean reads, in which 10 963 733 (PA64s), 12 717 274 (93-11), and 11 462 878 (LYP9) reads were from flag leaves, and 8 474 866 (PA64s), 9 147 347 (93-11), and 10 590 535 (LYP9) reads were from the panicles, respectively; these corresponded to 2 982 601, 3 652 768, 3 214 069, 3 407 271, 3 534 763, and 3 854 243 unique reads in each library (Supplementary Table S1 and Fig. S1 at JXB online). Of the sRNA reads in these libraries, 74.1–89.1% of total reads, corresponding to 48.2–81.4% unique reads, matched the indica rice 93-11 genome perfectly (Supplementary Table S1).
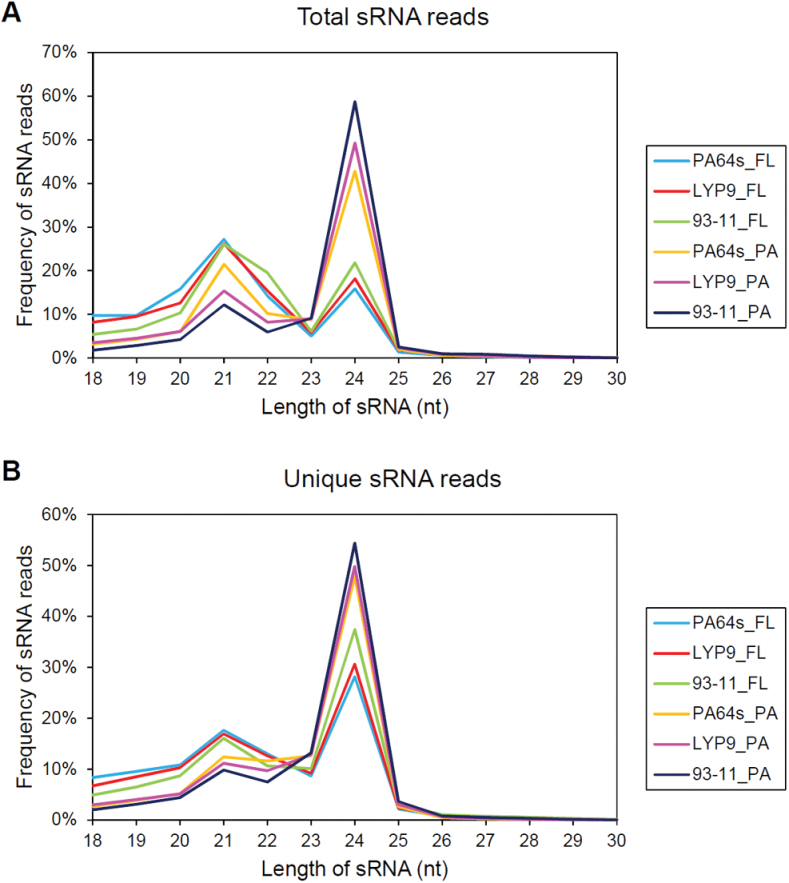
In these libraries, 84.6% on average of the total sRNAs were 20–24 nt (a typical size of Dicer-derived sRNAs; Henderson et al., 2006), and of these the 21 and 24 nt sRNAs formed the majority. In the libraries of flag leaves, numbers of 21 nt sRNAs slightly exceeded those of 24 nt sRNAs, whereas in panicles, 24 nt sRNAs were much more numerous than 21 nt sRNAs (Fig. 1A). Of the unique reads, the 24 nt sRNAs were most abundant in all six libraries (Fig. 1B).
Fig. 1.
Length distribution of sRNAs. (A) Total sRNAs in different libraries. (B) Unique sRNAs in different libraries. FL, flag leaves. PA, panicles.
To annotate and classify the total sRNAs, the sRNA clean reads were first aligned with the known ncRNAs, including rRNAs, tRNAs, snRNAs, and snoRNAs, in the GenBank and Rfam databases (Supplementary Table S1). Then the remaining sRNAs reads were aligned with the known miRNA precursors of rice in miRBase for miRNA annotation. The results showed that 1 859 743, 1 876 542, and 2 654 408 miRNA reads were detected in flag leaves of PA64s, LYP9, and 93-11, respectively, with much lower corresponding numbers of 844 637, 682 363, and 452 645 in panicles (Supplementary Table S1).
The remaining sRNA reads were in turn aligned with the rice TAS3 genes, NATs, and the repeat sequences in the rice genome, for ta-siRNA, nat-siRNA, and ra-siRNA annotations, respectively (Supplementary Table S1), for the classifications of the three types of endogenous siRNAs with different biogenesis and functions (Vazquez et al., 2010). There were more ta-siRNAs in panicles than in flag leaves, and the four rice TAS3 genes showed differential expression with each other in the different tissues. In flag leaves, TAS3a1 had the highest expression level followed by TAS3b1, with TAS3a2 and TAS3b2 showing very low expression. The order of expression levels in panicles from high to low was TAS3b1, TAS3b2, TAS3a1, and TAS3a2 (Supplementary Table S1). The ra-siRNAs were the second most abundant sRNAs in flag leaves after miRNAs, and were the most abundant sRNAs in panicles – they were followed in both tissues by nat-siRNAs, of which most were trans-nat-siRNAs (Supplementary Table S1).
Expression profiling of known miRNAs and ta-siARFs in hybrid rice LYP9 and its parents
We focused on the two kinds of sRNAs, miRNAs and ta-siRNAs (ta-siARFs, the functional ta-siRNAs), which usually direct the mRNA cleavage of target protein-coding genes (Mallory and Bouche, 2008).
Through mapping to the 547 distinct mature miRNA sequences of the 591 known rice miRNA genes in miRBase version 19 and five distinct mature ta-siARF sequences of the four TAS3 genes (Zhu et al., 2008), a total of 355 sRNAs comprising 350 miRNAs and five ta-siARFs were detected in either flag leaves or panicles. Of these, 244 sRNAs, including 241 miRNAs and three ta-siARFs, were detected in both tissues (Supplementary Table S2 and Fig. S2A at JXB online). Based on the identified significance (Robinson et al., 2010) of the expressed sRNAs, 45 and 47 DES between hybrid and MPV were identified in flag leaves and panicles, respectively (Supplementary Tables S3 and S4). This yielded in total 69 DES, comprising 67 miRNAs and two ta-siARFs, in both tissues. This result indicated the obvious expression changes of sRNAs between the F1 hybrid and its parents. Among them, 23 DES were detected in both tissues, but 22 of 45 (48.9%) unique DES in flag leaves and 24 of 47 (51.1%) in panicles, respectively, which exhibited the tissue-specificity of DES (Supplementary Fig. S2B).
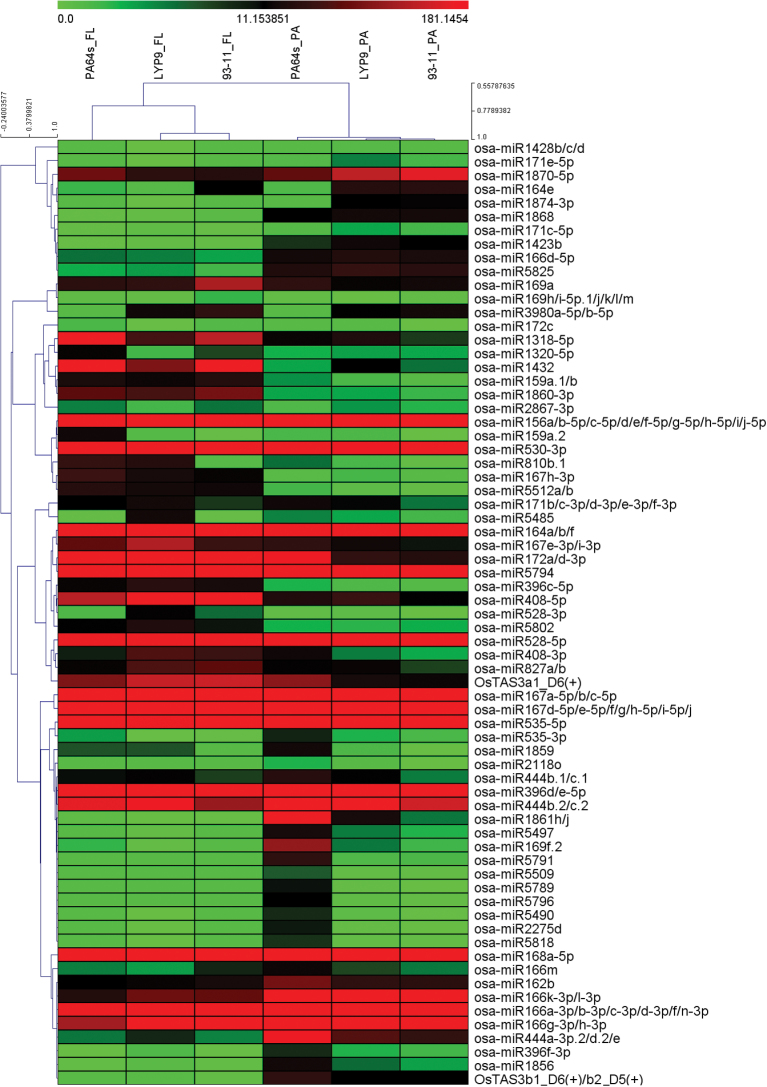
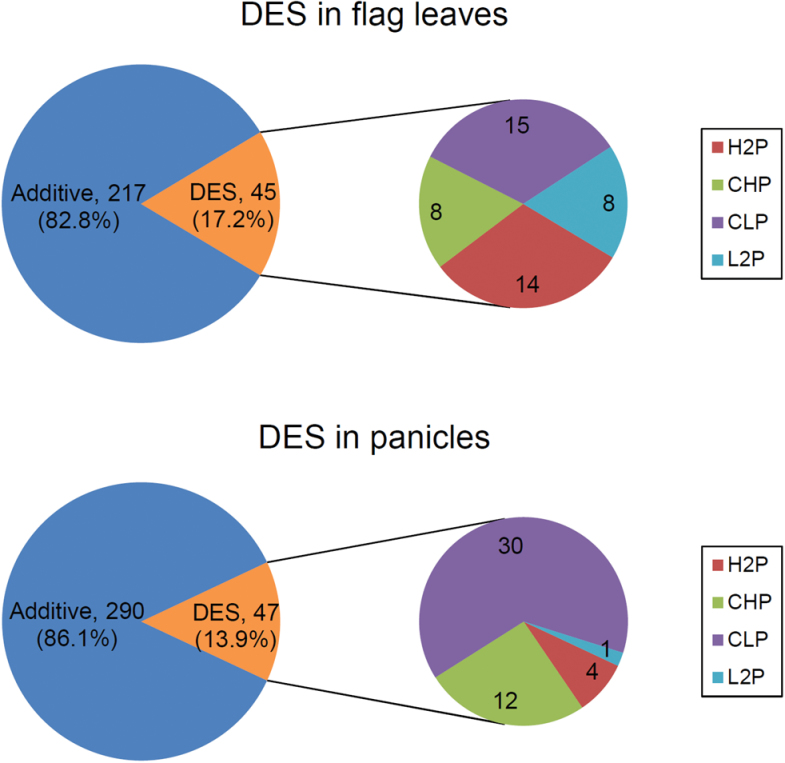
Hierarchical cluster analysis was used to investigate the similarity of DES expression profiles, and showed that libraries from different cultivars of the same tissue formed the primary groups. The DES profiles of LYP9 were much closer to paternal line 93-11 than to maternal line PA64s, both in flag leaves and in panicles at the grain-filling stage. This was consistent with our previous study (Wei et al., 2009) on the transcriptional profiles of the LYP9 hybrid combination (Fig. 2). Comparison of DES expression levels between the F1 hybrid and its parents enabled classification of the DES into four patterns: higher than both parents (H2P), close to higher parent (CHP), close to lower parent (CLP), and lower than both parents (L2P) (Fig. 3). Similar to our previous result for transcriptional profiling of the LYP9 hybrid rice combination (Wei et al., 2009), the CHP and CLP expression patterns of DES in panicles accounted for the majority of all DES (89.4%, Fig. 3).
Fig. 2.
Hierarchical cluster analysis of DES. The hierarchical clustering tree of 69 DES in different libraries of flag leaves (FL) and panicles (PA) was generated by MultiExperiment View version 4.9 using the average linkage clustering method. Read abundance (RPM) is denoted by colour; red and green represent high and low expression levels, respectively.
Fig. 3.
Expression patterns of DES in flag leaves and panicles. Additive represents additively expressed sRNAs without significant differential expression.
RNA-Seq analysis of super-hybrid rice LYP9 and its parental lines
To correlate DES with their target genes, we also compiled transcript profiles of flag leaves and panicles of LYP9 hybrid rice and its parental lines (the same materials used in the sRNA analysis) by RNA-Seq. Mapping the RNA-Seq reads to the gene models in Rice Genome Annotation release 6.1 (Ouyang et al., 2007) gave 11 107 734, 11 526 473, and 11 709 849 unique-matched reads of PA64s, LYP9, and 93-11 from flag leaves, respectively; and corresponding values of 11 448 322, 11 949 274 and 12 333 163 from panicles (Supplementary Table S5 at JXB online). Of the detected 40 754 transcripts either in flag leaves or in panicles of the LYP9 hybrid rice combination (Supplementary Fig. S2C), 7782 (23.2%) and 7629 (19.7%) were differentially expressed between the hybrid and MPV in flag leaves and panicles, respectively (Supplementary Tables S6 and S7 at JXB online). Of these, 3229 transcripts were differentially expressed in both tissues (Supplementary Fig. S2D). Among the four expression patterns of DGs, the CHP and CLP patterns were dominant in panicles (69.1%, Supplementary Fig. S3B at JXB online), which was similar to the DES patterns.
We employed KEGG annotations to reveal the potential functions of DGs in the hybrid rice LYP9 combination. Functional category analysis by KOBAS (Xie et al., 2011) showed that DGs were involved in 18 functional categories including two of the mostly enriched categories – carbohydrate metabolism and energy metabolism (Supplementary Fig. S3C), which was consistent with our previous result (Wei et al., 2009).
Correlation analysis between DES and their potential target genes
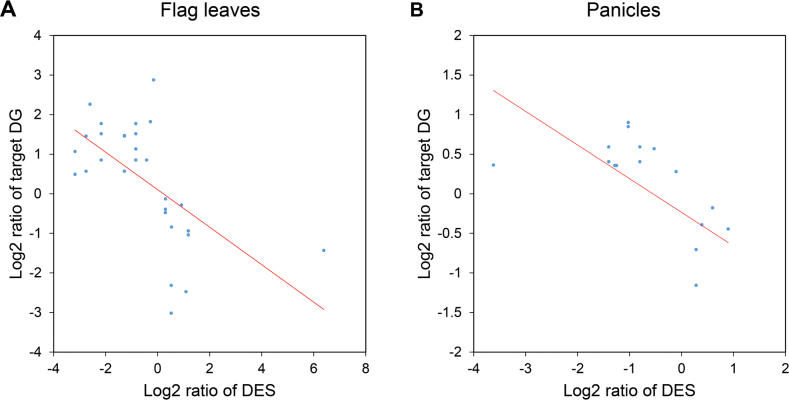
The target genes of DES were predicted from the gene models in Rice Genome Annotation release 6.1 (Ouyang et al., 2007) via psRobot (Wu et al., 2012) analysis. The result indicated that 34 of 69 DES had recognized target genes (Table 1), and a total of 176 target gene transcripts were predicted (Supplementary Tables S8 and S9 at JXB online), among which 123 (69.9%) transcripts were common to both tissues (Table 1). We further investigated the correlations between DES and DG targets by the Pearson correlation coefficient method (Rodgers and Nicewander, 1988). There were significant negative correlations between the expression levels of 23 (67.6%) DES either in flag leaves or in panicles and corresponding target DGs (Fig. 4 and Supplementary Table S9).
Table 1.
Annotation of DES target genes
| Sample name | DESa | Target transcripts | Target loci | TF transcripts (%)b | TFc | TF family |
|---|---|---|---|---|---|---|
| Totald | 34 | 176 | 110 | 112 (63.6) | 94 | 13 |
| Flag leaves | 25 | 145 | 94 | 102 (70.3) | 86 | 13 |
| Panicles | 25 | 154 | 92 | 103 (66.9) | 86 | 11 |
| Common parte | 16 | 123 | 76 | 93 (69.6) | 78 | 11 |
a DES with predicted target genes by psRobot analysis (Wu et al., 2012).
b TF annotations of DES targets were according to PlantTFDB (Zhang et al., 2011). Results in parentheses show the percentage of TF transcripts in total predicted DES target transcripts.
c TFs encoded by the DES target transcripts according to PlantTFDB.
d Union of flag leaves and panicles.
e Intersection of flag leaves and panicles
Fig. 4.
Expression correlation analysis between DES and target DGs in flag leaves (A) and in panicles (B). The Pearson correlation coefficient method (Rodgers and Nicewander, 1988) was used in the correlation analysis.
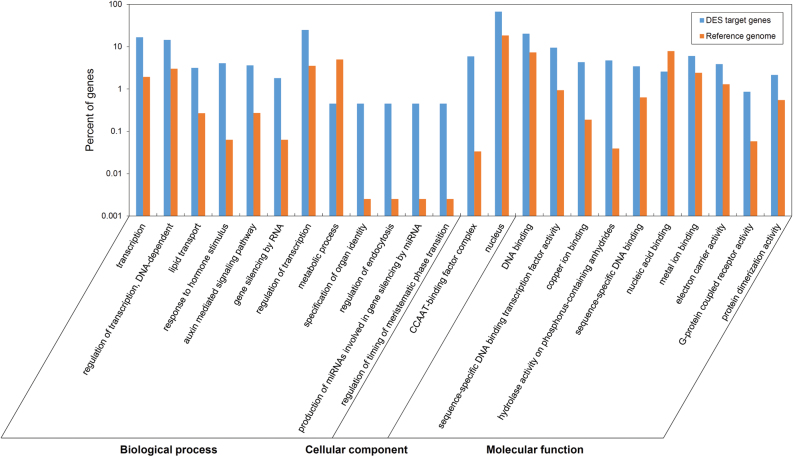
According to the plant transcription factor database PlantTFDB (Zhang et al., 2011), the DES targets annotation showed that 112 (63.6%) of the target genes encoded 94 TFs from 13 families (Table 1). GO enrichment analysis by ROAD (Cao et al., 2012) showed that the target genes of DES either in flag leaves or in panicles were significantly enriched in 11 biological processes including the auxin-mediated signalling pathway and regulation of transcription, two cellular components including the nucleus and the CCAAT-binding factor complex, and nine molecular functions including sequence-specific DNA-binding TF activity, compared with total genes in the rice genome (P≤0.01, Fig. 5).
Fig. 5.
Significantly enriched biological processes, cellular components, and molecular functions identified by GO analysis of DES target genes. The significant GO terms (P≤0.01) of target genes were plotted, with the whole genome as the background reference.
Validation of the DES and target genes expression
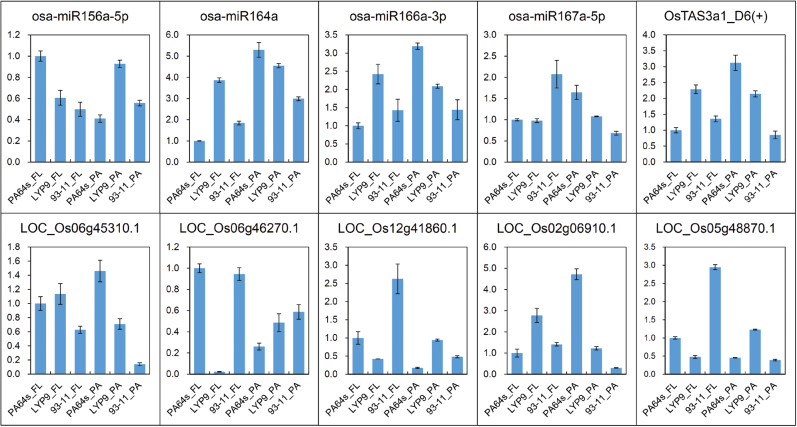
To validate the expression levels of DES identified in the sRNA sequencing and their potential targets, five DES and five of their target genes were randomly selected for examination by real-time qRT-PCR in flag leaves and panicles of the LYP9 hybrid rice combination. qRT-PCR revealed that all five sRNA mature sequences in flag leaves [osa-miR156a-5p, osa-miR164a, osa-miR166a-3p, osa-miR167a-5p, and OsTAS3a1_D6(+)], as well as three of four DES in panicles (osa-miR156a-5p, osa-miR166a-3p, and osa-miR167a-5p), showed similar expression patterns with the sequencing data (Fig. 6), demonstrating the high quality of sRNA sequencing. Moreover, expression of the potential target genes generally had opposite trends to the corresponding DES (Fig. 6), which confirmed the negative regulation by DES.
Fig. 6.
qRT-PCR analysis of DES and target genes. U6 snRNA and ACT1 were used as internal references for the qRT-PCR of DES mature sequences and target gene transcripts (corresponding vertical lower figure), respectively. The y-axis represents the relative gene expression level in different samples. Error bars indicate the standard deviation (SD) for three biological replicates. FL, flag leaves. PA, panicles.
Regulatory network between DES and their target genes
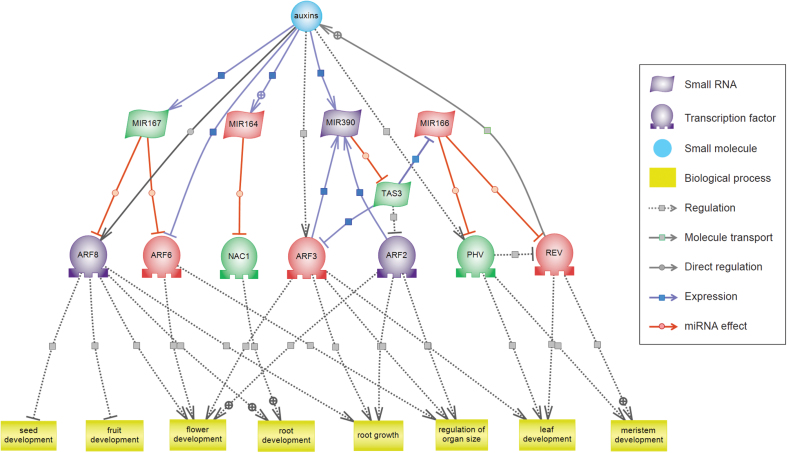
Genes often interact with other genes to accomplish complex biological processes, as do miRNAs, ta-siRNAs, and their target genes. Using Pathway Studio software (Nikitin et al., 2003), the ‘gold standard’ of pathway analysis, we investigated the roles of the interaction network between DES and their target genes in heterosis of hybrid rice (Fig. 7). The results showed that the regulatory factors of the auxin signalling pathway were regulated by DES. Of these, auxin response factor (ARF) 2 and ARF3 were regulated by ta-siARFs from miR390-directed TAS3 cleavage, and ARF6 and ARF8 were regulated by miR167. Additionally, pathway analysis showed that the TF NAM/ATAF/CUC 1 (NAC1) was regulated by miR164, and that TFs PHAVOLUTA (PHV) and REVOLUTA (REV) were regulated by miR166. The resulting expression changes of ARFs, NAC1, PHV, and REV facilitated the auxin signalling pathway, which plays important regulatory roles in plant development and growth of important tissues including roots, stems, leaves, flowers, fruit, and seeds (Fig. 7).
Fig. 7.
Construction of the auxin signalling regulatory network. DES and their target genes were used for direct interaction analysis by Pathway Studio 9.0. Upregulated, downregulated and unchanged sRNAs and targets in hybrid rice were represented with red, green and purple, respectively. TAS3, trans-acting siRNA 3; ARF2, auxin response factor 2; ARF3, auxin response factor 3; ARF6, auxin response factor 6; ARF8, auxin response factor 8; NAC1, NAM/ATAF/CUC 1; PHV, PHAVOLUTA; REV, REVOLUTA.
Mapping DES and target genes to QTL in the rice genome
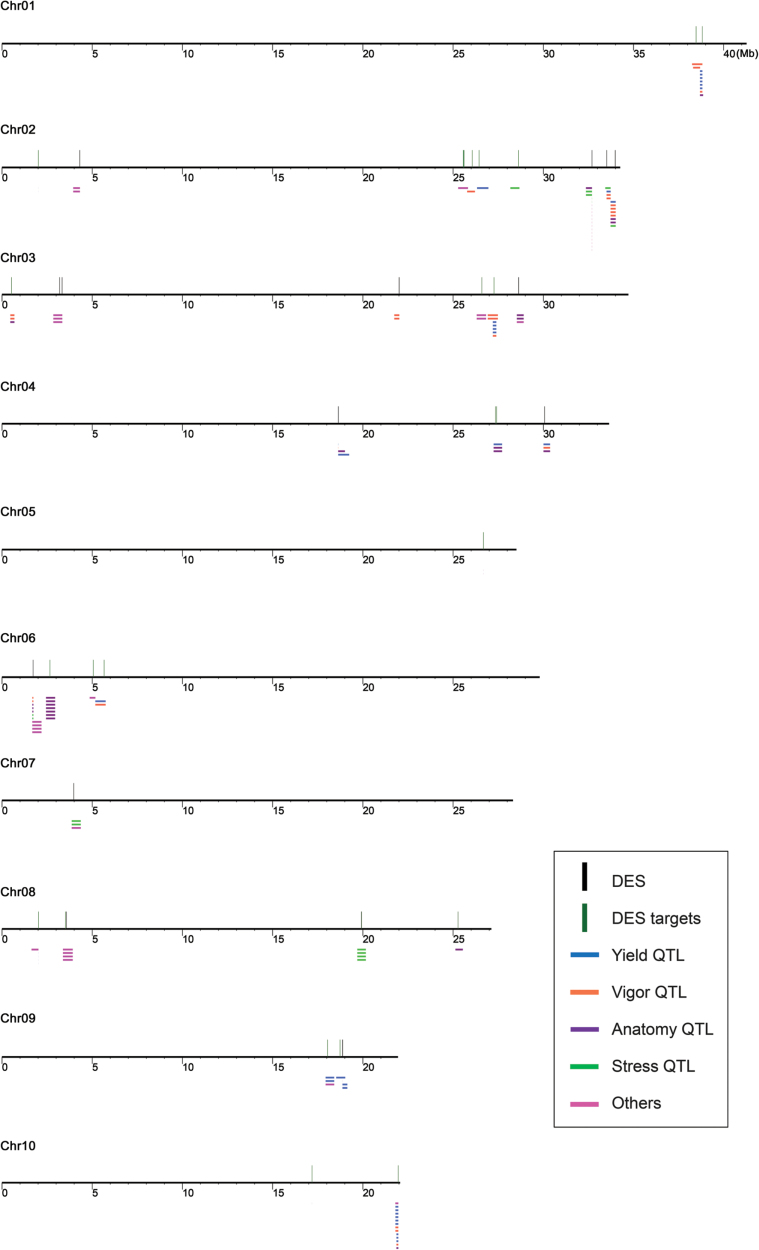
QTL are identified within genomic interval regions across chromosomes, containing one or more genes associated with the trait being measured. To investigate the association of sRNA expression variation with phenotypic changes in hybrid rice, we mapped the loci of DES and their potential targets to rice QTL in the Gramene database (Youens-Clark et al., 2011), which includes QTL identified for numerous agronomic traits, with genome coordinates. We mapped 211 of 212 (99.5%) loci (comprising 102 DES loci and 110 target loci) to 3214 QTL with 208 traits, which could be classified into nine categories including yield, vigour, and quality (Table 2 and Supplementary Table S10 at JXB online). Furthermore, by using all the gene loci in the rice genome as reference, we found 44 (20.8%) DES and target loci significantly enriched in 141 QTL of small intervals spanning less than 100 genes (P=0.0008, Supplementary Table S11 at JXB online), with 54 traits of nine categories and five classes (Table 2 and Fig. 8). Hybrid rice LYP9 is advantageous compared with its parents mainly in terms of grain yield, growth vigour, and plant architecture (Lu and Zou, 2000; Yu et al., 2002). As expected, 18 (40.9%), 14 (31.8%), and 16 (36.4%) DES and target loci could be significantly mapped to 38 yield-related, 25 vigour-related, and 26 anatomy-related QTL of small intervals, respectively, among which four DES target loci were mapped to nine yield- or anatomy-related QTL that spanned only one gene (Table 3).
Table 2.
QTL classification of DES and target genes
| Trait category | All QTL in Gramene database | QTL of small intervalsa | ||||
|---|---|---|---|---|---|---|
| Traitb | QTL | Genec | Traitb | QTL | Genec | |
| Abiotic stress | 44 | 237 | 180 | 9 | 12 | 7 |
| Anatomy | 43 | 528 | 204 | 10 | 26 | 16** |
| Biochemical | 20 | 95 | 161 | 2 | 2 | 4 |
| Biotic stress | 8 | 143 | 169 | 2 | 3 | 2 |
| Development | 11 | 348 | 206 | 3 | 6 | 5 |
| Quality | 34 | 248 | 190 | 3 | 11 | 6 |
| Sterility or fertility | 8 | 104 | 134 | 2 | 18 | 3 |
| Vigour | 14 | 753 | 208 | 8 | 25 | 14* |
| Yield | 26 | 758 | 209 | 15 | 38 | 18* |
| Total | 208 | 3,214 | 211 | 54 | 141 | 44** |
* and ** denote significant enrichment at P≤0.05 and P≤0.01, respectively, according to the hypergeometric test in Table S11.
a QTL contains gene number ≤100.
b Trait classified in Gramene database (Youens-Clark et al., 2011).
c Loci number of DES and target genes enriched in the QTL trait.
Fig. 8.
QTL mapping of DES and target genes. The rice QTL from the Gramene QTL database of small intervals (number of genes <100) that harbour DES and the target genes were aligned with the gene coordinates in MSU rice genome release 6. The long horizontal lines represent the rice chromosomes with the scale in Mb, the short horizontal lines represent QTL intervals of different trait categories in different colours, and the short vertical lines represent DES and targets in black and green, respectively. Five classes of QTL traits wee determined: yield, vigour, anatomy, stress (biotic and abiotic stress), and others (biochemical, development, quality, and sterility or fertility).
Table 3.
DES target genes in yield- or anatomy-related QTL that spanned only one gene
| QTL IDa | Chrb | Startb | Endb | Trait category | Trait name | Gene IDc | Corresponding DES |
|---|---|---|---|---|---|---|---|
| AQE036 | Chr2 | 2105542 | 2105840 | Yield | Grain yield per panicle | LOC_Os02g04680 | osa-miR156/535 |
| AQE049 | Chr2 | 2105542 | 2105840 | Yield | 100-Seed weight | LOC_Os02g04680 | osa-miR156/535 |
| AQFJ005 | Chr8 | 2114843 | 2115100 | Yield | Grain yield per plant | LOC_Os08g04310 | osa-miR528 |
| AQFJ015 | Chr8 | 2114843 | 2115100 | Yield | Panicle tiller ratio | LOC_Os08g04310 | osa-miR528 |
| AQGS037 | Chr8 | 2114843 | 2115100 | Yield | Grain yield | LOC_Os08g04310 | osa-miR528 |
| AQGS069 | Chr8 | 2114843 | 2115100 | Yield | Grain yield | LOC_Os08g04310 | osa-miR528 |
| AQFP005 | Chr5 | 27976107 | 27977104 | Anatomy | Culm thickness | LOC_Os05g48870 | OsTAS3-ta-siARF |
| AQFP027 | Chr5 | 27976107 | 27977104 | Anatomy | Culm thickness | LOC_Os05g48870 | OsTAS3-ta-siARF |
| AQGM008 | Chr10 | 18014265 | 18014633 | Anatomy | Culm length | LOC_Os10g33960 | osa-miR166 |
a QTL in the Gramene database (Youens-Clark et al., 2011).
b Genomic coordinates of QTL. Chr, Chromosome.
c DES target gene loci mapped in the QTL.
Discussion
Interaction and functional analysis between DES and their target genes
Recent studies (Ha et al., 2009; He et al., 2010; Groszmann et al., 2011; Barber et al., 2012) have analysed the relationships between sRNA variations and phenotypic changes in types of plant hybrids, providing some information for exploring the role of sRNAs in the heterosis mechanism. However, the role of the interaction between miRNAs/ta-siRNAs and their targets in heterosis is still poorly described in hybrid rice. Previously, we performed a comparative analysis of the transcriptional profiles on the super-hybrid rice LYP9 combination to approach the regulatory mechanism of heterosis in hybrid rice (Wei et al., 2009). Obvious changes of gene expression were detected in flag leaves and panicles of hybrid rice at the grain-filling stage, which play important roles in grain yield. In this study, we used the same materials to investigate the expression profiles of miRNAs and ta-siRNAs, two class of naturally occurring non-coding sRNAs with clear functions of regulating target mRNAs by causing their cleavage (Chen, 2009), and probed genome-wide for interactions between DES and predicted target genes, and their influence on the vigorous phenotypes of hybrid rice.
Pearson correlation analysis (Rodgers and Nicewander, 1988) indicated that 23 of the 34 (67.6%) DES in two tissues (Supplementary Table S9) displayed opposite expression patterns to the corresponding target DGs (Fig. 4). This result was consistent with the negatively regulating action of plant miRNAs and ta-siRNAs guiding Argonaute 1 (AGO1)- or AGO7-mediated cleavage of the partially complementary target mRNAs (Mallory and Bouche, 2008).
TF annotation analysis indicated that 112 of 176 (63.6%) DES target genes encoded TFs that belonged to 13 families (Table 1), including SBP, NAC, HD-ZIP, ARF, and AP2 (Nakano et al., 2006; Ariel et al., 2007; Guilfoyle and Hagen, 2007; Fang et al., 2008; Guo et al., 2008) (Supplementary Table S9). Intriguingly, all 11 TF families of target genes in panicles were also found in flag leaves (Table 1), suggesting a common transcriptional regulatory mechanism mediated by sRNAs shared in both tissues (Chen, 2009; Jeong et al., 2011).
To further investigate the biological functions of DES target genes, we analysed the GO classifications of the potential target genes of DES by ROAD (Cao et al., 2012) (Fig. 5). The results indicated that the DES target genes were enriched in the biological processes of regulatory functions including regulation of transcription, the response to hormone stimulus, the auxin-mediated signalling pathway, gene silencing by RNA, and regulation of timing of meristematic phase transition, but less in the metabolic process than the reference (Fig. 5) – consistent with the above result that most of the DES targets are TFs.
DES and their target genes are involved in the auxin signalling regulatory network
To elucidate the potential role of DES and their target genes, we performed interaction network analysis using Pathway Studio, and the result showed that a regulatory network in the auxin signalling pathway was correlated with some of our identified DES and their potential target genes (Fig. 7).
Auxin is a critical factor in plant development that has important influences on the final shape and functions of cells and tissues in all higher plants (Ljung, 2013). In this network, miR164, miR167, and miR390 (Guo et al., 2005; Yang et al., 2006; Yoon et al., 2010), as well as the target genes ARF6 and ARF8 of miR167 (Faivre-Rampant et al., 2004; Yang et al., 2006), the target gene ARF3 of ta-siRNA (Pekker et al., 2005), and the target gene PHV of miR166 (Weijers and Jurgens, 2005), could be directly regulated by the plant hormone auxin to execute biological functions in plant development (Ljung, 2013; Pierre-Jerome et al., 2013). REV is another target gene of miR166, which is antagonized by PHV, and plays a role in altering the auxin polar transport (Zhong and Ye, 1999, 2001; Prigge et al., 2005) (Fig. 7).
The upregulation of miR164 led to a downregulated change in NAC1, which transmits auxin signals from the auxin receptor transport inhibitor response 1 (TIR1) to downstream auxin-responsive genes to promote root development (Xie et al., 2000; Guo et al., 2005). NAC1 is a negative regulator of drought tolerance in rice (Fang et al., 2014), and therefore downregulation of NAC1 might increase the stress resistance of hybrid rice. ARF6 and ARF8, targeted by miR167, have effects on timing of flower maturation, development of seed and fruit, root elongation, and phosphate homeostasis (Goetz et al., 2006; Wang et al., 2014). Downregulation of ta-siARFs generated from miR390-dependent TAS3 acted on its target genes ARF2 and ARF3, modulating lateral root growth, flowering, leaf senescence and floral organ abscission, and leaf longevity (Ellis et al., 2005; Lim et al., 2010; Marin et al., 2010). The expression changes of miR166 target genes PHV and REV could direct shoot apical meristem development, lateral organ polarity, and leaf adaxial identity (Huang et al., 2006a ; Nagasaki et al., 2007; Grigg et al., 2009) (Fig. 7). The downregulation of TAS3 and resulting upregulation of miR166 might promote the growth of hybrid rice, according to a study of TAS3 ta-siRNA overexpression transgenic rice with a retarded growth phenotype (Wang et al., 2010). Furthermore, most of the DES in this network were validated, and exhibited opposite trends in expression to the corresponding target genes according to qRT-PCR (Fig. 6). Our results suggest that these DES have a potential role in heterosis in rice, but how exactly the sRNA expression changes contribute to heterosis has yet to be understood.
The auxin signalling network has interactions with the light-signalling network via phytochrome-interacting factors (PIFs) (Hornitschek et al., 2012; Sun et al., 2013). Sentandreu et al. (2011) revealed that PIF3 negatively regulated the Arabidopsis mitogen-activated protein kinase MPK12, which was proposed to be a negative regulator in auxin signalling (Lee et al., 2009). Our previous study on comparative transcriptional profiling of a hybrid rice combination (Song et al., 2010) revealed significantly differential expression for the rice PIF3 homologous gene Os01g18290 (Nakamura et al., 2007). Likewise, Os01g18290 was also identified as DG in the present study (Supplementary Tables S6 and S7). In Arabidopsis, PIF3 interacts with photoreceptors phytochrome A (phyA) and phyB, receiving the light signal at the first step to mediate light signalling (Castillon et al., 2007). The role of PIF3 in rice has not yet been identified completely; it might execute similar biological functions as AtPIF3 (Nakamura et al., 2007). PIF3 regulated the expression of the core circadian-rhythm regulatory gene late elongated hypocotyl (LHY) by binding to its promoter (Martinez-Garcia et al., 2000), which further facilitates the integration of light-signalling and the circadian-rhythm regulatory network. As a result, PIF3 might play an important role in regulating the downstream genes involved in photosynthesis and carbohydrate metabolic pathways, which will increase the carbon fixation and photosynthetic efficiency in the F1 hybrid (Ni et al., 2009; Song et al., 2010; Chen, 2013). LHY (Os08g06110) and GIGANTEA (GI) (Os01g08700), two important members in the circadian regulatory network, were also identified as DGs in both this study (Supplementary Tables S6 and S7) and a previous study (Song et al., 2010). Shen et al. (2012) analysed the genome-wide DNA methylation and transcriptional profiles in Arabidopsis and found that the differential expression of the auxin-related genes, including NAC1 and vacuolar H + -pyrophosphatase 1 (AVP1), led to an increase of auxin signalling, which might contribute to the growth vigour of Arabidopsis F1 hybrids. In the present study, the DES miR164 target gene NAC1 displayed obvious expression changes and had negatively correlated expression with miR164 (Supplementary Table S9 and Fig. 6), and the AVP1 homologous gene Os02g09150 was also identified as a DG (Supplementary Tables S6 and S7). The above results indicated that changes in the complex auxin signalling regulatory network by the interaction between DES and their target genes might contribute to the growth vigour and grain-yield heterosis of hybrid rice.
DES and their targets were associated with heterosis-related QTL
As an effective tool to investigate the relationship between genotype and phenotype, QTL analysis has been widely used in the crop breeding as well as heterosis studies (Miura et al., 2011; Qu et al., 2012; Fang et al., 2013).
In the present study, we further investigated the correlation of DES and their target genes with heterosis phenotypes using QTL analysis. There were 211 DES and target loci mapped to known rice QTL that were related to nine categories of agronomic traits (Table 2). For example, we mapped the DES gene osa-MIR156f and its target gene Os08g39890, which encodes SBP-box family protein OsSPL14, to the QTL CQAW25 for tiller number trait in the vigour category (Supplementary Table S10). It has been reported that a point mutation in OsSPL14 impairs the regulation of OsmiR156 on OsSPL14, resulting in ideal rice plant architecture related to tiller number and grain yield, by QTL fine-mapping analysis (Jiao et al., 2010). Moreover, Os02g04680, another target gene of miR156, was significantly mapped to the QTL AQE036 and AQE049, which only spanned one gene, for grain yield per panicle and 100-seed weight trait of yield, respectively (Table 3). These confirmed the strong correlations between expression variation of miR156 and yield-related phenotypes in the hybrid rice LYP9 (Lu and Zou, 2000; Yu et al., 2002).
Furthermore, the anatomy-related QTL of small intervals was the most significant category that DES and their target genes were enriched in (P=0.0037, Supplementary Table S11), suggesting the contributions of DES and targets to the plant architecture of hybrid rice. For instance in this category, the ta-siARF target gene Os05g48870 and miR166 target gene Os10g33960 could be localized in the QTL AQFP005 for culm thickness trait and AQGM008 for culm length trait that spanned only one locus, respectively (Table 3), implying correlations between DES and the thick culm architecture of the hybrid rice LYP9 (Lu and Zou, 2000). In addition, the differential expression of miR166 and miR167, which were enriched in the auxin regulatory network in this study, were observed in the hybrid rice SY63 and its parental lines, ZS97A and MH63. These two miRNAs were predicted in the QTL intervals of the yield heterotic trait, RG653-G342 and C9098B-RM17 (Fang et al., 2013). These results suggested that the DES-enriched auxin pathway contributed to high-yield traits in different hybrid rice combinations.
In summary, QTL analysis indicated the critical roles of DES and their interactions with target genes in agronomic phenotypic changes of hybrid rice. Our findings provide strong molecular evidence that sRNAs are important regulators that contribute to heterosis in hybrid rice, and open up a new line of deciphering the molecular mechanism of heterosis. Such an understanding of the molecular basis of heterosis can be useful for breeders to take advantage of heterosis in the near future for crop improvement.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Common and specific sRNA reads in different libraries and tissues of LYP9 hybrid rice combination.
Supplementary Fig. S2. Common and specific detected miRNAs/ta-siARFs, DES, detected protein-coding genes, and DGs in different tissues.
Supplementary Fig. S3. Expression patterns and functions of DGs in flag leaves and panicles at the grain-filling stage.
Supplementary Table S1. Statistics of annotations and classifications of sRNA reads in different libraries.
Supplementary Table S2. Expression profiling of the known miRNAs and ta-siARFs in different libraries.
Supplementary Table S3. DES in flag leaves of hybrid rice and its parents.
Supplementary Table S4. DES in panicles of hybrid rice and its parents.
Supplementary Table S5. Statistics of RNA-Seq reads in different libraries.
Supplementary Table S6. DG in flag leaves of hybrid rice and its parents.
Supplementary Table S7. DG in panicles of hybrid rice and its parents.
Supplementary Table S8. Predicted target genes of DES with degradome data support.
Supplementary Table S9. List of target genes of DES in flag leaves and panicles.
Supplementary Table S10. List of QTL mapped with DES and target genes.
Supplementary Table S11. Enrichment analysis of DES and target genes in QTL of small intervals.
Supplementary Table S12. Primers used in qRT-PCR.
Acknowledgments
We thank Dr Guisheng Song for thoughtful suggestions and comments on the manuscript, Professor Xiujie Wang and Dr Huajun Wu for helpful advice in the use of psRobot software and data analysis, and Dr Gang Wei for kindly helping in the QTL mapping analysis. This work was supported by the National Natural Science Foundation of China (31100931 and 91335111), the Ministry of Science and Technology of China (2012AA10A304 and 2013AA102701), the Ministry of Agriculture of China (2014ZX08012-002), the State Key Laboratory of Plant Genomics of China (SKLPG2011B0404), and the ‘Strategic Priority Research Program’ of the Chinese Academy of Sciences (XDA08010402).
Glossary
Abbreviations:
- AGO
Argonaute
- ARF
auxin response factor
- CHP
close to higher parent
- CLP
close to lower parent
- DES
differentially expressed sRNA
- DG
differentially expressed gene
- FDR
false discovery rate
- GO
Gene Ontology
- H2P
higher than two parents
- L2P
lower than two parents
- miRNA
microRNA;
- MPV
mid-parental value
- NAT
natural antisense transcripts
- nat-siRNA
natural antisense transcripts-generated siRNA
- ncRNA
non-coding RNA
- PIF
phytochrome interacting factor
- qRT-PCR
quantitative reverse transcription-PCR
- QTL
quantitative trait loci
- ra-siRNA
repeat-associated siRNA
- RPKM
reads per kilobase transcriptome per million mapped reads
- RPM
reads per million mapped reads
- rRNA
ribosomal RNA;
- snoRNA
small nucleolar RNA
- snRNA
small nuclear RNA
- siRNA
small interfering RNA
- sRNA
small RNA
- ta-siRNA
trans-acting siRNA
- TF
transcription factor.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221 [DOI] [PubMed] [Google Scholar]
- Ariel FD, Manavella PA, Dezar CA, Chan RL. 2007. The true story of the HD-Zip family. Trends in Plant Science 12, 419–426 [DOI] [PubMed] [Google Scholar]
- Baranwal VK, Mikkilineni V, Zehr UB, Tyagi AK, Kapoor S. 2012. Heterosis: emerging ideas about hybrid vigour. Journal of Experimental Botany 63, 6309–6314 [DOI] [PubMed] [Google Scholar]
- Barber WT, Zhang W, Win H, Varala KK, Dorweiler JE, Hudson ME, Moose SP. 2012. Repeat associated small RNAs vary among parents and following hybridization in maize. Proceedings of the National Academy of Sciences, USA 109, 10444–10449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- Bruce AB. 1910. The Mendelian theory of heredity and the augmentation of vigor. Science 32, 627–628 [DOI] [PubMed] [Google Scholar]
- Cao P, Jung KH, Choi D, Hwang D, Zhu J, Ronald PC. 2012. The Rice Oligonucleotide Array Database: an atlas of rice gene expression. Rice 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E. 2007. Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends in Plant Science 12, 514–521 [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, et al. 2005. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Research 33, e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Yuan C, Zhang J, Zhang Z, Bai L, Meng Y, Chen LL, Chen M. 2012. PlantNATsDB: a comprehensive database of plant natural antisense transcripts. Nucleic Acids Research 40, D1187–D1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, He G, He H, Chen W, Zhu X, Liang M, Chen L, Deng XW. 2010. Expression analysis of miRNAs and highly-expressed small RNAs in two rice subspecies and their reciprocal hybrids. Journal of Integrative Plant Biology 52, 971–980 [DOI] [PubMed] [Google Scholar]
- Chen X. 2009. Small RNAs and their roles in plant development. Annual Review of Cell and Developmental Biology 25, 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ. 2013. Genomic and epigenetic insights into the molecular bases of heterosis. Nature Reviews: Genetics 14, 471–482 [DOI] [PubMed] [Google Scholar]
- Cheng SH, Zhuang JY, Fan YY, Du JH, Cao LY. 2007. Progress in research and development on hybrid rice: a super-domesticate in China. Annals of Botany 100, 959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. 1876. The effects of cross and self fertilisation in the vegetable kingdom. London, UK: John Murray Press [Google Scholar]
- Ding D, Wang Y, Han M, Fu Z, Li W, Liu Z, Hu Y, Tang J. 2012. MicroRNA transcriptomic analysis of heterosis during maize seed germination. PLoS One 7, e39578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East EM. 1936. Heterosis. Genetics 21, 375–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW. 2005. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana . Development 132, 4563–4574 [DOI] [PubMed] [Google Scholar]
- Faivre-Rampant O, Cardle L, Marshall D, Viola R, Taylor MA. 2004. Changes in gene expression during meristem activation processes in Solanum tuberosum with a focus on the regulation of an auxin response factor gene. Journal of Experimental Botany 55, 613–622 [DOI] [PubMed] [Google Scholar]
- Fang R, Li L, Li J. 2013. Spatial and temporal expression modes of microRNAs in an elite rice hybrid and its parental lines. Planta 238, 259–269 [DOI] [PubMed] [Google Scholar]
- Fang Y, Xie K, Xiong L. 2014. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. Journal of Experimental Botany 65, 2119–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, You J, Xie K, Xie W, Xiong L. 2008. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Molecular Genetics and Genomics 280, 547–563 [DOI] [PubMed] [Google Scholar]
- Gale MD, Devos KM. 1998. Comparative genetics in the grasses. Proceedings of the National Academy of Sciences, USA 95, 1971–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Vivian-Smith A, Johnson SD, Koltunow AM. 2006. AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis . Plant Cell 18, 1873–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Zhang Q. 2013. Heterosis in elite hybrid rice: speculation on the genetic and biochemical mechanisms. Current Opinion in Plant Biology 16, 221–227 [DOI] [PubMed] [Google Scholar]
- Grigg SP, Galinha C, Kornet N, Canales C, Scheres B, Tsiantis M. 2009. Repression of apical homeobox genes is required for embryonic root development in Arabidopsis . Current Biology 19, 1485–1490 [DOI] [PubMed] [Google Scholar]
- Groszmann M, Greaves IK, Albertyn ZI, Scofield GN, Peacock WJ, Dennis ES. 2011. Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proceedings of the National Academy of Sciences, USA 108, 2617–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. 2007. Auxin response factors. Current Opinion in Plant Biology 10, 453–460 [DOI] [PubMed] [Google Scholar]
- Guo AY, Zhu QH, Gu X, Ge S, Yang J, Luo J. 2008. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 418, 1–8 [DOI] [PubMed] [Google Scholar]
- Guo HS, Xie Q, Fei JF, Chua NH. 2005. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17, 1376–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Lu J, Tian L, Ramachandran V, Kasschau KD, Chapman EJ, Carrington JC, Chen X, Wang XJ, Chen ZJ. 2009. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proceedings of the National Academy of Sciences, USA 106, 17835–17840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T. 2008. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44, 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbritter F, Vaidya HJ, Tomlinson SR. 2012. GeneProf: analysis of high-throughput sequencing experiments. Nature Methods 9, 7–8 [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952 [DOI] [PubMed] [Google Scholar]
- He G, He H, Deng XW. 2013. Epigenetic variations in plant hybrids and their potential roles in heterosis. Journal of Genetics and Genomics 40, 205–210 [DOI] [PubMed] [Google Scholar]
- He G, Zhu X, Elling AA, et al. , 2010. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22, 17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE. 2006. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nature Genetics 38, 721–725 [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, et al. , 2012. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant Journal 71, 699–711 [DOI] [PubMed] [Google Scholar]
- Huang W, Pi L, Liang W, Xu B, Wang H, Cai R, Huang H. 2006a. The proteolytic function of the Arabidopsis 26S proteasome is required for specifying leaf adaxial identity. Plant Cell 18, 2479–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Zhang L, Zhang J, Yuan D, Xu C, Li X, Zhou D, Wang S, Zhang Q. 2006b. Heterosis and polymorphisms of gene expression in an elite rice hybrid as revealed by a microarray analysis of 9198 unique ESTs. Plant Molecular Biology 62, 579–591 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing P. 2005. The map-based sequence of the rice genome. Nature 436, 793–800 [DOI] [PubMed] [Google Scholar]
- Jeong DH, Park S, Zhai J, Gurazada SG, De Paoli E, Meyers BC, Green PJ. 2011. Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23, 4185–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, et al. , 2010. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nature Genetics 42, 541–544 [DOI] [PubMed] [Google Scholar]
- Jones DF. 1917. Dominance of linked factors as a means of accounting for heterosis. Genetics 2, 466–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Research 12, 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Wang S, Sritubtim S, Chen JG, Ellis BE. 2009. Arabidopsis mitogen-activated protein kinase MPK12 interacts with the MAPK phosphatase IBR5 and regulates auxin signaling. Plant Journal 57, 975–985 [DOI] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. 2009a. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 [DOI] [PubMed] [Google Scholar]
- Li X, Wei Y, Nettleton D, Brummer EC. 2009b. Comparative gene expression profiles between heterotic and non-heterotic hybrids of tetraploid Medicago sativa . BMC Plant Biology 9, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Lee IC, Kim J, Kim HJ, Ryu JS, Woo HR, Nam HG. 2010. Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. Journal of Experimental Botany 61, 1419–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Chen Z, Song X, et al. , 2007. Oryza sativa Dicer-like4 reveals a key role for small interfering RNA silencing in plant development. Plant Cell 19, 2705–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- Ljung K. 2013. Auxin metabolism and homeostasis during plant development. Development 140, 943–950 [DOI] [PubMed] [Google Scholar]
- Lu C, Zou J. 2000. Breeding and utilization of two-line intersubspecific hybrid rice Liangyou Peijiu. Hybrid Rice 15, 4–5 [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. 2011. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Research 39, D52–D57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Bouche N. 2008. MicroRNA-directed regulation: to cleave or not to cleave. Trends in Plant Science 13, 359–367 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Vaucheret H. 2006. Functions of microRNAs and related small RNAs in plants. Nature Genetics 38 (Suppl. ), S31–S36 [DOI] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A. 2010. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22, 1104–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH. 2000. Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863 [DOI] [PubMed] [Google Scholar]
- Miura K, Ashikari M, Matsuoka M. 2011. The role of QTLs in the breeding of high-yielding rice. Trends in Plant Science 16, 319–326 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods 5, 621–628 [DOI] [PubMed] [Google Scholar]
- Nagasaki H, Itoh J, Hayashi K, et al. , 2007. The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proceedings of the National Academy of Sciences, USA 104, 14867–14871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Kato T, Yamashino T, Murakami M, Mizuno T. 2007. Characterization of a set of phytochrome-interacting factor-like bHLH proteins in Oryza sativa . Bioscience, Biotechnology, and Biochemistry 71, 1183–1191 [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. 2006. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology 140, 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DW, Lu J, Chen ZJ. 2012. Big roles for small RNAs in polyploidy, hybrid vigor, and hybrid incompatibility. Current Opinion in Plant Biology 15, 154–161 [DOI] [PubMed] [Google Scholar]
- Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ. 2009. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457, 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin A, Egorov S, Daraselia N, Mazo I. 2003. Pathway studio--the analysis and navigation of molecular networks. Bioinformatics 19, 2155–2157 [DOI] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, et al. , 2007. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Research 35, D883–D887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y. 2005. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17, 2899–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre-Jerome E, Moss BL, Nemhauser JL. 2013. Tuning the auxin transcriptional response. Journal of Experimental Botany 64, 2557–2563 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. 2005. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17, 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z, Li L, Luo J, Wang P, Yu S, Mou T, Zheng X, Hu Z. 2012. QTL mapping of combining ability and heterosis of agronomic traits in rice backcross recombinant inbred lines and hybrid crosses. PLoS One 7, e28463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JL, Nicewander WA. 1988. Thirteen ways to look at the correlation coefficient. American Statistician 42, 59–66 [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. 2006. TM4 microarray software suite. Methods in Enzymology 411, 134–193 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Springer NM. 2013. Progress toward understanding heterosis in crop plants. Annual Review of Plant Biology 64, 71–88 [DOI] [PubMed] [Google Scholar]
- Schnell FW, Cockerham CC. 1992. Multiplicative vs. arbitrary gene action in heterosis. Genetics 131, 461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentandreu M, Martin G, Gonzalez-Schain N, Leivar P, Soy J, Tepperman JM, Quail PH, Monte E. 2011. Functional profiling identifies genes involved in organ-specific branches of the PIF3 regulatory network in Arabidopsis . Plant Cell 23, 3974–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, He H, Li J, et al. , 2012. Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell 24, 875–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Dunn RM, Santos BA, Bassett A, Baulcombe DC. 2012. Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO Journal 31, 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull GH. 1908. The composition of a field of maize. Journal of Heredity 4, 296–301 [Google Scholar]
- Song GS, Zhai HL, Peng YG, et al. , 2010. Comparative transcriptional profiling and preliminary study on heterosis mechanism of super-hybrid rice. Molecular Plant 3, 1012–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, Stupar RM. 2007. Allelic variation and heterosis in maize: how do two halves make more than a whole? Genome Research 17, 264–275 [DOI] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Zhai Q, Li C. 2013. PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis . Plant Cell 25, 2102–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Jia Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS. 2006. All possible modes of gene action are observed in a global comparison of gene expression in a maizeF1 hybrid and its inbred parents. Proceedings of the National Academy of Sciences, USA 103, 6805–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Legrand S, Windels D. 2010. The biosynthetic pathways and biological scopes of plant small RNAs. Trends in Plant Science 15, 337–345 [DOI] [PubMed] [Google Scholar]
- Wang J, Gao X, Li L, Shi X, Zhang J, Shi Z. 2010. Overexpression of Osta-siR2141 caused abnormal polarity establishment and retarded growth in rice. Journal of Experimental Botany 61, 1885–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tian L, Lee HS, et al. , 2006. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172, 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhang S, Sun C, Xu Y, Chen Y, Yu C, Qian Q, Jiang DA, Qi Y. 2014. Auxin response factor (OsARF12), a novel regulator for phosphate homeostasis in rice (Oryza sativa). New Phytologist 201, 91–103 [DOI] [PubMed] [Google Scholar]
- Wei G, Tao Y, Liu G, et al. , 2009. A transcriptomic analysis of superhybrid rice LYP9 and its parents. Proceedings of the National Academy of Sciences, USA 106, 7695–7701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Jurgens G. 2005. Auxin and embryo axis formation: the ends in sight? Current Opinion in Plant Biology 8, 32–37 [DOI] [PubMed] [Google Scholar]
- Wu HJ, Ma YK, Chen T, Wang M, Wang XJ. 2012. PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucleic Acids Research 40, W22–W28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. 2011. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Research 39, W316–W322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua NH. 2000. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes & Development 14, 3024–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Han SJ, Yoon EK, Lee WS. 2006. Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Research 34, 1892–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Dogra Gray A, Auger DL, Birchler JA. 2013. Genomic dosage effects on heterosis in triploid maize. Proceedings of the National Academy of Sciences, USA 110, 2665–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon EK, Yang JH, Lim J, Kim SH, Kim SK, Lee WS. 2010. Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Research 38, 1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youens-Clark K, Buckler E, Casstevens T, et al. , 2011. Gramene database in 2010: updates and extensions. Nucleic Acids Research 39, D1085–D1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, et al. , 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92 [DOI] [PubMed] [Google Scholar]
- Yu SB, Li JX, Xu CG, Tan YF, Gao YJ, Li XH, Zhang Q, Saghai Maroof MA. 1997. Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proceedings of the National Academy of Sciences, USA 94, 9226–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai R, Feng Y, Wang H, et al. 2013. Transcriptome analysis of rice root heterosis by RNA-Seq. BMC Genomics 14, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Jin J, Tang L, Zhao Y, Gu X, Gao G, Luo J. 2011. PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Research 39, D1114–D1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Peng Y, Dong Y, Li H, Wang W, Zhu Z. 2013. Progress of Genomics and Heterosis Studies in Hybrid Rice. In: Chen ZJ, Birchler JA, eds. Polyploid and hybrid genomics. New York: Wiley-Blackwell Press, 117–135 [Google Scholar]
- Zhong R, Ye ZH. 1999. IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11, 2139–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. 2001. Alteration of auxin polar transport in the Arabidopsis ifl1 mutants. Plant Physiology 126, 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, Helliwell C. 2008. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Research 18, 1456–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.