Summary
Ammonium nutrition is toxic to many plants. Arabidopsis displays high intraspecific variability in ammonium tolerance (shoot biomass), and ammonium accumulation seems to be an important player in this variability.
Key words: Ammonium, Arabidopsis thaliana, glutamate dehydrogenase, glutamine synthetase, natural variation, nitrate, nitrogen.
Abstract
Plants are dependent on exogenous nitrogen (N) supply. Ammonium (NH4 +), together with nitrate (NO3 –), is one of the main nitrogenous compounds available in the soil. Paradoxically, although NH4 + assimilation requires less energy than that of NO3 –, many plants display toxicity symptoms when grown with NH4 + as the sole N source. However, in addition to species-specific ammonium toxicity, intraspecific variability has also been shown. Thus, the aim of this work was to study the intraspecific ammonium tolerance in a large panel of Arabidopsis thaliana natural accessions. Plants were grown with either 1mM NO3 – or NH4 + as the N source, and several parameters related to ammonium tolerance and assimilation were determined. Overall, high variability was observed in A. thaliana shoot growth under both forms of N nutrition. From the parameters determined, tissue ammonium content was the one with the highest impact on shoot biomass, and interestingly this was also the case when N was supplied as NO3 –. Enzymes of nitrogen assimilation did not have an impact on A. thaliana biomass variation, but the N source affected their activity. Glutamate dehydrogenase (GDH) aminating activity was, in general, higher in NH4 +-fed plants. In contrast, GDH deaminating activity was higher in NO3 –-fed plants, suggesting a differential role for this enzyme as a function of the N form supplied. Overall, NH4 + accumulation seems to be an important player in Arabidopsis natural variability in ammonium tolerance rather than the cell NH4 + assimilation capacity.
Introduction
Plants have a fundamental dependence on inorganic nitrogen (N), and intensive agriculture requires the use of N compounds to supplement the natural supply from the soil. Indeed, >100 Mt of nitrogenous fertilizers are added to the soil worldwide annually (Good and Beatty, 2011). In part because of the intense use of fertilizers, agriculture is now a dominant force behind many environmental threats, including climate change and degradation of land and fresh water (Foley et al., 2011; Tilman et al., 2011). Moreover, recent studies suggest that agricultural output would need to roughly double to meet the expected demand associated with the increase in the world’s population (FAO, 2009).
Nitrate (NO3 −) and ammonium (NH4 +) are the main forms of N available for plants. There is a serious concern regarding NO3 − loss in the field, giving rise to soil and water pollution. Moreover, incomplete capture and poor conversion of nitrogen fertilizer also causes global warming through emissions of nitrous oxide. Due to these detrimental effects of adding high NO3 − concentrations to ecosystems (Gruber and Galloway, 2008), the potential of NH4 + as an N source for agriculture has been reconsidered alongside the search to improve N use efficiency (NUE) while mitigating the impact of agriculture (IPCC, 2007). Similarly, lowering fertilizer input and breeding plants with better NUE without affecting yield is a main goal for research in plant nutrition (Xu et al., 2012).
Plants have differential N source preference, but this depends not only on their genetic background but also on a wide and dynamic range of environmental variables including soil pH, temperature, etc. Thus, a robust classification of plants species adapted to NO3 − or NH4 + does not exist. However, it appears that most non-bred plants preferentially take up NH4 + (Bloom et al., 1993; Kronzucker et al., 2001). Moreover, crop species have traditionally been bred under nitric or combined N nutrition, provoking a negative selection pressure towards NH4 + assimilation, and this undoubtedly is one of the reasons they prefer NO3 −, although NO3 − must be taken up against an electrochemical gradient and then be reduced to NH4 + with the consequent energy cost (Kronzucker et al., 2001). In this sense, NH4 + nutrition has been generally considered as toxic for plants, particularly when NH4 + is supplied as the sole N source. Indeed, NH4 + is also toxic to animals and fungi when present in excess amounts (Britto and Kronzucker, 2002).
Ammonium toxicity syndrome in plants includes several symptoms, among others leaf chlorosis, ion imbalance, hormone deregulation, disorder in pH regulation, decrease in net photosynthesis, and changes in metabolite levels including amino acids, organic acids, and carbohydrates. At the whole-plant level, a reduction in plant growth with increasing external NH4 + concentrations, as compared with NO3 − nutrition, is a common effect of NH4 + nutrition (Cruz et al., 2006). Biomass reduction has been associated with carbohydrate limitation for growth due to excessive sugar consumption for NH4 + assimilation and to the energy costs of futile transmembrane NH3/NH4 + cycling in root cells (Coskun et al., 2013). Indeed, plant growth is probably the best indicator of NH4 + stress as it is a comprehensive measure of the physiology of the plant as a whole (Cruz et al., 2006; Dominguez-Valdivia et al., 2008; Ariz et al., 2011).
Substantial variations in NH4 + tolerance have been observed amongst closely related species (Monselise and Kost, 1993) and even within species (Rauh et al., 2002; Cruz et al., 2011; Li et al., 2011), suggesting the evolution of highly distinct mechanisms to cope with this stress. The strategies plants deploy to avoid NH4 + toxicity include enhancing NH4 + assimilation and increasing the efflux outside the cell or into the vacuole. Nevertheless, at present there is no consensus as to which traits confer NH4 + tolerance or sensitivity to plants. Ammonium assimilation mainly occurs via the glutamine synthetase/glutamate synthase cycle (GS/GOGAT). However, it seems that other alternative pathways could be involved in ammonium assimilation when NH4 + is supplied as the sole source of N. Although controversial, under these conditions, glutamate dehydrogenase (GDH), that catalyses the reversible deamination of glutamate to 2-oxoglutarate, might be collaborating in NH4 + assimilation (Skopelitis et al., 2006; Setien et al., 2013).
Arabidopsis thaliana and the Brassicaceae family are considered to be a species, and a family, sensitive to NH4 +. Most of the works focused on NH4 + toxicity in Arabidopsis have compared plants fed with NO3 − versus plants fed with a combined nutrition of NO3 − supplemented with increasing concentrations of NH4 +. Studies where Arabidopsis has been grown under long-term ammonium supply as the sole N source are scarce and have shown how NH4 + causes a retardation of seedling growth or a dramatic reduction in plant biomass (Rauh et al., 2002; Hoffmann et al., 2007; Helali et al., 2010). Also, recent genetic approaches have been useful to identify new molecular players involved in the signalling pathways that lead to NH4 + sensitivity, for example a GDP-mannosepyrophosphorylase enzyme (Qin et al., 2008) or the ammonium transporter AMT1:3 (Lima et al., 2010).
Overall, the evolutionary trade-off between high productivity, adaptation to low-nutrient environments, and the use of ammonium as fertilizer is a challenge to most plant cultivars that have been selected under non-limiting NO3 − or combined NH4 +/NO3 − fertilization (Presterl et al., 2003; Xu et al., 2012). Approaches based on natural variation have become an important means to study plant adaptation to the environment. In Arabidopsis, it has already been reported that a plant’s response to N availability is dependent on both the genotype and the N fertilization level (Loudet et al., 2003), and natural variation has been observed for N remobilization during seed filling, among others (Masclaux-Daubresse and Chardon, 2011). Thus, the present work compares the natural intraspecific variability of A. thaliana grown under a low NO3 − or NH4 + supply, focusing on the importance of N assimilation mechanisms in relation to the differential N source provided.
Materials and methods
Experimental procedures and growth conditions
Forty-seven A. thaliana world natural accessions lines (http://publiclines.versailles.inra.fr/naturalAccession/index) were used throughout the study. Seeds were directly sown in 37cm3 pots containing autoclaved perlite:vermiculite substrate mixture (1:1, v/v).
Seeds were cold-treated during 4 d in the dark at 4 ºC and then transferred into a controlled-conditions phytotron: 14h, 200mol m–2 s–1 light intensity, 60% relative humidity (RH), and 22 ºC day conditions, and 10h, 70% RH, and 18ºC night conditions. Pots were initially misted with a modified Murashige and Skoog (MS) solution containing 0.5mM NH4NO3. Nine days after transfer into the growth chamber, a single seedling was retained per pot and treatment was initiated. Plants were irrigated three times a week with a modified MS solution (3mM CaCl2, 1.25mM KH2PO4, 1.5mM MgSO4, 5mM KCl, 0.085mM Na2EDTA, 0.5mM MES, 5 μM KI, 0.1 μM CuSO4, 100 μM MnSO4, 100 μM H3BO3, 0.1 μM CoCl2, 100 μM FeSO4, 30 μM ZnSO4, and 0.1 μM Na2MoO4) with 0.5mM Ca(NO3)2 or 0.5mM (NH4)2SO4 as N source. NH4 +-fed plants were supplemented with 0.5mM CaSO4 to compensate the Ca2+ supplied together with the NO3 –.
Thirty days after transfer into the growth chamber, rosette biomass was recorded and leaves were immediately frozen in liquid nitrogen and stored at –80 ºC.
Determination of ammonium and total amino acids content
Aliquots of ~25mg of frozen material were ground to powder with liquid nitrogen and homogenized with 800 μl of ultrapure water. Samples were then incubated at 80 ºC during 5min and centrifuged at 4000 g and 4 ºC for 20min, and supernatants were recovered.
Total free amino acids were determined by the ninhydrin method (Yemm and Cocking, 1955). Ammonium content was determined by using the colorimetric method based on the phenol hypochlorite assay (Berthelot reaction).
Protein extraction
Proteins were extracted as described in Gibon et al. (2004). Briefly, leaves (~40mg per sample) were homogenized using a mortar and pestle with 0.8ml of extraction buffer [10mM MgCl2, 1mM EDTA, 1mM EGTA, 10mM dithiothreitol (DTT), 0.1% Triton X-100, 10% glycerol, 0.05% bovine serum albumin (BSA), 0.5% polyvinylpolypyrrolidone (PVPP), 50mM HEPES pH 7.5] in the presence of a cocktail of proteases inhibitors [1mM phenylmethylsulfonyl fluoride (PMSF), 1mM ε-aminocaproic acid, 10 μM leupeptin, 1mM benzamidine]. Samples were then centrifuged at 4000 g for 30min at 4 ºC and the supernatants recovered. Protein content of the supernatants was quantified by the Bradford assay (Bradford, 1976).
Enzyme activities
The GS reaction was measured at 30 ºC in a reaction buffer containing: 50mM TRIS-HCl (pH 7,6), 20mM MgSO4, 8mM sodium glutamate, 6mM hydroxylamine, 4mM Na2-EDTA, and 8mM ATP. The reaction was stopped by adding 0.12M FeCl3, 0.5M trichloroacetic acid (TCA), and 2 N HCl. Samples were centrifuged at 13 200 g for 5min, and the absorbance of γ-glutamyl monohydroxamate (γ-GHM) was measured at 540nm.
GDH activity was determined in the aminating direction in a reaction buffer containing 100mM TRIS-HCl (pH 8), 1mM CaCl2, 13mM 2-oxoglutarate, 50mM (NH4)2SO4, and 0.25mM NADH, and in the deaminating direction in 100mM TRIS-HCl (pH 9), 1mM CaCl2, 30mM glutamic acid, and 0.25mM NAD. Both kinetic activities were monitored spectrophotometrically at 30 ºC by consumption of NADH (amination) or appearance of NADH (deamination) at 340nm.
Nitrate reductase (NR) activity was measured at 30 ºC. The reaction medium consisted of 50mM HEPES-KOH, pH 7.6, 5mM KNO3, 0.2mM NADH, 10 μM FAD phosphate, 1mM DTT, 20mM EDTA. The reaction was started by adding 50 μl of protein extract to 250 μl of reaction medium and stopped by adding 32 μl of 50mM zinc acetate. Then, samples were centrifuged, 100 μl of supernatant was recovered, 8 μl of 50mM phenacin metosulphate added, and the samples incubated for 20min at room temperature. Finally, 80 μl of 1% sulphanilamide in 3M HCl and 80 μl of 0.02% N-(1-naphthyl)ethylenediamine dihydrochloride were added and the absorbance determined at 546nm.
Western blotting
Sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) was performed in a 1.5mm thick 10% (w/v) resolving gel and a 4.6% acrylamide (w/v) stacking gel in a vertical electrophoresis cell (Mini- Protean III; Bio-Rad) at 150V for 150min. Gels were electroblotted onto nitrocellulose membrane for 75min at 100V in a Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad). Blots were blocked in 5% (w/v) skim milk in 20mM TRIS-buffered saline at 4 °C for 1h. α-GDH (1:5000), α-GS (1:2000), and α-NR (1:1000; Agrisera, Sweeden) were used as primary antibodies. The secondary antibody was goat anti-rabbit horseradish peroxidase conjugate (1:50 000, Sigma-Aldrich, St. Louis, MO, USA). Immunoreactive bands were visualized with a highly sensitive chemiluminescent substrate for peroxidase detection (GE Healthcare Europe GmbH, Freiburg, Germany).
Data analysis
Data analyses were carried out using SPSS 17.0 (Chicago, IL, USA). Statistical differences between nitrate and ammonium nutrition for each accession and variable were assessed comparing the mean values by paired t-test. To test the connectivity between variables, Pearson’s correlation coefficient was calculated for P≤0.05. Multiple regressions provided a view of the relationship between a trait and shoot biomass independent of other correlated traits. Multiple regression estimations can suffer from multicollinearity wherein highly correlated traits might act redundantly. Thus, to help in interpretation, Akaike’s information criterion (AIC) was also used to determine the ‘best’ model by rewarding added explanatory power but penalizing the inclusion of additional terms. This provides the simplest model with the least collinearity and, thus, supposedly, the best estimation of selection (Shaw and Geyer, 2010).
Results
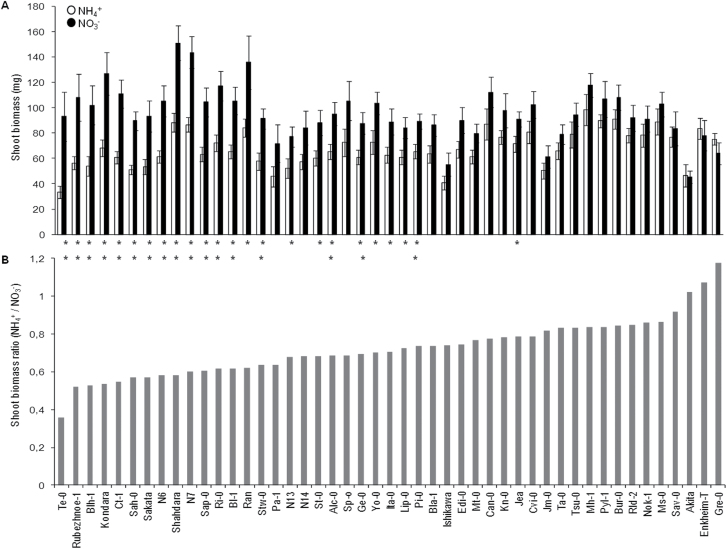
To evaluate NUE with ammonium as the sole N source, Arabidopsis rosette biomass was compared after 3 weeks of growth under 1mM NH4 + [0.5mM (NH4)2SO4] or 1mM NO3 – [0.5mM Ca(NO3)2], and the ratio between shoot biomass under NH4 + and NO3 – conditions (SB NH4 +/NO3 –) was used to estimate ammonium tolerance as it has been previously used in other studies (Cruz et al., 2006; Ariz et al., 2011). In general, Arabidopsis is a species sensitive to NH4 + and nearly every ecotype analysed showed shoot biomass inhibition in response to NH4 +. Twenty-four out of the forty-seven accessions analysed experienced a significant growth inhibition upon NH4 + nutrition (Fig. 1A). The accession Te-0 was the one showing the lowest SB NH4 +/NO3 – ratio (<0.4), which was significantly lower than that of the next most sensitive accession to NH4 + (Rubenzhnoe-1; SB NH4 +/NO3 – 0.56). Only three accessions had an SB NH4 +/NO3 – ratio >1, but without significant differences between both types of nutrition (Akita, Enkheim-T, and Gre-0; Fig. 1B). Overall, intraspecific shoot growth variability under a contrasting N source is evident by the use of this collection of accessions (Fig. 1B).
Fig. 1.
Natural variation of Arabidopsis thaliana growth under nitrate or ammonium as N source. (A) Shoot biomass. (B) Ratio between shoot biomass under NH4 + and NO3 – nutrition. Means and standard errors were calculated from 8–12 plants. Significant differences between shoot biomass under ammonium compared with nitrate nutrition are indicated for each accession (*P<0.05; **P>0.01).
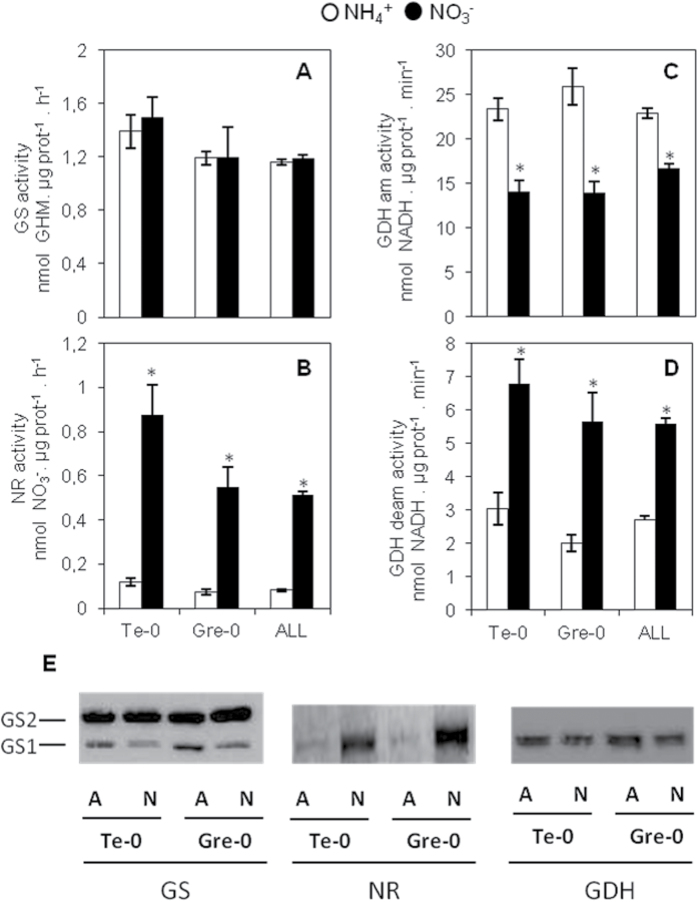
The content of ammonium and free amino acids (Supplementary Table S1 available at JXB online) as well as NR, GS, and GDH enzyme activities (Supplementary Table S1) were determined. GDH activity was measured both in the aminating (GDHam) and in the deaminating (GDHdeam) direction. Regarding NH4 + content, overall, plants under NH4 + nutrition contained significantly more NH4 + compared with plants fed with NO3 –. Eight accessions (Enkheim-T, Gre-0, Ishikawa, Jea, Ms-0, Ran, Ta-0, and Tsu-0) did not show significant differences between both treatments (Supplementary Table S1). Amino acid content followed a similar trend to NH4 + content (Supplementary Fig. S1) and every accession under NH4 + nutrition contained significantly more amino acids compared with under NO3 – nutrition (Supplementary Table S1, Fig. S1). Concerning the enzyme activities, as expected, every accession under NO3 – nutrition had a higher NR activity (Fig. 2B; Supplementary Table S1). GS activity was similar for every accession under both forms of nutrition, except for Mt-0 and Ct-1 that showed a slightly higher GS activity under NO3 – nutrition and for Rld-2, N7, and N14 that experienced a small increase under NH4 + nutrition (Fig. 2A; Supplementary Table S1). GDHam activity was higher under NH4 + nutrition in 35 out of the 47 accessions. In contrast, GDHdeam activity was higher under NO3 – nutrition in every accession except for Akita, Ishikawa, Rld-2, Pa-1, and Sah-0, which did not show significant differences between both forms of nutrition (Fig. 2C, D; Supplementary Table S1).
Fig. 2.
Enzyme activities of Te-0 and Gre-0 accessions and the mean of every accession (ALL) for (A) GS, (B) NR, (C) GDHam, and (D) GDH deam, and (E) western blot of GS, GDH, and NR for Te-0 and Gre-0 accessions grown under ammonium or nitrate nutrition. An asterisk indicates a significant difference for P<0.05 (n=6).
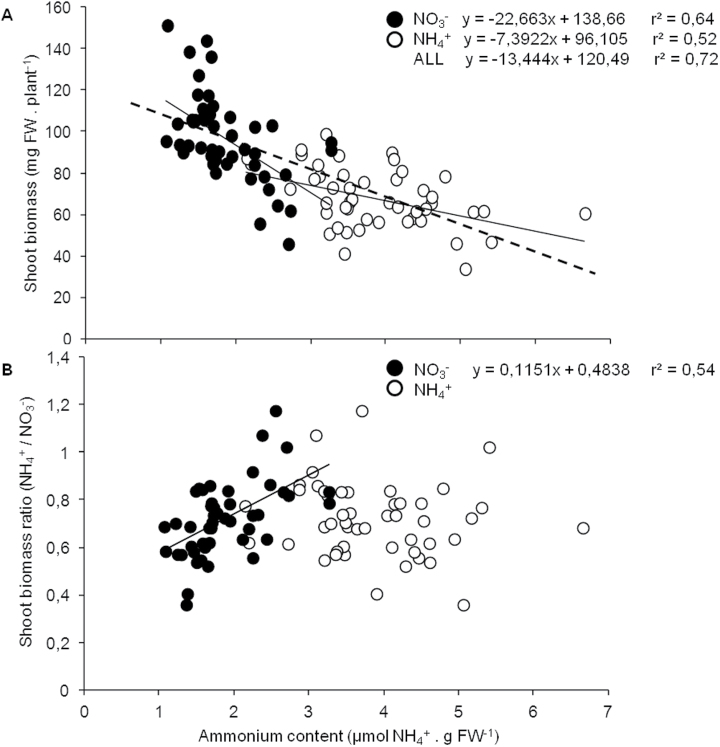
To investigate the connectivity between the different parameters, a Pearson correlation analysis was performed for each pair of parameters. Values are given for the correlation coefficient (r 2) and the significance (P). The results are presented separately for the plants grown under NH4 + (Table 1) and NO3 – nutrition (Table 2). Shoot biomass under both ammonium and nitrate nutrition was negatively correlated with NH4 + and free amino acid content (Tables 1, 2; Fig. 3A), which is reasonable because it could mean that part of the absorbed N is not being used for growth, and ammonium accumulation inside plant tissues is known to be deleterious for plant performance (Britto and Krontzuker, 2002; Ludewig et al., 2007). None of the parameters determined in NH4 +-fed plants showed any correlation with the SB NH4 +/NO3 – ratio (Table 1). In contrast, in NO3 –-fed plants, NH4 + and amino acid content, together with GDHam activity, were positively correlated with the SB NH4 +/NO3 – ratio (Table 2).
Table 1.
Pearson correlations between the determined parameters in Arabidopsis thaliana plants under NH4 + nutritionSB indicates the shoot biomass, and SB NH4 +/NO3 – denotes the shoot biomass ratio between NH4 +- and NO3 –-fed plants.
| SB NH4 +/NO3 – | SB | NH4 + | Amino acids | NR activity | GS activity | GDHam activity | GDHdeam activity | ||
|---|---|---|---|---|---|---|---|---|---|
| SB NH4 +/NO3 – | r 2 | 1 | |||||||
| P | |||||||||
| SB | r 2 | 0.427** | 1 | ||||||
| P | 0.002 | ||||||||
| NH4 + | r 2 | –0.144 | –0.447** | 1 | |||||
| P | 0.328 | 0.001 | |||||||
| Amino acids | r 2 | –0.001 | –0.405** | 0.554** | 1 | ||||
| P | 0.997 | 0.004 | 0.000 | ||||||
| NR activity | r 2 | –0.014 | –0.192 | 0.099 | 0.048 | 1 | |||
| P | 0.926 | 0.192 | 0.505 | 0.744 | |||||
| GS activity | r 2 | 0.105 | –0.016 | –0.118 | 0.002 | 0.248 | 1 | ||
| P | 0.476 | 0.913 | 0.426 | 0.988 | 0.089 | ||||
| GDHam activity | r 2 | 0.212 | 0.011 | 0.327* | 0.321* | 0.056 | 0.054 | 1 | |
| P | 0.149 | 0.940 | 0.023 | 0.026 | 0.704 | 0.717 | |||
| GDHdeam activity | r 2 | 0.136 | –0.052 | 0.265 | 0.305* | 0.139 | 0.010 | 0.687** | 1 |
| P | 0.355 | 0.723 | 0.069 | 0.035 | 0.346 | 0.948 | 0.000 |
Table 2.
Pearson correlations between the determined parameters in Arabidopsis thaliana plants under NO3 – nutritionSB indicates the shoot biomass, and SB NH4 +/NO3 – denotes the shoot biomass ratio between NH4 +- and NO3 –-fed plants.
| SB NH4 +/NO3 – | SB | NH4 + | Amino acids | NR activity | GS activity | GDHam activity | GDHdeam activity | ||
|---|---|---|---|---|---|---|---|---|---|
| SB NH4 +/NO3 – | r 2 | 1 | |||||||
| P | |||||||||
| SB | r 2 | –0.524** | 1 | ||||||
| P | 0.000 | ||||||||
| NH4 + content | r 2 | 0.547** | –0.566** | 1 | |||||
| P | 0.000 | 0.000 | |||||||
| Amino acid content | r 2 | 0.478** | –0.544** | 0.496** | 1 | ||||
| P | 0.001 | 0.000 | 0.000 | ||||||
| NR activity | r 2 | 0.124 | –0.138 | 0.340* | 0.085 | 1 | |||
| P | 0.403 | 0.349 | 0.018 | 0.567 | |||||
| GS activity | r 2 | 0.029 | –0.105 | 0.139 | –0.014 | 0.335* | 1 | ||
| P | 0.845 | 0.478 | 0.346 | 0.927 | 0.020 | ||||
| GDHam activity | r 2 | 0.438** | –0.238 | 0.389* | 0.326* | 0.162 | 0.066 | 1 | |
| P | 0.002 | 0.103 | 0.006 | 0.024 | 0.271 | 0.655 | |||
| GDHdeam activity | r2 | 0.048 | 0.146 | 0.078 | –0.187 | 0.220 | 0.489** | 0.156 | 1 |
| p | 0.744 | 0.321 | 0.596 | 0.204 | 0.133 | 0.000 | 0.288 |
Fig. 3.
Scatter plots of ammonium content (horizontal axis) versus (A) shoot biomass and (B) the ratio between shoot biomass under NH4 + and NO3 –. Linear regression and Pearson r 2 are given only if P was <0.05.
Regarding the enzyme activities, in NH4 +-fed plants, neither GS nor NR activity showed any correlation with any of the parameters determined (Table 1). GDHam and GDHdeam activities were positively correlated with each other, suggesting that when a genotype shows high GDH activity, it occurs in both the aminating and deaminating directions. Both GDHam and GDHdeam activities were positively correlated with amino acid content; however, only GDHam activity was positively correlated with NH4 + content (Table 1). In NO3 –-fed plants, NR activity was positively correlated with NH4 + content and with GS activity (Table 2). In addition, GS activity was also correlated with GDHdeam activity. Interestingly, and similarly to NH4 +-fed plants, in NO3 –-fed plants GDHam activity was also correlated with ammonium and amino acid content (Table 2).
In order to better understand the relationships between the SB NH4 +/NO3 – ratio and the different determined parameters, a multiple regression full model and AIC best model (AIC-selected) were applied. The full model only indicated a significant selection for the ammonium content in NO3 –-fed plants (Table 3) and explained 23% of the variance in SB NH4 +/NO3 –. In the best model, the percentage of the variance in SB NH4 +/NO3 – explained increased up to 38%. From the four traits retained in the best model (ammonium content in both NH4 +- and NO3 –-fed plants; amino acid content in NO3 –-fed plants; and NR activity under NH4 + nutrition), NH4 + and amino acid accumulation in NO3 –-fed plants were significantly retained. Interestingly, NH4 + content explained 53% of the best model.
Table 3.
Full and Akaike’s information criterion (AIC)-selected best multiple regression models of Arabidopsis thaliana ammonium tolerance based on the ratio of the rosette biomass between plants grown under NH4 + or NO3 – nutritionSelection gradients (β) and standard errors (SE) are presented along with P-values.
| Trait | Treatment | SB NH4 +/NO3 – | |||
|---|---|---|---|---|---|
| Full model | AIC-selected best model | ||||
| β ±SE | P-value | β ±SE | P-value | ||
| NH4 + | A | –0.037±0.029 | 0.214 | –0.033±0.020 | 0.108 |
| NH4 + | N | 0.155±0.045 | 0.002 | 0.106±0.041 | 0.002 |
| NO3 – | A | –0.001±0.003 | 0.848 | – | – |
| NO3 – | N | –0.001±0.002 | 0.651 | – | – |
| Amino acids | A | 0.001±0.002 | 0.630 | – | – |
| Amino acids | N | 0.010±0.005 | 0.081 | 0.008±0.004 | 0.040 |
| NR activity | A | –0.882±1.064 | 0.412 | –1.266±0.795 | 0.119 |
| NR activity | N | –0.031±0.183 | 0.867 | – | – |
| GS activity | A | –0.053±0.172 | 0.760 | – | – |
| GS activity | N | –0.042±0.143 | 0.773 | – | – |
| GDHam activity | A | –0.005±0.009 | 0.594 | – | – |
| GDHam activity | N | 0.010±0.008 | 0.215 | – | – |
| GDHdeam activity | A | 0.021±0.048 | 0.669 | – | – |
| GDHdeam activity | N | –0.004±.0.022 | 0.852 | – | – |
| r 2 0.23 | r 2 0.38 | ||||
Significant selection gradients are presented in bold.
A, ammonium-fed plants; N, nitrate-fed plants.
The same analysis was performed for the shoot biomass under both forms of nutrition. For NH4 +-fed plants, the models only indicated selection for ammonium content, and both the full and best models only explained 19% of the variance in shoot biomass (Supplementary Table S3 at JXB online). For NO3 –-fed plants, both the full and the best model explained 39% of the variance in shoot biomass. The full model indicated selection for ammonium and amino acid content (Supplementary Table S2), and both models significantly retained the ammonium and amino acid content (Supplementary Table 2).
According to the importance given by both Pearson correlations and the multiple regression models, the correlation of ammonium content both with shoot biomass and with SB NH4 +/NO3 – was illustrated (Fig. 3). As shown by Pearson analysis (Tables 1, 2), ammonium content was negatively correlated with shoot biomass under both NH4 + and NO3 – nutrition (Fig. 3A). Interestingly, and as suggested by the multiple regression model, only the ammonium content in NO3 –-fed plants was correlated with the SB NH4 +/NO3 – ratio (Fig. 3B).
To understand further the behaviour of the N-assimilating enzymes determined, the enzyme activities were illustrated and western blotting analysis was performed for the accessions Te-0 and Gre-0, the most sensitive and tolerant accessions to ammonium, respectively (Fig. 3). This analysis did not show any difference for any of the three enzymes under both forms of nutrition. However, it was useful to ascertain that although there were no significant differences in GS activity, the GS1 isoform content was clearly accumulated upon ammonium nutrition (Fig. 2E). NR protein content, in agreement with NR activity, was dramatically induced in NO3 –-fed Te-0 and Gre-0 plants. Finally, GDH content increased in NH4 +-fed plants, according to the increase in GDHam activity (Fig. 2C). In contrast, although GDH was induced upon ammonium nutrition, as described above, GDHdeam activity increased in NO3 –-fed plants (Fig. 2D). However, it must be noted that under NH4 + nutrition, the average GDHam activity was around eight times higher than the GDHdeam activity, whilst under NO3 – nutrition GDHam activity was about three times higher than GDHdeam activity.
Discussion
Plant response to N availability depends on the genotype, the N source, and N fertilization level, and the limiting steps in N metabolism are different at low and high N supply (Chardon et al., 2012; Xu et al., 2012). Overall, NUE is higher when N supply is limiting. In general, adaptation to low N environments is challenging to most cultivars, because they have been selected under high-nutrient environments but plants in natural field conditions are faced with environmental changes where N availability varies and the better NUE under low N conditions is a competitive advantage (Kant et al., 2011). Moreover, reducing N fertilizer input in the soil while maintaining productivity is an unavoidable strategy to reduce agricultural impact on the environment. Thus, and taking into account that Arabidopsis and the Brassicaceae family have been described as very susceptible to ammonium nutrition, in this work, a low N dose (1mM) was used. Because of this high sensitivity, most of the studies related to ammonium toxicity in Arabidopsis have been performed with mixed nutrition, and thus long-term ammonium-based nutrition studies involve the use of a low ammonium concentration.
Approaches based on intraspecific natural variation have become an important means to study plants adaptation. Regarding nitrate nutrition, studies based on natural variation have already been used in several species including maize (Coque and Gallais, 2007) and rice (Namai et al., 2009). Arabidopsis natural variation has also been studied under limiting and ample nitrate supply (North et al., 2009; Chardon et al., 2010) and to evaluate the capacity of different genotypes for N remobilization during seed filling (Masclaux-Daubresse and Chardon, 2011). In contrast, studies focused in intraspecific variation of N use with ammonium as the sole N source are more scarce, although examples exist, studying, among others, four maize cultivars (Schortemeyer et al., 1997), a collection of rice inbred lines (Obara et al., 2010), and four pea cultivars (Cruz et al., 2011). In this work, data from 47 natural accessions of Arabidopsis were collected and several traits related to N metabolism were measured to determine the natural variation of Arabidopsis growth and N metabolism (ammonium and amino acid content, and NR, GS, and GDH enzyme activities) under two different N sources (nitrate or ammonium). Biomass is considered as the best indicator of plant performance because it integrates every aspect of plant metabolism, from nutrient uptake to its assimilation, and the ratio of the shoot biomass under ammonium versus nitrate nutrition was considered here as an indicator of the plant’s tolerance/sensitivity to ammonium, as it has previously been used in other works (Cruz et al., 2006; Ariz et al., 2011). Arabidopsis accession N1438 grown under 2.5mM NH4 + for 21 d showed three times less biomass compared with plants grown under NO3 –, and the authors suggested ionic imbalance as a major cause of this toxicity (Helali et al., 2010). Similarly, Hoffman et al. (2007) reported a retardation of Arabidopsis Col-0 seedling growth under NH4 + nutrition compared with NO3 – nutrition. The present study confirms an overall sensitivity of Arabidopsis to ammonium, since, out of the 47 genotypes, 44 had a ratio <1 (23 accessions showing significant differences in shoot biomass between both forms of nutrition). However, this study highlights large intraspecific variation of ammonium tolerance expressed as SB NH4 +/NO3 –, which varied between 0.36 and 1.18. These values are in agreement with the values registered by Ariz et al. (2011) working with seven different species and ammonium concentrations. Thus, the present study, working with a low ammonium concentration, reveals a similar degree of intraspecific Arabidopsis ammonium tolerance variability to the interspecific degree of ammonium tolerance variability. This underscores the high variability within a single species and the power of natural variation approaches for plant adaptation studies.
Ammonium accumulation affects plant growth
‘Excessive’ ammonium accumulation is toxic to cells. However, the concept of ‘excessive’ is extremely variable depending on the plant species and on the soil NH4 + concentration. In fact, ammonium toxicity is considered to be ‘universal’ even in species labelled as ‘NH4 + specialists’ (Li et al., 2014). Excess ammonium causes an imbalance in, among others, pH homeostasis, ionic equilibrium, and primary metabolism (Britto and Kronzucker, 2002). Ammonium accumulation might derive from its direct uptake but also from amino acid deamination, protein degradation, and photorespiration. To prevent the cytosol from ammonium overload, plants deploy different strategies including AMT-type ammonium transporter regulation (Lanquar et al., 2009) or increasing ammonium assimilation (Setien et al., 2013). In the present work, as expected, NH4 +-fed plants accumulated more NH4 + and amino acids than NO3 –-fed plants and this NH4 + accumulation was negatively correlated with Arabidopsis rosette biomass (Fig. 3A). Interestingly, this correlation was found for plants grown under both forms of nutrition, suggesting that ammonium accumulation negatively influences plant growth even under nitric nutrition. NH4 + accumulation under low N supply might be due to a lack of proper carbohydrate supply for ammonium assimilation or to the toxicity caused by the excess NH4 + as stated above. To the authors’ knowledge, this is the first time that a correlation between plant shoot growth under NO3 – as sole N source and the accumulation of NH4 + in leaves has been reported, which provides evidence of the extreme sensitivity of Arabidopsis to ammonium.
Regarding the SB NH4 +/NO3 – ratio, of the parameters determined, only ammonium, amino acid content, and GDHam activity from NO3 –-fed plants showed a significant correlation (Table 2). Multiple regression full and best models retained ammonium and amino acid content, which both show a strong correlation (Supplementary Fig. S1 at JXB online), as significant factors explaining the variation in the SB NH4 +/NO3 – ratio (Table 3). Interestingly, the NH4 + content of NH4 +-fed plants did not show any significant correlation with the SB NH4 +/NO3 – ratio. Thus, the fact that NO3 –-fed plants with a higher NH4 + content present a smaller rosette biomass (Fig. 3A) could explain the relationship between ammonium content of NO3 –-fed plants and the SB NH4 +/NO3 – ratio (Fig. 3B). Alternatively, it can be speculated that evolutionarily a plant that under NO3 – nutrition is able to accumulate more ammonium could be genetically better adapted to an ammonium-based nutrition.
Role of NR, GS, and GDH in Arabidopsis response to ammonium
NO3 – absorbed from the nutrient solution is reduced to ammonium, whereas in NH4 +-fed plants this step is bypassed and ammonium is directly assimilated for plant growth. As expected, NR activity was induced upon NO3 – exposure but it was not related to differential plant growth. Indeed, NR or nitrite reductase overexpression in tobacco, potato, or Arabidopsis did not increase plant biomass, thus nitrate reduction does not seem to be a limiting step for plant growth (Pathak et al; 2008; Masclaux-Daubresse et al., 2010). Ammonium assimilation in normal conditions in plants mainly occurs via the GS/GOGAT cycle. There are two different GS isoforms. GS1 is encoded by five genes in Arabidopsis and functions primarily in assimilating ammonia during nitrogen remobilization. GS2 is encoded by a single gene in Arabidopsis and has been involved in assimilating the ammonia coming from nitrate reduction or photorespiration (Xu et al., 2012). In general, plants with higher GS activities are considered more tolerant to ammonium, and Cruz et al. (2006) showed a relationship between GS activity in the dark and ammonium tolerance. In this work, no difference in GS activity was found in almost every accession between NH4 +- and NO3 –-fed plants (Fig. 2A; Supplementary Table S2 at JXB online) and there was no correlation between GS activity and shoot biomass in plants under both forms of nutrition (Tables 1, 12). A western blot analysis was performed in two accessions with contrasting ammonium tolerance (Te-0 and Gre-0), and in both cases there was a clear accumulation of the GS1 isoform in response to ammonium nutrition. Overall, total GS activity does not seem to be crucial for ammonium tolerance in Arabidopsis; however, GS1 could have an important role when ammonium is supplied as the N source. Moreover, out of the five genes encoding GS1 in Arabidopsis GS1;2 is the most highly expressed in leaves and it is induced by ammonium (Lothier et al., 2011). Indeed, an Arabidopsis mutant lacking GS1;2 expression exhibited reduced growth under a 7 d ammonium treatment compared with the wild type (Lothier et al., 2011). Similarly, a rice mutant in the GS1;1 gene was also more sensitive upon ammonium nutrition (Kusano et al., 2011). Thus, it remains to be determined whether GS1;2 and the rest of the GS isozymes are related to Arabidopsis variability under ammonium nutrition. Also, very recently root NADH-GOGAT has been suggested to play an important role in ammonium assimilation under ammonium nutrition (Konishi et al., 2014s).
GDH is able to catalyse the in vitro reversible amination of 2-oxoglutarate to glutamate. In vivo, the existence of the N assimilating capacity of GDH is controversial and in the last years evidence has been accumulating in favour of the major role of GDH deamination, for example by the use of 15N-nuclear magnetic resonance (NMR) labelling studies showing that there was no direct incorporation of ammonia into glutamate when GS was inhibited (Labboun et al., 2009; Tercé-Laforgue et al., 2013). However, although in unstressed plants GDH ammonia assimilating capacity seems to be negligible, it appears that under stress conditions and under ammonium nutrition, GDH could incorporate NH4 + (Skopelitis; 2006; Setien et al., 2013). In the present study, a contrasting behaviour of GDH activity was found. GDHam activity was generally induced upon NH4 + exposure whereas GDHdeam activity was repressed (Fig. 2C, D; Supplementary Table S1 at JXB online). Moreover, in both NH4 +- and NO3 –-fed plants ammonium and amino acid contents were positively correlated with GDHam activity, and not with GDHdeam activity (Tables 1, 12). Thus, the present data suggest that NH4 + accumulation might be stimulating the ammonium-incorporating capacity of GDH rather than being a consequence of NH4 + release associated with GDH glutamate deamination. Nevertheless, experiments designed to ascertain the actual GDHam activity in conditions of plant growth under an exclusive ammoniacal nutrition, such as by 15N-NMR labelling, are necessary.
GDH is traditionally accepted to form seven isoenzymes composed of α and β homo- or heterodimers. Recently, the existence in Arabidopsis of a third gene encoding a γ subunit has been shown (Fontaine et al., 2012). However, the activity of this γ isoenzyme was exclusively from root (Fontaine et al., 2012), which is in line with the hypothesis that each of the GDH subunits may have specific biological functions (Purnell et al., 2005; Tercé-Laforgue et al., 2013). In the present work, after SDS–PAGE, GDH was accumulated under ammonium nutrition (Fig. 2E). An accumulation of GDH polypeptides has already been reported in several species including wheat (Setien et al. 2013), pea (Ariz et al., 2013), and tomato (Setien et al. 2014). The overall data indicate a key role for GDH in Arabidopsis under NH4 + nutrition.
Concluding remarks and future prospects
Overall, the results obtained in this work reveal that there exists high natural variation in A. thaliana growth as a function of the N source. This variation was partially due to the differential tissue NH4 + and amino acid accumulation in both NO3 –-fed and NH4 +-fed plants. Similarly, significant natural variability was detected in NH4 + tolerance expressed as the SB NH4 +/NO3 – ratio, and, interestingly, NH4 + accumulation in NO3-fed plants was the parameter showing the highest relevance, which may indicate an evolutionary adaptation suggesting that plants that under NO3 – nutrition are able to accumulate more ammonium could be genetically better adapted to an ammonium-based nutrition. Although plant NH4 + assimilation capacity is known to be a key aspect for ammonium tolerance, GS and GDH activity does not seem to be responsible for the variability shown in A. thaliana. However, the modulation of GDH activity as a function of the supplied N source was clearly observed, which suggests an important role for this enzyme in NH4 + assimilation. Similarly, the observed higher content of the GS1 isoform in NH4 +-fed plants could also contribute to NH4 + assimilation. The quality of the root system has also been suggested partly to explain the differences in nitrogen uptake and NUE (Loudet et al., 2005). Furthermore, several works have highlighted the importance of the root in NH4 + tolerance (Setien et al., 2013, 2014, Kojima et al., 2014). Thus, future works dealing with root metabolism will be useful to ascertain whether N assimilation in this organ could be related to the natural variability in NH4 + tolerance in A. thaliana. Also, approaches using larger A. thaliana natural populations in combination with genome-wide association studies (Atwell et al., 2010) will no doubt be very helpful in elucidating the genetic basis underlying the Arabidopsis intraspecific variability in ammonium tolerance.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Scatter plots of amino acids versus ammonium content of leaves of Arabidopsis thaliana grown under NH4 + and NO3 –.
Table S1. Ammonium and amino acid content and enzyme activities: whole data set.
Table S2. Full and Akaike’s information criterion (AIC)-selected best multiple regression models of Arabidopsis thaliana rosette biomass grown under NH4 + or NO3 – nutrition.
Acknowledgements
AS holds a PhD Grant from the Basque Government. The research leading to these results has received funding from the Basque Government (IT526-10) and the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme (FP7/2007–2013) under REA grant agreement number 334019. We thank Anabel Robredo for technical assistance. We are also grateful to Dr J.F. Moran and Dr K.A. Roubelakis-Angelakis for providing GS and GDH antibodies, respectively.
Glossary
Abbreviations:
- GDH
glutamate dehydrogenase
- GDHam
glutamate dehydrogenase aminating
- GDHdeam
glutamate dehydrogenase deaminating
- GS
glutamine synthetase
- NR
nitrate reductase
- NUE
nitrogen use efficiency
- SB
shoot biomass.
References
- Ariz I, Asensio AC, Zamarreno AM, Garcia-Mina JM, Aparicio-Tejo P, Moran JF. 2013. Changes in the C/N balance caused by increasing external ammonium concentrations are driven by carbon and energy availabilities during ammonium nutrition in pea plants: the key roles of asparagine synthetase and anaplerotic enzymes. Physiologia Plantarum 148, 522–537 [DOI] [PubMed] [Google Scholar]
- Ariz I, Cruz C, Moran JF, Gonzalez-Moro MB, Garcia-Olaverri C, Gonzalez-Murua C, Martins-Loucao MA, Aparicio-Tejo PM. 2011. Depletion of the heaviest stable N isotope is associated with NH4+/NH3 toxicity in NH4+-fed plants. BMC Plant Biology 11, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjalmsson BJ, et al. 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465, 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom AJ, Jackson LE, Smart DR. 1993. Root-growth as a function of ammonium and nitrate in the root zone. Plant, Cell and Environment 16, 199–206 [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry 72, 248–254 [DOI] [PubMed] [Google Scholar]
- Britto DT, Kronzucker HJ. 2002. NH4+ toxicity in higher plants: a critical review. Journal of Plant Physiology 159, 567–584 [Google Scholar]
- Chardon F, Barthelemy J, Daniel-Vedele F, Masclaux-Daubresse C. 2010. Natural variation of nitrate uptake and nitrogen use efficiency in Arabidopsis thaliana cultivated with limiting and ample nitrogen supply. Journal of Experimental Botany 61, 2293–2302 [DOI] [PubMed] [Google Scholar]
- Chardon F, Noel V, Masclaux-Daubresse C. 2012. Exploring NUE in crops and in Arabidopsis ideotypes to improve yield and seed quality. Journal of Experimental Botany 63, 3401–3412 [DOI] [PubMed] [Google Scholar]
- Coque M, Gallais A. 2007. Genetic variation for nitrogen remobilization and postsilking nitrogen uptake in maize recombinant inbred lines: heritabilities and correlations among traits. Crop Science 47, 1787–1796 [Google Scholar]
- Coskun D, Britto DT, Li MY, Becker A, Kronzucker HJ. 2013. Rapid ammonia gas transport accounts for futile transmembrane cycling under NH3/NH4+ toxicity in plant roots. Plant Physiology 163, 1859–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz C, Bio AFM, Dominguez-Valdivia MD, Aparicio-Tejo PM, Lamsfus C, Martins-Loucao MA. 2006. How does glutamine synthetase activity determine plant tolerance to ammonium? Planta 223, 1068–1080 [DOI] [PubMed] [Google Scholar]
- Cruz C, Dominguez-Valdivia MD, Aparicio-Tejo PM, Lamsfus C, Bio A, Martins-Loucao MA, Moran JF. 2011. Intra-specific variation in pea responses to ammonium nutrition leads to different degrees of tolerance. Environmental and Experimental Botany 70, 233–243 [Google Scholar]
- Dominguez-Valdivia MD, Aparicio-Tejo PM, Lamsfus C, Cruz C, Martins-Loucao MA, Moran JF. 2008. Nitrogen nutrition and antioxidant metabolism in ammonium-tolerant and -sensitive plants. Physiologia Plantarum 132, 359–369 [DOI] [PubMed] [Google Scholar]
- FAO. 2009. Global agriculture towards 2050. High Level Expert Forum—how to feed the world in 2050. Rome: Food and Agriculture Organization (United Nations) [Google Scholar]
- Foley JA, Ramankutty N, Brauman KA, et al. 2011. Solutions for a cultivated planet. Nature 478, 337–342 [DOI] [PubMed] [Google Scholar]
- Fontaine JX, Terce-Laforgue T, Armengaud P, et al. 2012. Characterization of a NADH-dependent glutamate dehydrogenase mutant of Arabidopsis demonstrates the key role of this enzyme in root carbon and nitrogen metabolism. The Plant Cell 24, 4044–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Hohne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M. 2004. A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. The Plant Cell 16, 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good AG, Beatty PH. 2011. Fertilizing nature: a tragedy of excess in the commons. PLoS Biology 9, e1001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber N, Galloway JN. 2008. An earth-system perspective of the global nitrogen cycle. Nature 451, 293–296 [DOI] [PubMed] [Google Scholar]
- Helali SM, Nebli H, Kaddour R, Mahmoudi H, Lachaal M, Ouerghi Z. 2010. Influence of nitrate–ammonium ratio on growth and nutrition of Arabidopsis thaliana . Plant and Soil 336, 65–74 [Google Scholar]
- Hoffmann A, Milde S, Desel C, et al. 2007. N form-dependent growth retardation of Arabidopsis thaliana seedlings as revealed from physiological and microarray studies. Journal of Plant Nutrition and Soil Science 170, 87–97 [Google Scholar]
- IPCC. 2007. Climate change 2007: mitigation of climate change. In: Metz B, Davidson O, Bosch P, Dave R, Meter L, eds. Contribution of Working Group III to the Fourth Assessment Report of the Intergovernmental Panel of Climate Change. Cambridge: Cambridge University Press [Google Scholar]
- Kant S, Bi YM, Rothstein SJ. 2011. Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. Journal of Experimental Botany 62, 1499–1509 [DOI] [PubMed] [Google Scholar]
- Konishi N, Ishiyama K, Matsuoka K, Muru I, Hayakawa T, Yamaya T, Kojima S. 2014. NADH-dependent glutamate synthase plays a crucial role in assimilating ammonium in the Arabidopsis root. Physiologia Plantarum 152, 138–151 [DOI] [PubMed] [Google Scholar]
- Kronzucker HJ, Britto DT, Davenport RJ, Tester M. 2001. Ammonium toxicity and the real cost of transport. Trends in Plant Science 6, 335–337 [DOI] [PubMed] [Google Scholar]
- Kusano M, Tabuchi M, Fukushima A, et al. 2011. Metabolomics data reveal a crucial role of cytosolic glutamine synthetase 1;1 in coordinating metabolic balance in rice. The Plant Journal 66, 456–466 [DOI] [PubMed] [Google Scholar]
- Labboun S, Terce-Laforgue T, Roscher A, et al. 2009. Resolving the role of plant glutamate dehydrogenase. I. In vivo real time nuclear magnetic resonance spectroscopy experiments Plant and Cell Physiology 50, 1994–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanquar V, Loque D, Hormann F, Yuan LX, Bohner A, Engelsberger WR, Lalonde S, Schulze WX, von Wiren N, Frommer WB. 2009. Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. The Plant Cell 21, 3610–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BH, Li GJ, Kronzucker HJ, Baluska F, Shi WM. 2014. Ammonium stress in Arabidopsis: signaling, genetic loci, and physiological targets. Trends in Plant Science 19, 107–114 [DOI] [PubMed] [Google Scholar]
- Li B, Shi W, Su Y. 2011. The differing responses of two Arabidopsis ecotypes to ammonium are modulated by the photoperiod regime. Acta Physiologiae Plantarum 33, 325–334 [Google Scholar]
- Lima JE, Kojima S, Takahashi H, von Wiren N. 2010. Ammonium triggers lateral root branching in Arabidopsis in an AMMONIUM TRANSPORTER1;3-dependent manner. The Plant Cell 22, 3621–3633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lothier J, Gaufichon L, Sormani R, Lemaitre T, Azzopardi M, Morin H, Chardon F, Reisdorf-Cren M, Avice JC, Masclaux-Daubresse C. 2011. The cytosolic glutamine synthetase GLN1;2 plays a role in the control of plant growth and ammonium homeostasis in Arabidopsis rosettes when nitrate supply is not limiting. Journal of Experimental Botany 62, 1375–1390 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Merigout T, Talbotec J, Daniel-Vedele F. 2003. Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis. Plant Physiology 131, 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Gaudon V, Trubuil A, Daniel-Vedele F. 2005. Quantitative trait loci controlling root growth and architecture in Arabidopsis thaliana confirmed by heterogeneous inbred family. Theoretical and Applied Genetics 110, 742–753 [DOI] [PubMed] [Google Scholar]
- Ludewlg U, Neuhduser B, Dynowski M. 2007. Molecular mechanisms of ammonium transport and accumulation in plants. FEBS Letters 581, 2301–2308 [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Chardon F. 2011. Exploring nitrogen remobilization for seed filling using natural variation in Arabidopsis thaliana . Journal of Experimental Botany 62, 2131–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A. 2010. Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Annals of Botany 105, 1141–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monselise EBI, Kost D. 1993. Different ammonium-ion uptake, metabolism and detoxification efficiencies in 2 Lemnaceae—a N-15-nuclear magnetic-resonance study. Planta 189, 167–173 [Google Scholar]
- Namai S, Toriyama K, Fukuta Y. 2009. Genetic variations in dry matter production and physiological nitrogen use efficiency in rice (Oryza sativa L.) varieties. Breeding Science 59, 269–276 [Google Scholar]
- North KA, Ehlting B, Koprikova A, Rennenberg H, Kopriva S. 2009. Natural variation in Arabidopsis adaptation to growth at low nitrogen conditions. Plant Physiology and Biochemistry 47, 912–918 [DOI] [PubMed] [Google Scholar]
- Obara M, Tamura W, Ebitani T, Yano M, Sato T, Yamaya T. 2010. Fine-mapping of qRL6.1, a major QTL for root length of rice seedlings grown under a wide range of NH4 (+) concentrations in hydroponic conditions. Theoretical and Applied Genetics 121, 535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak RR, Ahmad A, Lochab S, Raghuram N. 2008. Molecular physiology of plant nitrogen use efficiency and biotechnological options for its enhancement. Current Science 94, 1394–1403 [Google Scholar]
- Presterl T, Seitz G, Landbeck M, Thiemt EM, Schmidt W, Geiger HH. 2003. Improving nitrogen-use efficiency in European maize: estimation of quantitative genetic parameters. Crop Science 43, 1259–1265 [Google Scholar]
- Purnell MP, Skopelitis DS, Roubelakis-Angelakis KA, Botella JR. 2005. Modulation of higher-plant NAD(H)-dependent glutamate dehydrogenase activity in transgenic tobacco via alteration of beta subunit levels. Planta 222, 167–180 [DOI] [PubMed] [Google Scholar]
- Qin C, Qian WQ, Wang WF, Wu Y, Yu CM, Jiang XH, Wang DW, Wu P. 2008. GDP-mannose pyrophosphorylase is a genetic determinant of ammonium sensitivity in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 105, 18308–18313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauh BL, Basten C, Buckler ES, IV. 2002. Quantitative trait loci analysis of growth response to varying nitrogen sources in Arabidopsis thaliana . Theoretical and Applied Genetics 104, 743–750 [DOI] [PubMed] [Google Scholar]
- Schortemeyer M, Stamp P, Feil B. 1997. Ammonium tolerance and carbohydrate status in maize cultivars. Annals of Botany 79, 25–30 [Google Scholar]
- Setien I, Fuertes-Mendizabal T, Gonzalez A, Aparicio-Tejo PM, Gonzalez-Murua C, Gonzalez-Moro MB, Estavillo JM. 2013. High irradiance improves ammonium tolerance in wheat plants by increasing N assimilation. Journal of Plant Physiology 170, 758–771 [DOI] [PubMed] [Google Scholar]
- Setien I, Vega-Mas I, Celestino N, Calleja-Cervantes ME, Gonzalez-Murua C, Estavillo JM, Gonzalez-Moro MB. 2014. Root phosphoenolpyruvate carboxylase and NAD-malic enzymes activity increase the ammonium-assimilating capacity in tomato. Journal of Plant Physiology 171, 49–63 [DOI] [PubMed] [Google Scholar]
- Shaw RG, Geyer CJ. 2010. Inferring fitness landscapes. Evolution 64, 2510–2520 [DOI] [PubMed] [Google Scholar]
- Skopelitis DS, Paranychianakis NV, Paschalidis KA, Pliakonis ED, Delis ID, Yakoumakis DI, Kouvarakis A, Papadakis AK, Stephanou EG, Roubelakis-Angelakis KA. 2006. Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. The Plant Cell 18, 2767–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terce-Laforgue T, Bedu M, Dargel-Grafin C, Dubois F, Gibon Y, Restivo FM, Hirel B. 2013. Resolving the role of plant glutamate dehydrogenase: II. Physiological characterization of plants overexpressing the two enzyme subunits individually or simultaneously. Plant and Cell Physiology 54, 1635–1647 [DOI] [PubMed] [Google Scholar]
- Tilman D, Balzer C, Hill J, Befort BL. 2011. Global food demand and the sustainable intensification of agriculture. Proceedings of the National Academy of Sciences, USA 108, 20260–20264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GH, Fan XR, Miller AJ. 2012. Plant nitrogen assimilation and use efficiency. Annual Review of Plant Biology 63, 153–182 [DOI] [PubMed] [Google Scholar]
- Yemm EW, Cocking EC. 1955. The determination of amino-acids with ninhydrin. Analyst 80, 209–213 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.