Abstract
Allergic asthma is characterized by airway inflammation caused by infiltration and activation of inflammatory cells that produce cytokines. Many studies have revealed that c-kit, a proto-oncogene, and its ligand, stem cell factor (SCF), play an important role in the development of asthmatic inflammation. Intranasal small interference RNA (siRNA) nanoparticles targeting specific viral gene could inhibit airway inflammation. In this study, we assessed whether silencing of c-kit with intranasal small interference RNA could reduce inflammation in allergic asthma. A mouse model of experimental asthma was treated with intranasal administration of anti-c-kit siRNA to inhibit the expression of the c-kit gene. We assessed the inflammatory response in both anti-c-kit siRNA-treated and control mice. Local administration of siRNA effectively inhibited the expression of the c-kit gene and reduced airway mucus secretion and the infiltration of eosinophils in bronchoalveolar lavage fluid. Moreover, c-kit siRNA reduced the production of SCF, interleukin-4 (IL-4), and IL-5, but had no effect on interferon-γ (IFN-γ) generation. These results show that intranasal siRNA nanoparticles targeting c-kit can decrease the inflammatory response in experimental allergic asthma.
Keywords: c-kit, small interference RNA, gene silencing, inflammation, asthma
Introduction
Asthma is a chronic inflammatory disease of the airways, characterized by infiltration of mast cells, eosinophils, and T lymphocytes that produce cytokines and proinflammatory lipid mediators, which contribute to induction of downstream inflammation processes such as mucous production and airway hyperresponsiveness (AHR) [1,2]. Many research studies have shown that allergic asthma is mainly mediated by Th2 immune response [3]. The pathophysiological features are the result of an imbalance in the Th1/Th2 paradigm, with the Th2 response being predominant. Antigen-presenting cells (APCs) present allergens that are taken up and processed by Th2 cells, causing Th2 cell activation. Activated Th2 cells produce a series of cytokines, such as interleukin-4 (IL-4), IL-5, and IL-13, which are named Th2 cytokines and play a pivotal role in asthmatic pathogenesis by promoting the survival and recruitment of eosinophils and mast cells through induction of goblet cell hyperplasia and bronchial hyperreactivity [4].
The white-spotting (w) and steel (sl) loci in mice encode the transmembrane protein tyrosine kinase receptor c-kit and its ligand, stem cell factor (SCF) [5,6]. c-kit is critical for the proliferation, survival, and differentiation of hematopoietic stem and progenitor cells and several nonhematopoietic tissues [7]. Several studies have demonstrated that SCF mediates eosinophil-induced degranulation, cytokine production, and survival [8,9]. In addition, SCF is important in the development of AHR, airway inflammation, and IL-4 production [10,11]. A recent study has shown that inflammation in allergic asthma is alleviated in c-kit-defective mice, which indicates that c-kit plays a role in the development of allergic inflammation [12].
RNA interference (RNAi) is an evolutionarily conserved process of sequence-specific, posttranscriptional gene silencing that uses double-stranded RNA (dsRNA) as a signal to trigger the degradation of homologous mRNA in most eukaryotic cells and inhibit gene expression. To date, many studies have shown that small interference RNA (siRNA) could effectively silence target gene expression in vitro or in vivo. Silencing of c-kit with small interference RNA has been achieved in vivo [13]. However, whether intranasal siRNA nanoparticles can attenuate the inflammation of experimental allergic asthma mediated by c-kit remains to be determined. In this study, we used specific siRNA to silence the expression of the c-kit gene and investigated the effect of c-kit on allergic airway inflammation in a murine model of asthma. We found that intranasal siRNA nanoparticles targeting c-kit reduces airway inflammation in experimental allergic asthma.
Materials and methods
Mice and murine model of experimental asthma
SPF-grade male C57BL/6 mice (8-10 wk old) were obtained from The Laboratory Animal Center of Xi’an Medical University. Experimental protocols were approved by The Institutional Animals Ethics Committee of Xi’an Medical University.
The experimental model of asthma was generated as previously described [14]. As shown in Figure 1, mice were given ovalbumin (OVA, 20 µg, grade V; Sigma-Aldrich) with aluminum hydroxide (alum, 2.25 mg; Imject Alum; Pierce Biotechnology, Inc.) in a 100-µl total volume via intraperitoneal (i.p.) injection on days 0, 7, and 14. Mice were challenged with five doses of aerosolized 1% OVA, each for 30 min, on days 19 to 23. Mice were sacrificed with i.p. injection of a lethal dose of pentobarbital on day 26.
Figure 1.
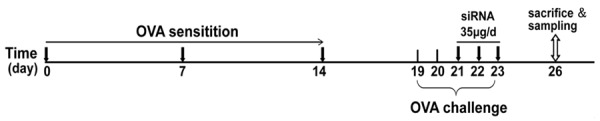
Experimental protocol for the induction of allergic asthma. Eight- to ten-week-old male C57BL/6 mice were grouped, sensitized, and challenged. The first group of asthmatic mice was administered 20 μl of PBS vehicle in each nostril. Similarly, the second and third groups of mice were administered 35 µg of scrambled siRNA and specific murine c-kit siRNA for 3 consecutive days from days 21 to 23. The mice were sacrificed and sampled on day 26, as described in Materials and methods.
Screening of the siRNA sequence
The siRNA sequence used for targeted silencing of c-kit [GenBank accession no. NM_021011] was designed by Qiagen, and siRNA sequences were selected according to the method of Sikarwar and Reddy [13]. The siRNA sequences were as follows: c-kit siRNA-1, sense: 5’-CUGUCUAGAAUUUACUCAAdTd-3’, antisense: 5’-UUGAUGAAAUUCUAGACAGdTdG-3’; c-kit-siRNA-2, sense: 5’-CCGUGACAUUCAACGUUUAdTdT-3’, antisense: 5’-UAAACGUUGAAUGUCACGGdAdA-3’.
Target siRNA was chemically modified by methylation to avoid or lessen its destruction in the tissue [15]. We selected two c-kit-specific siRNA duplexes on the basis of a previous in vitro study and used them as a mixture of an equal amount of each siRNA duplex [16]. We also designed a pair of control siRNAs, namely, scrambled siRNA and the sequence of scrambled siRNA with no complementation with any other genes in mice, to avoid specific gene silencing. The scrambled siRNA sequences were as follows: sense, 5’-UAGGCGCAGCUCCGGAUCGdTT-3’; antisense, 5’-CGAUCCGGAGCUGCGCCUAdTT-3’. All siRNAs were synthesized by Qiagen. Briefly, for each oligonucleotide, the two individual complementary strands of the siRNA were synthesized separately by the solid-phase method and then purified separately by ion-exchange chromatography. The complementary strands were annealed to form the double-stranded siRNA (duplex). The duplex was then ultrafiltered and lyophilized to form the solid drug substance.
Nasal administration of siRNA
Mice were divided into three groups and each group (n=5) was named according to treatment: PBS/vehicle, scrambled siRNA, and c-kit siRNA. On days 21 to 23, the mice were anesthetized with 0.2 ml of 2.5% Avertin (1 g/ml of tribromoethyl alcohol in tertiary amylalcohol) in PBS injected i.p. before receiving treatment. The first group of asthmatic mice was intranasally administered with 20 μl of PBS vehicle on each nostril. Similarly, the second and third groups of mice were administered with 35 µg of scrambled siRNA and specific murine c-kit siRNA, respectively, for 3 consecutive days from days 21 to 23. Lung tissue and bronchoalveolar lavage fluid (BALF) were collected 72 h after the last intranasal administration.
Histology
Lung tissue blocks from the right upper and middle lobes were rinsed in normal saline and fixed in 10% formalin for 24 h. The tissue blocks were sectioned at 4 µm with a Leica rotary microtome. The paraffin sections were stained with hematoxylin and eosin and periodic acid-Schiff reagent (Sigma). The sections were scored blindly by two histopathologists on a scale of 0 to 4 for inflammation and 0 to 4 for epithelial thickening. Inflammation was scored as follows: 0, no inflammation; 1, inflammatory cells present; 2, a few (< 3) loci of inflammation; 3, multiple (≥ 3) loci of inflammation; 4, inflammatory cells present throughout the lung. The scores for inflammation were added together and divided by 2 [0 (no disease) to 4 (maximal disease)]. The results are presented as means ± S.E.
Detection of inflammatory cytokines in the BALF
Bronchoalveolar lavage (BAL) samples were obtained, and histological evaluations were performed as previously described [17]. Briefly, mice were anesthetized, and the tracheas were isolated by blunt dissection. A small caliber tube was inserted and secured in the airway. Two successive volumes of 0.75 ml of PBS were instilled and gently aspirated, and these two volumes were pooled. Each BAL sample was centrifuged, and the supernatants were stored at -70°C until use.
Quantitation of eosinophils in the BALF
BAL with 1 ml of HBSS, instilled bilaterally with a syringe, provided lavage fluid, which was harvested by gentle aspiration three times and then centrifuged. The cells were identified as eosinophils by standard morphology, and at least 200 cells were counted under ×400 magnification. The percentage and absolute numbers of each cell type were then calculated.
RT-PCR for c-kit
For RNA isolation, immediately after removal the whole left lungs were frozen in liquid nitrogen and stored at -80°C. Total RNA was extracted from frozen lung tissue using TRIzol reagent (Gibco-BRL, Gaithersburg, MD) and amplified using the Promega PCR single-step kit (Promega, Madison, WI) according to the manufacturer’s instructions. Total RNA was transcribed into cDNA with a reverse transcriptase kit at 50°C for 30 min. The following primers for murine c-kit were used: forward, 5’-ACCCACAGGTGTCCAATTATTC-3’, and reverse, 5’-TGGCGTTCATAATTGAAGTCAC-3’. β-Actin served as an internal control. The PCR products were electrophoresed on a 3% agarose gel and visualized by ethidium bromide staining.
Western blotting
Lung tissues were taken on day 26 and immediately immersed in liquid nitrogen. The tissues were ground and then lysed in RIPA lysis buffer. Lysate (40 µg) was loaded per well and resolved on a 10% polyacrylamide gel (SDS-PAGE) under denaturing conditions; the proteins were transferred onto 0.45-µm nitrocellulose membranes. The blots were probed for murine c-kit with a murine c-kit antibody (Santa Cruz Biotechnology, Inc.) and with GAPDH antibody as a loading control (Santa Cruz Biotechnology, Inc.). Bands were visualized with an HRP-conjugated anti-rabbit IgG secondary antibody and the ECL western blotting detection system (GE Healthcare, UK).
Statistical analysis
The results are expressed as means ± SD, unless indicated otherwise. The differences between mean values were estimated using either an ANOVA followed by Fisher’s protected least significant difference tests or a Student’s t test for unpaired data. A value of P < 0.05 was considered statistically significant.
Results
c-kit siRNA regulates the production of cytokines in the BALF
SCF production decreased to a greater extent in the c-kit siRNA group than in the control and scrambled siRNA groups (Figure 2A); the same pattern was observed in the production of the Th2 cytokines IL-4 and IL-5 (Figure 2B and 2C). The production of interferon-γ (IFN-γ), a Th1 cytokine, did not increase with the decrease in Th2 cytokines (Figure 2D).
Figure 2.
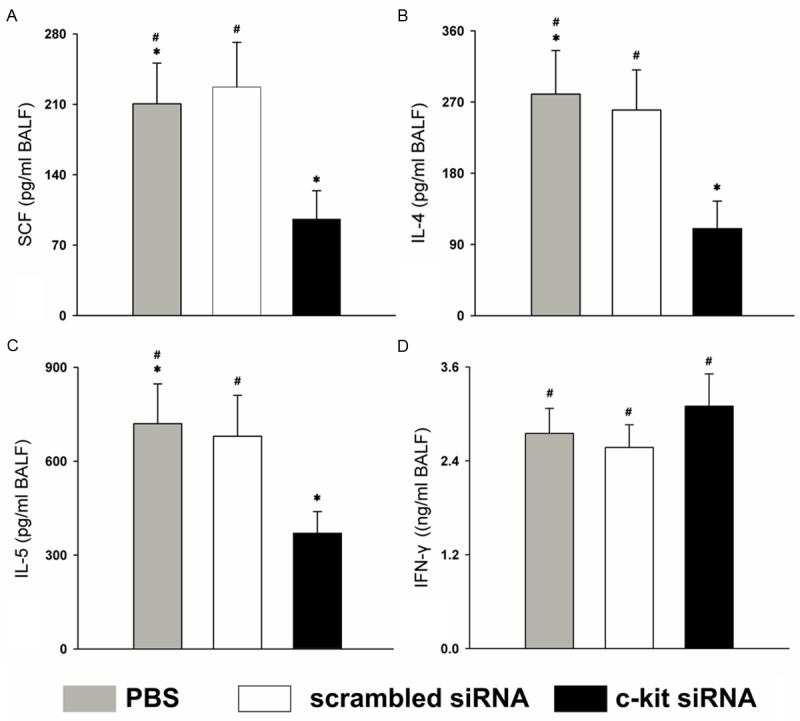
c-kit siRNA selectively regulates cytokines in the BALF. Samples were obtained 3 days after the injection of siRNA or PBS vehicle, on day 26. The levels of (A) SCF, (B) IL-4, (C) IL-5, and (D) IFN-γ in the BALF in the three groups were measured by ELISA. BALF was collected at 72 h after the last administration. *P < 0.05; #P > 0.05 (Student’s t test). Data are expressed as mean cytokine concentration ± SEM from n ≥ 5 mice and are representative of three independent experiments.
siRNA reduces eosinophil infiltration in the BALF
All BALF samples were obtained 3 days after the intranasal administration of siRNA or PBS vehicle on day 26. The number of eosinophils in the BALF showed a significantly greater decrease in the c-kit siRNA group than in the control and scrambled siRNA groups (P < 0.05) (Figure 3A). The percentage of eosinophils in the BALF in the c-kit-siRNA group was lower than those in the control or the scrambled siRNA group (P < 0.05) (Figure 3B).
Figure 3.
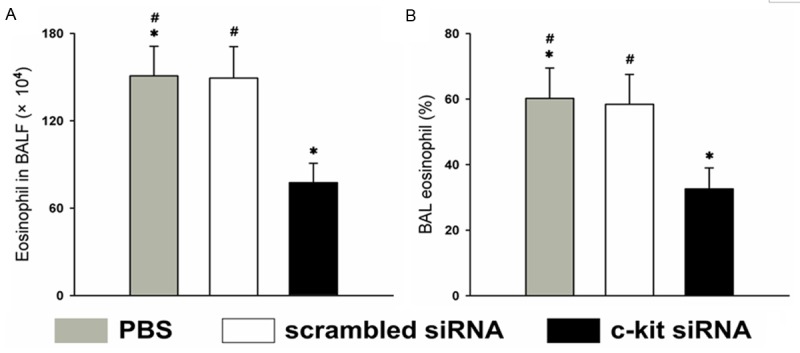
Characterization of pulmonary eosinophilic infiltration in the BALF. BALF was collected at 72 h after the last administration of siRNA or PBS vehicle, on day 26. A. Number of eosinophils in the BALF; B. Percentage of eosinophils in the BALF. *P < 0.05 vs. the control and scrambled siRNA groups. Data (means ± SEM) are representative of three independent experiments with ≥ 5 mice per group.
siRNA attenuates the inflammatory response in lung tissue
Histological examination of lung specimens showed that c-kit siRNA attenuated the inflammatory response in the lung tissue compared with the control and scrambled siRNA groups. Compared with the control group, the number of inflammatory cells that infiltrated the lung tissue was lower in the c-kit siRNA group (Figure 4).
Figure 4.
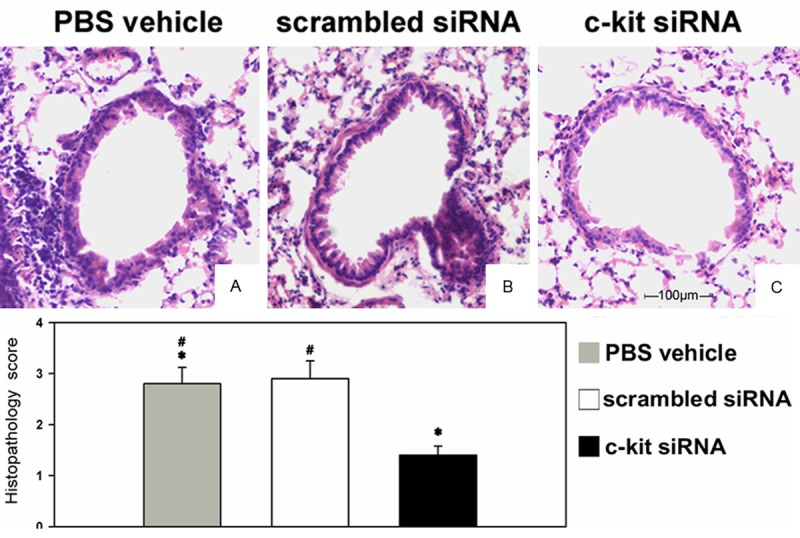
Histological characterization of the asthmatic lung and airway inflammation and its suppression by c-kit siRNA. Lung tissue sections at day 26 from mice given siRNA or vehicle; sections were obtained from fixed, paraffin-embedded lung tissue stained with H&E and scored for pathology as described in Materials and Methods. Original magnification, ×200. The mice were nasally administered (A) PBS vehicle, (B) scrambled siRNA, or (C) c-kit siRNA. The experiment was carried out three times; representative images are shown.
siRNA reduces the secretion of mucus in the airways of asthmatic mice
Periodic acid-Schiff staining showed that treatment with c-kit siRNA decreased the production of airway epithelial mucus, compared with the control and scrambled siRNA groups. The mucus was less thick in the c-kit siRNA group compared with the control group (Figure 5).
Figure 5.
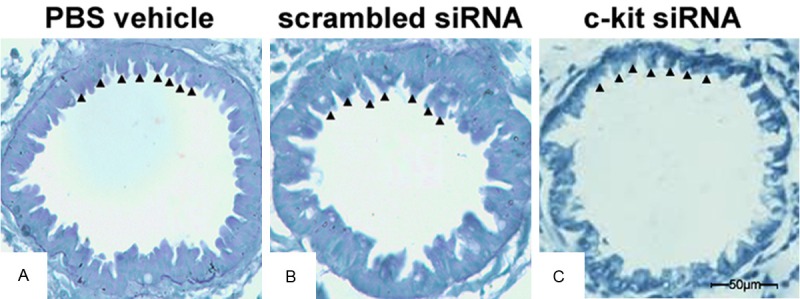
c-kit siRNA improves airway mucus clearance. Lung tissue sections at day 26 from mice administered PBS vehicle (A), scrambled siRNA (B), or c-kit siRNA (C); sections were obtained from fixed, paraffin-embedded lung tissue stained with periodic acid-Schiff reagent. Black arrowheads indicate mucus-containing goblet cells. Original magnification, ×400. The experiment was carried out three times; representative images are shown.
c-kit siRNA down-regulates the expression of c-kit gene in lung tissue
To investigate whether c-kit-specific siRNA could silence the expression of the c-kit gene, we measured the levels of c-kit mRNA by reverse transcription-polymerase chain reaction (RT-PCR). In comparison with the control and scrambled siRNA groups, the expression of c-kit mRNA was down-regulated in the anti-c-kit siRNA group at 72 h after the last administration, indicating that anti-c-kit siRNA effectively inhibited the expression of the c-kit gene (Figure 6).
Figure 6.
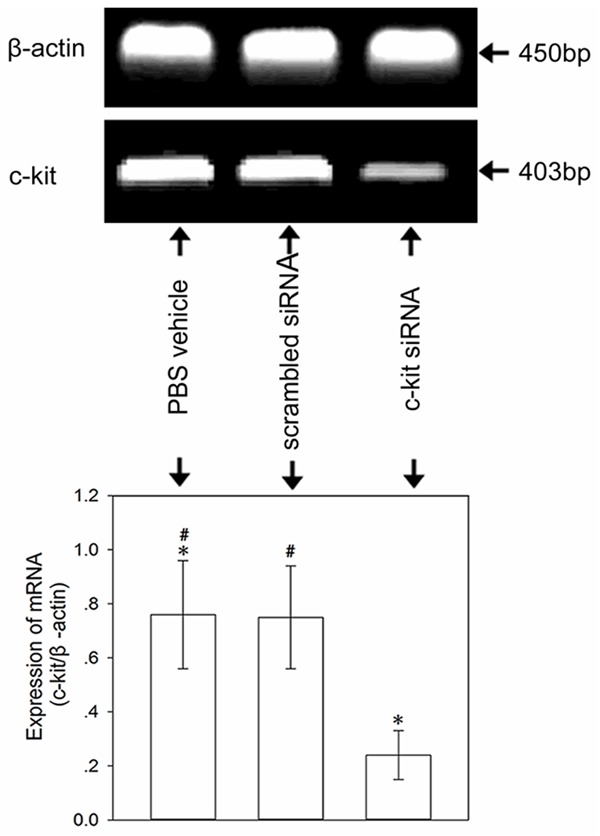
Representative RT-PCR analysis of c-kit mRNA expression in lung tissue. Note that c-kit mRNA expression was down-regulated at 72 h after the last administration in the anti-c-kit siRNA group. The bar graph represents the mean relative intensity (± SD) of the c-kit bands normalized to β-actin. *P < 0.05 compared with the control and scrambled siRNA groups. #P > 0.05 between the control and scrambled siRNA groups. The experiment was carried out three times; representative images are shown.
c-kit siRNA inhibits the expression of c-kit protein
Western blot analysis showed that anti-c-kit siRNA could effectively inhibit the expression of c-kit protein at 72 h after transfection in the c-kit siRNA group. In contrast, the control and scrambled siRNA groups showed expression of c-kit protein in lung tissue (Figure 7).
Figure 7.
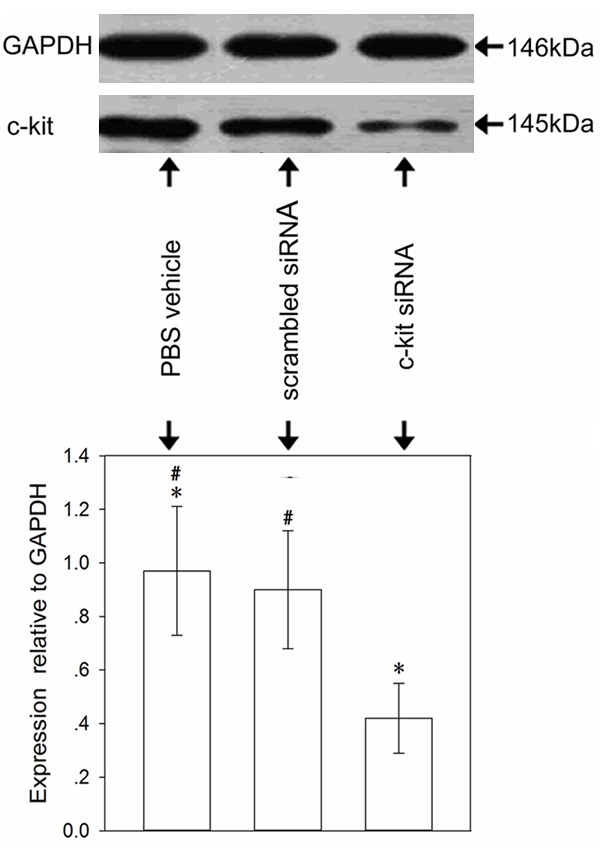
Expression of c-kit protein in lung tissue. Histograms represent the mean relative amount (± SD) of the c-kit bands normalized to GAPDH. There was a difference between the anti-c-kit siRNA group and the other groups. *P < 0.05 vs. the control and scrambled siRNA groups. #P > 0.05 between the control and scrambled siRNA groups. The experiment was carried out three times; representative images are shown.
Discussion
The term RNAi, first coined by Fire and Mello in 1998, describes the posttranslational silencing of gene expression that occurs in response to the introduction of dsRNA into a cell. This phenomenon can result in highly specific suppression of gene expression [18]. To date, many studies have been conducted on a variety of animal models to investigate the efficacy of siRNAs as therapeutic agents; their findings showed that specific siRNAs could suppress the expression of target genes when administered locally or systemically [16,19]. In our study, the experimental asthmatic mice were locally administered c-kit siRNA through the nose. We obtained evidence that specific c-kit siRNA acts as an anti-inflammatory mediator of allergic airway inflammation.
c-kit was first found to be expressed on the surface of mast cells in 1989 [20]. At present, c-kit protein expression has been found in a wide range of nonhemopoietic cell types including interstitial cells of Cajal, astrocytes, renal tubules, breast glandular epithelial cells, human airway epithelia, mast cells, eosinophils, and lymphocytes [21-25]. SCF-also termed Kit ligand, steel factor, or mast cell growth factor-is the ligand of the c-kit proto-oncogene product. SCF and its receptor, Kit, are normally present in both cell surface and soluble forms. Both forms of Kit can bind SCF [26]. In this study, we used specific siRNA to inhibit the expression of c-kit, and observed reduced production of SCF, IL-4, and IL-5 in the BALF. The number of eosinophils markedly decreased in the BALF from the mice administered c-kit siRNA. Histopathologic examination of the lung showed that treatment with c-kit siRNA reduced the inflammation of lung tissue and mucus secretion in the airways. Hartman et al. reported that human peripheral blood eosinophils expressed and released SCF, and blocking c-kit/c-kit ligand interaction could significantly decrease eosinophils in the BALF [27,28]. Moreover, SCF also induced the adhesion of eosinophils to fibronectin and to vascular cell adhesion molecule VCAM-1 in vitro. This effect was completely inhibited by α4-integrin and β1-integrin antibodies and was therefore dependent on very late antigen VLA-4 (α4β1) [29]. Intranasal administration of SCF antisense oligonucleotides decreased lung SCF protein expression, as well as AHR, IL-4 production, and eosinophil number in the BALF, in a murine model of asthma [10]. Treatment of mice with imatinib, a tyrosine kinase inhibitor, to inhibit the signal pathway of SCF/c-kit, which could significantly attenuate airway hyperreactivity and peribronchial eosinophil accumulation, significantly reduced the Th2 cytokines IL-4 and IL-13. In addition, chemokines associated with allergen-induced pulmonary disease, such as CCL2, CCL5, and CCL6, were significantly reduced in the lungs of imatinib-treated mice [30]. In this study, we found that the number and percentage of eosinophils in the BALF showed a significantly greater decrease in the c-kit siRNA group than in the control and scrambled siRNA groups.
IFN-γ, a Th1 cytokine, has been considered to attenuate the inflammatory response in allergic asthma. Previous studies have shown that treatment with antisense phosphorothioate oligonucleotides to the c-kit ligand suppresses airway inflammation, IL-4 production, and eosinophil infiltration, which are attributable to the SCF effects [10]. On the one hand, IL-4 promotes IgE synthesis, contributes to early recruitment of eosinophils, and is detected as early as 3h after challenge [31,32]. On the other hand, IL-4 can boost the production of IL-5, which is the most important chemokine in the recruitment of eosinophils into the airway [33]. Vladislav et al. found that the production of IL-25 (IL-17E) and IL-17RB myeloid cell-derived Th2 cytokines was dependent on SCF-induced responses during chronic allergic pulmonary disease, thus indicating that SCF contributes to the Th2 response in allergic asthma [34]. Our results showed that silencing of c-kit inhibited the Th2 reaction and led to depression of the airway inflammation in asthmatic mice. On the other hand, inhibition of Th2 response resulted in decreased IL-4 and IL-5 production.
In our study, we found that the levels of IL-4, IL-5, and SCF decreased in the BALF from the mice treated with c-kit siRNA, which suggests that activation of c-kit could promote the production of IL-4. Recent studies have shown that SCF antibody administered into the airways attenuates Th2 cytokine responses in allergic asthma, implying that SCF is linked to the production of IL-4 and IL-5 [34,35]. As a result, a lower production of IL-5 leads to diminished recruitment of eosinophils into the airway and, ultimately, to a decrease in the number of eosinophils in the BALF. In addition, IL-4 plays a pivotal role in the production of mucus in the airway [36,37]. In this study, reduced mucus in the airway was attributed to lower production of IL-4.
Multiple signaling pathways including the Ras/ERK, phosphatidylinositol 3-kinase (PI3K), phospholipase C and D, Src kinase, and JAK/STAT are activated downstream of c-kit after ligand binding [38,39]. Therefore, the c-kit-PI3K kinase signaling axis plays a pivotal role in the development of allergic asthma. The c-kit-PI3K positively regulates the production of IL-6; moreover, c-kit is associated with Th2 responses through the Notch ligand Jagged-2 signaling pathway [12,40]. c-kit up-regulation in dendritic cells led to immune skewing toward the Th2 and Th17 subsets and away from Th1 responses [41]. However, data on signal transduction induced by SCF demonstrate that MEK-mediated signaling pathways are crucial for SCF-induced CCL6 chemokine activation and eosinophil survival. CCL6 gene activation and production induced by SCF in the presence or absence of rather specific pathway inhibitors demonstrated that the MEK/MAPK pathway, but not the PI3K pathway, was crucial for the SCF-induced CCL6 gene activation. These same signaling pathways were shown to initiate antiapoptotic events and promote eosinophil survival, including up-regulation of BCL-2 and BCL-3 [42].
In conclusion, we have shown that siRNA targeting c-kit decreases the recruitment of eosinophils into the airways and attenuates allergic inflammation in asthma. Our findings may serve as an impetus for further research on inflammation control in allergic asthma.
Acknowledgements
This study was supported by grants from The Nature Science Foundation of The Education Department of Shaanxi Provincial Government (No. 11JK0662).
Disclosure of conflict of interest
None.
References
- 1.Holgate ST. The epidemic of allergy and asthma. Nature. 1999;402:B2–B4. doi: 10.1038/35037000. [DOI] [PubMed] [Google Scholar]
- 2.Haczku A, Takeda K, Redai I, Hamelmann E, Cieslewicz G, Joetham A, Loader J, Lee JJ, Irvin C, Gelfand EW. Anti-CD86 (B7.2) treatment abolishes allergic airway hyperresponsiveness in mice. Am J Respir Crit Care Med. 1999;159:1638–1643. doi: 10.1164/ajrccm.159.5.9711040. [DOI] [PubMed] [Google Scholar]
- 3.Bucchieri F, Puddicombe SM, Lordan JL, Richter A, Buchanan D, Wilson SJ, Ward J, Zummo G, Howarth PH, Djukanović R, Holgate ST, Davies DE. Asthmatic bronchial epithelium is more susceptible to oxidant-induced apoptosis. Am J Respir Cell Mol Biol. 2002;27:179–185. doi: 10.1165/ajrcmb.27.2.4699. [DOI] [PubMed] [Google Scholar]
- 4.Hammad H, Lambrecht BN. Dendritic cells and epithelial cells: linking innate and adaptive immunity in asthma. Nat Rev Immunol. 2008;8:193–204. doi: 10.1038/nri2275. [DOI] [PubMed] [Google Scholar]
- 5.Chabot B, Stephensson DA, Chapwen VM, Besmer P, Bernstein A. The proto-oncogene c-kit encodes a transmembrane tyrosine kinase receptor maps to the mouse W-locus. Nature. 1988;335:88–90. doi: 10.1038/335088a0. [DOI] [PubMed] [Google Scholar]
- 6.Williams DE, Eisenman J, Baird A, Rauch C, Ness KV, March CJ, Park LS, Martin U, Mochizukl DY, Boswell HS, Burgess GS, Cosman D, Lyman SD. Identification of a ligand for the c-kit proto-oncogene. Cell. 1990;63:167–179. doi: 10.1016/0092-8674(90)90297-r. [DOI] [PubMed] [Google Scholar]
- 7.Edling CE, Hallberg B. c-kit-a hematopoietic cell essential receptor tyrosine kinase. Int J Biochem Cell Biol. 2007;39:1995–1998. doi: 10.1016/j.biocel.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Oliveira SH, Taub DD, Nagel J, Smith R, Hogaboam CM, Berlin A, Lukacs NW. Stem cell factor induces eosinophil activation and degranulation: mediator release and gene array analysis. Blood. 2002;100:4291–4297. doi: 10.1182/blood.V100.13.4291. [DOI] [PubMed] [Google Scholar]
- 9.Berlin AA, Lincoln P, Tomkinson A, Lukacs NW. Inhibition of stem cell factor reduces pulmonary cytokine levels during allergic airway responses. Clin Exp Immunol. 2004;136:15–20. doi: 10.1111/j.1365-2249.2004.02404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finotto S, Buerke M, Lingnau K, Schmitt E, Galle PR, Neurath MF. Local administration of antisense phosphorothioate oligonucleotides to the c-kit ligand, stem cell factor, suppresses airway inflammation and IL-4 production in a murine model of asthma. J Allergy Clin Immunol. 2001;107:279–286. doi: 10.1067/mai.2001.113049. [DOI] [PubMed] [Google Scholar]
- 11.Oliveira SH, Hogaboam CM, Berlin A, Lukacs NW. SCF-induced airway hyperreactivity is dependent on leukotriene production. Am J Physiol Lung Cell Mol Physiol. 2001;280:L1242–1249. doi: 10.1152/ajplung.2001.280.6.L1242. [DOI] [PubMed] [Google Scholar]
- 12.Krishnamoorthy N, Oriss TB, Paglia M, Fei M, Yarlagadda M, Vanhaesebroeck B, Ray A, Ray P. Activation of c-kit in dendritic cells regulates T helper cell differentiation and allergic asthma. Nat Med. 2008;14:565–573. doi: 10.1038/nm1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sikarwar AP, Reddy KV. siRNA-mediated silencing of c-kit in mouse primary spermatogonial cells induces cell cycle arrest. Oligonucleotides. 2008;18:145–160. doi: 10.1089/oli.2008.0108. [DOI] [PubMed] [Google Scholar]
- 14.Paveglio SA, Allard J, Mayette J, Whittaker LA, Juncadella I, Anguita J, Poynter ME. The tick salivary protein, Salp15, inhibits the development of experimental asthma. J Immunol. 2007;178:7064–7071. doi: 10.4049/jimmunol.178.11.7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strapps WR, Pickering V, Muiru GT, Rice J, Orsborn S, Polisky BA, Sachs A, Bartz SR. The siRNA sequence and guide strand overhangs are determinants of in vivo duration of silencing. Nucleic Acids Res. 2010;38:4788–4797. doi: 10.1093/nar/gkq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pushparaj PN, H’ng SC, Melendez AJ. Refining siRNA in vivo transfection: silencing SPHK1 reveals its key role in C5a-induced inflammation in vivo. Int J Biochem Cell Biol. 2008;40:1817–1825. doi: 10.1016/j.biocel.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Zhu Z, Homer RJ, Wang Z, Chen Q, Geba GP, Wang J, Zhang Y, Elias JA. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest. 1999;103:779–788. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1988;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 19.Li BJ, Tang Q, Cheng D, Qin C, Xie FY, Wei Q, Xu J, Liu Y, Zheng BJ, Woodle MC, Zhong N, Lu PY. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat Med. 2008;11:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nocka K, Majumder S, Chabot B, Ray P, Cervone M, Bernstein A, Besmer P. Expression of c-kit gene products in known cellular targets of W mutations in normal and W mutant mice-evidence for an impaired c-kit kinase in mutant mice. Genes Dev. 1989;3:816–826. doi: 10.1101/gad.3.6.816. [DOI] [PubMed] [Google Scholar]
- 21.Broudy VC, Kovach NL, Bennett LG, Lin N, Jacobsen FW, Kidd PG. Human umbilical vein endothelial cells display high-afinity c-kit receptors and produce a soluble form of the c-kit receptor. Blood. 1994;83:2145–2152. [PubMed] [Google Scholar]
- 22.Torihashi S, Ward SM, Nishikawa S, Nishi K, Kobayashi S, Sanders KM. c-kit-dependent development of interstitial cells and electrical activity in the murine gastrointestinal tract. Cell Tissue Res. 1995;280:97–111. doi: 10.1007/BF00304515. [DOI] [PubMed] [Google Scholar]
- 23.Huizinga JD, Thuneberg L, Klüppel M, Malysz J, Mikkelsen HB, Bernstein A. W/kit gene required for interstitial cells of Cajal and for intestinal pacemaker activity. Nature. 1995;373:347–349. doi: 10.1038/373347a0. [DOI] [PubMed] [Google Scholar]
- 24.Al-muhsen SZ, Shablovsky G, Olivenstein R, Mazer B, Hamid Q. The expression of stem cell factor and c-kit receptor in human asthmatic airways. Clin Exp Allergy. 2004;34:911–916. doi: 10.1111/j.1365-2222.2004.01975.x. [DOI] [PubMed] [Google Scholar]
- 25.Broudy VC. Stem cell factor and hematopoiesis. Blood. 1997;90:1345–1364. [PubMed] [Google Scholar]
- 26.Langley KE, Bennett LG, Wypych J, Yancik SA, Liu XD, Westcott KR, Chang DG, Smith KA, Zsebo KM. Soluble stemcell factor in human serum. Blood. 1993;81:656–660. [PubMed] [Google Scholar]
- 27.Hartman M, Piliponsky AM, Temkin V, Levi-Schaffer F. Human peripheral blood eosinophils express stem cell factor. Blood. 2001;97:1086–1091. doi: 10.1182/blood.v97.4.1086. [DOI] [PubMed] [Google Scholar]
- 28.Lukacs NW, Strieter RM, Lincoln PM, Brownell E, Pullen DM, Schock HJ, Chensue SW, Taub DD, Kunkel SL. Stem cell factor (c-kit ligand) influences eosinophil recruitment and histamine levels in allergic airway inflammation. J Immunol. 1996;156:3945–3951. [PubMed] [Google Scholar]
- 29.Yuan Q, Austen KF, Friend DS, Heidtman M, Boyce JA. Human peripheral blood eosinophils express a functional c-kit receptor for stem cell factor that stimulates very late antigen 4 (VLA-4)-mediated cell adhesion to fibronectin and vascular cell adhesion molecule 1 (VCAM-1) J Exp Med. 1997;186:313–323. doi: 10.1084/jem.186.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aaron AB, Nicholas WL. Treatment of Cockroach Allergen Asthma Model with Imatinib Attenuates Airway Responses. Am J Respir Crit Care Med. 2005;171:35–39. doi: 10.1164/rccm.200403-385OC. [DOI] [PubMed] [Google Scholar]
- 31.Corry DB, Folkesson HG, Warnock ML, Erle DJ, Matthay MA, Wiener-Kronish JP, Locksley RM. Interleukin 4, but not interleukin 5 or eosinophils, is required in a murine model of acute airway hyperreactivity. J Exp Med. 1996;183:109–117. doi: 10.1084/jem.183.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukacs NW, Strieter RM, Chensue SW, Kunkel SL. Interleukin-4-dependent pulmonary eosinophil infiltration in a murine model of asthma. Am J Respir Cell Mol Biol. 1994;10:526–532. doi: 10.1165/ajrcmb.10.5.8179915. [DOI] [PubMed] [Google Scholar]
- 33.Wills-Karp M. Murine models of asthma in understanding immune dysregulation in human asthma. Immunopharmacology. 2000;48:263–268. doi: 10.1016/s0162-3109(00)00223-x. [DOI] [PubMed] [Google Scholar]
- 34.Vladislav D, Bryan CP, Alison LB, Aaron AB, Nicholas W. Pulmonary IL-17E (IL-25) production and il-17rb myeloid cell-derived th2 cytokine production are dependent upon stem cell factor-induced responses during chronic allergic pulmonary disease. J Immunol. 2009;183:5705–5715. doi: 10.4049/jimmunol.0901666. [DOI] [PubMed] [Google Scholar]
- 35.Petersen B, Dolgachev V, Lukacs NW. SCF and IL-25 induced Th2 associated pathophysiology in chronic cockroach antigen induced asthma. J Immunol. 2009;182:79–19. [Google Scholar]
- 36.Ronnstrand L. Signal transduction via the stem cell factor receptor/c-Kit. Cell Mol Life Sci. 2004;61:2535–2548. doi: 10.1007/s00018-004-4189-6. [DOI] [PubMed] [Google Scholar]
- 37.Temann UA, Prasad B, Gallup MW, Basbaum C, Ho SB, Flavell RA, Rankin JA. A novel role for murine IL-4 in vivo: induction of MUC5AC gene expression and mucin hypersecretion. Am J Respir Cell Mol Biol. 1997;16:471–478. doi: 10.1165/ajrcmb.16.4.9115759. [DOI] [PubMed] [Google Scholar]
- 38.Dabbagh K, Takeyama K, Heung ML, Iris FU, James AL, Jay AN. IL-4 Induces Mucin Gene Expression and Goblet Cell Metaplasia In Vitro and In Vivo. J Immunol. 1997;162:6233–6237. [PubMed] [Google Scholar]
- 39.Lennartsson J, Jelacic T, Linnekin D, Shivakrupa R. Normal and oncogenic forms of the receptor tyrosine kinase kit. Stem Cells. 2005;23:16–43. doi: 10.1634/stemcells.2004-0117. [DOI] [PubMed] [Google Scholar]
- 40.Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–526. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- 41.Ray P, Krishnamoorthy N, Oriss TB, Ray A. Signaling of c-kit in dendritic cells influences adaptive immunity. Ann NY Acad Sci. 2010;1183:104–122. doi: 10.1111/j.1749-6632.2009.05122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vladislav D, Molly T, Aaron B, Nicholas WL. Stem cell factor-mediated activation pathways promote murine eosinophil CCL6 production and survival. J Leukoc Biol. 2007;81:1111–1119. doi: 10.1189/jlb.0906595. [DOI] [PubMed] [Google Scholar]


