Abstract
miR-183, a member of an evolutionarily conserved miRNA cluster (miR-96, miR-182, and miR-183), has been demonstrated to act as both a tumor suppressor and oncogene in various type of human cancer. However, the biological role of miR-183 in gastric cancer (GC) still remains unclear. In the present study, miR-183 expression was significantly decreased in gastric cancer tissues compared with its’ adjacent normal tissues, and down-regulation of miR-183 was significantly associated with lymph node metastasis and pathological TNM stage. Furthermore, Erzin, which was reported to be up-regulated in gastric cancer, was identified as an efficient target of miR-183. Overexpression of miR-183 markedly suppressed cells invasion by downregulation of Ezrin expression. However, miR-183 expression didn’t affect cells proliferation and cell cycle distribution of GC. In conclusion, our study demonstrated that miR-183 acts as a tumor suppressor in GC, partially at least via regulation of Ezrin. Therefore, miR-183 may be a potential target for the treatment of gastric cancer.
Keywords: MicroRNA-183, gastric cancer (GC), Ezrin, metastasis
Introduction
After lung cancer, gastric cancer (GC) is the second most frequent cause of cancer-related deaths, leading to approximately 738,000 (10%) deaths worldwide [1]. Although the incidence of GC has substantially declined due to the increased availability of fresh fruits and vegetables and reductions in chronic H. pylori infection, some 400,000 new cases are diagnosed every year in China, accounting for 42% of the total cases reported worldwide [2]. Clinical data have shown that most GC patients eventually suffer metastasis after the curative resection (R0) of the cancer [3]. During metastasis, the invasion of GC into the surrounding tissue is a crucial early step [4]. However, the mechanisms of invasion are not yet fully understood.
miRNAs are a class of short, non-coding RNA molecules that negatively regulate gene expression and play important roles in various biological processes. In most mammals, mature miRNAs, typically comprise 21-24 nucleotides, generated from pri-miRNAs and pre-miRNAs through a series of enzymatic reactions. A large number of mature miRNAs have been recently implicated in cancer metastasis, including miR-99a, miR-107, miR-200a, miR-375, miR-484, miR-520c, and miR-205 in breast cancer [5-9]; miR-21, miR-31, miR-126, miR-141, and miR-145 in colorectal cancer [10-12]; miR-132, miR-138, and miR-182 in lung cancer [13-15]; miR-200b and miR-361 in prostate cancer [16,17]; and miR-7, miR-10a, miR-133a, miR-133b and miR-145 in gastric cancer [18-20]. Emerging evidence has revealed that miR-183 plays an oncogenic role in the development and metastasis of tumors. miR-183 is up-regulated in human hepatocellular carcinoma and inhibit apoptosis in HCC cells through the suppression of programmed cell death 4 (PDCD4) expression [21]. It has also been reported that miR-183 is significantly overexpressed and promotes cell migration though the negative regulation of two tumor suppressor genes (EGR1 and PTEN) [22]. In addition, increasing evidence has demonstrated that miR-183 could act as a tumor suppressor gene in the metastasis of several types of tumors. The over-expression of miR-183 inhibited cell migration and invasion through the targeting of Ezrin both in lung and breast cancers [23,24]. In osteosarcoma, the dysregulation of miR-183 significantly impacts tumor metastasis via Ezrin targeting [25,26]. These studies indicate the important roles of miR-183 in tumorigenesis and metastasis. However, there are few studies concerning miR-183 in GC, and the biological role of miR-183 in GC pathogenesis remains unknown.
In the present study, we investigated the potential role of miR-183 in the development and progression of GC. Using quantitative RT-PCR, we observed that miR-183 was remarkably down-regulated in GC tissues compared with adjacent normal tissues, and the down-regulation of miR-183 was significantly associated with lymph node metastasis and the pathological stage of TNM. In addition, functional assays showed that miR-183 over-expression in highly metastatic cells could inhibit cell invasion, but does not affect cell proliferation and cell cycle distribution, through increased Ezrin expression. Furthermore, using the luciferase reporter system, we demonstrated that Ezrin is a direct target of miR-183. Altogether, these results suggest that miR-183 plays an efficient regulatory role in gastric cancer metastasis, suggesting that miR-183 might be a novel diagnostic and prognostic marker of GC.
Materials and methods
Primary reagents
Ezrin antibodies were purchased from Abcam (ab4069, Abcam, Cambridge, MA). GAPDH antibodies were purchased from Cell Signaling Technology. Horseradish peroxidase-conjugated goat anti-rabbit IgG and goat anti-mouse IgG were obtained from Sigma. The hsa-miR-183 pre-miR miRNA (Ambion cat. no. AM17100) and the Pre-miR™ miRNA precursor molecule, as a negative control (Ambion, cat. no. AM17110), were obtained from Ambion. The mirVanaTM miRNA Isolation Kit (Ambion, cat. no. 1560, UK) and the RecoverAllTM Total Nucleic Acid Isolation Kit (Ambion, cat. no. 1975, UK) were obtained from Ambion. The miRCURY LNATM Universal cDNA Synthesis Kit II (Exiqon, cat. no. 203301, Vedbaek, Denmark) and ExiLENT SYBR® Green master mix were purchased from Exiqon (catalog no. 203402, Vedbaek, Denmark). Primers for miR-183 (catalog no. 204652) and U6 snRNA (catalog no. 203907) were designed and synthetized at Exiqon.
Clinical tumor tissues
Samples of GC and adjacent normal tissues were obtained from 55 patients at FuJian Medical University Union Hospital and the detailed Clinicopathological parameters were assessed. All gastric cancer patients were diagnosed and gastrectomized with lymph node dissection in the Department of Gastric Surgery of the Union Hospital from 2006 to 2014. A total of 5 tissue samples from 3 patients (gastrectomized during 2014) were collected to detect mature miR-183 expression using quantitative RT-PCR, and Ezrin protein expression was analyzed through western blotting. A total of 104 specimens from 52 patients (gastrectomized from 2006 to 2008) were obtained to determine mature miR-183 and Ezrin protein expression through quantitative RT-PCR and immunohistochemistry, respectively. All patients had a well-documented clinical history and detailed follow-up information. None of the patients underwent preoperative chemotherapy and radiation therapy. The ethics committee of Fujian Medical University Union Hospital approved this study, and written consent was obtained from all patients involved.
Immunohistochemistry analysis
Immunohistochemical staining for Ezrin was performed on formalin-fixed, paraffin-embedded gastric tissue sections (3 μm thick, tumor or normal). The Ezrin protein expression was immunohistochemically demonstrated as yellowish to brown staining in the cytoplasm and membrane of gastric glandular cells. By Two pathologists, blinded to the clinical data, reviewed the immunoreactivity for Ezrin protein under a light microscope, and the protein expression was scored independently according to the intensity of cellular staining and the proportion of stained tumor cells. The staining intensity was scored as 0 (no staining), 1 (weak staining, light yellow), 2 (moderate staining, yellow brown), and 3 (strong staining, brown), and the proportion of stained tumor cells was classified as 0 (≤5% positive cells), 1 (6% to 25% positive cells), 2 (26% to 50% positive cells), and 3 (≥51% positive cells). The product of the scores for intensity and proportion was used to signify the level of protein expression. A score of 3 or less was considered low Ezrin expression, and a score of 4 or more was considered high Ezrin expression.
Western blot analysis
Fresh tissues and cells were homogenized in 100~200 μl of radioimmunoprecipitation assay lysis buffer (RIPA), containing protease inhibitors, at 4°C for 30 minutes, followed by centrifugation at 16,000 g for 15 minutes at 4°C. The supernatants, containing whole-cell lysates, were prepared for use in subsequent experiments. The protein concentration was measured using the BCA Protein Assay Kit (Thermo). A total of 40 mg of protein from each sample was denatured and loaded onto each well, separated through SDS-PAGE, and transferred to a polyvinylidene difluoride membrane (Millipore, Billerica, MA). Subsequently, the membrane was blocked with 5% nonfat milk at room temperature for 1 hour. The membrane was incubated with mouse anti-Ezrin (1:500; ab4069, Abcam, Cambridge, MA) or rabbit anti-GAPDH (Cell Signaling Technology) primary antibodies overnight at 4°C. After washing with wash buffer (10 mmol/L Tris-HCl, 150 mmol/L NaCl, and 0.1% Tween 20), the membrane was further incubated with horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse IgG (Sigma, St. Louis, MO) at a 1:3000 dilution for 1 hour at room temperature. Subsequently, the membrane was washed for 30 minutes with wash buffer and detected using enhanced chemiluminescence (Amersham Corporation, Arlington Heights, IL, USA).
RNA extraction and quantitative RT-PCR
MicroRNA was extracted from cells and frozen tissues using the mirVanaTM miRNA Isolation Kit (Ambion, cat. no. 1560, UK) and from FFPE cells using the RecoverAllTM Total Nucleic Acid Isolation Kit (Ambion, cat. no. 1975, UK) according to the manufacturer’s instructions. Total RNA was extracted from the cells and frozen tissues using TRIZOL Reagent (Invitrogen). The miRNA mRNA expression levels were detected through quantitative RT-PCR. For miRNA detection, reverse transcription was performed using the miRCURY LNATM Universal cDNA Synthesis Kit II, and quantitative RT-PCR was performed using ExiLENT SYBR® Green master mix. U6 snRNA levels were used as an endogenous control. PCR reactions were performed using the ABI Prism 7900 Sequence Detection System (Applied Biosystems). The PCR amplification conditions included polymerase activation/denaturation at 95°C for 10 min, followed by 40 amplification cycles at 95°C for 10 s and 60°C for 1 min.
Cell lines and cell culture
Human GC cell lines MGC-803, SGC-7901, BGC-823, MKN-45, and MKN-28 and the normal gastric epithelial cell line, GES-1, were purchased from The Institute of Biochemistry and Cell Biology at The Chinese Academy of Sciences (Shanghai, China). The cells were routinely cultured at 37°C in a humidified atmosphere of 5% CO2 in RPMI 1640 medium containing 10% fetal bovine serum (Gibco).
Precursor microRNA transfection
For miR-183 overexpression, precursor molecules (pre-miR miRNA precursor and the negative control) were transfected into gastric cancer cell lines using Lipofectamine 2000 (Invitrogen) and OPTI-MEM I Reduced-Serum Medium (Invitrogen) according to manufacturer’s instructions. The culture medium was changed after 24 h of transfection. The efficiency of transfection and overexpression of miR-183 were evaluated at 72 hours post-transfection using quantitative RT-PCR. Each experiment was performed in triplicate.
Transwell invasion assay
A 24-well transwell plate (8 μm pore size, Corning, USA) was used to examine the invasion of each cell line. Briefly, serum-free medium was added into the upper chamber to rehydrate the Matrigel for 1 hour. Subsequently, the medium was removed and the lower chambers of the Transwell plate were filled with 500 μl of RPMI 1640 medium containing 20% FBS. The transfected cells were trypsinized, washed using serum-free RPMI 1640 medium and resuspended to a density of 5×105 cells/mL in serum-free medium. A total of 300 μl of the cell suspension was added to the upper chambers. After incubation at 37°C for 24 hours, the upper chambers were wiped with several cotton tips to completely remove non-invasive cells. The invading cells under the surface of the membrane were fixed in 100% methanol for 10 minutes, air-dried, and stained in 0.1% crystal violet for 30 minutes. The cells in 3 random fields of view (100× magnification) were counted and represented the average number of cells. Each experiment was performed in triplicate.
Cell viability assays
The cells were seeded onto 96-well plates and maintained in RPMI-1640 medium supplemented with 10% FBS. The cells were transiently transfected with pre-miR-183 and miR-control. At 5 days post-transfection, the cell viability was determined daily using a commercial Cell Counting Kit (CCK-8, Dojindo, Japan). A total of 10 ml of CCK-8 reagent was added to each well, and the cells were incubated at 37°C for 3 hours, until the media turned yellow. The absorbance was measured at 450 and 600 nm using a spectrophotometer. The cell viability was calculated using the following equation: cell viability = OD450 nm - OD600 nm. Each experiment was performed in triplicate and repeated at least three times.
Flow cytometry assay
To examine changes in the cell cycle, the cells were transfected with pre-miR-183 and miR-control, trypsinized and collected in 2 ml microcentrifuge tubes, followed by centrifugation at 500 g for 5 minutes at room temperature. After washing with 1X PBS, the cells were fixed in 70% ethanol and stained with 50 mg/ml of propidium iodide (BD Pharmingen, San Jose, CA, USA). Cells at different cell cycle stages were subsequently analyzed using a Beckman coulter flow cytometer, and the cell cycle profiles were analyzed using ModFit 3.0 software (Verity Software House, Topsham, ME, USA).
Luciferase activity assay
The 3’UTR segment of the Ezrin gene, containing the miR-183 binding site, was amplified through PCR and inserted into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, Madison, WI). A plasmid construct carrying Ezrin gene with a mutation in the miR-183 binding site was generated using the Quick Change Site-Directed Mutagenesis Kit (Agilent). MKN-45 cells were cotransfected with Ezrin 3’UTR or mutEzrin 3’UTR and Pre-miR-183 or miR-control using Lipofectamine 2000 (Invitrogen). The luciferase activity was analyzed at 48 hours post-transfection using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI). For each transfection, the luciferase activity was averaged from three replicates.
Statistical analysis
All measurement data are presented as the means ± SE and analyzed using SPSS 17.0 for Windows (SPSS, Chicago, IL) and Prism 5.0 software (GraphPad). The enumeration data were analyzed with the χ2 test using SPSS 17.0 software. Student’s t-test or one-way analysis of variance was used to analyze various clinicopathological parameters. The Kaplan-Meier method was used to analyze the survival curves. The relationships between miR-183 and Ezrin protein were compared through Spearman’s nonparametric analysis. P<0.05 was considered statistically significant, and all P values were 2-sided.
Results
Down-regulation of miR-183 in gastric cancer tissues and cells
The miR-183 level was detected in four gastric neoplasm tissue samples (indicated as T1, T2, T3 and T4) and one normal tissue sample (indicated as N) from three different gastric cancer patients using quantitative RT-PCR. Among these samples, T2, T3 and N were derived from the same patient, while T1 and T4 were derived from two other patients. The miR-183 expression levels in the tumor tissues were significantly lower than in the normal tissue (Figure 1A). We also detected the miR-183 expression levels in five gastric cancer cell lines (BGC-823, SGC-7901, MGC-803, MKN-45 and MKN-28) and one gastric epithelium cell line (GES-1), and the result showed that miR-183 expression was lower in four of the five cancer cell lines than in the epithelium cell line (Figure 1B).
Figure 1.
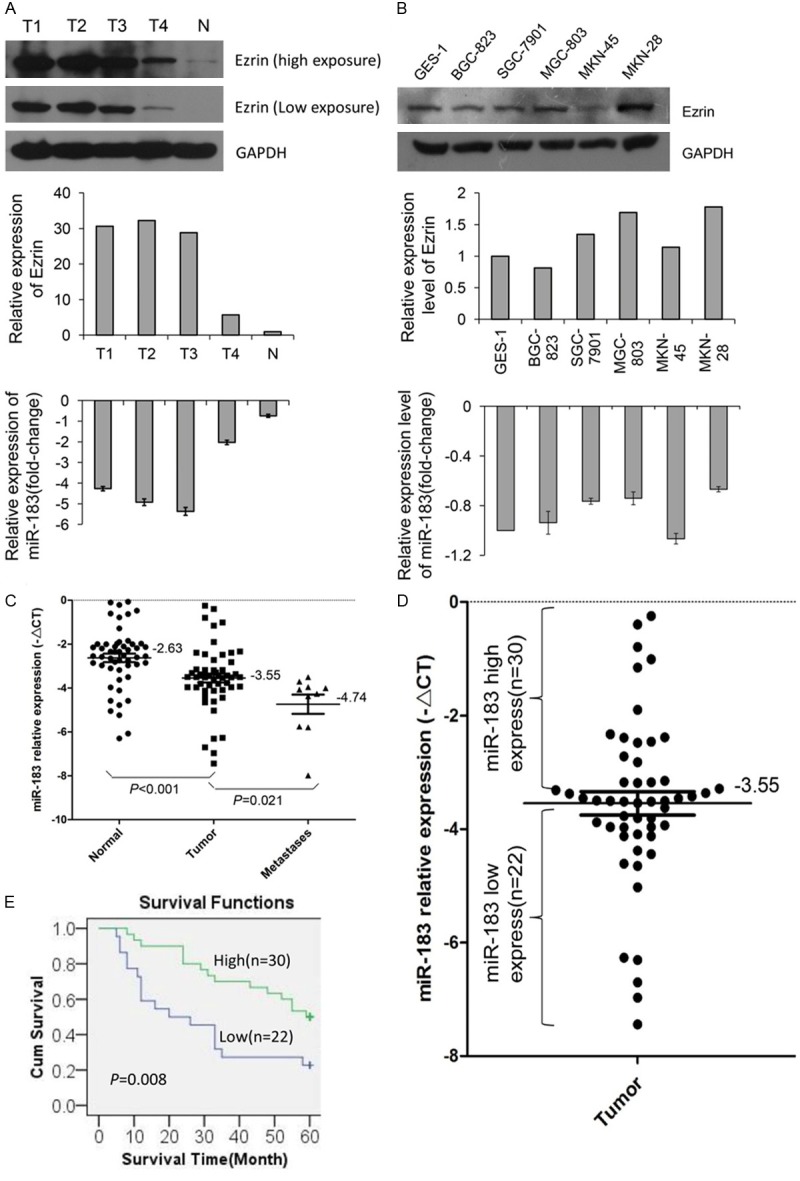
The miR-183 expression levels in gastric cancer tissues and cells. (A) The expression of miR-183 and Ezrin protein in five fresh tissue samples obtained from three patients. Top and middle: western blot analysis of Ezrin protein levels. Bottom: RT-qPCR analysis of miR-183 levels. T1: hepatic metastases; T2: peritoneal metastases; T3: primary tumor with lymph node metastases; T4: primary tumor without lymph node metastases; and N: normal gastric tissue. (B) The expression of miR-183 and Ezrin protein in gastric cell lines. Top and middle: western blot analysis of Ezrin protein levels. Bottom: quantitative RT-PCR analysis of miR-183 levels. (C) Quantitative RT-PCR analysis of miR-183 in 52 pairs of paraffin-embedded primary gastric tumor tissues and adjacent normal tissues and 10 peritoneal metastases. (D) The same primary gastric tumor tissue samples as used in (C) were divided into two groups according to the mean expression of miR-183. Cases with levels of miR-183 below the mean were considered as the low expression group (n=22), and those with levels above the mean were considered as the high expression group (n=30). (E) The results of the Kaplan-Meier survival curve and log-rank test between high and low miR-183 expression in gastric tumor patients.
To determine the relationship between the clinicopathological parameters and miR-183 expression, we examined the expression of miR-183 in paraffin-embedded sections of gastric tumor tissues and adjacent normal tissues (among which 10 had paired peritoneal metastases) from 52 patients using quantitative RT-PCR. The term ΔCt was used to describe the miR-183 expression level. The miR-183 expression level in tumor tissues (-3.546±0.205, mean ± SE) was significantly lower than that in adjacent normal tissues (-2.626±0.193, mean ± SE) (P<0.001, t=4.949, paired t-test) and significantly higher than that in peritoneal metastases (-4.74±0.438, mean ± SE) (P<0.021, t=2.363, unpaired t-test) (Figure 1C). The association of the miR-183 expression levels and clinicopathological parameters was evaluated through a comparison of tumor tissues with normal tissues. As shown in Table 1, the levels of miR-183 expression were associated with lymph node metastasis and pathological TNM stage (P=0.005 and P=0.007, respectively). The median expression of miR-183 was -2.518±0.352 in 12 cases without lymph node metastasis, whereas the median expression was -3.854±0.225 in 40 cases with lymph node metastasis. In addition, the expression of miR-183 was significantly lower in 36 cases with stage III or IV TNM than in 16 cases with stage I or II TNM. However, the expression of miR-183 was not correlated with age, sex, tumor location and size, grade of differentiation, depth of invasion and distant metastasis in the present study. Furthermore, the miR-183 expression in T3 and T4 tissue samples was lower than that in T1 and T2 samples, although this difference was not statistically significant (P=0.081, t=1.779). To accurately evaluate the clinical prognostic role of miR-183 in patients with GC, we examined the association of miR-183 expression with survival time in these patients using the Kaplan-Meier survival analysis. miR-183 expression level was divided into low expression (n=22) and high expression (n=30) based on the median value (-ΔCt=-3.55, Figure 1D). Overall, the survival for patients with low miR-183 expression was significantly lower than that for patients with high expression (P=0.008, log-rank test), as shown in the Kaplan-Meier survival curves (Figure 1E).
Table 1.
The relationship between clinicopathological parameters and miR-183 expression in primary gastric adenocarcinoma
| Clinicopathological Parameters | Patients (n) % | Expression level of miR-183 T/F Test P | |||
|---|---|---|---|---|---|
| Gender | 1.049 | 0.299 | |||
| Male | 40 | 77% | -3.428±0.215 | ||
| Female | 12 | 23% | -3.938±0.531 | ||
| Age (Y) | 0.143 | 0.887 | |||
| ≤60 | 25 | 48% | -3.576±0.227 | ||
| >60 | 27 | 52% | -3.517±0.339 | ||
| Location of tumor | 0.065 | 0.978 | |||
| Upper stomach | 16 | 31% | -3.447±0.415 | ||
| Middle stomach | 7 | 13% | -3.750±0.628 | ||
| Lower stomach | 24 | 46% | -3.554±0.279 | ||
| Mixed | 5 | 10% | -3.537±0.715 | ||
| Tumor size | 0.731 | 0.469 | |||
| ≤5 cm | 13 | 25% | -3.285±0.420 | ||
| >5 cm | 39 | 75% | -3.633±0.237 | ||
| Grade of differentiation | 0.286 | 0.776 | |||
| Well and moderate | 31 | 60% | -3.594±0.274 | ||
| Poor and not | 21 | 40% | -3.474±0.315 | ||
| Depth of invasion | 1.779 | 0.081 | |||
| T1+T2 | 12 | 23% | -2.893±0.235 | ||
| T3+T4 | 40 | 77% | -3.741±0.250 | ||
| Lymph node metastasis | 2.945 | 0.005* | |||
| Negative | 12 | 23% | -2.518±0.352 | ||
| Positive | 40 | 77% | -3.854±0.225 | ||
| Distant metastasis | 0.962 | 0.341 | |||
| Negative | 49 | 94% | -3.497±0.215 | ||
| Positive | 3 | 6% | -4.343±0.287 | ||
| TNM stage | 2.829 | 0.007* | |||
| I+II | 16 | 46 | -2.730±0.283 | ||
| III+IV | 36 | 33 | -3.908±0.247 | ||
P<0.05, statistical significance.
Taken together, these results suggested that gastric tumor tissues showed a dramatic decrease in miR-183 expression compared with adjacent normal tissues. Moreover, GC patients with less miR-183 expression had more lymphatic metastasis, a higher TNM stage, and a shorter survival time.
miR-183 inhibited gastric cancer cell invasion in vitro
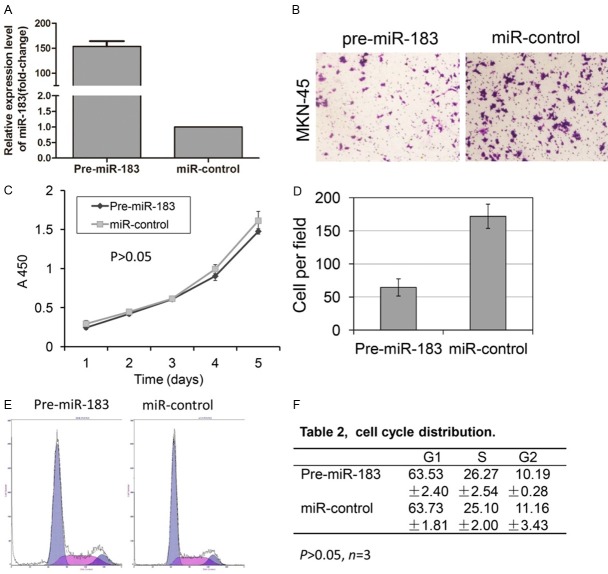
To further assess the functional role of miR-183 in GC, we used two gastric cancer cell lines, MKN-45 and MKN-28. The gastric cancer cells were transfected with hsa-miR-183 pre-miR miRNA precursor and pre-miR-negative control (Ambion catalog no. AM17100) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Notably, the miR-183 expression level was significantly higher in cells transfected with the miRNA precursor (Pre-miR-183 group) than in cells transfected with the pre-miR-negative control (miR-control group) (P<0.001, Figure 2A). Next, we examined the invasion of the gastric cancer cell lines using transwell membrane inserts. We observed that gastric cancer cells transfected with Pre-miR-183 showed a two- to three-fold decrease in invasion compared with the negative control (Figure 2B and 2D, Supplemental Figure 1B and 1D). Invasive cells were quantified in five fields of view on each membrane, and each experiment was repeated in triplicate.
Figure 2.
miR-183 inhibits MKN-45 cell invasion, but does not influence proliferation and cell cycle distribution. A. Quantitative RT-PCR analysis of miR-183 in MKN-45 cells transfected with Pre-miR-183 and miR-control. B, D. Representative images of invasive cells on polycarbonate transwell membranes, and the average number of invasive cells from three independent experiments are shown in D (P<0.01). C. The proliferation rates of the cell lines were detected using a CCK-8 assay (P>0.05). E, F. Cell cycle distribution was detected through flow cytometry analysis (P>0.05).
To explore the distinct mechanisms underlying the decrease in cell invasion after the over-expression of mature miR-183, we applied CCK-8 and flow cytometry assays to detect the proliferation and cell cycle distribution of GC cells, respectively. The results show that the over-expression of miR-183 did not affect the proliferation and cell cycle distribution of MKN-45 (Figure 2C, 2E and 2F) and MKN-28 (Supplemental Figure 1C, 1E and 1F) cells. Taken together, these results suggest that miR-183 inhibits metastasis without affecting cell proliferation and cell cycle distribution. These results are consistent with the results obtained in gastric tumor tissues.
Negative relationship of miR-183 and Ezrin expression in gastric cancer cells and tissues
To explore the mechanisms underlying the reduced invasion of gastric cancer cells through the over-expression of miR-183, we conducted a bioinformatics search for potential protein targets of miR-183 using four common databases: miRanda, Pictar, TargetScan and microRNA.org. At least three of these databases predicted the 3’UTR of Ezrin as a potential target of miR-183, suggesting that Ezrin might be associated with the inhibition of gastric cancer cell invasion through miR-183. To examine this hypothesis, the expression levels of miR-183 and Ezrin protein were assessed in five gastric cancer cell lines and one gastric epithelium cell line. The results showed that there was an inverse correlation between miR-183 and Ezrin protein expression in these cell lines (Figure 1B). Moreover, the transfection of Pre-miR-183 into MKN-45 and MKN-28 cells resulted in the obvious increase in the levels of mature miR-183 expression (Figure 2A, Supplemental Figure 1A). In contrast, Ezrin protein expression was significantly decreased (Figure 3A), further suggested that Ezrin was negatively regulated through miR-183 in GC cell lines.
Figure 3.
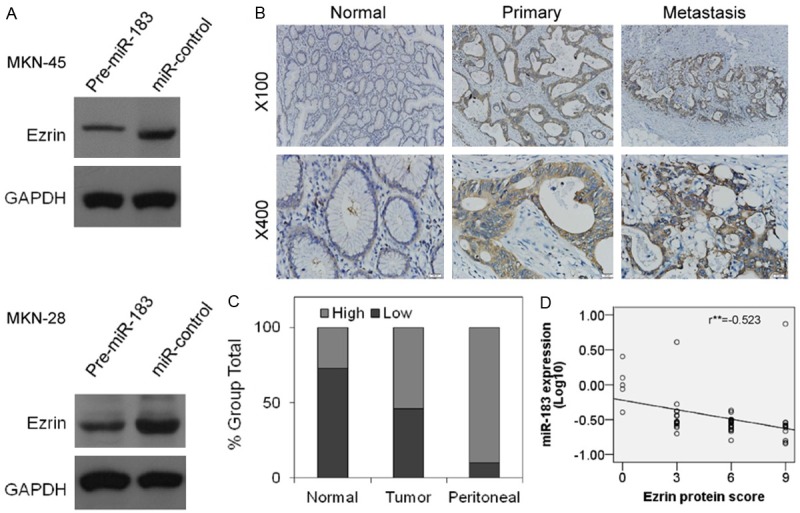
Inverse correlation of miR-183 with Ezrin protein expression levels in gastric tumor tissues and cell lines. A. Ezrin protein levels were decreased when miR-183 was up-regulated in MKN-45 (Top) and MKN-28 (Bottom) cells. B. Representative Ezrin immunostaining in normal gastric tissues, primary gastric tumors, and peritoneal metastases from the same patient. Ezrin protein was low or undetected in normal gastric tissues and high in primary gastric tumors, particularly in metastases. Picture magnification 100× (Top) and 400× (Bottom). C. Ezrin protein expression levels (low and high) in 52 pairs of normal gastric tissues and primary gastric tumors and 10 cases of peritoneal metastases. D. An inverse relationship between miR-183 and Ezrin protein expression is shown in the correlative analysis curve, r=-0.523, with a significant P<0.001.
The inverse relationship of miR-183 and Ezrin expression levels was further demonstrated through quantitative RT-PCR and immunohistochemistry. As previously demonstrated, the Ezrin protein expression in the tumor tissues, particularly in hepatic or peritoneal metastases, was significantly higher than that in normal tissue, whereas miR-183 showed a contrasting expression pattern (Figure 1A). Moreover, we previously examined miR-183 expression levels in 52 cases of gastric tumor tissues and adjacent normal tissues and 10 cases of peritoneal metastases using quantitative RT-PCR (Figure 1C). We also detected the Ezrin protein expression in these paraffin-embedded sections through immunohistochemistry. The pathological immunoreactivity scores were used to describe Ezrin protein expression. As expected, the expression of Ezrin in tumor tissues was significantly higher than that in adjacent normal tissues (P=0.005, χ2=7.828, χ2 test) and obviously lower than that in peritoneal metastases (p=0.04, Fisher’s Exact test) (Figure 3B and 3C). The inverse relationship between miR-183 and Ezrin protein expression was also observed in the correlative analysis curve (Figure 3D). These data suggested that the down-regulation of miR-183 might account for the Ezrin upregulation observed in gastric cancer.
3’UTR region of Ezrin mRNA is a direct target of miR-183
Three prediction programs, including TargetScan, miRBase, and PicTar, have predicted Ezrin 3’UTR as a potential target of miR-183 (Figure 4A). To confirm whether the 3’UTR region of Ezrin is a direct target of miR-183, we used the luciferase reporter system. The 3’UTR region of Ezrin mRNA, containing the miR-183 binding site, was inserted into a luciferase reporter gene. MKN-45 cells were used for this assay. The results showed that the luciferase activity in the pre-miR-183 group was significantly decreased compared with that in the miR-control and mock group (Figure 4B). However, the luciferase activity was not significantly different among the three groups cotransfected with mutant reporter genes (Figure 4C). Thus, these results showed that the Ezrin expression level was affected, at least in part, through the direct binding of miR-183 to the 3’UTR region of Ezrin mRNA and Ezrin is a direct target of miR-183.
Figure 4.
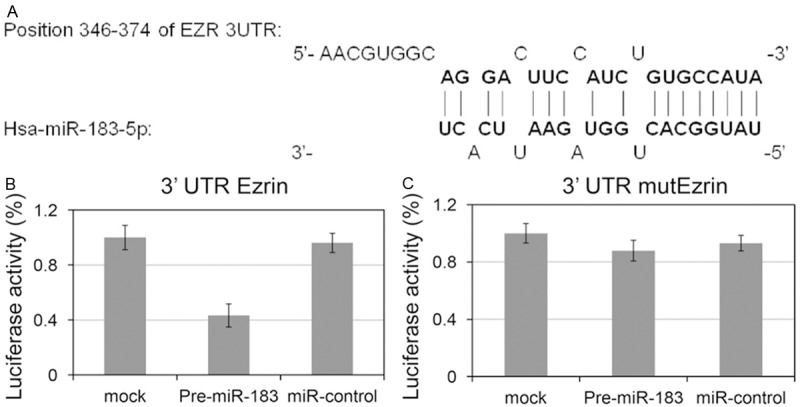
The 3’UTR region of Ezrin mRNA is a direct target of miR-183. A. The results from three prediction programs (TargetScan, miRBase, and PicTar) suggested that Ezrin 3’UTR is a potential target of miR-183. B. Luciferase activities of wild-type 3’UTR-Ezrin-luc constructs among three groups (Pre-miR-183, miR-control, and mock) of MKN-45 cells. C. Luciferase activities of mutant 3’UTR-Ezrin-luc constructs among three groups of MKN-45 cells. *P<0.05.
Discussion
Human gastric cancer is the second most common cause of cancer death, making this disease a major public health issue. Gastric cancer invasion and metastasis are the primary factors that affect patient survival time [27]. Many studies have shown that the estimated overall survival rate of patients with gastric cancer was 57.6% or less [28,29]. Gastric cancer metastasis is a multistep and multifactorial process, associated with various genetic and molecular alterations, including the activation of various oncogenes, inactivation of tumor suppressor genes, dysregulation of the cell cycle, etc. [30,31]. However, the detailed molecular mechanisms of gastric cancer metastasis remain unclear. With the identification and characterization of genes, molecules and associated pathways, an increasing number of studies have provided evidence to increase our understanding of gastric cancer metastasis. Recently, microRNAs have emerged as an important mechanism of gastric cancer metastasis.
miRNAs have been demonstrated to promote [21,22] or suppress [23,24] cancer development, metastasis and cell cycle progression, providing a new perspective on tumorigenesis. Nonetheless, the role of miR-183 in GC metastasis remains unclear. Preliminary studies have shown that miR-183 expression in stage II GC tissues is lower than that in adjacent normal tissues, and patients with lower miR-183 expression have more lymph node metastasis and a shorter survival time [32]. In the present study, we focused on the effect of miR-183 on GC metastasis and confirmed that miR-183 acts as a tumor suppressor in GC metastasis. We observed that miR-183 expression is dramatically decreased in gastric tumor tissues and cells compared with paired adjacent normal tissues and gastric epithelium cells. Statistical analyses revealed that the level of miR-183 expression is associated with lymph node metastasis and pathological TNM stage, and patients with less miR-183 expression have a shorter survival time, consistent with the results of a previous study. Moreover, we observed that the restoration of miR-183 expression in gastric cancer cells reduced cell invasion in vitro. Furthermore, we demonstrated that the miR-183-mediated suppression of GC cell invasion reflects the silencing of Ezrin expression. Thus, the results of the present study suggested that miR-183 could be a promising diagnostic and therapeutic target for the treatment of metastatic gastric cancer.
Extensive studies over the past decade have revealed that microRNAs play critical roles in regulating multiple aspects of biological processes, including cell proliferation, differentiation, metabolism and cell cycle progression. miR-142-3p acts as a regulator of the balance between proliferation and differentiation in lung mesenchymal cells. The downregulation of miR-142-3p expression inhibited the proliferation of parabronchial smooth muscle cells, resulting in an imbalance in lung development [33]. It has been reported that various miRNAs, including miR-19b, miR-33, and miR-27b, play key roles in liver cell development and metabolism [34-36]. These studies suggest that miRNAs acts as an effective regulator in normal cell proliferation, and the dysregulation of these miRNAs might lead to cell hyperplasia and eventual tumorigenesis. The data obtained in the present study showed that miR-183 expression is obviously decreased in GC tissues compared with paired adjacent normal tissues, suggesting that miR-183 might function as a tumor suppressor to inhibit gastric tumorigenesis. To test this hypothesis, we overexpressed miR-183 in GC cells and examined cell viability using a CCK-8 assay and detect cell cycle distribution through flow cytometry. The results suggested that miR-183 does not regulate gastric cancer proliferation and cell cycle progression. The results of a recent study demonstrated that miR-183 suppressed cancer cell proliferation in various types of cancer, including retinoblastoma [37], prostate cancer [38], etc. The different roles of miR-183 suggest that the same microRNAs might have different function in different tumor types.
Ezrin, forming the ERM protein family with radixin and moesin, acts as a molecular crosslinker between actin filaments and proteins anchored in the cell membrane. Studies have shown that Ezrin expression is organ specific and expressed at high levels in the small intestine, lung, pancreas and stomach [39]. Increasing evidence has shown that Ezrin is associated with the metastasis of various human cancers [40-42]. The in silico analysis of Ezrin and microRNAs, using at least three prediction programs, including TargetScan, miRBase, and PicTar, revealed that Ezrin is a target of miR-183. Lowery et al. demonstrated that miR-183 overexpression inhibited breast cancer cell migration, and this effect might reflect the downregulation of Ezrin [24]. Wang et al. provided evidence that miR-183 down-regulated Ezrin expression and repressed lung cancer metastasis [23]. Zhu et al. recently reported that miR-183 expression was down-regulated in osteosarcoma tissues and cells, and the down-regulation of miR-183 promoted metastasis of osteosarcoma through Ezrin targeting [25]. In the study, we showed the overexpression of Ezrin in primary gastric tumor tissues, particularly peritoneal metastases, whereas miR-183 showed an inverse expression pattern. The transfection of miR-183 into MKN-45 and MKN-28 cells showed opposing expression patterns between miR-183 and Ezrin. This result was further confirmed using luciferase activity assays. It is likely that the tumor suppression function of miR-183 reflects the down-regulation of Ezrin through miR-183 in gastric cancer. It has been reported that miR-183 could indirectly regulate MAPK/ERK signaling through Ezrin in osteosarcomas cell lines [25]. Thus, it would be interesting to determine whether there are signaling pathways downstream of miR-183 and Ezrin cross talk in gastric tumorigenesis. Taken together, the results of the present study suggest that miR-183 inhibits the invasion of gastric cancer primarily through the down-regulation of Ezrin.
Thus, these findings indicated that miR-183 plays a critical role in the aggressiveness of gastric cancer and functions as a new tumor suppressor in gastric cancer metastasis. These results might provide important information concerning new diagnostic and therapeutic targets for the treatment of metastatic gastric cancer.
Acknowledgements
We thank Drs. Yang YH and Kang DY at the Department of Pathology, Fujian Medical University Union Hospital for assessment of immunostaining score, and Tian J at statistic teaching and research section of Fujian Medical University for statistical analysis. This work was supported by The Natural Science Foundation of Fujian province, China (Grant: 2012J01350).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12:17–20. doi: 10.3748/wjg.v12.i1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet. 2009;374:477–490. doi: 10.1016/S0140-6736(09)60617-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein CA. Cancer. The metastasis cascade. Science. 2008;321:1785–1787. doi: 10.1126/science.1164853. [DOI] [PubMed] [Google Scholar]
- 5.Hu Y, Zhu Q, Tang L. MiR-99a antitumor activity in human breast cancer cells through targeting of mTOR expression. PLoS One. 2014;9:e92099. doi: 10.1371/journal.pone.0092099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li XY, Luo QF, Wei CK, Li DF, Li J, Fang L. MiRNA-107 inhibits proliferation and migration by targeting CDK8 in breast cancer. Int J Clin Exp Med. 2014;7:32–40. [PMC free article] [PubMed] [Google Scholar]
- 7.Yao J, Zhou E, Wang Y, Xu F, Zhang D, Zhong D. microRNA-200a inhibits cell proliferation by targeting mitochondrial transcription factor A in breast cancer. DNA Cell Biol. 2014;33:291–300. doi: 10.1089/dna.2013.2132. [DOI] [PubMed] [Google Scholar]
- 8.Ye XM, Zhu HY, Bai WD, Wang T, Wang L, Chen Y, Yang AG, Jia LT. Epigenetic silencing of miR-375 induces trastuzumab resistance in HER2-positive breast cancer by targeting IGF1R. BMC Cancer. 2014;14:134. doi: 10.1186/1471-2407-14-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zearo S, Kim E, Zhu Y, Zhao JT, Sidhu SB, Robinson BG, Soon P. MicroRNA-484 is more highly expressed in serum of early breast cancer patients compared to healthy volunteers. BMC Cancer. 2014;14:200. doi: 10.1186/1471-2407-14-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin J, Bai Z, Song J, Yang Y, Wang J, Han W, Zhang J, Meng H, Ma X, Yang Y, Wang T, Li W, Zhang Z. Differential expression of serum miR-126, miR-141 and miR-21 as novel biomarkers for early detection of liver metastasis in colorectal cancer. Chin J Cancer Res. 2014;26:95–103. doi: 10.3978/j.issn.1000-9604.2014.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y, Zhu J, Ou C, Deng Z, Chen M, Huang W, Li L. MicroRNA-145 inhibits tumour growth and metastasis in colorectal cancer by targeting fascin-1. Br J Cancer. 2014;110:2300–2309. doi: 10.1038/bjc.2014.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen T, Yao LQ, Shi Q, Ren Z, Ye LC, Xu JM, Zhou PH, Zhong YS. MicroRNA-31 contributes to colorectal cancer development by targeting factor inhibiting HIF-1alpha (FIH-1) Cancer Biol Ther. 2014;15:516–523. doi: 10.4161/cbt.28017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.You J, Li Y, Fang N, Liu B, Zu L, Chang R, Li X, Zhou Q. MiR-132 suppresses the migration and invasion of lung cancer cells via targeting the EMT regulator ZEB2. PLoS One. 2014;9:e91827. doi: 10.1371/journal.pone.0091827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Y, Fan X, Li W, Ping W, Deng Y, Fu X. miR-138-5p reverses gefitinib resistance in non-small cell lung cancer cells via negatively regulating G protein-coupled receptor 124. Biochem Biophys Res Commun. 2014;446:179–186. doi: 10.1016/j.bbrc.2014.02.073. [DOI] [PubMed] [Google Scholar]
- 15.Zhu YJ, Xu B, Xia W. Hsa-mir-182 downregulates RASA1 and suppresses lung squamous cell carcinoma cell proliferation. Clin Lab. 2014;60:155–159. doi: 10.7754/clin.lab.2013.121131. [DOI] [PubMed] [Google Scholar]
- 16.Williams LV, Veliceasa D, Vinokour E, Volpert OV. miR-200b inhibits prostate cancer EMT, growth and metastasis. PLoS One. 2013;8:e83991. doi: 10.1371/journal.pone.0083991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu D, Tao T, Xu B, Chen S, Liu C, Zhang L, Lu K, Huang Y, Jiang L, Zhang X, Huang X, Zhang L, Han C, Chen M. MiR-361-5p acts as a tumor suppressor in prostate cancer by targeting signal transducer and activator of transcription-6(STAT6) Biochem Biophys Res Commun. 2014;445:151–156. doi: 10.1016/j.bbrc.2014.01.140. [DOI] [PubMed] [Google Scholar]
- 18.Xie J, Chen M, Zhou J, Mo MS, Zhu LH, Liu YP, Gui QJ, Zhang L, Li GQ. miR-7 inhibits the invasion and metastasis of gastric cancer cells by suppressing epidermal growth factor receptor expression. Oncol Rep. 2014;31:1715–1722. doi: 10.3892/or.2014.3052. [DOI] [PubMed] [Google Scholar]
- 19.Jia H, Zhang Z, Zou D, Wang B, Yan Y, Luo M, Dong L, Yin H, Gong B, Li Z, Wang F, Song W, Liu C, Ma Y, Zhang J, Zhao H, Li J, Yu J. MicroRNA-10a is down-regulated by DNA methylation and functions as a tumor suppressor in gastric cancer cells. PLoS One. 2014;9:e88057. doi: 10.1371/journal.pone.0088057. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Qiu T, Zhou X, Wang J, Du Y, Xu J, Huang Z, Zhu W, Shu Y, Liu P. MiR-145, miR-133a and miR-133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancer. FEBS Lett. 2014;588:1168–1177. doi: 10.1016/j.febslet.2014.02.054. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Fu H, Xu C, Tie Y, Xing R, Zhu J, Qin Y, Sun Z, Zheng X. miR-183 inhibits TGF-beta1-induced apoptosis by downregulation of PDCD4 expression in human hepatocellular carcinoma cells. BMC Cancer. 2010;10:354. doi: 10.1186/1471-2407-10-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010;70:9570–9580. doi: 10.1158/0008-5472.CAN-10-2074. [DOI] [PubMed] [Google Scholar]
- 23.Wang G, Mao W, Zheng S. MicroRNA-183 regulates Ezrin expression in lung cancer cells. FEBS Lett. 2008;582:3663–3668. doi: 10.1016/j.febslet.2008.09.051. [DOI] [PubMed] [Google Scholar]
- 24.Lowery AJ, Miller N, Dwyer RM, Kerin MJ. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC Cancer. 2010;10:502. doi: 10.1186/1471-2407-10-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu J, Feng Y, Ke Z, Yang Z, Zhou J, Huang X, Wang L. Down-regulation of miR-183 promotes migration and invasion of osteosarcoma by targeting Ezrin. Am J Pathol. 2012;180:2440–2451. doi: 10.1016/j.ajpath.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 26.Zhao H, Guo M, Zhao G, Ma Q, Ma B, Qiu X, Fan Q. miR-183 inhibits the metastasis of osteosarcoma via downregulation of the expression of Ezrin in F5M2 cells. Int J Mol Med. 2012;30:1013–1020. doi: 10.3892/ijmm.2012.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hippo Y, Yashiro M, Ishii M, Taniguchi H, Tsutsumi S, Hirakawa K, Kodama T, Aburatani H. Differential gene expression profiles of scirrhous gastric cancer cells with high metastatic potential to peritoneum or lymph nodes. Cancer Res. 2001;61:889–895. [PubMed] [Google Scholar]
- 28.Huang CM, Lin JX, Zheng CH, Li P, Xie JW, Lin BJ. Effect of negative lymph node count on survival for gastric cancer after curative distal gastrectomy. Eur J Surg Oncol. 2011;37:481–487. doi: 10.1016/j.ejso.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Siewert JR, Bottcher K, Stein HJ, Roder JD. Relevant prognostic factors in gastric cancer: ten-year results of the German Gastric Cancer Study. Ann Surg. 1998;228:449–461. doi: 10.1097/00000658-199810000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson SM, Evers BM. Translational research in gastric malignancy. Surg Oncol Clin N Am. 2008;17:323–340. viii. doi: 10.1016/j.soc.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Kim K, Chun KH, Suh PG, Kim IH. Alterations in cell proliferation related gene expressions in gastric cancer. Crit Rev Eukaryot Gene Expr. 2011;21:237–254. doi: 10.1615/critreveukargeneexpr.v21.i3.20. [DOI] [PubMed] [Google Scholar]
- 32.Zheng WW, Huang CM, Xie JW, Zheng CH, Li P, Wang JB, Lin JX. [Expression of microRNA-183 in stage II (gastric cancer and its association with Ezrin protein] . Zhonghua Wei Chang Wai Ke Za Zhi. 2012;15:723–726. [PubMed] [Google Scholar]
- 33.Carraro G, Shrestha A, Rostkovius J, Contreras A, Chao CM, El Agha E, Mackenzie B, Dilai S, Guidolin D, Taketo MM, Gunther A, Kumar ME, Seeger W, De Langhe S, Barreto G, Bellusci S. miR-142-3p balances proliferation and differentiation of mesenchymal cells during lung development. Development. 2014;141:1272–1281. doi: 10.1242/dev.105908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allen RM, Marquart TJ, Albert CJ, Suchy FJ, Wang DQ, Ananthanarayanan M, Ford DA, Baldan A. miR-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol Med. 2012;4:882–895. doi: 10.1002/emmm.201201228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lakner AM, Steuerwald NM, Walling TL, Ghosh S, Li T, McKillop IH, Russo MW, Bonkovsky HL, Schrum LW. Inhibitory effects of microRNA 19b in hepatic stellate cell-mediated fibrogenesis. Hepatology. 2012;56:300–310. doi: 10.1002/hep.25613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vickers KC, Shoucri BM, Levin MG, Wu H, Pearson DS, Osei-Hwedieh D, Collins FS, Remaley AT, Sethupathy P. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2013;57:533–542. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Wang X, Li Z, Liu H, Teng Y. MicroRNA-183 suppresses retinoblastoma cell growth, invasion and migration by targeting LRP6. FEBS J. 2014;281:1355–1365. doi: 10.1111/febs.12659. [DOI] [PubMed] [Google Scholar]
- 38.Ueno K, Hirata H, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, Hinoda Y, Dahiya R. microRNA-183 is an oncogene targeting Dkk-3 and SMAD4 in prostate cancer. Br J Cancer. 2013;108:1659–1667. doi: 10.1038/bjc.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akisawa N, Nishimori I, Iwamura T, Onishi S, Hollingsworth MA. High levels of ezrin expressed by human pancreatic adenocarcinoma cell lines with high metastatic potential. Biochem Biophys Res Commun. 1999;258:395–400. doi: 10.1006/bbrc.1999.0653. [DOI] [PubMed] [Google Scholar]
- 40.Hunter KW. Ezrin, a key component in tumor metastasis. Trends Mol Med. 2004;10:201–204. doi: 10.1016/j.molmed.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 41.Li Q, Gao H, Xu H, Wang X, Pan Y, Hao F, Qiu X, Stoecker M, Wang E, Wang E. Expression of ezrin correlates with malignant phenotype of lung cancer, and in vitro knockdown of ezrin reverses the aggressive biological behavior of lung cancer cells. Tumour Biol. 2012;33:1493–1504. doi: 10.1007/s13277-012-0400-9. [DOI] [PubMed] [Google Scholar]
- 42.Jaroensong T, Endo Y, Lee SJ, Kamida A, Mochizuki M, Nishimura R, Sasaki N, Nakagawa T. Effects of transplantation sites on tumour growth, pulmonary metastasis and ezrin expression of canine osteosarcoma cell lines in nude mice. Vet Comp Oncol. 2012;10:274–282. doi: 10.1111/j.1476-5829.2011.00294.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.