Abstract
Recent studies have highlighted the role of microRNA-21 (miR-21) as a prognostic biomarker of breast cancer. However, controversy still remains. The present study aimed to summarize available evidences and obtain a more precise estimation of a prognostic role of miR-21 in breast cancer patients. All eligible studies were searched from PubMed and EMBASE through multiple search strategies. Data were extracted from studies comparing survival in breast cancer patients having higher miR-21 expression with those having lower expression. A meta-analysis was performed to clarify prognostic role of miR-21 in patients with breast cancer. Subgroup analysis was also performed according to patients’ ethnicity. A total of 6 eligible articles comprising 951 cases were selected for this meta-analysis. The combined hazard ratios (HRs) and 95% confidence intervals (95% CIs) for overall survival (OS) were 2.11 (1.09-4.08) and for disease free survival (DFS) was 1.6 (1.30-1.96). Subgroup analysis indicated high miR-21 expression was significantly associated with worse OS in Asian patients (HR = 4.39, 95% CI: 2.47-7.80) but not in non-Asian patients (HR = 1.18, 95% CI: 0.81-1.70). Sensitivity analysis revealed results of this meta-analysis were robust. Odds ratios (ORs) showed that miR-21 expression was closely associated with estrogen receptor (ER), progesterone receptor (PR), lymph node metastasis, histological grade, Her2/neu. The pooled ORs and 95% CIs were 0.53 (0.35-0.80), 0.49 (0.32-0.74), 2.32 (1.54-3.50), 2.44 (1.58-3.75), 4.29 (2.34-7.85), respectively. Our results indicated that elevated miR-21 expression could potentially predict poor survival in patients with breast cancer.
Keywords: MicroRNA-21, breast cancer, prognosis, biomarker, meta-analysis
Introduction
Breast cancer is one of the most common malignancies in women, which is a significant health problem ranking as a leading cause of cancer-related deaths among women around the world. The morbidity rate of breast cancer has gradually increased in recent decades [1-3] and 1.38 million individuals worldwide are affected every year [4]. Despite the extensive use of adjuvant systematic therapies such as radiotherapy, chemotherapy, hormone treatment, and biological therapy, the prognosis is still poor. More than 20% of patients with early breast cancer could develop incurable metastatic disease eventually [5,6]. Therefore, factors to effectively evaluate the patients’ survival outcome are needed urgently.
To date, there are a number of independent prognostic factors identified in clinical management of breast cancer, including tumor size, histological grade, nodal status, and patient age [7-9]. However, breast cancer is a heterogeneous disease with multiple factors involved in, and alterations in molecular mechanisms may affect tumor growth and progression, thereby the prognostic value of clinicopathological parameters is potentially limited [10]. Nowadays, a variety of potential prognostic biomarkers are being studied and applied in basic and clinical research, such as HER-2/neu, estrogen receptor (ER), progesterone receptor (PR), cyclin E, p53, bcl-2, vascular endothelial growth factor, urokinase-type plasminogen activator-1. However, no single biomarker was able to predict those patients with the best (or worst) prognosis due to its the discriminant value on the complex , heterogenetic disease [10].
MicroRNAs (miRNAs) are endogenous, small non-coding RNAs molecules with a length of 18-25 nucleotides. They were first reported in 1993 [11]. MicroRNA could identify post transcriptional gene regulators that paired to complementary sequences in the 3’untranslated region (3’UTR) of target mRNAs, and regulate protein-coding genes expression by mRNA degradation or translational repression [12]. These miRNAs may regulate the translation of specific protein-coding genes [13,14].
Recent studies have revealed that microRNA-21 (miR-21) is elevated in many kinds of cancer, including breast cancer [15,16], colorectal cancer [17,18], lung cancer [19,20], pancreatic cancer [21,22], prostate cancer [23], gastric cancer [23], glioblastoma [24], and is recognized a potential oncomicroRNA. Many studies have shown that overexpression of miR-21 could increase cell growth, migration, invasion, survival [25,26] and inhibit apoptosis [27]. Studies also demonstrate that miR-21 can modulate several tumor suppressor genes, including phosphatase and tensin homolog (PTEN) [28], tropomyosin 1 (TPM1) [29], programmed cell death 4 (PDCD4) [30], and may play a role in cancer invasion and metastasis.
Prognostic role of microRNA-21 (miR-21) in breast cancer has been highlighted in some studies. However, controversy still remains. In this study, we aim to perform a meta-analysis to evaluate and predict the overall risk of high miR-21 expression for survival prognosis and clinicopathological features in breast cancer patients and discuss the challenges of miR-21 as a possible prognosis biomarker in breast cancer.
Material and methods
This meta-analysis was carried out in accordance with the guidelines of the meta-analysis of the Observational Studies in Epidemiology group (MOOSE) [31].
Search strategy
PubMed and EMBASE were searched for the last time on May 15, 2014. The search strategy included the following keywords: miR-21, microRNA-21, breast Cancer, breast carcinoma, prognostic and prognosis. A manual review of the references of relevant publications was also performed to obtain additional studies.
Study selection
Studies were considered eligible if they met the following criteria: (i) they had to study the patients with breast cancer; (ii) they had to detect the expression of miR-21 in tissue, serum, or other clinical samples; (iii) they had to investigate the survival outcome or the correlation between miR-21 expression and the clinical variables. (iv) the method for detecting miR-21 must be same. Articles were excluded based on any of the following reasons: (i) review articles, laboratory articles or letters; (ii) They were described the survival outcome of other tumors or other markers; (iii) They lacked key information for calculation with methods developed by Parmar, Williamson , and Tierney [32-34]; (iv) They were not English; (v) the articles from one author and the studies brought into the repeated samples from the same patients when a study already included.
The information such as titles, abstracts, full texts and reference lists of all of the identified reports was carefully identified in duplicate by two investigators (Pan and Mao). These extracted articles were double checked by Geng. Disagreements were resolved by discussion among all authors in this paper. All the data were consensus. The references from the relevant literatures, including all the identified studies, reviews and editorials, were also reviewed manually.
Quality assessment
The quality of all the studies included was systematically evaluated according to a critical review checklist of the Dutch Cochrane Centre proposed by MOOSE [31,35]. The current checklist included the following key points: (i) clear definition of study population and origin of country; (ii) clear definition of type of carcinoma; (iii) clear definition of study design; (iv) clear definition of outcome assessment; (v) clear definition of measurement of miR-21; (vi) sufficient period of follow-up. Any study without mentioning these points is excluded so as not to compromise the quality of the meta-analysis.
Data extraction
Data were extracted from each study according to the before-mentioned selection criteria. The primary information was collected in a form: first author’s name, year of publication, country of origin, ethnicity, total number of cases, follow-up time, multivariate analysis, univariate analysis, kaplan-meier survival analysis, P value, 95% CI, and hazard ratios independently. Other extracted data elements were included as the following: age, gender, TNM stage, lymph status, histological classification, method of detecting miR-21, Estrogen Receptor (ER), Progesterone Receptor (PR), Her2/neu. An HR of > 1 was considered significant association with a poorer outcome. If only survival curves were available, data were extracted from the graphical survival plots and HR was then estimated according to the previously described method [32,34].
Statistical methods
Statistics were conducted as described previously [36]. Hazard Ratios (HRs) and 95% confidence intervals (95% CIs) were calculated for each study. But some studies did not list the HRs or 95% confidence intervals (95% CIs) directly, only giving Kaplan-Meier survival curves instead. The necessary statistics were calculated using software developed by Matthew Sydes and Jayne Tierney (Medical Research Council Clinical Trials Unit, London, UK) [34].
Forest plots were used to estimate the aggregation effect of miR-21 expression on survival outcome (OS and DFS) and the correlation between miR-21 expression and clinical variables. Subgroup analysis of pooled hazard ratios was performed subsequently by ethnicity. For each meta-analysis performed, Cochran’s Q test and Higgins I-squared statistic were carried out to assess the between-study heterogeneity, and heterogeneity was considered significant for P < 0.1. A random-effects model with DerSimonian and Laird method were then applied to calculate the poor HR, if heterogeneity was observed (P < 0.1), Otherwise, the fixed-effects model was used [37]. Pooled HR > 1 indicated poor prognosis for the groups with elevated miR-21 expression and was considered statistically significant if the 95% CI did not overlap 1. Sensitivity analysis was applied by excluding each single study. Finally, publication bias was evaluated using the funnel plot, Begg’s test [38], Egger’s test [39]. P > 0.05 was considered that there was no potential publication bias in the meta-analysis. All analyses were executed using the software Review Manager 5.1 (The Cochrane Collaboration, Oxford, United Kingdom) and Stata 12.0 (http://www.stata.com/; Stata Corporation, College Station, Texas, USA).
Results
Characteristics of the studies included in Meta-analysis
As shown in Figure 1, 130 records for miR-21 and breast cancer were identified in Pubmed and EMBASE. After reviewing these abstracts, 112 studies were excluded due to their irrelevance to the current analysis, letters, reviews and duplicate studies. Furthermore, 12 potential studies were excluded, due to laboratory studies or records without sufficient survival data for calculation. Therefore, 6 eligible articles were enrolled in this meta-analysis [40-45].
Figure 1.

Flow diagram of the study selection process.
These eligible studies were published between 2008 and 2013. One studies evaluated patients from China, one evaluated patients from Italy, one evaluated patients from Korea, one evaluated patients from Japan, one evaluated patients from Greek, and one evaluated patients from the United States of America. These studies included a total of 951 patients with a mean number of 158.5 patients per study. These six eligible studies were all retrospective cohort studies. The method of miR-21 detection was all quantitative real-time polymerase chain reaction (qRT-PCR). MiR-21 expression levels were measured in tumor tissue or bone marrow. The mean or medium length of follow-up ranged from 35.5 to 86.2 months. Characteristics of the eligible studies are summarized in Table 1.
Table 1.
Clinical characteristics of studies included in the meta-analysis
| First author | Year | Population | Study Design | Number | Materials | Stage | Method | Cut-off | Survival analysis | Hazard Ratios | Follow-up, months | Quality score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yan | 2008 | China | R | 113 | Tumor | I-III | qRT-PCR | Mean | OS | Reported | 66.2 (10.4-81.0) | 7 |
| Qian | 2008 | Italy | R | 301 | Tumor | I-IV | qRT-PCR | Median | OS | Reported | 86.2 (8.0-108) | 6 |
| Reported | 86.2 (8.0-108) | |||||||||||
| Lee | 2011 | Korea | R | 109 | Tumor | I-III | qRT-PCR | Mean | OS | Reported | median 54 | 7 |
| DFS | SC | median 54 | ||||||||||
| OTA | 2011 | Japan | R | 291 | Bone marrow | -- | qRT-PCR | 5.84 | OS | SC | 61 (2-90) | 7 |
| DFS | Reported | 61 (2-90) | ||||||||||
| Markou | 2013 | Greece | R | 112 | Tumor | I-IV | qRT-PCR | Median | OS | Reported | 84 (10-149) | 7 |
| DFI | Reported | 84 (10-149) | ||||||||||
| Walter | 2011 | the USA | R | 25 | Tumor | I-IV | qRT-PCR | Median | OS | SC | median 35.5 | 6 |
The studies included here are all retrospective cohort studies with different groups of patients. qRT-PCR, quantitative real-time PCR; --, not mentioned; OS, overall survival; DFS, disease-free survival; DFI, disease-free interval; SC, survival curve.
Methodological quality of the studies
The qualities of 6 eligible studies included in our meta-analysis were assessed according to the Newcastle-Ottawascale (NOS). The NOS contained eight items of methodology, which were categorized into the three dimensions of selection, comparability, and outcome. It was used to assess the quality of non-randomized studies in meta-analysis. For quality, scores ranged from 0 (lowest) to 9 (highest), and studies with scores of more than 5 were identified as high quality. All these 6 eligible studies gained more than 5 scores in methodological assessment, indicating that they were of high quality relatively (Table 1).
Correlation between miR-21 expression and survival outcome (OS and DFS)
The meta-analysis was performed on 6 studies [40-45] assessing the association of miR-21 expression with OS and DFS. As shown in Figure 2, the pooled HR for OS was 2.11 (95% CI: 1.09-4.08; Z = 2.22; P = 0.03) with heterogeneity (I 2 = 69.0%, P = 0.006). In the case of heterogeneity, a random-effects model was used. The pooled HR of four studies [41-43,45] for DFS was 1.60 (95% CI: 1.30-1.96; Z = 4.46; P < 0.00001) with no heterogeneity (I 2 = 0%, P = 0.88), and the fixed efforts model was used. It suggested that high miR-21 expression was statistically significant with the worse prognosis of breast cancer and high miR-21 expression was a valuable prognostic factor in breast cancer.
Figure 2.
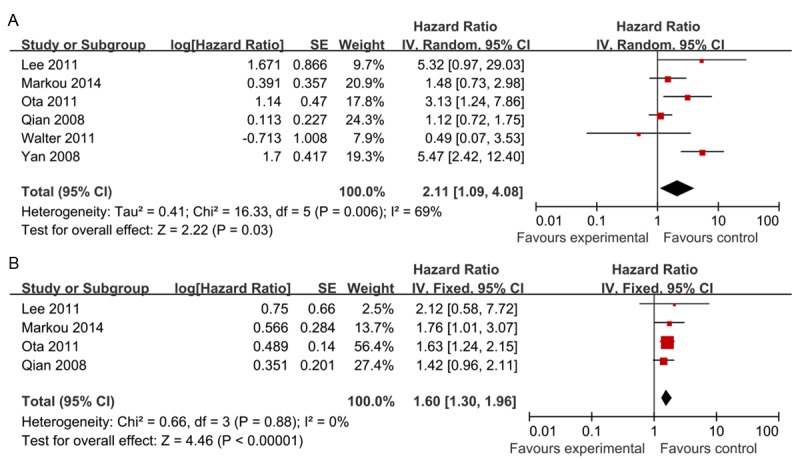
Forest plot of studies evaluating hazard ratio (HR) for the association of high miR-21 expression with overall survival (OS) (A) and disease-free survival (DFS) (B) in patients with breast cancer patients.
Subgroup analysis
Moreover, we carried out subgroup analysis by the patients’ ethnicity. The results showed that there was significant relation between high miR-21 expression and OS, especially in Asian countries (Figure 3) (Table 2). The combined HR of Asian studies [40,42,43] for OS was 4.39 (95% CI: 2.47-7.80; Z = 5.04; P < 0.00001), while the combined HR of the other yielded non-Asian studies [41,44,45] was 1.18 (95% CI: 0.81-1.70; Z = 0.86; P = 0.39). As a result of this subgroup analysis, the heterogeneity was both eliminated. We also tried to use the other grouping term to examine the prognostic role of miR-21, such as number of patients (Table 2). No results could give clinical significance.
Figure 3.
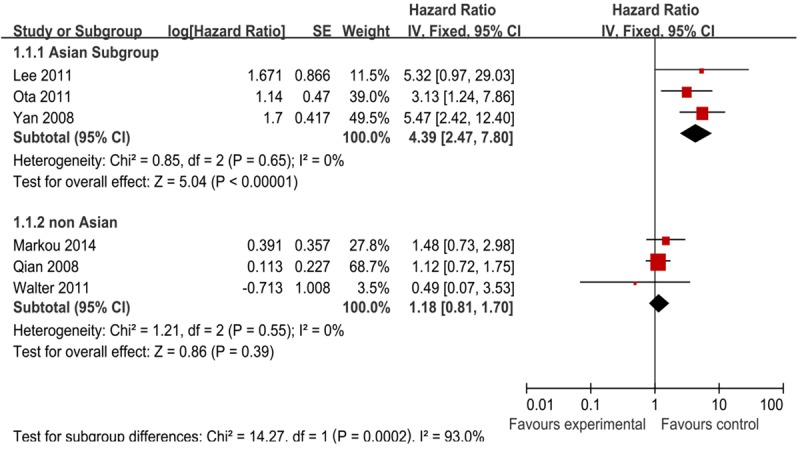
Forest plot of studies evaluating hazard ratio (HR) for the association of high miR-21 expression with overall survival (OS) stratified according to source of ethnicity.
Table 2.
Subgroup analysis of pooled HRs of breast cancer patients with high miR-21 expression for OS
| Heterogeneity | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Subgroup | Number of studies | Number of patients | HR (95% CI) | P value | I 2 (%) | P value |
| Patients’ ethnicity | ||||||
| Asian | 3 | 513 | 4.39 (2.47-7.80) | 0.00001 | 0% | 0.65 |
| non-Asian | 3 | 438 | 1.18 (0.81-1.70) | 0.39 | 0% | 0.55 |
| Number of patients | ||||||
| >200 | 2 | 592 | 1.72 (0.64-4.65) | 0.28 | 74% | 0.05 |
| <200 | 4 | 359 | 2.4 (0.89-6.44) | 0.08 | 67% | 0.03 |
Sensitivity analysis
The conclusions remained similar when a single study involved in the meta-analysis was removed each time to reflect the influence of the rest data-set on the pooled HRs. The summary HR for OS ranged from 1.42 (95% CI: 1.01-1.99) after excluding the study of Yan [40] to 2.60 (95% CI: 1.26-5.36) after excluding the study of Qian [41] (Table 3). The summary HR for DFS ranged from 1.56 (95% CI: 1.14-2.13) after excluding the study of OTA to 1.67 (95% CI: 1.31-2.13) after excluding the study of Qian (Table 4).
Table 3.
HRs (95% CI) of sensitivity analysis on OS for the meta-analysis
| Heterogeneity | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Study omitted | Estimated HR | Low value of 95% CI | High value of 95% CI | P value | I 2 (%) | P value |
| Yan 2008 | 1.42 | 1.01 | 1.99 | 0.04 | 46% | 0.12 |
| Qian 2008 | 2.6 | 1.26 | 5.36 | 0.01 | 57% | 0.05 |
| Lee 2011 | 1.91 | 0.96 | 3.82 | 0.07 | 73% | 0.006 |
| OTA 2011 | 1.94 | 0.9 | 4.18 | 0.09 | 72% | 0.006 |
| Walter 2011 | 2.39 | 1.2 | 4.74 | 0.01 | 73% | 0.005 |
| Markou 2013 | 2.32 | 0.97 | 5.54 | 0.06 | 75% | 0.003 |
| Combined | 2.11 | 1.09 | 4.08 | 0.03 | 69% | 0.006 |
Table 4.
HRs (95% CI) of sensitivity analysis on DFS for the meta-analysis
| Heterogeneity | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Study omitted | Estimated HR | Low value of 95% CI | High value of 95% CI | P value | I 2 (%) | P value |
| Qian 2008 | 1.67 | 1.31 | 2.13 | 0.0001 | 0% | 0.91 |
| Lee 2011 | 1.59 | 1.29 | 1.95 | 0.0001 | 0% | 0.79 |
| OTA 2011 | 1.56 | 1.14 | 2.13 | 0.006 | 0% | 0.74 |
| Markou 2013 | 1.57 | 1.26 | 1.96 | 0.0001 | 0% | 0.77 |
| Combined | 1.6 | 1.3 | 1.96 | 0.00001 | 0% | 0.88 |
Correlation between miR-21 expression and clinical characteristics
The studies which referred the association between miR-21 expression and some clinical characteristics (ER, PR, lymph node metastasis, histological grade, Her2/neu, and age) have been combined to calculate the Odds ratios (ORs). Four studies [40,42-44] evaluated the correlation between miR-21 expression and ER and PR. The pooled ORs were 0.53 (95% CI: 0.35-0.80, P = 0.002) with no heterogeneity (I 2 = 48%, P = 0.12) and 0.49 (95% CI: 0.32-0.74, P = 0.0007) with no heterogeneity (I 2 = 0%, P = 0.46) respectively, indicating that miR-21 expression was negatively related to ER and PR (Figure 4A and 4B) (Table 5). We also assessed the correlation between miR-21 expression and lymphnode node metastasis and Histologic grade. The pooled ORs were 2.32 (95% CI: 1.54-3.50, P = 0.0001) and 2.44 (95% CI: 1.58-3.75, P = 0.0001) respectively (Figure 4C and 4D) (Table 5), which suggested that high miR-21 expression was associated with metastasis and histologic grade of breast cancer. Moreover, the association between miR-21 expression and Her2/neu was identified, with the pooled OR 4.29 (95% CI: 2.34-7.85, P = 0.00001), suggesting that high miR-21 expression was related to Her2/neu. Meanwhile, there was no significant association between high miR-21 expression and age (Figure 4E). All these results could be reviewed in Table 5.
Figure 4.

Forrest plot of odds ratios (ORs) for the association of miR-21 expression with clinicopathological features in breast cancer. A. ORs with corresponding 95% CIs of miR-21 expression with Estrogen Receptor (ER). B. ORs with corresponding 95% CIs of miR-21 expression with Progesterone Receptor (PR). C. ORs with corresponding 95% CIs of miR-21 expression with lymphnode metastasis. D. ORs with corresponding 95% CIs of miR-21 expression with Histologic grade. E. ORs with corresponding 95% CIs of miR-21 expression with HER2/neu. F. ORs with corresponding 95% CIs of miR-21 expression with age.
Table 5.
Meta-analysis of miR-21 expression classified by clinical features
| Heterogeneity | |||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| Clinicopathological features | Number of studies | Number of patients | Model | Pooled OR (95% CI) | P value | I 2 (%) | P value |
| ER (postive vs. negative)* | 4 | 529 | Fixed | 0.53 (0.35-0.80) | 0.002 | 48% | 0.12 |
| PR (postive vs. negative)* | 4 | 524 | Fixed | 0.49 (0.32-0.74) | 0.0007 | 0% | 0.46 |
| Lymph node metastasis (postive vs. negative) | 4 | 536 | Fixed | 2.32 (1.54-3.50) | 0.0001 | 0% | 0.47 |
| Histologic grade (G3 vs. G1, 2) | 4 | 536 | Fixed | 2.44 (1.58-3.75) | 0.0001 | 0% | 0.61 |
| HER2/neu (postive vs. negative) | 3 | 294 | Fixed | 4.29 (2.34-7.85) | 0.00001 | 0% | 0.79 |
| Age (> median vs. < median) | 3 | 242 | Fixed | 1.52 (0.88-2.62) | 0.13 | 0% | 0.61 |
ER: Estrogen Receptor; PR: Progesterone Receptor.
Assessment of publication bias
In the funnel plot analysis, the shape of the funnel plot seemed symmetrical for both OS and DFS. Egger’s test and Begg’s test were used to examine publication bias. There were no significant publication biases in the meta-analysis of miR-21 prediction value for OS (Egger’s test, P = 0.429; Begg’s test, P = 0.999) and DFS (Egger’s test, P = 0.521; Begg’s test, P = 0.308) respectively (Figure 5).
Figure 5.
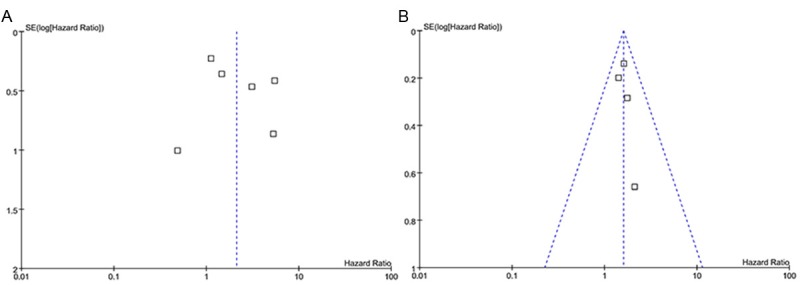
A: Funnel Plots of elevated miR-21 expression and OS in patients with breast cancer. B: Funnel Plots of elevated miR-21 expression and DFS in patients with breast cancer.
Discussion
Alteration of biological markers in tumor tissues plays an important role in predicting the prognostic value of the breast cancer patients [10]. Breast cancer is a malignant disease and is known to be quite complex and heterogeneous in development, progress and response to treatment. As a result, biomarkers available such as ER, PR, HER2, could not reflect the whole prognostic significance for breast cancer patients. Therefore, besides ER, PR, and HER2, it is important to find out new prognostic biomarkers for patients with breast cancer.
Recently, numbers of studies are emerging that microRNA could be considered as revolutionary sources of biomarkers for cancer prognosis [46]. MiR-21, known as a potential oncogenic role, is one of the most commonly observed aberrant miRNAs in a variety of cancers [15-24]. And it could be measured stably and easily in tumor tissues, formalin-fixed paraffin-embedded tissues and blood circulation [14,47]. In breast cancer, some studies found that miR-21 was significantly associated with patients’ survival [40,43]. However, insignificant results of miR-21 were also observed in other studies [41]. Meta-analysis is a systematical approach applied widely to the evaluation of prognostic indicators in different studies. There is no consensus on the association between high miR-21 expression and poor survival prognosis in patients with breast cancer currently, nevertheless the prognostic role of miR-21 has been discussed in head and neck squamous cell carcinoma (HNSCC), non-small-cell lung cancer (NSCLC) and colorectal cancer (CRC) [36,48,49]. Thus, a quantitative meta-analysis was performed to determine the association between miR-21 expression and the survival prognosis and clinicopathological features in breast cancer patients.
This meta-analysis revealed that elevated miR-21 expression did predict poor survival in patients of breast cancer. The pooled HR for OS was 2.11 (95% CI: 1.09-4.08, P = 0.03) with heterogeneity (I 2 = 69.0%, P = 0.006). The differences of HRs were found to be statistically significant, but significant heterogeneity was also observed among the studies. Then we used random effects model to analyze the data, however, heterogeneity was still a potential problem to affect meta-analysis results. Moreover, the result remained similar in a sensitivity analysis when a single study was removed each time. After subgroup analysis by the patients’ ethnicity, the heterogeneity was eliminated both in Asian studies and non-Asian studies. The HR for Asian countries was 4.39 (95% CI: 2.47-7.80, P < 0.00001). It suggested that the miR-21 expression played significant prognostic role on breast cancer in Asian group. Meanwhile, the combined HR for non-Asian group was 1.18 (95% CI: 0.81-1.70, P = 0.39), which suggested that the miR-21 had no significant effect to predict survival outcome in non-Asian group. As a result, the heterogeneity in this meta-analysis might be explained by the patients’ ethnicity and miR-21 expression could be racial different as a prognostic factor. The pooled HR for DFS was 1.60 (95% CI: 1.30-1.96, P < 00001) with no heterogeneity (I 2 = 0%, P = 0.88). The difference was statistically significant, though the HR was not so strong. As referred in Hayes [50], a prognostic factor with HR > 2 is considered strongly predictive. Combining the results for OS and DFS, it may suggest that miR-21 expression in breast cancer patients could predict their prognosis practically, especially in the Asian population.
Furthermore, the correlation between miR-21 expression and clinical characteristics was calculated. There was no significant association between high miR-21 expression and age, while ORs for ER, PR, lymph node metastasis, histological grade, Her2/neu, were significant. Thus, elevated miR-21 expression was closely associated with worse clinical characteristics.
Still some limitations existed in this meta-analysis. First, there are only six studies included in this meta-analysis and the pooled HR was calculated on the basis of these 6 studies with a small sample size of 951 patients. Second, the cut-off values of miR-21 were different in the selected studies. Median and mean values were often chosen as the cut-off values. However, there was no final conclusion to confirm how high was considered high. Third, miR-21 was detected in tumor tissue and bone marrow but few in serum or plasma in the selected studies. Circulating prognostic markers were more likely to be accepted than tissue markers and it could be detected before surgery and be monitored throughout the lives of the cancer patients. Fourth, this meta-analysis did not evaluate the prognostic value of a combination of miR-21 and other miRNA markers for breast cancer cases. Fifth, some studies didn’t provide HRs, and the data were extracted from survival curves, which might be of less credibility than direct analysis on HRs.
Publication bias is a major concern for meta-analysis. In our research, neither Egger’s test nor Begger’s test showed evidence of publication bias; however, bias might still have occurred. It should be noted that positive results tend to be accepted by journals, while negative results are often rejected or not even submitted.
In conclusion, our meta-analysis showed that elevated miR-21 expression was significantly associated with poor survival in breast cancer patients and might potentially predict the poor survival in patients with breast cancer. More multi-center clinical investigations with larger sample size should be conducted to further confirm these results.
Acknowledgements
We would like to thank Cancer Center, Division of Internal Medicine, Chinese PLA General Hospital and Chinese PLA Medical School.
Disclosure of conflict of interest
None.
References
- 1.Berger U, Bettelheim R, Mansi JL, Easton D, Coombes RC, Neville AM. The relationship between micrometastases in the bone marrow, histopathologic features of the primary tumor in breast cancer and prognosis. Am J Clin Pathol. 1988;90:1–6. doi: 10.1093/ajcp/90.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Morimoto T, Sasa M, Yamaguchi T, Kondo H, Akaiwa H, Sagara Y. Breast cancer screening by mammography in women aged under 50 years in Japan. Anticancer Res. 2000;20:3689–3694. [PubMed] [Google Scholar]
- 3.Westlake S, Cooper N. Cancer incidence and mortality: trends in the United Kingdom and constituent countries, 1993 to 2004. Health Stat Q. 2008;38:33–46. [PubMed] [Google Scholar]
- 4.Lee BL, Liedke PE, Barrios CH, Simon SD, Finkelstein DM, Goss PE. Breast cancer in Brazil: present status and future goals. Lancet Oncol. 2012;13:e95–e102. doi: 10.1016/S1470-2045(11)70323-0. [DOI] [PubMed] [Google Scholar]
- 5.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 6.Berry DA, Cronin KA, Plevritis SK, Fryback DG, Clarke L, Zelen M, Mandelblatt JS, Yakovlev AY, Habbema JD, Feuer EJ. Effect of screening and adjuvant therapy on mortality from breast cancer. N Engl J Med. 2005;353:1784–1792. doi: 10.1056/NEJMoa050518. [DOI] [PubMed] [Google Scholar]
- 7.Duraker N, Caynak ZC. Prognostic value of the 2002 TNM classification for breast carcinoma with regard to the number of metastatic axillary lymph nodes. Cancer. 2005;104:700–707. doi: 10.1002/cncr.21199. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson NL, Bell J, Heidel R, Lee S, Vanmeter S, Duncan L, Munsey B, Panella T, Orucevic A. Prognostic value of breast cancer subtypes, Ki-67 proliferation index, age, and pathologic tumor characteristics on breast cancer survival in Caucasian women. Breast J. 2013;19:22–30. doi: 10.1111/tbj.12059. [DOI] [PubMed] [Google Scholar]
- 9.Gebauer G, Fehm T, Lang N, Jager W. Tumor size, axillary lymph node status and steroid receptor expression in breast cancer: prognostic relevance 5 years after surgery. Breast Cancer Res Treat. 2002;75:167–173. doi: 10.1023/a:1019601928290. [DOI] [PubMed] [Google Scholar]
- 10.Coradini D, Daidone MG. Biomolecular prognostic factors in breast cancer. Curr Opin Obstet Gynecol. 2004;16:49–55. doi: 10.1097/00001703-200402000-00010. [DOI] [PubMed] [Google Scholar]
- 11.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Yang W, Lee DY, Ben-David Y. The roles of microRNAs in tumorigenesis and angiogenesis. Int J Physiol Pathophysiol Pharmacol. 2011;3:140–155. [PMC free article] [PubMed] [Google Scholar]
- 14.Ng EK, Wong CL, Ma ES, Kwong A. MicroRNAs as New Players for Diagnosis, Prognosis, and Therapeutic Targets in Breast Cancer. J Oncol. 2009;2009:305420. doi: 10.1155/2009/305420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 16.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 17.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 18.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 20.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696–1704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 21.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, Croce CM. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J. Clin. Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 22.Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12:2171–2176. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 25.Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PubMed] [Google Scholar]
- 26.Yang CH, Yue J, Pfeffer SR, Handorf CR, Pfeffer LM. MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J Biol Chem. 2011;286:39172–39178. doi: 10.1074/jbc.M111.285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 28.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 30.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 31.Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA, Thacker SB. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283:2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 32.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17:2815–2834. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 33.Williamson PR, Smith CT, Hutton JL, Marson AG. Aggregate data meta-analysis with time-to-event outcomes. Stat Med. 2002;21:3337–3351. doi: 10.1002/sim.1303. [DOI] [PubMed] [Google Scholar]
- 34.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong L, Han Y, Yang J, Zhang H, Jin Y, Brain L, Li M, Zhao Q. Prognostic value of epidermal growth factor receptor in patients with gastric cancer: a meta-analysis. Gene. 2013;529:69–72. doi: 10.1016/j.gene.2013.07.106. [DOI] [PubMed] [Google Scholar]
- 36.Ma XL, Liu L, Liu XX, Li Y, Deng L, Xiao ZL, Liu YT, Shi HS, Wei YQ. Prognostic role of microRNA-21 in non-small cell lung cancer: a meta-analysis. Asian Pac J Cancer Prev. 2012;13:2329–2334. doi: 10.7314/apjcp.2012.13.5.2329. [DOI] [PubMed] [Google Scholar]
- 37.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 39.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian B, Katsaros D, Lu L, Preti M, Durando A, Arisio R, Mu L, Yu H. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res Treat. 2009;117:131–140. doi: 10.1007/s10549-008-0219-7. [DOI] [PubMed] [Google Scholar]
- 42.Lee JA, Lee HY, Lee ES, Kim I, Bae JW. Prognostic Implications of MicroRNA-21 Overexpression in Invasive Ductal Carcinomas of the Breast. J Breast Cancer. 2011;14:269–275. doi: 10.4048/jbc.2011.14.4.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ota D, Mimori K, Yokobori T, Iwatsuki M, Kataoka A, Masuda N, Ishii H, Ohno S, Mori M. Identification of recurrence-related microRNAs in the bone marrow of breast cancer patients. Int J Oncol. 2011;38:955–962. doi: 10.3892/ijo.2011.926. [DOI] [PubMed] [Google Scholar]
- 44.Walter BA, Gomez-Macias G, Valera VA, Sobel M, Merino MJ. miR-21 Expression in Pregnancy-Associated Breast Cancer: A Possible Marker of Poor Prognosis. J Cancer. 2011;2:67–75. doi: 10.7150/jca.2.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Markou A, Yousef GM, Stathopoulos E, Georgoulias V, Lianidou E. Prognostic Significance of Metastasis-Related microRNAs in Early Breast Cancer Patients with a Long Follow-up. Clin Chem. 2014;60:197–205. doi: 10.1373/clinchem.2013.210542. [DOI] [PubMed] [Google Scholar]
- 46.Ferracin M, Veronese A, Negrini M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2010;10:297–308. doi: 10.1586/erm.10.11. [DOI] [PubMed] [Google Scholar]
- 47.Chen J, Wang X. MicroRNA-21 in breast cancer: diagnostic and prognostic potential. Clin Transl Oncol. 2014;16:225–33. doi: 10.1007/s12094-013-1132-z. [DOI] [PubMed] [Google Scholar]
- 48.Fu X, Han Y, Wu Y, Zhu X, Lu X, Mao F, Wang X, He X, Zhao Y. Prognostic role of microRNA-21 in various carcinomas: a systematic review and meta-analysis. Eur J Clin Invest. 2011;41:1245–1253. doi: 10.1111/j.1365-2362.2011.02535.x. [DOI] [PubMed] [Google Scholar]
- 49.Xia X, Yang B, Zhai X, Liu X, Shen K, Wu Z, Cai J. Prognostic Role of microRNA-21 in Colorectal Cancer: a Meta-Analysis. PLoS One. 2013;8:e80426. doi: 10.1371/journal.pone.0080426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayes DF, Isaacs C, Stearns V. Prognostic factors in breast cancer: current and new predictors of metastasis. J Mammary Gland Biol Neoplasia. 2001;6:375–392. doi: 10.1023/a:1014778713034. [DOI] [PubMed] [Google Scholar]


