Abstract
Hepatocellular carcinoma (HCC) is one of the most malignant tumors worldwide, especially in Eastern China where HBV infection confirmed as the most important pathological element. HBV X gene, extremely easy to mutate and integrate into hepatocytes, plays a significant role in HBV infection and HCC development. We deduced that mutations of integrated HBx gene make transformation more malignant. The aim of the study was to investigate whether there were different mutation patterns between the HCC tissues and the pericarcinoma liver tissues (PCLT) from patients with HCC in Eastern China. Methods: HBx genes extracted from 287 HCC tissue samples and 195 PCLT tissue samples were analyzed by sequence alignment and stratified analysis with the matched medical records. Results: Mutations occurred complicated and changeable in both HCC and PCLT. COOH-terminal truncation is more frequently found in HCC than PCLT (P < 0.05). There is no single site mutation of nucleic acid or amino acid makes distribution discrepancy between HCC and PCLT. Hydrophobic/hydrophilic character of amino acid of site 43, 47, 127, 131, 132 make distribution discrepancy between HCC and PCLT in men when stratified for gender (P < 0.05). Hydrophobic/hydrophilic character of amino acid of site 40 makes distribution discrepancy between HCC and PCLT in both male and female (P < 0.05). Hydrophobic/hydrophilic character of amino acid of site 47 and 127 make significant discrepancy among clinical stage I, II, III (P < 0.05). Conclusions: During the infection and replication of HBV, HBx mutates to adjust itself to the hepatocyte and increase the carcinogenesis. COOH-terminal truncated HBX may play a stimulative role in HBV-related HCC carcinogenesis as well as hydrophobic/hydrophilic character changes in some specific amino acid sites.
Keywords: Hepatitis B virus, X gene, hepatocellular carcinoma, pericarcinoma liver tissue, mutation
Introduction
Hepatocellular carcinoma (HCC) is one of the most malignant tumors worldwide. Annually newly reported HCC cases and death cases are both approximately 700 thousand, half of which occurred in China [1]. Chronic hepatitis B virus (HBV) infection has been confirmed as the leading pathological element for HCC development [2]. Over 80% Chinese HCC patients were infected by HBV, especially in Eastern China [3]. Evidence is accumulating that HBV X protein, the product of the fourth open reading frame (ORF), which is a multifunctional regulator, could modulate transcription, intracellular signal transduction, malignant cell proliferation, apoptosis, plays a significant role in HBV infection and HCC carcinogenesis [4-11]. Exhausting numbers of proteins were proved to interact with HBX. Just as Murakami said: “If there is a protein that is not among the list of HBX interactors, then this protein has probably not yet been tested” [2].
HBx is extremely easy to integrate into the chromosomal DNA of hepatocyte [12]. The clinical records that mutated and truncated HBx gene could often be detected from serum and tissue samples of HCC patients and cirrhotic patients were consistent with molecular mechanism research [13]. Although lacking of proofreading function of HBV polymerase results in frequent and multiple mutations in X gene during HBV infection and replication [14], integrated, even truncated HBx gene could encode functional X protein that still acts as a promoter for malignant transformation [2].
Furthermore, several mutation patterns have been documented. Research from Carman and Okamoto [15,16] documented the HBV double mutations 1762T/1764A (equally 389T and 391A in X gene region) of chronic HBV carriers from Greek and Japan, which result in a HBeAg (-) phenotype. More interestingly, Chen found a brand new mutation pattern of Hong Kong HCC patients, that insert mutation 204AGGCCC co-existed with two point mutations 260 (G→A) and 264 (G/C/T→A), therefore named “204 insert + 260/264 point” [17]. Muroyama reported a substitution mutation in X protein, S38, correlated significantly with an increased risk of hepatocellular carcinoma in Japan [18]. Whereas a different result was obtained by a following research of Japanese patients from another region that S38 substitution mutation did not show any correlation with HCC [19].
Hardly a research focus on the HBX mutation differences between HCC and PCLT samples of patients in Eastern China has been done due to the difficulty in samples obtaining, results in the unknown whether there are special mutation patterns required for HCC pathogenesis in Eastern China. The aim of our study was to fill in the blank.
Materials and methods
Subjects
287 hepatocellular carcinoma tissue samples and 195 pericarcinoma liver tissue samples were obtained from patients who underwent surgical resection of tumors in Zhongshan Hospital affiliated to Fudan University and were tested negative for antibody for hepatitis C virus and positive for hepatitis B virus surface antigen. Informed consent for the study was obtained from all these patients. PCR for the HBx coding region was successful in 192 HCC samples and 149 PCLT samples. These 341 tissue samples and matched medical records could be eligible for HBx sequence analysis.
DNA extraction and nested PCR
All tissue samples were frozen in liquid nitrogen immediately after surgical resection and stored at -80°C until the procedure for tissue DNA extraction. Tissue DNA was extracted using QIAamp® DNA Mini Kit (QIAGEN GmbH, Hilden, Germany). DNA yield was measured by absorbance at 260 nm with DanoDrop 2000 system (Thermo Fisher Scientific, Waltham, MA, USA), which was also applied for DNA purity determined and concentration dilution to 50 ng/μl. The primers for the first round PCR were 5’-ATCGTATCCATGGCTGCTAGGCT-3’ (forward 1) and 5’-CACAGCTTGGAGGCTTGAACA-3’ (reverse 1), and for the second round PCR were 5’-CATGGCTGCTAGGCTGTGCTG-3’ (forward 2), and 5’-GAGAGTATTAGGCAGAGGTGAAAAAG-3’ (reverse 2) [17]. 200 ng genome DNA was used as template in the first round PCR performed with Pyrobest PCR system (TaKaRa Bio, Japan) according to the instruction on the manufacturer’s manual with DNA Thermal Cycler (GeneAmp® PCR system 9600, Applied Biosystems, Foster City, CA, USA). The first round PCR was carried out as described [17]. 4 μl of the first round PCR product was used as template for the second round PCR, which was also proceeded as described [17]. The products of second round PCR were gel-purified using QIAquick® Gel Extraction Kit (QIAGEN GmbH, Hilden, Germany) according to the instruction.
Sequencing of X region
Sequencing primers were 5’-GACGTCCTTTGTCTACG-3’ (HBx-seq-forward), 5’-TATGCCTACAGCCTCC-3’ (HBx-seq-reverse). ABI PRISM® BigDyeTM Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA) was used for sequencing reactions. Briefly, 4 μl of Terminator Ready Reaction Mix, 1 μl of 10 pmol/μl primer (forward or reverse), 5 μl of 15 ng/μl gel purified PCR product were well mixed in a PCR reaction tube and placed in DNA Thermal Cycler (GeneAmp® PCR system 9600, Applied Biosystems, Foster City, CA, USA). The reaction was proceeded at 96°C for 20 s, 50°C for 10 s, 60°C for 4 min and was repeated for 30 cycles. Purification and denaturation processes were carried out as described [17]. Sequencing electrophoresis was performed on ABI PRISM 3730TM DNA Sequencer (Applied Biosystems, Foster City, CA, USA). Sequences obtained from each sample were compared with the HBx sequence in NCBI (HBVgp3, NC_003977.1) using software DNAStar 6.0.
Statistical analysis
Mutations between HCC and PCLT were analyzed with chi-square test (χ2 analysis) and P-Value by SPSS 11.5. Matched medical records were performed with stratified analysis. A threshold of P < 0.05 was defined as statistically significant.
Results
HBx amplification and sequencing
DNA was successfully amplified in 192 HCC samples and 169 HBx sequences (88%) were obtained after sequencing. Parallely, 117 HBx sequences (78.5%) were obtained from 149 PCLT samples sequencing.
Mutations in nucleic acid sequences
Point mutations, truncated mutations and insert mutations were found in both HCC samples and PCLT samples throughout the 465 nucleotide.
In HCC samples, there were 121 sites had pointed mutations; In PCLT samples, 133 sites with total of 154 sites. Some mutations, occurred in HCC samples, did not exhibit in PCLT samples, and vice versa. Mutation rate is less than 5% in approximately 68% (106/156) mutation sites. Mutation rate is higher than 20% in approximately 8% (12/156) sites, which is mainly allocated in the COOH-terminal of the HBx coding region, the region of transcriptional activity [20,21]. Unexpectedly, HBV double mutations 1762T/1764A (equally HBx 389T/391A), which located in an essential region of HBV for trans-activation function [22] and made significant discrepancy between serum from HCC patients and that from normal controls [15,16,23], did not make distribution discrepancy between HCC and PCLT in our study.
As shown in Figure 1A, the complete HBx sequence (white areas) should acquire UTRs (3’- and 5’-, gray areas) and two-sided primer regions (base areas) after nested PCR in our study according to the primer design [17]. Here, we exhibit the 68 truncated mutations in HCC samples (Figure 1B) and 24 truncated mutations (Figure 1C) in PCLT samples, black bars stand for the truncated parts. Truncated mutations were found to have a significant higher percentage in HCC samples than PCLT samples (Figure 1D) (χ2=12.75, P < 0.01). Moreover, as the COOH-terminal of HBx coding region is the region of transcriptional activity [20,21], we divided these sequences into COOH-terminal truncation sequences and Non-COOH-terminal truncation sequences. COOH-terminal truncation made distribution discrepancy between HCC samples and PCLT samples (Figure 1E) (χ2=4.23, P < 0.05).
Figure 1.
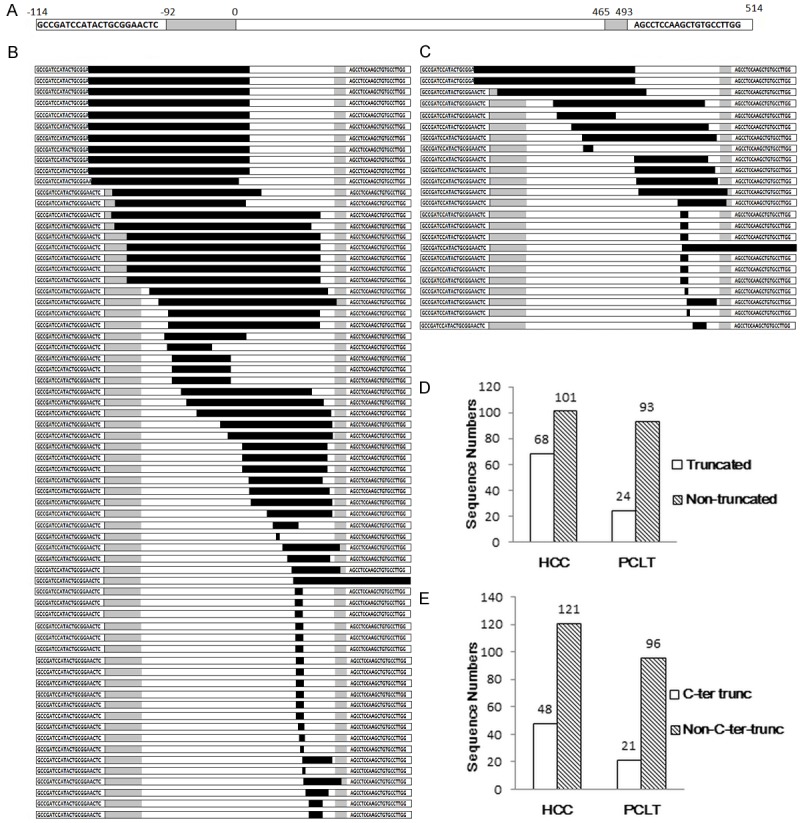
Basic situation of truncated mutations in the study. A: The sketch map of complete HBx sequence after nested PCR. the complete HBx sequence (white areas) should acquire UTRs (3’- and 5’-, gray areas) and two-sided primer regions (base areas), black bars stand for the truncated parts. B: 68 truncated mutant sequences in HCC samples. C: 24 truncated mutant sequences in PCLT samples. D: Statistics of truncated mutations in HCC and PCLT. Truncated mutations were found to have a significant higher percentage in HCC samples than PCLT samples. (χ2=12.75, P < 0.01). E: Statistics of COOH-terminal truncated mutations in HCC and PCLT. COOH-terminal truncated mutations were also found to have a significant higher percentage in HCC samples than PCLT samples. (χ2=4.23, P < 0.05).
Insert mutations existed in 15 sequences from HCC samples and 9 sequences from PCLT samples, which did not make distribution discrepancy. But it was noteworthy that, in HCC samples, insertion mutations only existed in sequences that from patients whose serum alpha fetoprotein (AFP) level > 20 ng/ml.
Mutations in deduced amino acid sequences
We did not find any type of amino acid made distribution discrepancy between HCC samples and PCLT samples (Figure 2A). Furthermore, neither the physicochemical properties (Figure 2B, 2C) nor the chemical constructions (Figure 2D, 2E) of amino acids made significant difference between HCC and PCLT. Hardly any accumulation of changes in physicochemical properties or chemical constructions results in the distinction between HCC and PCLT.
Figure 2.
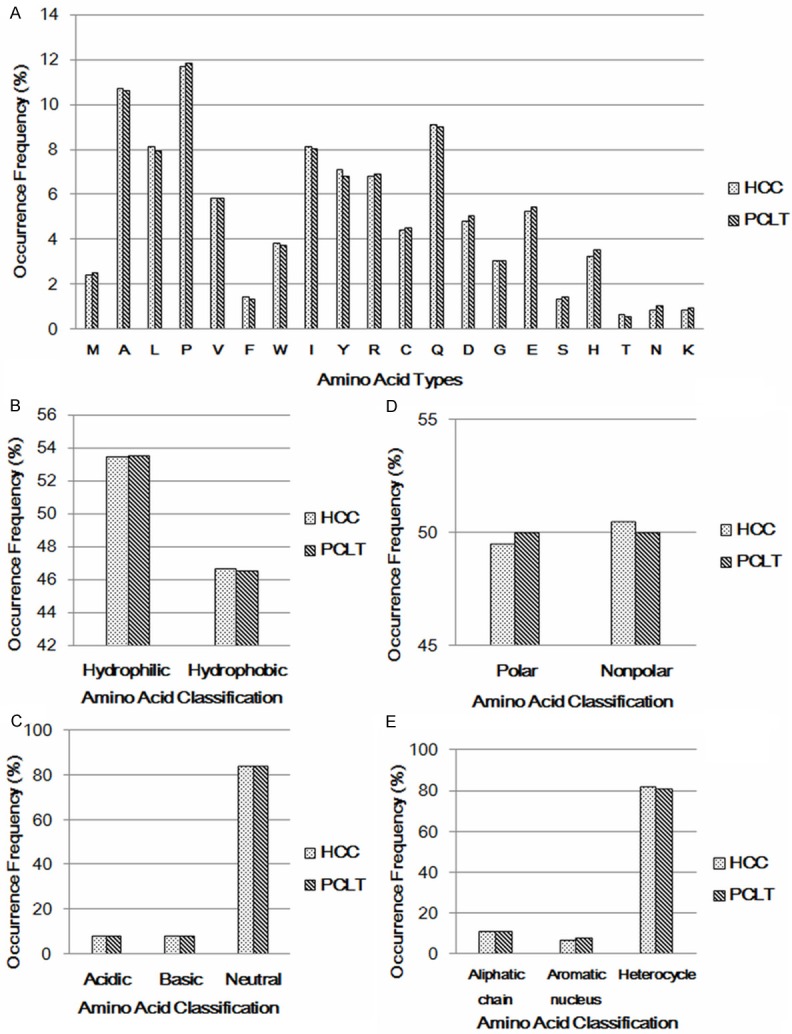
Statistics of types and properties of amino acids in HCC and PCLT. A: Statistics of amino acids types. There was no type of amino acid made distribution discrepancy between HCC samples and PCLT samples. B: Statistics of hydrophobic and hydrophilic amino acids. Numbers of total hydrophobic and hydrophilic amino acids did not make distribution discrepancy between HCC samples and PCLT samples. C: Statistics of acidic, basic and neutral amino acids. Numbers of total acidic, basic and neutral amino acids did not make distribution discrepancy between HCC samples and PCLT samples. D: Statistics of polar and nonpolar amino acids. Numbers of total polar and nonpolar amino acids did not make distribution discrepancy between HCC samples and PCLT samples. E: Statistics of amino acids with special side chains. Numbers of amino acids with special side chains did not make distribution discrepancy between HCC samples and PCLT samples.
So we considered another approaches: compare the physicochemical properties changes in every amino acid position instead of the overall situation between HCC and PCLT.
Not surprisingly, we found position 40 makes significant difference between HCC and PCLT when taken the hydrophobic/hydrophilic character into consideration (χ2=5.04, P < 0.05).
Stratified analysis with medical records
Epidemical investigation showed that, and was well-known that, when infected by HBV, males are more likely to become carriers and to develop chronic liver disease and even primary liver cancer. The ratio of males to females with hepatocellular carcinoma was approximately 2.7:1 in China [24], which was even higher in the Eastern China. So we performed the stratified analysis focused on the male group.
There were significant differences in 5 positions in male group between HCC and PCLT when taken the hydrophobic/hydrophilic character of amino acid into consideration besides position 40 (Table 1).
Table 1.
Positions related to the hydrophobic/hydrophilic character of amino acid made significant differences between HCC and PCLT in male group
| Position | Amino acid change hydrophobic/hydrophilic | HCC | PCLT | χ2 | P-value |
|---|---|---|---|---|---|
| 40 | P + A/S + T | 82/10 | 77/2 | 4.63 | 0.033 |
| 43 | P + A/S | 11/83 | 2/77 | 5.78 | 0.016 |
| 47 | A/T + S | 77/14 | 75/3 | 6.75 | 0.009 |
| 127 | I + V/N + T + S | 60/14 | 39/25 | 6.91 | 0.009 |
| 130 | M + I + L/K | 23/27 | 38/19 | 4.67 | 0.031 |
| 132 | F/Y | 78/4 | 60/10 | 4.06 | 0.044 |
Note: Amino acids were exhibited in single letter abbreviations.
Furthermore, position 47 and 127 made significant discrepancy in TNM staging related to the hydrophobic/hydrophilic character of amino acid in both male and female HCC samples. Analogously, position 43 made significant discrepancy in TNM staging related to the hydrophobic/hydrophilic character of amino acid only in male HCC samples (Table 2).
Table 2.
Positions related to the hydrophobic/hydrophilic character of amino acid made significant differences in TNM staging in HCC samples
| Position | Group(s) | TNM Staging hydrophobic percentage (%) | χ2 | P-value | ||
|---|---|---|---|---|---|---|
|
| ||||||
| I | II | III | ||||
| 43 | Male | 27.78 | 10.00 | 3.13 | 7.09 | 0.029 |
| 47 | Male & Female | 66.67 | 84.62 | 100 | 7.00 | 0.030 |
| 127 | Male & Female | 100 | 76.92 | 0.00 | 12.64 | 0.002 |
Hydrophobic/hydrophilic character change in specific regions
Interestingly, as we could see, these mutation hot positions were concentrated in two regions around the positions 40 and 120, which were relative to the hydrophobic/hydrophilic character of amino acid. Therefore, we referred to the Kyte-Doolittle Hydropathy Plots (http://gcat.davidson.edu/DGPB/kd/kyte-doolittle.htm) to investigate whether there was special hydrophobic area or hydrophilic area around the positions 40 and 120. In Kyte-Doolittle Hydropathy Plots, hydropathy score represented hydrophobic/hydrophilic character of amino acid. The higher the score, the greater the hydrophobicity is, the lower the score, the greater the hydrophilic is. As shown in Figure 3A, these mutated positions were located in two regions of special hydrophobic/hydrophilic character, neutral area around position 40 and hydrophilic area around position 120. The result indicated that mutations mainly affected the hydrophobic/hydrophilic character of HBX and may alter the pathology feature of HBX.
Figure 3.
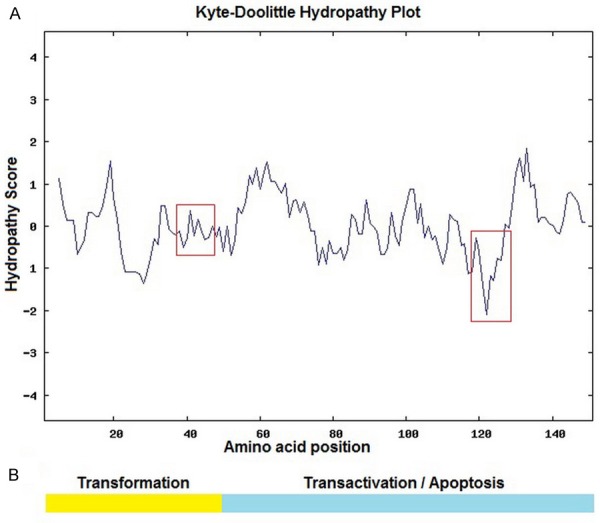
Kyte-Doolittle Hydropathy Plots of HBX and functional domain of HBX. A: Kyte-Doolittle Hydropathy Plots of HBX. Mutations found in our study were located in two concentrated regions (red boxes) around position 40 and position 120, position 40 is located in a neutral region in the N-terminal, position 120 is the most hydrophilic part of X protein. B: The functional domain of HBX. The N-terminal part of HBX (yellow part) is the transformation functional part of X protein. The COOH-terminal part (green part) is regulating region for transactivation and apoptosis.
Discussion
Hepatocellular carcinoma (HCC), with poor prognosis and high recurrence rate, is one of the most malignant tumors worldwide and occupy the third in the cancer death in China. It is generally accepted that chronic hepatitis B virus (HBV) infection plays a major pathological role in HCC development in China. Over 80% Chinese HCC patients were infected by HBV, especially in Eastern China [3]. The HBX protein, product of HBV X gene, a 16-18 kDa protein, has been reported to be a transcriptional transactivator and can modulate the transcription of an extensive variety of viral and cellular promoters [25]. More importantly functional X protein, usually mutated or truncated due to the lacking of proofreading function of HBV polymerase, is maintained and expressed in most of the integrated subviral genomes [26], could induce the cell proliferation [11], deregulate the cell cycle check point [11], induce HCC in certain transgenic mice [27] and modify several cellular pathways [25].
The correlation between HBX protein mutations and HCC development, reported distinguished behavior in different geographic regions, has not been clearly elucidated. Furthermore, due to the difficulties in tissue samples obtaining, most research were focused on the HBx mutations in serum samples or a small amount of tissue samples.
In our study, we focused on the different mutation patterns between hepatocellular carcinoma (HCC) samples and pericarcinoma liver tissues (PCLT) samples. In terms of the overall situation, we obtained 169 HBx sequences from 287 HCC samples and 117 HBx sequences from 195 PCLT samples. Stratified analysis was performed with corresponding medical records to investigate the potential mutation positions that related to the HCC carcinogenesis or malignant transformation and special mutation patterns that exclusive to the inhabitants in Eastern China.
In our study, point mutations, truncated mutations and insert mutations were found in both HCC samples and PCLT samples, which were in accordance with some classic studies [22,28] and demonstrated the occurrence of mutations in X-ORF is independent with geographic regions. Furthermore, some mutations, occurred in HCC samples, did not exhibit in PCLT samples, and vice versa, which were consistent with Chen’s study [17]. Whereas we did not found the special mutation pattern “204 insert + 260/264 point”, which was inconsistent with Chen’s study as Hong Kong has relatively isolated ethic Chinese population, which has their own special living styles in many aspects [17]. It’s worth mentioning that HBV double mutations 1762T/1764A (equally 389T and 391A in X gene region) of chronic HBV carriers from Greek and Japan [15,16] and two substitution mutations in X protein in Japan, S38 [18] and Y94 [29], did not make significant difference between HCC samples and PCLT samples, which might be because of the different geographic regions or non-functioning of these mutations in HCC carcinogenesis or malignant transformation.
The COOH-terminal region of X protein is pivotal to the transactivational function and pro-apoptosis effect of the protein and for mediating cell proliferation (Figure 3B) [30]. HBV DNA sequences from HCC patients, especially those isolated from integrated viral DNA, very common, have a deletion in the COOH-terminal region of the X-ORF, this indicates that the deletion parts may be implicated in the HCC carcinogenesis or malignant transformation [11]. Undoubtedly, in our study COOH-terminal truncations were found in both HCC samples and PCLT samples (Figure 1B, 1C). Furthermore, our study showed that COOH-terminal truncation made distribution discrepancy between HCC samples and PCLT samples (Figure 1E), which validated the potential pathological function of COOH-terminal of X protein in HBV-related HCC carcinogenesis from another point of view.
The most important finding of our study was the identification of two mutation-concentrated regions (Tables 1, 2), particularly in male, related to hydrophobic/hydrophilic character’s switch. To our knowledge, these two special regions have never been reported before. One of the regions was located around position 40 (Figure 3A), a neutral region in the N-terminal, the regulating region for transformation. Another is located around position 120 (Figure 3A), the most hydrophilic part of X protein, also in the pro-apoptosis functional part of X protein. These two special regions changes may cause structure change and conformation alteration of X protein and hence alter the regulatory and transactivational functions. However, further experiments need to be done to confirm the inference.
Another feature of our study is that we discovered insert mutations only existed in sequences that from patients whose serum AFP level > 20 ng/ml. Clinically, serum AFP level > 20 ng/ml is the symbol of chronic liver disease. AFP is a classic liver cancer marker in adults and is reported to have a promotive effect in HCC carcinogenesis [31,32]. However, the molecular mechanism behind the connection between AFP and HBX insert mutations is complicated and difficult to interpret. On the other hand, the correlation between AFP and HBX insert mutations observed in our study might be because the coincidence in samples obtaining or the sample size was not large enough.
In summary, we conducted a stratified analysis with corresponding medical records between HCC samples and PCLT samples based on the advantages in tissue samples obtaining and found some novel phenomenon that exclusive to the inhabitants in Eastern China. Combined with the reviewed studies, our research might represent a possible strategy of HBV subviral genome to escape the immunological surveillance of parasitifer and experience some co-evolutionary process with the host cells so that they could contribute to the process of multi-step hepatocellular carcinoma deterioration.
Acknowledgements
The study is supported by The National Natural Science Fund (31071193) and The Major National Science and Technology Projects (2013ZX10002010). We wish to thank Zhongshan hospital affiliated to Fudan University for furnishing the hepatocellular carcinoma samples and the pericarcinoma liver tissues samples.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Murakami S. Hepatitis B virus X protein: a multifunctional viral regulator. J Gastroenterol. 2001;36:651–660. doi: 10.1007/s005350170027. [DOI] [PubMed] [Google Scholar]
- 3.Szmuness W. Hepatocellular carcinoma and the hepatitis B virus: evidence for a causal association. Prog Med Virol. 1978;24:40–69. [PubMed] [Google Scholar]
- 4.Miyaki M, Sato C, Gotanda T, Matsui T, Mishiro S, Imai M, Mayumi M. Integration of region X of hepatitis B virus genome in human primary hepatocellular carcinomas propagated in nude mice. J Gen Virol. 1986;67:1449–1454. doi: 10.1099/0022-1317-67-7-1449. [DOI] [PubMed] [Google Scholar]
- 5.Slagle BL, Lee TH, Medina D, Finegold MJ, Butel JS. Increased sensitivity to the hepatocarcinogen diethylnitrosamine in transgenic mice carrying the hepatitis B virus X gene. Mol Carcinog. 1996;15:261–269. doi: 10.1002/(SICI)1098-2744(199604)15:4<261::AID-MC3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 6.Lara-Pezzi E, Armesilla AL, Majano PL, Redondo JM, Lopez-Cabrera M. The hepatitis B virus X protein activates nuclear factor of activated T cells (NF-AT) by a cyclosporin A-sensitive pathway. Embo J. 1998;17:7066–7077. doi: 10.1093/emboj/17.23.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin EC, Shin JS, Park JH, Kim H, Kim SJ. Expression of fas ligand in human hepatoma cell lines: role of hepatitis-B virus X (HBX) in induction of Fas ligand. Int J Cancer. 1999;82:587–591. doi: 10.1002/(sici)1097-0215(19990812)82:4<587::aid-ijc19>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 8.Carretero M, Gomez-Gonzalo M, Lara-Pezzi E, Benedicto I, Aramburu J, Martinez-Martinez S, Redondo JM, Lopez-Cabrera M. The hepatitis B virus X protein binds to and activates the NH(2)-terminal trans-activation domain of nuclear factor of activated T cells-1. Virology. 2002;299:288–300. doi: 10.1006/viro.2002.1526. [DOI] [PubMed] [Google Scholar]
- 9.Schaefer S. Hepatitis B virus: significance of genotypes. J Viral Hepat. 2005;12:111–124. doi: 10.1111/j.1365-2893.2005.00584.x. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka Y, Kanai F, Ichimura T, Tateishi K, Asaoka Y, Guleng B, Jazag A, Ohta M, Imamura J, Ikenoue T, Ijichi H, Kawabe T, Isobe T, Omata M. The hepatitis B virus X protein enhances AP-1 activation through interaction with Jab1. Oncogene. 2006;25:633–642. doi: 10.1038/sj.onc.1209093. [DOI] [PubMed] [Google Scholar]
- 11.Kew MC. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J Gastroenterol Hepatol. 2011;26(Suppl 1):144–152. doi: 10.1111/j.1440-1746.2010.06546.x. [DOI] [PubMed] [Google Scholar]
- 12.Wei Y, Neuveut C, Tiollais P, Buendia MA. Molecular biology of the hepatitis B virus and role of the X gene. Pathol Biol (Paris) 2010;58:267–272. doi: 10.1016/j.patbio.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Girones R, Miller RH. Mutation rate of the hepadnavirus genome. Virology. 1989;170:595–597. doi: 10.1016/0042-6822(89)90455-8. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Lau SH, Sham JS, Wu MC, Wang T, Guan XY. Characterization of HBV integrants in 14 hepatocellular carcinomas: association of truncated X gene and hepatocellular carcinogenesis. Oncogene. 2004;23:142–148. doi: 10.1038/sj.onc.1206889. [DOI] [PubMed] [Google Scholar]
- 15.Carman WF, Jacyna MR, Hadziyannis S, Karayiannis P, McGarvey MJ, Makris A, Thomas HC. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet. 1989;2:588–591. doi: 10.1016/s0140-6736(89)90713-7. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto H, Tsuda F, Akahane Y, Sugai Y, Yoshiba M, Moriyama K, Tanaka T, Miyakawa Y, Mayumi M. Hepatitis B virus with mutations in the core promoter for an e antigen-negative phenotype in carriers with antibody to e antigen. J Virol. 1994;68:8102–8110. doi: 10.1128/jvi.68.12.8102-8110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen GG, Li MY, Ho RL, Chak EC, Lau WY, Lai PB. Identification of hepatitis B virus X gene mutation in Hong Kong patients with hepatocellular carcinoma. J Clin Virol. 2005;34:7–12. doi: 10.1016/j.jcv.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Muroyama R, Kato N, Yoshida H, Otsuka M, Moriyama M, Wang Y, Shao RX, Dharel N, Tanaka Y, Ohta M, Tateishi R, Shiina S, Tatsukawa M, Fukai K, Imazeki F, Yokosuka O, Shiratori Y, Omata M. Nucleotide change of codon 38 in the X gene of hepatitis B virus genotype C is associated with an increased risk of hepatocellular carcinoma. J Hepatol. 2006;45:805–812. doi: 10.1016/j.jhep.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 19.Shinkai N, Tanaka Y, Ito K, Mukaide M, Hasegawa I, Asahina Y, Izumi N, Yatsuhashi H, Orito E, Joh T, Mizokami M. Influence of hepatitis B virus X and core promoter mutations on hepatocellular carcinoma among patients infected with subgenotype C2. J Clin Microbiol. 2007;45:3191–3197. doi: 10.1128/JCM.00411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu H, Bonura C, Giannini C, Mouly H, Soussan P, Kew M, Paterlini-Brechot P, Brechot C, Kremsdorf D. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res. 2001;61:7803–7810. [PubMed] [Google Scholar]
- 21.Lee SW, Lee YM, Bae SK, Murakami S, Yun Y, Kim KW. Human hepatitis B virus X protein is a possible mediator of hypoxia-induced angiogenesis in hepatocarcinogenesis. Biochem Biophys Res Commun. 2000;268:456–461. doi: 10.1006/bbrc.2000.2093. [DOI] [PubMed] [Google Scholar]
- 22.Arii M, Takada S, Koike K. Identification of three essential regions of hepatitis B virus X protein for trans-activation function. Oncogene. 1992;7:397–403. [PubMed] [Google Scholar]
- 23.Li W, Chen G, Yu X, Shi Y, Peng M, Wei J. Accumulation of the mutations in basal core promoter of hepatitis B virus subgenotype C1 increase the risk of hepatocellular carcinoma in Southern China. Int J Clin Exp Pathol. 2013;6:1076–1085. [PMC free article] [PubMed] [Google Scholar]
- 24.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossner MT. Review: hepatitis B virus X-gene product: a promiscuous transcriptional activator. J Med Virol. 1992;36:101–117. doi: 10.1002/jmv.1890360207. [DOI] [PubMed] [Google Scholar]
- 26.Robinson WS. Molecular events in the pathogenesis of hepadnavirus-associated hepatocellular carcinoma. Annu Rev Med. 1994;45:297–323. doi: 10.1146/annurev.med.45.1.297. [DOI] [PubMed] [Google Scholar]
- 27.Kim CM, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 28.Kumar V, Jayasuryan N, Kumar R. A truncated mutant (residues 58-140) of the hepatitis B virus X protein retains transactivation function. Proc Natl Acad Sci U S A. 1996;93:5647–5652. doi: 10.1073/pnas.93.11.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poussin K, Dienes H, Sirma H, Urban S, Beaugrand M, Franco D, Schirmacher P, Brechot C, Paterlini BP. Expression of mutated hepatitis B virus X genes in human hepatocellular carcinomas. Int J Cancer. 1999;80:497–505. doi: 10.1002/(sici)1097-0215(19990209)80:4<497::aid-ijc3>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Zhang W, Liu Q, Zhang X, Lv N, Ye L, Zhang X. A mutant of hepatitis B virus X protein (HBxDelta127) promotes cell growth through a positive feedback loop involving 5-lipoxygenase and fatty acid synthase. Neoplasia. 2010;12:103–115. doi: 10.1593/neo.91298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng L, Gong W, Liang P, Huang X, You N, Han KQ, Li YM, Li J. Effects of AFP-activated PI3K/Akt signaling pathway on cell proliferation of liver cancer. Tumour Biol. 2014;35:4095–9. doi: 10.1007/s13277-013-1535-z. [DOI] [PubMed] [Google Scholar]
- 32.Cameron AM. AFP in OLT for HCC?: Another shadow on the cave wall: Comment on “Evaluation of absolute serum alpha-fetoprotein levels in liver transplant for hepatocellular cancer”. Arch Surg. 2011;146:33–34. doi: 10.1001/archsurg.2010.280. [DOI] [PubMed] [Google Scholar]


