Abstract
MicroRNA-218 (miR-218) acts as a tumor suppressor in numerous types of cancer by regulation of the expression of target genes. The aim of this study was to investigate whether polymorphisms in miR-218 LAMB3 pathway were associated with the risk and prognosis of esophageal squamous cell carcinoma (ESCC). Pri-mir-218 rs11134527 and LAMB3 rs2566 were genotyped in ESCC patients and 745 controls to assess their associations with cancer risk and overall survival. Pri-mir-218 rs11134527 was significantly associated with a decreased risk of ESCC under codominant, recessive and additive models. Although there was a significant association between rs11134527 and better survival of ESCC patients under codominant, recessive and additive models, the association disappeared after adjustment for TNM and LNM. However, further stratified analysis revealed that the association remained significant in patients with TNM stages I and II or non-LNM. Our data suggest that pri-miR-218 rs11134527 may contribute to the genetic susceptibility and prognosis for ESCC in Chinese Han population.
Keywords: Esophageal squamous cell carcinoma, miR-218, laminin 5, single nucleotide polymorphism, susceptibility
Introduction
Esophageal cancer is the third most common form of cancer of the digestive tract and the fifth leading cause of cancer-related death worldwide [1]. The incidence rates vary greatly worldwide, with the highest being in Southern and Eastern Africa and Eastern Asia [1]. Esophageal squamous cell carcinomas (ESCC) is the most common type, accounting for over 90% of esophageal cancer [2]. The prognosis of ESCC is uniformly poor despite the significant improvements in diagnosis and therapy in last ten years [3]. Although environmental risk factors for ESCC have been identified, the molecular mechanisms underlying carcinogenesis remain poorly understood.
Dysregulation of microRNAs (miRNAs) has been implicated in the development of various types of cancer [4-6]. miRNAs are a class of small, single-stranded, non-coding RNAs that normally negatively regulate expression of approximately 90% human genes at the post-transcriptional level [7]. A single miRNA can regulate hundreds of target genes, whereas one gene can be targeted by multiple miRNAs [7,8]. Much evidence has clearly demonstrated that miRNAs are involved in various diseases, including cancer [9]. miRNAs function as oncogenes and tumor suppressors in cancer initiation, progression and metastasis [5,6]. Therefore, changes in miRNA expression or miRNA dysfunction may affect cancer initiation and progression. Accumulating evidence reveals that genetic variation in miRNA genes affect the processing and expression of mature miRNAs and target mRNA-binding activity, leading to the changed expression of target genes [10-13]. Accordingly, functional single nucleotide polymorphisms (SNPs) not only contribute to the susceptibility to human diseases [10,13-19], but also can affect prognosis and response to treatment [11,14,20].
Many studies have demonstrated that miRNA SNPs are associated with the risk and prognosis of cancer [11,21]. So far, few studies have mentioned the relationship between miRNA SNPs and the risk and prognosis of ESCC. Previous studies found that pri-miR-218 rs11134527 was related to the risk of cervical carcinoma [17,18]. In the present study, we assessed the relationship between Pri-mir-218 rs11134527 and rs2566 in miR-218 target gene laminin 5 β3 (LAMB3), and the risk and prognosis of ESCC in Chinese Han.
Materials and methods
Patients
The study protocol was approved by the ethics committee of Taizhou People’s Hospital. A total of 706 pathologically confirmed ESCC patients and 745 controls were recruited at Taizhou People’s Hospital. All subjects were genetically-unrelated ethnic Han Chinese and lived within the same geographic region (Jiangsu Province). Patients with a prior diagnosis of cancer, other than ESCC, were excluded from this study. Furthermore, no individual had a blood transfusion in the last 6 months. Three ml of peripheral blood was collected from each participant after written informed consent was obtained from all subjects.
Genotyping
Genomic DNA was extracted from the peripheral leucocytes using the Universal Genomic DNA Extraction Kit Version 3.0 (Takara, Dalian, China) according to the manufacturer’s instructions. The concentration and quality of genomic DNA was measured by NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, DE, USA) and then stored at -20°C for later use. The genotypes of rs11134527 and rs2566 were determined using polymerase chain reaction-ligation detection reaction (PCR-LDR) method as described previously [22].
Statistical analyses
The differences between cases and controls were assessed using Pearson’s χ2 test and t-tests, as appropriate. The Hardy-Weinberg equilibrium (HWE) was tested by a χ2test to compare the expected genotype frequencies with observed genotype frequencies in controls. The differences in frequencies of SNPs between cases and controls were analyzed using χ2 test or Fisher’s exact test. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated using univariate and multivariable logistic regression model. Survival analyses were carried out using the Kaplan-Meier method, and the differences in overall survival were examined using log-rank tests. Hazard ratios (HRs) were estimated by Cox proportional hazard regression model. The SPSS 19.0 software (IBM Corporation, Armonk, NY, USA) was used to perform all statistical analyses. A P < 0.05 was considered statistically significant.
Results
Characteristics of ESCC patients and controls
The demographic characteristics of the 704 ESCC cases and 745 cancer-free controls were summarized in Table 1. The mean age of cases and controls were 61.2 and 60.6 years, respectively. There were no significant differences between cases and controls in terms of age and sex.
Table 1.
Clinical characteristics of cases and controls
| Characteristics | Cases (%) | Controls (%) | P values |
|---|---|---|---|
| Total | 706 | 745 | |
| Age (years) | 61.2±9.2 | 60.6±9.9 | 0.177 |
| Sex, % male | 495 (70.1) | 502 (67.4) | 0.282 |
| Pathologic type | |||
| medullary | 268 (38.0) | ||
| ulcerative | 278 (39.4) | ||
| others | 113 (16.0) | ||
| unknown | 47 (6.7) | ||
| Histologic grade | |||
| well | 60 (8.5) | ||
| moderate | 378 (53.5) | ||
| poor | 247 (35.0) | ||
| unknown | 21 (3.0) | ||
| Tumor size (cm) | |||
| > 4.5 | 235 (33.3) | ||
| ≤ 4.5 | 340 (48.2) | ||
| unknown | 131 (18.6) | ||
| TNM | |||
| I | 35 (5.0) | ||
| II | 304 (43.1) | ||
| III | 252 (35.7) | ||
| IV | 78 (11.0) | ||
| unknown | 37 (5.2) | ||
| LNM | |||
| yes | 379 (53.7) | ||
| no | 294 (41.6) | ||
| unknown | 33 (4.7) |
Association of rs11134527 and rs2566 with the risk of ESCC
The genotype and allele frequency distributions for rs11134527 and rs2566 in 704 cases and 745 controls were presented in Table 2. The genotype distributions of these two SNPs were in agreement with the Hardy-Weinberg equilibrium among controls (P > 0.05). We carried out association analysis with codominant, dominant, recessive and additive models using conditional logistic regression model. In the codominant model, rs11134527 GG genotype was significantly associated with a decreased risk of ESCC compared with AA genotype (OR=0.677, 95% CI: 0.493-0.931, P=0.016) (Table 2). The difference remained significant even after adjusted for age and sex (adjusted OR=0.669, 95% CI: 0.480-0.932, P=0.018). Furthermore, rs11134527 was significant protect factor for ESCC under both recessive (adjusted OR=0.678, 95% CI: 0.500-0.919, P=0.012) and additive models (adjusted OR=0.853, 95% CI: 0.729-0.997, P=0.046). In the dominant model, rs11134527 showed no significant association with ESCC risk (P > 0.05). There was no significant association between rs2566 and the risk of ESCC in our study population.
Table 2.
Associations between 2 SNPs and ESCC risk
| SNP | Genotype | Case | Control | Crude OR (95% CI) | P value | Adjusted OR (95% CI) | P value |
|---|---|---|---|---|---|---|---|
| rs11134527 | AA | 273 (38.7) | 268 (36.0) | 1 | 1 | ||
| AG | 344 (48.7) | 348 (46.7) | 0.970 (0.775-1.215) | 0.794 | 0.976 (0.773-1.234) | 0.842 | |
| GG | 89 (12.6) | 129 (17.3) | 0.677 (0.493-0.931) | 0.016 | 0.669 (0.480-0.932) | 0.018 | |
| Dominant model | 0.891 (0.720-1.103) | 0.289 | 0.894 (0.716-1.115) | 0.321 | |||
| Recessive model | 0.689 (0.514-0.923) | 0.012 | 0.678 (0.500-0.919) | 0.012 | |||
| Additive model | 0.855 (0.736-0.993) | 0.041 | 0.853 (0.729-0.997) | 0.046 | |||
| Allele A | 890 63.0) | 884 (59.3) | 1 | 1 | |||
| Allele G | 522 (37.0) | 606 (40.7) | 0.856 (0.737-0.994) | 0.041 | 0.866 (0.744-1.007) | 0.062 | |
| rs2566 | GG | 305 (43.2) | 310 (41.6) | 1 | 1 | ||
| AG | 290 (41.1) | 327 (43.9) | 1.109 (0.887-1.387) | 0.363 | 1.105 (0.881-1.386) | 0.386 | |
| AA | 111 (15.7) | 108 (14.5) | 0.957 (0.703-1.303) | 0.781 | 0.940 (0.687-1.286) | 0.698 | |
| Dominant model | 1.067 (0.867-1.314) | 0.54 | 1.060 (0.835-1.294) | 0.591 | |||
| Recessive model | 0.909 (0.682-1.212) | 0.515 | 0.894 (0.668-1.197) | 0.451 | |||
| Additive model | 0.993 (0.858-1.148) | 0.922 | 1.000 (0.863-1.160) | 0.995 | |||
| Allele A | 512 | 543 | 1 | 1 | |||
| Allele G | 900 | 947 | 0.992 (0.853-1.154) | 0.919 | 0.997 (0.855-1.162) | 0.971 | |
Effects of rs11134527 and rs2566 on ESCC survival
In the univariate analysis of various clinicopathological parameters for overall survival, LNM (HR=1.883, 95% CI: 1.500-2.364, P < 0.001), TNM stages III and IV (HR=2.055, 95% CI: 1.644-2.569, P < 0.001), rs11134527 GG genotype (GG vs. AA, HR=0.630, 95% CI: 0.434-0.914, P=0.015; GG vs. AA+AG, HR=0.652, 95% CI: 0.461-0.923, P=0.016) were significantly associated with prognosis in ESCC patients (Table 3, Figure 1). Furthermore, rs11134527 was also associated with better survival in ESCC patients under additive model (HR=0.838, 95% CI: 0.713-0.985, P=0.032). In the multivariate analysis, TNM was the only independent prognostic factor for ESCC (adjusted HR=1.775, 95% CI: 1.136-2.773, P=0.012). The rs2566 did not show any significant correlation with overall survival (Table 3).
Table 3.
Univariate and multivariate Cox regression analysis of overall survival in ESCC patients
| Features | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age (years), > 60 vs. ≤ 60 | 1.029 (0.830-1.275) | 0.797 | ||
| Sex, male vs. female | 0.870 (0.686-1.104) | 0.252 | ||
| Tumor size (cm), > 4.5 vs. ≤ 4.5 | 0.813 (0.639-1.034) | 0.092 | ||
| Tumor differentiation, poor vs. well, moderate | 0.937 (0.834-1.053) | 0.274 | ||
| Pathologic type | 0.984 (0.848-1.142) | 0.829 | ||
| LNM, yes vs. no | 1.883 (1.500-2.364) | < 0.001 | 1.118 (0.713-1.754) | 0.626 |
| TNM stage, III+IV vs. I+II | 2.055 (1.644-2.569) | < 0.001 | 1.775 (1.136-2.773) | 0.012 |
| rs11134527 | ||||
| AA | 1 | |||
| AG | 0.943 (0.749-1.186) | 0.614 | 0.987 (0.783-1.243) | 0.910 |
| GG | 0.630 (0.434-0.914) | 0.015 | 0.725 (0.497-1.057) | 0.095 |
| Dominant model | 0.870 (0.697-1.086) | 0.219 | ||
| Recessive model | 0.652 (0.461-0.923) | 0.016 | 0.731 (0.514-1.039) | 0.080 |
| Additive model | 0.838 (0.713-0.985) | 0.032 | 0.893 (0.758-1.052) | 0.177 |
| rs2566 | ||||
| GG | 1 | |||
| AG | 1.205 (0.953-1.523) | 0.119 | ||
| AA | 1.279 (0.934-1.750) | 0.125 | ||
| Dominant model | 1.224 (0.984-1.523) | 0.070 | ||
| Recessive model | 1.166 (0.873-1.557) | 0.298 | ||
| Additive model | 0.873 (0.753-1.013) | 0.073 | ||
Figure 1.
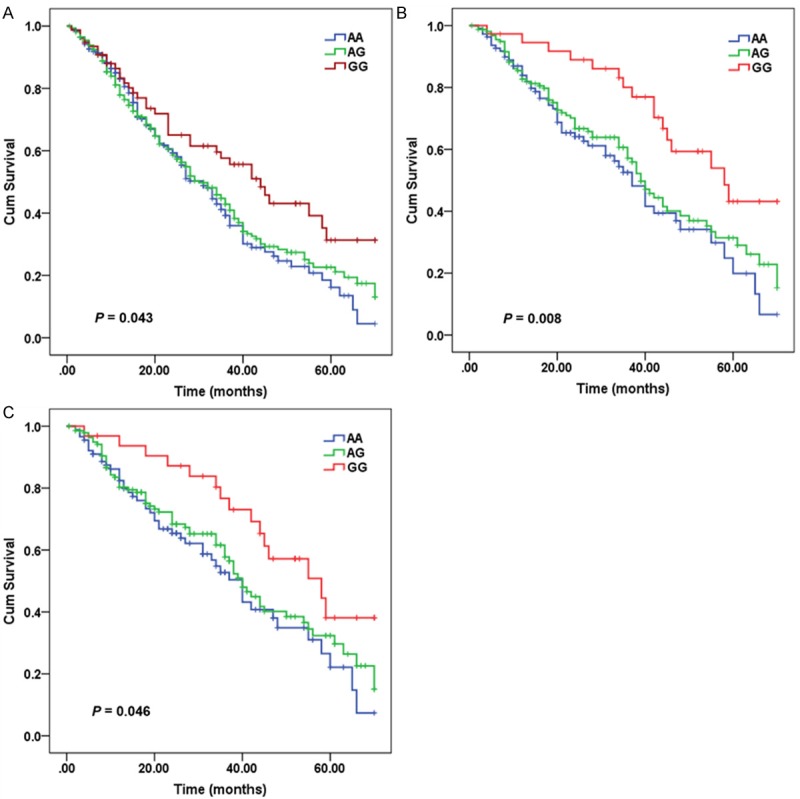
Kaplan-Meir curve for overall survival of ESCC patients according to pri-mir-218 rs11134527 genotypes. A. All patients; B. Patients with TNM stages I and II; C. Lymph node-negative patients.
The association between rs11134527 and overall survival was further evaluated by stratification of clinicopathological parameters. We found that when compared with rs11134527 AA genotype, GG genotype was more prominentin patients with TNM stages I and II (adjusted HR=0.419, 95% CI: 0.238-0.738, P=0.003) or non-LNM (adjusted HR=0.532, 95% CI: 0.295-0.960, P=0.036) (Figure 1). Compared with subjects with rs11134527 AA+GG genotypes and with TNM stages III and IV or LNM, patients with GG genotype and with TNM stages I and II or non-LNM had a significantly lower mortality rate (adjusted HR=0.580, 95% CI: 0.338-0.996, P=0.048).
Discussion
In the present study, we found that rs11134527 GG genotype was related to a significantly decreased risk of ESCC. Furthermore, a significantly increased survival time was observed in ESCC patients with GG genotypes. The effect was even stronger in those with TNM stages I and II or non-LNM. This is the first report to evaluate the relationship between SNPs in the LAMB3-miR-218 pathway and clinical outcome of ESCC patients.
miR-218 functions as a tumor suppressor [23-25] and is downregulated in various types of cancer, such as colorectal cancer [25], cervical squamous cell carcinoma [26] and head and neck squamous cell carcinoma [27]. Overexpression of miR-218 inhibits cancer cell proliferation, migration and invasion [24,26,27], and promotes colon cancer cell apoptosis [28]. In addition, miR-218 can enhance the sensitivity of cervical cancer cell to cisplatin [29] and radiotherapy [23]. The expression level of miR-218 is associated with cancer progression and poor prognosis [25,30,31]. Previous study has demonstrated that rs11134527 G allele in pri-miR-218 region leads to hairpin structure change, which increases the expression of mature miR-218 [12]. Therefore, tissue with rs11134527 GG genotype might have a higher level of miR-218, leading to the decreased risk of carcinogenesis. In this study, we found that rs11134527 GG genotype was relate to a reduced risk of ESCC, which was in agreement with previous observations that rs11134527 GG genotype was associated with decreased risk of cervical carcinoma [17,18]. However, Zhang et al. [32] found no association between rs11134527 and the risk of ESCC. One of the reasons for the contradiction may be genetic heterogeneity among different population samples. Moreover, ESCC patients with rs11134527 GG genotype had a better prognosis, especially in those with early-stage disease. This indicated that the level of miR-218 in early-stage ESCC may be higher than that in later stage. Further studies are required to confirm involvement of pri-miR-218 rs11134527 with the risk and prognosis of ESCC.
Several target genes of miR-218 have been identified, including BMI1 [28], LAMB3 [26,27] and LASP1 [24]. LAMB3 encodes β3 chain of laminin-5 that is abundant in the basal membrane and regulates important biological processes including cell differentiation, migration, adhesion and angiogenesis [33]. The level of LAMB3 is upregulated in ESCC and high LAMB3 expression is correlated with invasion and worse prognosis [34]. These imply that LAMB3-miR-218 pathway may be involved in the development of ESCC. Zhou et al. [17] reported that rs2566 CT and TT genotypes in the 3’UTR of LAMB3 was associated with increased risk of cervical carcinoma. However, Shi et al. [18] found no association between rs2566 and the risk of cervical carcinoma. Additionally, in this study, rs2566 was not related to the risk as well as prognosis of ESCC. Rs2566 may not affect miRNA binding and mRNA splicing.
In conclusion, our study revealed that pri-miR-218 rs11134527 GG genotype might decrease the risk of ESCC and be also associated with better survival in ESCC patients, especially in those with early-stage disease. Larger well-designed epidemiological studies and functional evaluations are warranted to confirm these initial findings.
Acknowledgements
This work was supported by the Health Scientific Research Foundation, Jiangsu, China (grant No. H201260).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X, He C, He C, Chen B, Liu Y, Kong M, Wang C, Lin L, Dong Y, Sheng H. Nuclear PKM2 expression predicts poor prognosis in patients with esophageal squamous cell carcinoma. Pathol Res Pract. 2013;209:510–515. doi: 10.1016/j.prp.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Han G, Tian Y, Duan B, Sheng H, Gao H, Huang J. Association of nuclear annexin A1 with prognosis of patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:751–759. [PMC free article] [PubMed] [Google Scholar]
- 4.Wu C, Cao Y, He Z, He J, Hu C, Duan H, Jiang J. Serum levels of miR-19b and miR-146a as prognostic biomarkers for non-small cell lung cancer. Tohoku J Exp Med. 2014;232:85–95. doi: 10.1620/tjem.232.85. [DOI] [PubMed] [Google Scholar]
- 5.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014 doi: 10.1016/j.molmed.2014.06.005. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 6.Huang J, Zhang SY, Gao YM, Liu YF, Liu YB, Zhao ZG, Yang K. MicroRNAs as oncogenes or tumour suppressors in oesophageal cancer: potential biomarkers and therapeutic targets. Cell Prolif. 2014;47:277–286. doi: 10.1111/cpr.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 8.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Hesse M, Arenz C. MicroRNA maturation and human disease. Methods Mol Biol. 2014;1095:11–25. doi: 10.1007/978-1-62703-703-7_2. [DOI] [PubMed] [Google Scholar]
- 10.Lin J, Huang Y, Zhang X, Chen J, Sheng H. Association of miR-146a rs2910164 with childhood IgA nephropathy. Pediatr Nephrol. 2014 Apr 30; doi: 10.1007/s00467-014-2818-3. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 11.Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, Zeng Y, Miao R, Jin G, Ma H, Chen Y, Shen H. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao X, Yang L, Ma Y, Yang J, Zhang G, Huang G, Huang Q, Chen L, Fu F, Chen Y, Su D, Dong Y, Ma X, Lu C, Peng X. No association of functional variant in pri-miR-218 and risk of congenital heart disease in a Chinese population. Gene. 2013;523:173–177. doi: 10.1016/j.gene.2013.03.119. [DOI] [PubMed] [Google Scholar]
- 13.Ding SL, Wang JX, Jiao JQ, Tu X, Wang Q, Liu F, Li Q, Gao J, Zhou QY, Gu DF, Li PF. A pre-microRNA-149 (miR-149) genetic variation affects miR-149 maturation and its ability to regulate the Puma protein in apoptosis. J Biol Chem. 2013;288:26865–26877. doi: 10.1074/jbc.M112.440453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhi H, Wang L, Ma G, Ye X, Yu X, Zhu Y, Zhang Y, Zhang J, Wang B. Polymorphisms of miRNAs genes are associated with the risk and prognosis of coronary artery disease. Clin Res Cardiol. 2012;101:289–296. doi: 10.1007/s00392-011-0391-3. [DOI] [PubMed] [Google Scholar]
- 15.Wang N, Li Y, Zhou RM, Wang GY, Wang CM, Chen ZF, Liu W. Hsa-miR-196a2 functional SNP is associated with the risk of ESCC in individuals under 60 years old. Biomarkers. 2014;19:43–48. doi: 10.3109/1354750X.2013.866164. [DOI] [PubMed] [Google Scholar]
- 16.Guo H, Wang K, Xiong G, Hu H, Wang D, Xu X, Guan X, Yang K, Bai Y. A functional varient in microRNA-146a is associated with risk of esophageal squamous cell carcinoma in Chinese Han. Fam Cancer. 2010;9:599–603. doi: 10.1007/s10689-010-9370-5. [DOI] [PubMed] [Google Scholar]
- 17.Zhou X, Chen X, Hu L, Han S, Qiang F, Wu Y, Pan L, Shen H, Li Y, Hu Z. Polymorphisms involved in the miR-218-LAMB3 pathway and susceptibility of cervical cancer, a case-control study in Chinese women. Gynecol Oncol. 2010;117:287–290. doi: 10.1016/j.ygyno.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 18.Shi TY, Chen XJ, Zhu ML, Wang MY, He J, Yu KD, Shao ZM, Sun MH, Zhou XY, Cheng X, Wu X, Wei Q. A pri-miR-218 variant and risk of cervical carcinoma in Chinese women. BMC Cancer. 2013;13:19. doi: 10.1186/1471-2407-13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C, Hong H, Chen L, Shi X, Chen Y, Weng Q. Association of microRNA Polymorphisms with the Risk of Myocardial Infarction in a Chinese Population. Tohoku J Exp Med. 2014;233:89–94. doi: 10.1620/tjem.233.89. [DOI] [PubMed] [Google Scholar]
- 20.Wu C, Li M, Hu C, Duan H. Prognostic role of microRNA polymorphisms in patients with advanced esophageal squamous cell carcinoma receiving platinum-based chemotherapy. Cancer Chemother Pharmacol. 2014;73:335–341. doi: 10.1007/s00280-013-2364-x. [DOI] [PubMed] [Google Scholar]
- 21.Yuan Z, Zeng X, Yang D, Wang W, Liu Z. Effects of common polymorphism rs11614913 in Hsa-miR-196a2 on lung cancer risk. PLoS One. 2013;8:e61047. doi: 10.1371/journal.pone.0061047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Chen Q, He C, Mao W, Zhang L, Xu X, Zhu J, Chen B. Polymorphisms on 8q24 are associated with lung cancer risk and survival in Han Chinese. PLoS One. 2012;7:e41930. doi: 10.1371/journal.pone.0041930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan W, Xiaoyun H, Haifeng Q, Jing L, Weixu H, Ruofan D, Jinjin Y, Zongji S. MicroRNA-218 enhances the radiosensitivity of human cervical cancer via promoting radiation induced apoptosis. Int J Med Sci. 2014;11:691–696. doi: 10.7150/ijms.8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishikawa R, Goto Y, Sakamoto S, Chiyomaru T, Enokida H, Kojima S, Kinoshita T, Yamamoto N, Nakagawa M, Naya Y, Ichikawa T, Seki N. Tumor-suppressive microRNA-218 inhibits cancer cell migration and invasion via targeting of LASP1 in prostate cancer. Cancer Sci. 2014;105:802–811. doi: 10.1111/cas.12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu H, Gao G, Jiang L, Guo L, Lin M, Jiao X, Jia W, Huang J. Decreased expression of miR-218 is associated with poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2013;6:2904–2911. [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto N, Kinoshita T, Nohata N, Itesako T, Yoshino H, Enokida H, Nakagawa M, Shozu M, Seki N. Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion by targeting focal adhesion pathways in cervical squamous cell carcinoma. Int J Oncol. 2013;42:1523–1532. doi: 10.3892/ijo.2013.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinoshita T, Hanazawa T, Nohata N, Kikkawa N, Enokida H, Yoshino H, Yamasaki T, Hidaka H, Nakagawa M, Okamoto Y, Seki N. Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion through targeting laminin-332 in head and neck squamous cell carcinoma. Oncotarget. 2012;3:1386–1400. doi: 10.18632/oncotarget.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He X, Dong Y, Wu CW, Zhao Z, Ng SS, Chan FK, Sung JJ, Yu J. MicroRNA-218 inhibits cell cycle progression and promotes apoptosis in colon cancer by downregulating BMI1 polycomb ring finger oncogene. Mol Med. 2012;18:1491–1498. doi: 10.2119/molmed.2012.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Ping Z, Ning H. MiR-218 Impairs Tumor Growth and Increases Chemo-Sensitivity to Cisplatin in Cervical Cancer. Int J Mol Sci. 2012;13:16053–16064. doi: 10.3390/ijms131216053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xin SY, Feng XS, Zhou LQ, Sun JJ, Gao XL, Yao GL. Reduced expression of circulating microRNA-218 in gastric cancer and correlation with tumor invasion and prognosis. World J Gastroenterol. 2014;20:6906–6911. doi: 10.3748/wjg.v20.i22.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Z, Xu Y, Du J, Tan J, Jiao H. Expression of microRNA-218 in human pancreatic ductal adenocarcinoma and its correlation with tumor progression and patient survival. J Surg Oncol. 2014;109:89–94. doi: 10.1002/jso.23475. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Huang X, Xiao J, Yang Y, Zhou Y, Wang X, Liu Q, Yang J, Wang M, Qiu L, Zheng Y, Zhang P, Li J, Wang Y, Wei Q, Jin L, Wang J, Wang M. Pri-miR-124 rs531564 and pri-miR-34b/c rs4938723 polymorphisms are associated with decreased risk of esophageal squamous cell carcinoma in chinese populations. PLoS One. 2014;9:e100055. doi: 10.1371/journal.pone.0100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bosman FT, Stamenkovic I. Functional structure and composition of the extracellular matrix. J Pathol. 2003;200:423–428. doi: 10.1002/path.1437. [DOI] [PubMed] [Google Scholar]
- 34.Kita Y, Mimori K, Tanaka F, Matsumoto T, Haraguchi N, Ishikawa K, Matsuzaki S, Fukuyoshi Y, Inoue H, Natsugoe S, Aikou T, Mori M. Clinical significance of LAMB3 and COL7A1 mRNA in esophageal squamous cell carcinoma. Eur J Surg Oncol. 2009;35:52–58. doi: 10.1016/j.ejso.2008.01.025. [DOI] [PubMed] [Google Scholar]


