Abstract
ERp27 (endoplasmic reticulum protein 27.7 kDa) is a homologue of PDI (protein disulfide-isomerase) localized to the endoplasmic reticulum. ERp27 is predicted to consist of two thioredoxin-fold domains homologous with the non-catalytic b and b′ domains of PDI. The structure in solution of the N-terminal b-like domain of ERp27 was solved using high-resolution NMR data. The structure confirms that it has the thioredoxin fold and that ERp27 is a member of the PDI family. 15N-NMR relaxation data were obtained and ModelFree analysis highlighted limited exchange contributions and slow internal motions, and indicated that the domain has an average order parameter S2 of 0.79. Comparison of the single-domain structure determined in the present study with the equivalent domain within full-length ERp27, determined independently by X-ray diffraction, indicated very close agreement. The domain interface inferred from NMR data in solution was much more extensive than that observed in the X-ray structure, suggesting that the domains flex independently and that crystallization selects one specific interdomain orientation. This led us to apply a new rapid method to simulate the flexibility of the full-length protein, establishing that the domains show considerable freedom to flex (tilt and twist) about the interdomain linker, consistent with the NMR data.
Keywords: endoplasmic reticulum, endoplasmic reticulum protein 27.7 kDa (ERp27), NMR, protein disulfide-isomerase (PDI), protein dynamics, thioredoxin-fold
Abbreviations: 2D, two-dimensional; 3D, three-dimensional; ER, endoplasmic reticulum; ERp27, ER protein 27.7 kDa; HSQC, heteronuclear single-quantum coherence; NOE, nuclear Overhauser effect; PDI, protein disulfide-isomerase; RMSD, root mean square deviation; Trx, thioredoxin
INTRODUCTION
The lumen of the ER (endoplasmic reticulum) is the subcellular compartment responsible for folding and quality control of proteins exported to the cell surface or extracellular spaces. This compartment contains many resident proteins which carry out these folding and quality assurance functions. One prominent group of such resident proteins is the PDI (protein disulfide-isomerase) family of proteins [1–3]. There are 20 known members of the human PDI family and their best-known role is in facilitating formation of the correct pairing of disulfide bonds, a key aspect of the folding process for most extracellular or cell-surface proteins. Proteins of the PDI family have a range of functions in addition to disulfide bond isomerization, and not all members of the family are catalytically active. All proteins of the PDI family have, or are thought to have, at least one domain with significant structural similarity to Trx (thioredoxin). These Trx-like domains have a mixed β/α fold with the typical secondary structure topology β1-α1-β2-α2-β3-α3-β4-β5-α4 [3]. Those proteins which are catalytically active in disulfide bond isomerization act through a conserved Cys-Xaa-Xaa-Cys active-site motif in a Trx-like domain. This motif is located in the N-terminus of α2 and incorporates the loop immediately preceding this helix. There is typically also a proline residue in the cis conformation in the loop between α3 and β4. This loop is adjacent to the active site in the tertiary structure and the cis-proline residue is thought to play a role in substrate binding [4]. Those members of the PDI family which are not catalytically active possess at least one Trx-like domain which lacks the active-site motif and may or may not possess the conserved cis-proline residue.
PDI is composed of four Trx-like domains, a, b, b′ and a′, with an acidic α-helical extension on the C-terminus [2,5]. The a and a′ domains contain the catalytic sites involved in thiol-disulfide oxidoreduction and show high sequence similarity to one another and to Trx. The b and b′ domains are non-catalytic; the b domain is of unknown function, whereas the b′ domain plays a major role in substrate binding [6]. This domain contains a hydrophobic pocket which is thought to act as the principal substrate-binding site on the protein [7,8], with the other domains making smaller contributions to substrate binding.
ERp27 is a 27.7 kDa ER-resident protein of unknown function and a putative member of the PDI family [9]. On the basis of sequence analysis, ERp27 is thought to be composed of two non-catalytic PDI-like domains, b and b′. The protein contains two cysteine residues, one in each domain. These cysteine residues have been shown to exist as free thiols and to be buried within each domain [9]. The protein is thus unlikely to be involved in thiol-disulfide oxidoreduction. The C-terminal domain of ERp27 shows sequence similarity to the b′ domain of PDI, and the similarity includes residues involved in the substrate-binding site of PDI [7,8], suggesting that ERp27 may possess a similar binding site. Indeed, this domain of ERp27 has been shown to bind the 14-residue peptide Δ-somatostatin [9]. ERp27 has also been shown to interact with ERp57, another member of the PDI family, both in vitro and in vivo [9]. The binding site for ERp57 has been mapped to the C-terminal (b′) domain of ERp27.
In the present paper we describe the solution structure and backbone dynamics of the N-terminal (b) domain of human ERp27. Structural data have been deposited in the Protein Data Bank (PDB code 2L4C). NMR relaxation data were collected for this domain, indicating regions of increased mobility. A comparison of the HSQC (heteronuclear single-quantum coherence) spectrum for this isolated domain and that for full-length ERp27 identified a large contiguous surface on the b domain affected by connection to the b′ domain. This putative interface region was considerably more extensive than that found in the X-ray structure of full-length ERp27 (PDB code 4F9Z, H. Schindelin and F.-X. Kober) and stimulated an exploration of the flexibility of ERp27 about the interdomain linker. We found that the domains show great interdomain flexibility making extensive transient interdomain contacts, consistent with the NMR data. This implies that crystallization selected a specific interdomain orientation from an ensemble of possible orientations.
EXPERIMENTAL
Protein expression and purification
Mature human ERp27 (Glu26–Leu273) and the individual b domain (Glu26–Leu141) were expressed in Escherichia coli BL21 (DE3) pLysS cells with an N-terminal His tag (MHHHHHHM) as described previously [9]. 15N- and 15N/13C-labelled protein was expressed in M9 medium containing [15N]ammonium chloride (1 g/l) or [15N]ammonium chloride (1 g/l) and [13C]glucose (2 g/l). The recombinant protein was purified by immobilized metal-affinity chromatography followed by ion-exchange chromatography. Purified protein was concentrated and buffer-exchanged into 25 mM monosodium phosphate and 100 mM sodium chloride (pH 6.5). Sodium azide (0.01%) was added as a preservative.
Multidimensional NMR spectroscopy
Protein samples were prepared to a concentration of 1–1.5 mM for structural determination experiments and 0.5 mM for relaxation studies in 25 mM monosodium phosphate and 100 mM sodium chloride (pH 6.5). The samples were placed in 5 mm Shigemi BMS-005V tubes with 10% 2H2O. All NMR experiments were carried out on four-channel Varian Unity INOVA NMR spectrometers with 1H resonance frequencies of 600 MHz, at the University of Kent, or 800 MHz at the Medical Research Council National Institute for Medical Research, The Ridgeway, Mill Hill, London, U.K. All spectra were acquired at 25°C. Referencing for the 1H chemical shift was based on the position of the water 1H resonance, according to its known relationship with temperature [10] and 13C/15N referencing was achieved using gamma ratios. NMR experiments used WATERGATE [11] for solvent suppression to attenuate the 1H water signal. Indirect dimensions were acquired using the hypercomplex method [12]. Data were processed on a Linux platform (SuSe 10.0) using NMRPipe [13], and analysed using the CcpNmr Analysis package [14].
NMR resonance assignment
1H-15N HSQC, CBCA(CO)NH and CBCANH standard triple-resonance NMR experiments were used to assign the backbone Cα, Cβ and amide 15N and 1H resonances. 15N-edited TOCSY-HSQC and HCCH-TOCSY experiments, with mixing times of 60 ms and 18 ms respectively, were used to assign the aliphatic side-chain 1H and 13C resonances. Where necessary, 3D (three-dimensional) 15N- and 13C-edited NOESY spectra were used to aid in the assignments. Side-chain NH2 resonances of asparagine and glutamine were assigned using the 1H-15N HSQC spectrum and a 15N-edited NOESY spectrum. Aromatic side-chain 1H and 13C resonances were assigned using 2D (two-dimensional) 1H-1H TOCSY with 60 ms mixing time, 2D 1H-1H NOESY with 100 ms mixing time, 2D aromatic 1H-1H NOESY with 80 ms mixing time and a 2D selective aromatic 1H-13C HSQC spectrum.
15N NMR relaxation measurements
15N T1 and T2 experiments were acquired using developed pulse sequences comparable with those described elsewhere [15]. T1 and T2 delay times were set as 128, 256 (×2), 384, 512, 640 (×2), 769 and 897 ms, and 20, 40, 60 (×2), 80, 100, 120 (×2), 140 and 160 ms respectively. 15N Heteronuclear NOE (nuclear Overhauser effect) experiments were collected with a relaxation delay of 5 s with and without saturation of the amide protons that was achieved using 120° high-power pulses [16]. Relaxation times were calculated as the exponential fit of single exponential decays to peak intensity values: I=I0exp(−t/Tx) where Tx=T1 or T2, and I=resonance intensity at time t. Heteronuclear NOEs were calculated using the expression η=I/I0. ModelFree analysis of the relaxation data was carried out using the ModelFree 4.0 suite of programs [17–19] to obtain the optimal parameter fits of S2 (order parameter of motion), τe (internal correlation time) and Rex (exchange broadening). All ModelFree optimization used the most appropriate model with the lowest number of parameters possible to provide an optimal fit to the relaxation data.
Structure calculations
NOE distance restraints were obtained from 3D 15N-edited NOESY-HSQC and 13C-edited NOESY-HSQC spectra. Both experiments were recorded at 800 MHz with a mixing time of 80 ms. Dihedral angle (φ/ϕ) restraints were generated from backbone chemical-shift data using TALOS [20]. Hydrogen-bond donors were determined from 15N-HSQC experiments involving H/D (hydrogen/deuterium) exchange and hydrogen bond donor–acceptor pairs within secondary structure elements were identified from preliminary structure calculations. Structure calculations initially used only NOE restraints and were refined by subsequent inclusion of the dihedral angle and hydrogen-bond restraints. Structures were calculated using ARIA version 2.1 [21] and CNS version 1.2 [22]. For assignment of ambiguous NOESY cross-peaks with ARIA, the frequency window sizes were left at the default settings of 0.02 p.p.m. for direct and 0.04 p.p.m. for indirect proton dimensions, and 0.5 p.p.m. for heteronuclear dimensions. For each round of structure calculations, ten iterations were performed. For the first iteration, 50 structures were calculated and the 20 lowest-energy structures were used for calibration and violation analysis. For the next seven iterations, 20 structures were calculated, with the seven lowest-energy structures used for calibration and violation analysis. For the ninth iteration, 50 structures were calculated and the 20 lowest-energy structures were used for calibration and violation analysis. For the final (tenth) iteration, 100 structures were calculated. For iterations 0–9, the violation tolerances were set to 1000.0, 5.0, 3.0, 1.0, 1.0, 1.0, 0.1 0.1, 0.1 and 0.1 Å (1 Å=0.1 nm) respectively and the partial assignment ambiguity cut-off values were set to 1.0, 0.9999, 0.999, 0.99, 0.98, 0.96, 0.93, 0.9, 0.85 and 0.8 respectively. For each iteration, the violation threshold was set to 0.5 Å and the maximum number of contributions for partial assignment was 20. Structure calculations with CNS used torsion angle dynamics. Each iteration used 18000 cooling steps in the simulated annealing protocol fixed in the ratio of 5:4 for the first and second cooling stages respectively.
Mapping of the b/b′ domain interface on the b domain of ERp27
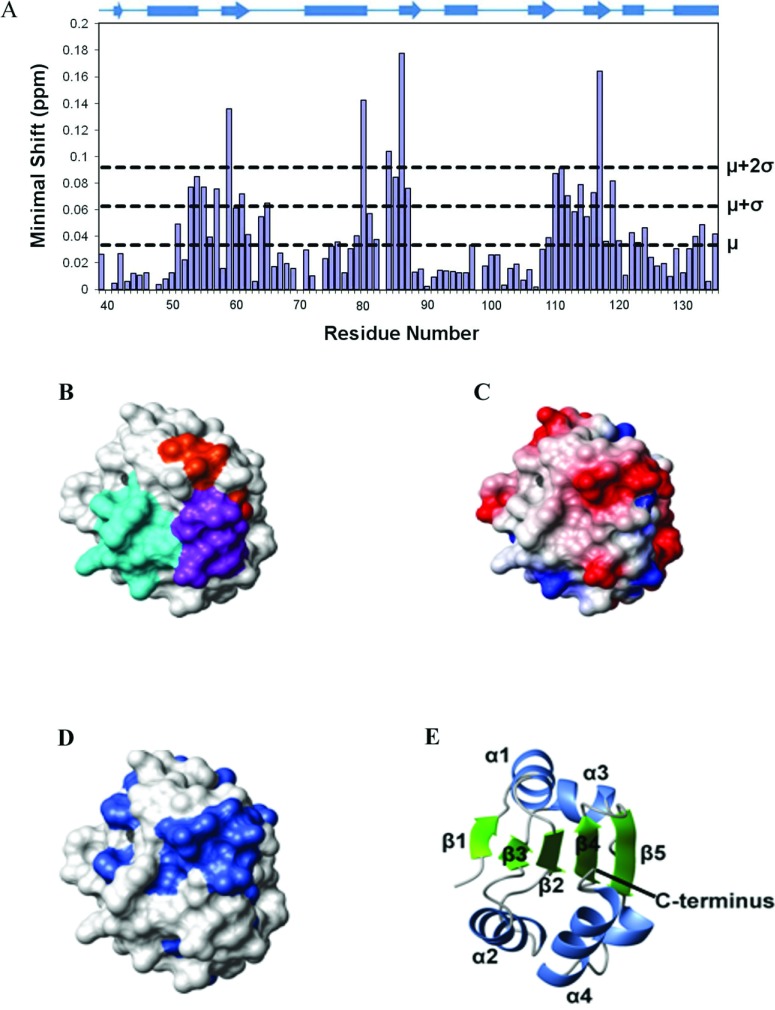
Residues from the b domain of ERp27 potentially forming the interface with the b′ domain were identified using the minimal shift mapping approach [23,24]. Minimal shift mapping provides a measure of the chemical-shift difference, Δshift, that corresponds to the minimal, or closest distance, for each b peak to any peak in an equivalent full-length ERp27 (bb′) spectrum. Therefore a peak in the spectrum of b with a corresponding peak in bb′ in an identical position would score a Δshift of zero with the value of Δshift increasing as the bb′ peak moves further away from being coincident to the b peak. These changes in chemical shift are indicative of a change in chemical environment in the full-length protein compared with the single-domain protein. Hence, a comparison of the 15N-HSQC spectra for the b domain and the full-length protein provides identification of residues with NH chemical shifts that are different in the b construct compared with the bb′ construct. For each peak in the HSQC spectrum for the b domain, the combined 1H and 15N chemical-shift difference was calculated for all peaks in the HSQC spectrum for the full-length protein. The following equation was used:
where Δ1HN is the chemical-shift difference in the 1H dimension and Δ15N is the chemical-shift difference in the 15N dimension. The Δ15N shift difference was reduced by a factor of 6 to account for the difference in 15N-HSQC amide chemical-shift ranges of approximately 30 p.p.m. and 5 p.p.m. for 15N and 1H respectively.
Simulation of the flexibility of full-length ERp27
The atomic co-ordinates of full-length ERp27 (PDB code 4F9Z) were kindly provided by F.-X. Kober and H. Schindelin in advance of publication and were used to simulate and represent the relative motion of the domains. Simulation was by a novel rapid method which analyses the protein structure as a network of rigid clusters and flexible linkers using the program FIRST [25], separately calculates the normal modes of motion of the structure by coarse-grained elastic network modelling using ElNemo [26] and then generates trajectories of motion for the network by geometric simulation using the FRODA algorithm [27] in a calculation in which the normal mode eigenvectors are used to bias the motion of the network [28]. This method of simulation of flexible motion retains an all-atom representation of the protein and uses a simplified physical model [29] to maintain the covalent bond geometry and the network of hydrogen bonds and hydrophobic tethers found in the input structure, and to enforce steric exclusion, while neglecting long-range interactions. This simplification allows for a systematic exploration of motion along multiple normal mode directions, achieving large amplitudes, using modest computational resources. The retention of all-atom detail and of the essential local covalent and noncovalent interactions, meanwhile, makes the generated structures physically realistic and useful for the interpretation of experimental data [30].
Normal modes of motion were generated for both full-length ERp27 (molecule B) and for the central bb′ domains of full-length yeast PDI (from PDB code 2B5E). For each structure, trajectories of flexible motion were generated using the ten lowest-frequency non-trivial normal modes; these are modes 7–16, as modes 1–6 are combinations of trivial rigid body motions. Biases were applied both parallel and antiparallel to the mode eigenvectors generating two opposed trajectories for each normal mode, indicated as 7− and 7+. The bias step size in each iteration of the simulation was 0.01 Å. The constraint network included all hydrophobic tethers identified by FIRST in the input structures. Hydrogen bonds identified in the input structure are assigned a (negative) energy in FIRST on the basis of their geometry, and the set of bonds retained in the simulations is determined by setting a cut-off value. Simulations were carried out using cut-off values of −1, −2 and −3 kcal/mol (1 kcal=4.184 kJ), with essentially identical results. Results are presented for the cut-off of −2 kcal/mol. In total 2000 conformations were generated along each trajectory with every 100th conformation being recorded as a PDB file for subsequent analysis. All simulations were carried out overnight on a dual-core 2.4 GHz desktop workstation running Linux.
In this case our principal interest was in the relative orientation of the two domains, and the variation of this orientation in the course of large-scale flexible motion. To represent the relative orientations of the two domains, a plane was calculated as representative of the β-sheet plane in each domain, by selecting the Cα atoms of four ‘central’ residues, namely alternating central residues in each of the adjacent antiparallel strands β2 and β4. These four atoms in each domain give a quadrilateral, from which the plane normals and interplane axis are calculated. The ‘tilt’ angle between adjacent domains (θ) is defined using the scalar product of the vectors corresponding to the respective plane normals. The dihedral ‘twist’ angle (ω) makes use of the vector between the ‘average’ position of the four selected Cα atoms in each domain. This vector and the plane normal for each domain define a plane; the dihedral angle between these two planes is the ‘twist’ angle (ω). Tilt and twist values were extracted for the input structures and for the structures generated in the simulations of flexible motion.
RESULTS
Resonance assignment of the ERp27 b domain
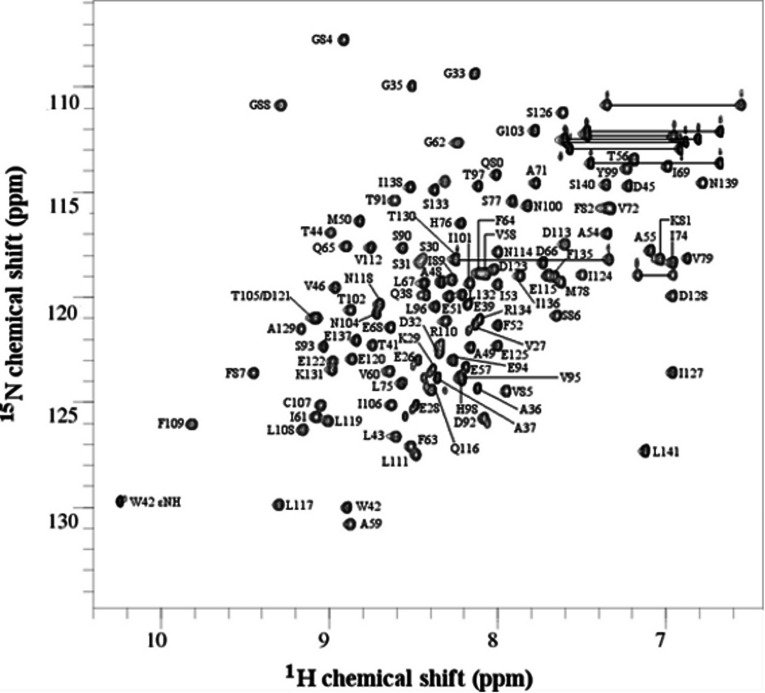
Excluding the eight residues of the His tag (MHHHHHHM), chemical-shift assignments were made for 100% of the backbone amide 15N and 1H resonances, 100% of the 13Cα and 13Cβ resonances, 96.7% of the 1Hα resonances, 94.5% of the 1Hβ resonances, 88.8% of the other side-chain 1H resonances (1Hγ, 1Hδ, 1Hϵ, 1Hζ and 1Hη), 70.6% of the side-chain 13C resonances (13Cγ, 13Cδ, 13Cϵ, 13Cζ and 13Cη) and 26.1% of the side-chain 15N resonances (15Nδ, 15Nϵ, 15Nζ and 15Nη). Six residues had no side-chain assignments other than 13Cβ; Glu26, Glu28, Ser30, Ser31, Glu122 and Glu125, of which four (Glu26, Glu28, Ser30 and Ser31) are located at the N-terminus and, following structure determination, were found to be in a flexible extension outside of the core Trx fold (see below). The assigned backbone amide resonances are shown in the 2D 15N-1H HSQC spectrum in Figure 1. The well-dispersed pattern of peaks indicates that the protein is folded. A total of 130 HSQC peaks were identified. Of these, 18 were identified as asparagine and glutamine side-chain peaks, one was assigned to the indole amide of the single tryptophan residue, one to the His-tag backbone and the remainder were assigned to the backbone amides of the 110 non-proline residues.
Figure 1. 2D 15N-1H HSQC spectrum of the ERp27 b domain.
Peaks are annotated to show the backbone amide assignments. All 110 non-proline residues have been assigned. The indole ϵ1 amide of the single tryptophan residue (W42) has also been assigned. Side-chain amide NH2 resonances are connected with horizontal lines.
NMR relaxation analysis
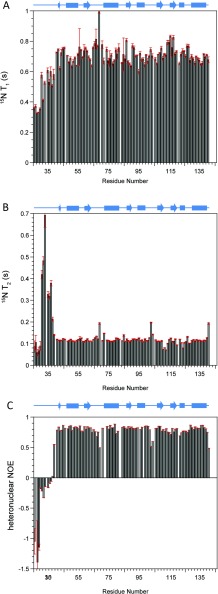
15N-NMR relaxation parameters T1, T2 and heteronuclear NOE were obtained for the b domain of ERp27 at 14.1 T (600 MHz 1H) and 25°C (Figure 2). The average values for 15N T1 and T2 were recorded as 704±46 ms and 122±64 ms respectively; these data were used to estimate the global correlation time for the domain of 7.13±0.71 ns using χ2 optimization of the theoretical dipolar and practical T1/T2 ratios. This estimate of global correlation time confirms that the domain is monomeric under the experimental conditions used. NMR relaxation parameters for each individual 15N were interrogated further using ModelFree analysis which provides motional information in terms of order parameters S2, internal correlation times τe and contributions to chemical exchange Rex, for individual NH vectors across the domain backbone (Supplementary Figure S1 at http://www.biochemj.org/bj/450/bj4500321add.htm). An unstructured N-terminal region was identified from combined low S2 and high τe values for residues 26–38. The average S2 value across the structured region (residues 39–141) was 0.79±0.04 where S2 values of 1.0 support a completely rigid domain backbone and 0.01 support an entirely random flexible backbone.
Figure 2. 15N NMR relaxation parameters of the ERp27 b domain.
Spin–lattice relaxation times T1 (A), spin–spin relaxation times T2 (B) and 1H-15N heteronuclear NOE data (C) were collected at 14.1 T and 25°C and are plotted as a function of residue number. A schematic diagram of the domain secondary structure as solved by NMR is shown above each plot. Cylinders represent α-helices and arrows represent β-strands.
This value of S2 can be compared with those of oxidized and reduced ERp18 (0.81 and 0.90 respectively) [31] and implies a relatively flexible backbone which may provide capacity for diverse interactions with partner proteins. Average internal correlation times across the structured region were 85±18 ps, a typical value for a well-behaved globular domain. Within the structured region, five residues were estimated to have τe values above 300 ps as seen in Supplementary Figure S1(B): Leu43, Val72, Val95, Gly103 and Leu141. These residues are all associated with loops or are the first residues of secondary structure elements and these higher τe are due to either depressed T1 or elevated T2 values as seen in Figures 2(A) and 2(B). These unusual τe values were all obtained with low errors and, according to ModelFree analysis, they indicate significant areas of the domain that have abnormally slow internal motions. Additional ModelFree contributions to Rex were only observed for two residues, Leu111 and Asp113 within the loop between strands β4 and β5 (Supplementary Figure S1C), and are indicative of low T2 observed for the residues in Figure 2(B).
Structure calculations
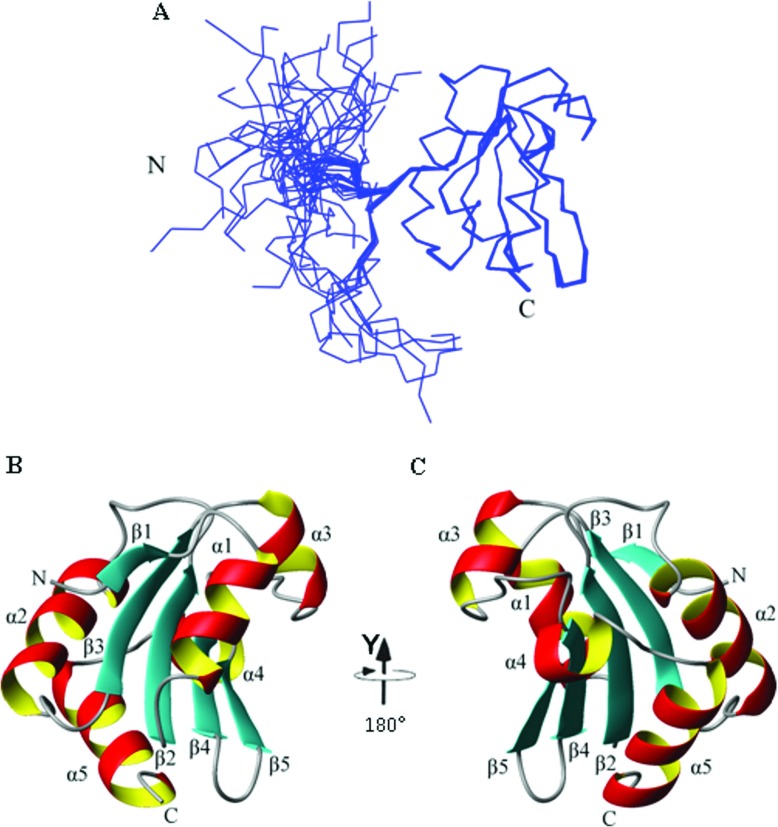
In the final round of structure calculations, 1939 NOE-derived distance restraints, 117 dihedral angle restraints and 42 hydrogen-bond restraints were used. In total 100 structures were generated, of which 92 had converged satisfactorily. The converged structures had no distance violations greater than 0.5 Å and no dihedral angle restraint violations greater than 5°. Of the 92 converged structures, the representative structure ensemble was chosen as the 50 lowest-energy structures; superimposed backbone tracings of these structures are shown in Figure 3(A). None of the structures in the ensemble had NOE or hydrogen-bond restraint violations greater than 0.3 Å. The NMR restraints and structural statistics are summarized in Table 1. NOE distribution, structural calculations and NMR relaxation data analysis confirmed that the structured region of the b domain extends from Glu39 to Leu141. Figure 3(A) illustrates the diversity of conformations of the N-terminal region in the individual structures and the structural limits of the domain are also confirmed by comparing the RMSD (root mean square deviation) values between residues 26 and 141 and those between residues 39 and 141 (Table 1). Description and analysis of the structure below is restricted to this structured region (Glu39–Leu141) and is based on the individual structure in the ensemble closest to the mean structure; this is illustrated in Figures 3(B) and 3(C) showing the assigned secondary structure.
Figure 3. Solution structure of the ERp27 b domain.
(A) Superimposition of the ensemble of 50 representative structures, residues 26–141. Each structural model in the ensemble was superimposed over the mean structure for the ordered region 39–141. The mean structure was calculated using MOLMOL [46]. The Cα traces are shown. The positions of the N- and C-termini are labelled N and C respectively. (B) and (C) show the single representative structure of ERp27 b, residues 39–141 and are related by a 180° rotation about the y-axis. Secondary structure elements are labelled.
Table 1. Structural statistics and RMSD values for the ERp27 b domain ensemble (50 structures).
For (A), (B), (D) and (E), values are reported ±S.D. (A) The energy was obtained using GROMOS96 within DEEPVIEW. (B) The RMSD to the mean structure was calculated using MOLMOL [46]. The mean structure was calculated independently for residues 26–141 and residues 39–141. The RMSD over the secondary structure regions used the mean structure calculated for residues 39–141. The secondary structure was as assigned using DSSP-CONT. (C) The total number of restraints was calculated using the total number of restrained hydrogen bonds, rather than the total number of hydrogen-bond restraints. This is because each restrained hydrogen bond had a set of two distance restraints. (D) The average number of all restraint violations (NOE, hydrogen bond and dihedral restraint violations) was calculated for all NOE and hydrogen bond violations greater than 0.1 Å and all dihedral angle violations greater than 5°. The data in (D) and (E) were generated using CNS. (F) Procheck Ramachandran plot statistics calculated for non-glycine residues from 39 to 141 for the ensemble of 50 structures.
| Parameter | Measurement |
|---|---|
| (A) Average energy (kJ/mol) | −2165±121 |
| (B) Average RMSD to mean structure (Å) | |
| Residues 26–141 | |
| Backbone | 3.27±0.82 |
| Heavy | 3.48±0.81 |
| Residues 39–141 | |
| Backbone | 0.10±0.02 |
| Heavy | 0.54±0.03 |
| Secondary structure | |
| Backbone | 0.09±0.02 |
| Heavy | 0.52±0.03 |
| (C) Number of restraints | |
| Total | 2098 |
| Average per residue | 18.1 |
| NOE | |
| Total | 1939 |
| Ambiguous | 1059 |
| Unambiguous | 880 |
| Intra (i−j=0) | 296 (33.6%) |
| Sequential (|i−j|=1) | 232 (26.4%) |
| Medium-range (1<|i−j|<5) | 146 (16.6%) |
| Long-range (|i−j|>5) | 206 (23.4%) |
| Hydrogen bond | |
| Total constrained | 42 |
| Total constraints | 84 |
| Long-range constraints (|i−j|>5) | 38 |
| Dihedral angle (ϕ/ψ) | |
| Total | 117 |
| (D) Restraint violations (mean values per structure) | |
| All restraints | 16.0±1.7 |
| NOE | |
| Violations>0.5 Å | 0 |
| Violations>0.3 Å | 0 |
| Violations>0.1 Å | 13.1±1.7 |
| Hydrogen bond | |
| Violations>0.5 Å | 0 |
| Violations>0.3 Å | 0 |
| Violations>0.1 Å | 2.9±0.3 |
| Dihedral angle | |
| Violations>5° | 0 |
| (E) RMSD from experimental restraints | |
| NOEs (Å) | 0.020 |
| Unambiguous (Å) | 0.018±0.001 |
| Ambiguous (Å) | 0.022 |
| Hydrogen bonds (Å) | 0.031±0.001 |
| Dihedral angles (°) | 0.110±0.015 |
| (F) Procheck statistics (residues 39–141, 50 structures) | |
| Most favoured regions | 3403 (74%) |
| Additional allowed regions | 1197 (26%) |
| Generously allowed regions | 0 |
| Disallowed regions | 0 |
The ordered region of the protein (residues 39–141) consists of a five-stranded mixed β-sheet surrounded by five α-helices (Figure 3). The secondary structure topology is β1-α1-β2-α2-β3-α3-β4-β5-α4-α5. Strands 1, 2 and 3 are parallel with each other, whereas strands 2, 4 and 5 are antiparallel. This corresponds to the variant of the canonical Trx fold (SCOP reference number 52832) commonly found in the PDI family [3], but with an α-helical insert (α4) before the C-terminal α-helix of the fold (α5). The β1 strand is short (two residues), but this is common in the family (e.g. in the various domains of yeast PDI [5] this strand comprises two, three, zero and four residues). This structural analysis is supported by chemical-shift data (Supplementary Figure S2 at http://www.biochemj.org/bj/450/bj4500321add.htm).
The structure ensemble was analysed using PROCHECK-NMR [32,33]. Over residues 39–141 of the ensemble, 74% of the non-glycine dihedral angles were in the most-favoured regions and 26% were in the additionally allowed regions. Gly103 was the only residue with dihedral angles in a generously allowed region of the plot. This residue is located on the loop between α3 and β4. This corresponds to the cis-proline loop found in many domains in the PDI family. Although there is no conserved proline residue in this loop in ERp27, Gly103 is in the equivalent position as the residue preceding the cis-proline in the catalytic PDI domains.
Structural homology
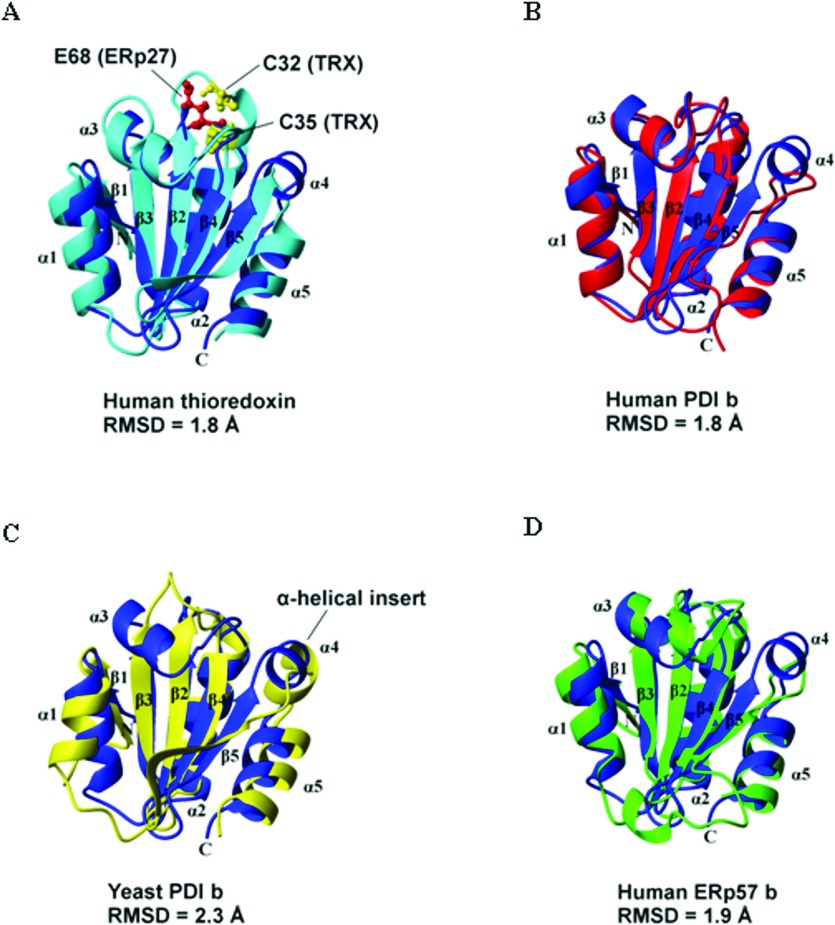
Structural homologues of the b domain of ERp27 were identified by a search of the PDB database using DALI [34]. In total 591 structure matches with a Z-score greater than 2.0 were found. Matches were detected to each of the five classes of Trx-fold proteins: Trx, glutaredoxin, glutathione transferase, DsbA and glutathione peroxidase. The highest-scoring matches were the structures of the b domains of ERp57 [35], human PDI [36], calsequestrin [37], ERp72 [38], yeast PDI [5], ERp29 [39] and ERp44 [40]. ERp27 b also showed significant structural similarity to Trx despite lacking the active-site residues and the conserved cis-proline residue. Ten out of the 20 highest-scoring matches were to Trxs.
The structure of ERp27 b domain is notable in having an α-helical insert between β5 and α4 of the standard PDI fold. This helix is short, consisting of only four residues, and is situated adjacent in the tertiary structure to the loop which contains the active site (in a-type PDI domains). A similar non-canonical helix is found in the yeast PDI b domain [5]. However, the structure of yeast PDI b differs significantly from that of ERp27 b in that it does not contain α3 of the canonical fold. Both ERp57 b and ERp72 b differ from the structure of ERp27 b most significantly in the loop region between β4 and β5, which is extended in these two proteins. Structure superimpositions between ERp27 b and selected homologous proteins are shown in Figure 4. A structure-based multiple sequence alignment between ERp27 b and selected homologues is shown in Supplementary Figure S3 (at http://www.biochemj.org/bj/450/bj4500321add.htm).
Figure 4. Structure superimpositions between ERp27 b and homologous proteins.
Structures were aligned using DALI [34]. The single structure of ERp27 b with the lowest backbone RMSD to the mean structure over residues 39–141 was used. The structure of ERp27 b is shown in blue. The secondary structure elements of ERp27 b are labelled. The N- and C-termini of ERp27 b are labelled N and C. ERp27 b is aligned with (A) human Trx (PDB code 1ERT, residues 1–105, RMSD=1.8 Å over 95 residues), (B) human PDI b domain (PDB code 2BJX, residues 119–216, RMSD=1.8 Å over 98 residues), (C) yeast PDI b domain (PDB code 2B5E, residues 142–237, RMSD=2.3 Å over 90 residues) and (D) human ERp57 b′ domain (PDB code 2H8L, residues 135–240, RMSD=1.9 Å over 97 residues). In (A) the catalytic cysteine residues of Trx are displayed in ball-and-stick representation. Glu68 of ERp27 b is also shown. This residue is located in the active-site loop and is strictly conserved in ERp27. In (C) the α-helical insertion in ERp27 b (α4) and yeast PDI b is indicated.
Sequence conservation
The sequence of ERp27 has only been found in vertebrate genomes. The sequence alignment in Supplementary Figure S4 (at http://www.biochemj.org/bj/450/bj4500321add.htm) shows a range of ERp27 b sequences from mammalian, bird, reptile and fish genomes. The sequence of ERp27 b is well conserved across species. The majority of conserved residues are located in the hydrophobic core of the protein, predominantly on β-strands 2, 3 and 4 and on the interior faces of the surrounding α-helices. In the alignment in Supplementary Figure S4 there are 13 positions that are invariant between ERp27 sequences. These correspond to Leu43, Glu57, Val58, Gly62, Phe63, Glu68, Val85, Val95, Leu108, Phe109, Arg110, Asp113 and Phe135 of human ERp27. Of these residues, nine (Leu43, Glu58, Val58, Glu68, Val85, Phe109, Arg110, Asp113 and Phe135) are surface-exposed. Gly62 and Phe63 are buried within the hydrophobic core of the protein and are situated at the C-terminal end of β2. These two residues are highly conserved in all b domains of the human PDI family (Supplementary Figure S3), with the exception of ERp29. Glu68 is located on the loop between β2 and α2. This is the location of the active site in the catalytically active domains of the PDI family. Glu57 and Val58 are situated at the N-terminus of β2, Arg110 is situated at the C-terminus of β4 and Asp113 is situated on the loop between β4 and β5. These residues are surface-exposed on a common face of the protein. The side chains of Arg110 and Asp113 are positioned so as to form a salt bridge. These residues are also present in the b domains of human PDI, ERp57 and ERp29, although in human PDI the arginine residue is replaced by a lysine (Supplementary Figure S3).
The b to b′ domain interface on the b domain
1H-15N HSQC spectra have been collected for both full-length ERp27 and the individual b domain (Supplementary Figure S5 at http://www.biochemj.org/bj/450/bj4500321add.htm). Signals in these spectra are highly sensitive to the local chemical environment of the backbone amide groups. A comparison of these spectra may therefore allow identification of those residues in b which are in contact with b′ in full-length ERp27. Residues in b potentially forming the interface between b and b′ in full-length ERp27 were therefore identified using the minimal shift approach [23,24]. The distance between each assigned cross-peak in the spectrum for the b domain to the nearest cross-peak in the spectrum for the full-length protein was measured. The minimal shift values per residue are plotted in the histogram in Figure 5(A). Residues 136–141 at the C-terminus of the b domain are inevitably close to the subsequent b′ domain; omitting these from further consideration, the average minimal shift of the remaining residues was 0.0333±0.0292. Figure 5(A) shows that the residues with large minimal shifts are clustered in three regions of sequence, namely residues 53–65, 80–87 and 110–119; within these regions, 19 out of 31 residues have minimal shifts >1 S.D. above the mean and most of the others have shifts above the mean. To interpret the shifts in relation to structure, all residues within these regions were included in a map of the possible interdomain surface; these were plotted on to the domain structure in Figure 5(B), which shows clearly that the regions identified in this way are located on one face of the protein. Similar regions of the fold make up the b to b′ domain interfaces in PDI, ERp57, ERp44, calsequestrin and ERp72 [5,35,37,38,40]. Calculation of the electrostatic potential shows the postulated interface region to be predominantly hydrophobic with charged residues around the edge (Figure 5C). There are patches of negative charge created, for example, by the surface-exposed side chains of Glu57, Asp113 and Glu137. There is considerable sequence conservation on the surface of the postulated b to b′ interface (Figure 5D). In particular, Glu57, Val58, Val85, Arg110, Asp113 and Phe135 are strictly conserved in ERp27 (Supplementary Figure S4), whereas Ala59, Val112, Asn139 and Leu141 in the interface are conserved in over 80% of ERp27 sequences.
Figure 5. Mapping of the b to b′ domain interface on the b domain of ERp27.
(A) Histogram showing the minimal chemical shifts per residue for ERp27 b (residues 39–135). The secondary structure is indicated above the histogram. Cylinders represent α-helices and arrows represent β-strands. The minimal shift values corresponding to the mean (μ), mean+1 S.D. (μ+σ) and mean+2 S.D. (μ+2σ) are marked by horizontal lines. (B–D) The contact molecular surface generated using MOLMOL with default settings [46]. In (B), residues are coloured according to the regions of structure showing the highest minimal shift values; residues 53–65 are coloured cyan, residues 80–87 are coloured orange and residues 110–119 are coloured purple. In (C), the electrostatic potential generated using MOLMOL is shown; blue represents positive charge, red represents negative charge and white represents neutral charge. In (D), sequence conservation is mapped on to the structure; residues with >80% identity (in the alignment in Supplementary Figure S3 at http://www.biochemj.org/bj/450/bj4500321add.htm) are coloured. (E) Cartoon representation of the protein backbone showing the orientation of the protein used for (B–D).
We were provided with the atomic co-ordinates of an X-ray structure of full-length ERp27 in advance of publication (PDB code 4F9Z) allowing a direct comparison of our inferred interface with that determined by X-ray diffraction.
The NMR minimal shift data in the present paper predict that the interdomain interface is homologous with that between the b and b′ domains in other multidomain members of the PDI family and this is broadly confirmed by the X-ray structure of the full-length protein, although the X-ray structure indicates an unusual interdomain angle. For a precise comparison, we selected ‘domain contact residues’ on the b domain by choosing the residues showing the greatest ‘minimal shifts’ in 1H-15N resonances in the NMR data and the closest interdomain atom contacts in the crystal structure data, excluding the residues at the C-terminus of the b domain (residues 136–141) which are inevitably in contact with or close to the b′ domain. Both approaches highlighted the same three regions on the b domain: I53AATEVAVIGFFQ65, Q80KFPGVSF87 and R110LVDNEQLNL119. Within these regions, eight residues were identified as contact residues by both criteria (Ile53, Ala54, Ala55, Glu57, Gly84, Leu111, Val112 and Asn114), 11 were identified by the NMR criterion only (Ala59, Ile61, Gln65, Gln80, Val85, Ser86, Phe87, Arg110, Gln116, Leu117 and Leu119), and four by the X-ray criterion only (Thr56, Val58, Asp113 and Glu115). Hence the contact surface highlighted by NMR is more extensive than that indicated in the crystal structure.
The X-ray data represent a static structure whereas shifts in NMR resonances could represent indirect effects, but will certainly also reflect perturbations that arise from solution dynamics. We therefore attempted to explore the potential flexibility of the molecule using a new rapid method [28].
Simulation of the flexibility of full-length ERp27 reconciles NMR and X-ray analyses of the interdomain interface
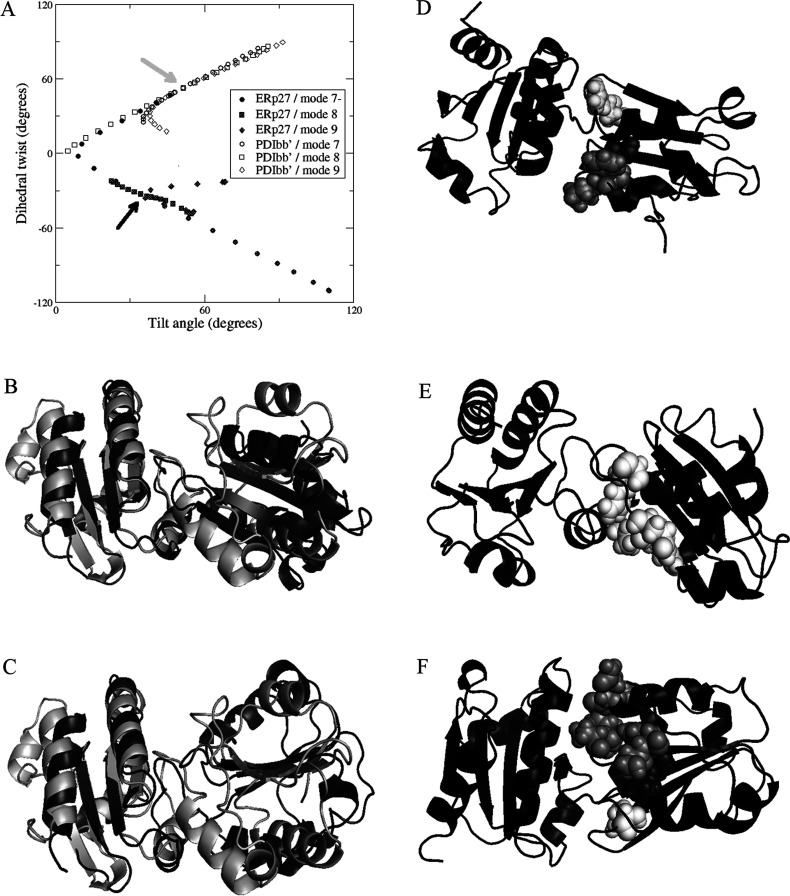
Examination of the trajectories of flexible motion showed that, for both ERp27 and PDI bb′, the largest amplitudes of flexible motion are achieved by the three lowest-frequency non-trivial normal modes (modes 7, 8 and 9), which are combinations of rotation about the interdomain axis, and tilting around the interdomain interface. Further discussion of results is focussed on these modes. To analyse these motions, we have defined the planes represented by the core β-sheet in each domain and extracted the ‘tilt’ and ‘twist’ angles between these planes (see the Materials and methods section). Figure 6(A) is a ‘tilt/twist’ plot of the interdomain orientations and confirms that the relative orientation of the domains in ERp27 is distinct from that in yeast PDI (arrows); it shows that the two structures differ in twist angle as well as in tilt angle (ERp27, tilt +34o and twist −33o; compared with PDI bb′, tilt +51o and twist +53o).
Figure 6. Domain orientation, flexibility and interdomain contacts in ERp27.
(A) Relative orientation of the β-sheet planes in adjacent Trx-fold domains for ERp27 and for the b-b′ moiety of yeast PDI. The planes are defined by the positions of four Cα atoms in each domain, as follows: for ERp27 domain b, Val60, Gly62, Ile106 and Leu108; for ERp27 domain b′, Leu164, Leu166, Leu222 and Ile224; for PDI b domain, Ile163, Gln165, Leu202 and Ile204; and for PDI b′ domain, Gly260, Leu262, Phe314 and Ile316. Orientation is described by a tilt angle between the plane normals, and by a dihedral twist angle formed by the plane normals and the interplane vector. The orientation in the PDB code 2B5E crystal structure of PDI is indicated by a grey arrow and that in the ERp27 crystal structure (PDB code 4F9Z [43]) by a black arrow. Symbols (open for PDI, closed for ERp27) show flexible motion along the three lowest-frequency non-trivial elastic network modes (modes 7, 8 and 9), in positive and negative directions, for each structure. Motion of ERp27 along mode 7− leads to PDI-like interdomain orientations. (B) Backbone cartoon overlay of ERp27 (black) with PDI b-b′ (white) crystal structures, aligned on the b′ domain (left-hand side) only, showing very different b-b′ orientations. (C) Corresponding backbone cartoon overlay of ERp27 after projection along mode 7− (black) with PDI b-b′ (white) crystal structure; the b domain β-sheets of ERp27 and PDI are now coplanar. (D) Backbone cartoon of ERp27 after projection along mode 7− (black). Residues 59 (dark grey), 82, 83 and 86 (all light grey), and 110 (white) are shown as spheres. These residues make new contacts with domain b′, not found in the input crystal structure. (E) Backbone cartoon of ERp27 after projection along mode 7+ (black), showing residues 110, 116, 117 and 118 as white spheres, forming new contacts with domain b′. (F) Backbone cartoon of ERp27 after projection along mode 8− (black), showing residues 59 and 60 (dark grey spheres), 80–83, 86 and 87 (light-grey spheres), and 110 (white spheres) forming new contacts with domain b′.
Trajectories of flexible motion are shown by calculating and plotting tilt and twist angles for every 100th structure in a simulated trajectory. In the case of ERp27, modes 8 and 9 correspond to tilting motions with the interdomain link acting as a hinge (as shown by a clear change in tilt angle but a very limited change in twist angle); PDI bb′ shows similar behaviour in modes 7 and 9 although the change in twist angle is slightly greater. For ERp27, mode 7, the lowest frequency non-trivial normal mode, shows the most striking results; this mode generates a pronounced twist, effectively a counter-rotation of the two domains around the axis joining the two domains. A similar motion in PDI bb′ is generated by its normal mode 8. What is notable is that this flexibility is much greater for ERp27 (a total amplitude of twist from −120o to +60o) than for PDI bb′, and that the extent of this motion for ERp27 allows it to take up a relative orientation very close to that found initially in PDI bb′. This reorientation of ERp27 to a more PDI-like structure is illustrated in Figures 6(B) and 6(C). Furthermore, in the course of facile motion along modes 7, 8 and 9, many residues in the loops identified by NMR as ‘contact loops’ move to make new contacts with the b′ domain (Figures 6D–6F). Using as the contact criterion that any backbone atom of a residue in the b domain is at a distance <7 Å from any non-hydrogen atom in the b′ domain, we find that Ile53–Val58, Gly84–Val85 and Leu111–Glu115 initially make contact with the b′ domain in at least one of the five molecules of ERp27 in the crystal unit cell. Flexible motion along the trajectories of modes 7–9 brings about new contacts, and almost all of the residues identified as being ‘contact’ residues by the NMR criterion are now seen to make contact with the b′ domain. Flexible twisting motion along the mode 7− trajectory (towards the PDI bb′ conformation) generates contacts between the b′ domain residues Ala59, Phe82, Pro83, Ser86 and Arg110, whereas motion along trajectory 7+ generates new contacts with Pro83, Phe87, Arg110, Gln116, Leu117 and Asn118. Similarly, tilting motion along mode 8− generates new contacts with Ala59, Val60, Gln80, Lys81, Phe82, Pro83, Ser86, Phe87 and Arg110 and that along mode 8+ generates new contacts with Arg110, Gln116, Leu117 and Asn118. New contacts made by motion along trajectories 9− and 9+ are all drawn from the same set of residues. Of the 11 non-proline residues in this set, four (Ala59, Gln80, Ser86 and Leu117) are the residues showing the greatest resonance shifts in the NMR analysis (Figure 5A) and these are four out of five residues showing shifts of greater than the mean+2 S.D., whereas a further three of the ‘new contact’ set (Phe87, Arg110 and Gln116) have shifts greater than the mean+S.D. Hence our simulations of flexible motion in ERp27 reconcile the apparent discrepancy between the interdomain contact region identified by NMR and that identified in the crystal structure
DISCUSSION
The N-terminal (b) domain of ERp27 was expressed in high yield, was highly soluble and very stable in solution, allowing the collection of high-quality 2D and 3D NMR data. By contrast, the C-terminal domain (b′) could not be expressed in isolation as a soluble protein and we did not succeed in strategies aimed at deriving it from cleavage of a fusion construct. Full-length ERp27 was readily expressed and gave a well-dispersed HSQC spectrum [9], but did not remain soluble through long-term NMR data collection at temperatures >30°C, preventing the collection of usable 3D data sets for full assignment and structure calculations. It is now well-established that the ligand-binding b′-type domains of the PDI family are the most difficult to generate and study in isolation; high-resolution structures of these domains have only been determined in the presence either of an adjacent domain or of a C-terminal extension sequence which is capable of occupying the hydrophobic cleft which constitutes the substrate-binding site [41]. Without one or other of these extensions in place, the isolated b′ domains have a tendency to oligomerize and aggregate [42], presumably as a result of the exposure of their hydrophobic ligand-binding sites.
The solution structure of the b domain reported in the present paper is highly congruent with the structure of this domain in full-length ERp27, determined by X-ray diffraction [43]. The two structures superimpose extremely closely with RMSD values of 0.96 Å over 102 Cα atoms and of 1.02 Å over the corresponding 408 backbone atoms (taking molecule D of the crystal structure as representative). The residues showing the greatest RMSD are in loops at either end of β3 (Gly103 and Val112) and in the loop containing the ‘additional’ helix (Glu122). The structural context of Gly103 has been noted above as corresponding to the residue preceding the conserved cis-proline residue in catalytic redox-active Trx domains, and this residue also shows up in the relaxation data as having an unusually slow internal correlation time.
It was noted (above) that ERp27 b has an α-helical insert between β5 and α4 of the conventional PDI domain fold. The equivalent region in the catalytic a and a′ domains of PDI, the loop between β5 and α4, contains a conserved arginine residue that has been implicated in the catalytic mechanism [44]. Short α-helical inserts also exist in analogous positions in yeast PDI b [4] and ERp44 b′ [40]. There is some sequence similarity between these inserts, with the C-terminus of each helix ending in the sequence Asp-X, where X is a hydrophobic branched-chain aliphatic residue. The side chain of the aspartate residue is surface-exposed and that of the hydrophobic residue is directed inwards, towards the core of the protein. The b′ domain of yeast PDI also contains an α-helical insert at this position, but this is part of a larger insert between β5 and α4 of the fold. There is no structural information available for the b domains of PDIp or PDILT, but these proteins appear to have insertions in the protein sequence at a similar position to the helical inserts in ERp27 b and yeast PDI b (Supplementary Figure S3). It is therefore possible that PDIp and PDILT also possess the structural feature of an additional α-helix at this location.
We found that ERp27 has a large capacity for flexible motion owing to its two-domain structure with relatively few interdomain constraints. The lowest-frequency non-trivial normal modes of motion for ERp27 represent a twisting motion around the interdomain axis (mode 7) and tilting motions where the interdomain interface acts as a hinge (modes 8 and 9). The b-b′ moiety of PDI has a similar structure with slightly more interdomain constraints and a similar (although slightly smaller) range of flexible variation. Twist motion of ERp27 along mode 7− carries its interdomain orientation from the distinctive state identified in the crystal structure to a PDI-like state. The inference that ERp27 is a highly flexible molecule is corroborated by interdomain contact information from NMR, which also suggests considerable interdomain motion (including motion towards a more PDI-like orientational state). Simulations of flexible motion along low-frequency modes generate multiple new interdomain contacts which correlate well with the interdomain contacts suggested by NMR. A further implication is that both ERp27 and PDI have considerable conformational flexibility in solution and that the crystal structures have captured particular orientations from a much wider ensemble of flexible variation. The crystallization of full-length ERp27 in a novel interdomain orientation has revealed the extent of this variation, not previously revealed by other PDI family crystal structures.
The paucity of information on the function of the b domain of ERp27 makes it difficult to interpret these results in functional terms. If the function of the b domain is purely passive (e.g. to confer solubility on the functional ligand-binding b′ domain), then the flexibility of the interdomain linker could arise negatively from the lack of any functional constraints determining relative orientation; this would contrast with PDI and other multidomain family members, where the b-b′ domain pair must partially constrain the positions and orientations of the adjacent functional a and a′ domains [4,45]. Alternatively, if the b domain plays a role in interactions between ERp27 and other chaperones or folding factors within the ER lumen, then the interdomain flexibility may be a positive factor in enabling alternative orientations for the domains in interactions with different partners. A recent publication confirmed that many members of the PDI family, including PDI, PDIr, ERp57, ERp72, P5, ERdj5 and ERp29, make multiple interactions with partner proteins within the ER ([45] see, for example, Figure 2A in this reference). Unfortunately, ERp27 was not included as a subject in that study, and in any case we lack structural information to underpin the majority of these inferred protein–protein interactions. In the absence of more information on the cellular role of ERp27, further comments on the significance of the flexible interdomain hinge between b and b′ domains are speculative.
Online data
AUTHOR CONTRIBUTION
The project was devised and supervised by Robert Freedman, Mark Howard and Richard Williamson. Nader Amin performed the experimental work and most of the data analysis, with guidance and hands-on support from Katrine Wallis and Michelle Rowe. Stephen Wells carried out the simulations of flexible motion. Nader Amin, Katrine Wallis and Robert Freedman drafted the paper, and all authors contributed to generating the final version of the paper.
ACKNOWLEDGEMENTS
We thank Franz-Xaver Kober and Hermann Schindelin (Rudolf Virchow Center, University of Würzburg, Würzburg, Germany) for communicating their structure of full-length ERp27 and for very helpful discussions. The majority of NMR data were collected at the University of Kent Biomolecular NMR facility which is supported by the Wellcome Trust. We are also grateful to Dr Geoff Kelly and Dr Tom Frenkiel (MRC National Institute for Medical Research, Mill Hill, London, U.K.) for assistance in running 800 MHz NMR datasets.
FUNDING
This work was supported by the Leverhulme Trust (Early Career Research Fellowship to S.A.W.) and the Wellcome Trust (‘Value in People’ Fellowship to A.K.W.).
References
- 1.Ellgaard L., Ruddock L. W. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freedman R. B., Klappa P., Ruddock L. W. Protein disulfide isomerases exploit synergy between catalytic and specific binding domains. EMBO Rep. 2002;3:136–140. doi: 10.1093/embo-reports/kvf035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozlov G., Määttänen P., Thomas D. Y., Gehring K. A structural overview of the PDI family of proteins. FEBS J. 2010;277:3924–39364. doi: 10.1111/j.1742-4658.2010.07793.x. [DOI] [PubMed] [Google Scholar]
- 4.Guddat L. W., Bardwell J. C., Zander T., Martin J. L. The uncharged surface features surrounding the active site of Escherichia coli DsbA are conserved and are implicated in peptide binding. Protein Sci. 1997;6:1148–11565. doi: 10.1002/pro.5560060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian G., Xiang S., Noiva R., Lennarz W. J., Schindelin H. The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell. 2006;124:61–736. doi: 10.1016/j.cell.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 6.Klappa P., Ruddock L. W., Darby N. J., Freedman R. B. The b’ domain provides the principal peptide-binding site of protein disulfide isomerase but all domains contribute to binding of misfolded proteins. EMBO J. 1998;17:927–935. doi: 10.1093/emboj/17.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrne L. J., Sidhu A., Wallis A. K., Ruddock L. W., Freedman R. B., Howard M. J., Williamson R. A. Mapping of the ligand-binding site on the b′ domain of human PDI: interaction with peptide ligands and the x-linker region. Biochem. J. 2009;423:209–217. doi: 10.1042/BJ20090565. [DOI] [PubMed] [Google Scholar]
- 8.Denisov A. Y., Määttänen P., Dabrowski C., Kozlov G., Thomas D. Y., Gehring K. Solution structure of the bb’ domains of human protein disulfide isomerase. FEBS J. 2009;276:1440–1449. doi: 10.1111/j.1742-4658.2009.06884.x. [DOI] [PubMed] [Google Scholar]
- 9.Alanen H. I., Williamson R. A., Howard M. J., Hatahet F. S., Salo K. E., Kauppila A., Kellokumpu S., Ruddock L. W. ERp27, a new non-catalytic endoplasmic reticulum-located human protein disulfide isomerase family member, interacts with ERp57. J. Biol. Chem. 2006;281:33727–33738. doi: 10.1074/jbc.M604314200. [DOI] [PubMed] [Google Scholar]
- 10.Wishart D. S., Sykes B. D. Chemical shifts as a tool for structure determination. Methods Enzymol. 1994;239:363–392. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- 11.Piotto M., Saudek V., Sklenář V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 12.States D. J., Haberkorn R. A., Ruben D. J. A two-dimensional nuclear Overhauser experiment with pure absorption phase in 4 quadrants. J. Magn. Reson. 1982;48:286–292. [Google Scholar]
- 13.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 14.Wim F. V., Wayne B., Tim J. S., Rasmus H. F., Anne P., Miguel L., Eldon L. U., John L. M., John I., Ernest D. L. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins: Struct., Funct., Bioinf. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 15.Barbato G., Ikura M., Kay L. E., Pastor R. W., Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992;31:5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- 16.Neuhaus D., Vanmierlo C. P. M. Measurement of heteronuclear NOE enhancements in biological macromolecules: a convenient pulse sequence for use with aqueous solutions. J. Magn. Reson. 1992;100:221–228. [Google Scholar]
- 17.Lipari G., Szabo A. Model-free approach to the interpretation of nuclear magnetic-resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 18.Lipari G., Szabo A. Model-free approach to the interpretation of nuclear magnetic-resonance relaxation in macromolecules. 2. Analysis of experimental results. J. Am. Chem. Soc. 1982;104:4559–4570. [Google Scholar]
- 19.Palmer A. G., Rance M., Wright P. E. Intramolecular motions of a zinc finger DNA-binding domain from Xfin characterized by proton-detected natural abundance C-12 heteronuclear NMR spectroscopy. J. Am. Chem. Soc. 1991;113:4371–4380. [Google Scholar]
- 20.Cornilescu G., Delaglio F., Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 21.Rieping W., Habeck M., Bardiaux B., Bernard A., Malliavin T. E., Nilges M. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- 22.Brünger A., Adams P., Clore G., DeLano W., Gros P., Grosse-Kunstleve R., Jiang J., Kuszewski J., Nilges M., Pannu N., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1998;1:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Waters L. C., Veverka V., Bohm M., Schmedt T., Choong P. T., Muskett F. W., Klempnauer K. H., Carr M. D. Structure of the C-terminal MA-3 domain of the tumour suppressor protein Pdcd4 and characterization of its interaction with eIF4A. Oncogene. 2007;26:4941–4950. doi: 10.1038/sj.onc.1210305. [DOI] [PubMed] [Google Scholar]
- 24.Williamson R. A., Carr M. D., Frenkiel T. A., Feeney J., Freedman R. B. Mapping the binding site for matrix metalloproteinase on the N-terminal domain of the tissue inhibitor of metalloproteinases-2 by NMR chemical shift perturbation. Biochemistry. 1997;36:13882–13889. doi: 10.1021/bi9712091. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs D. J., Rader A. J., Kuhn L. A., Thorpe M. F. Protein flexibility predictions using graph theory. Proteins. 2001;44:150–165. doi: 10.1002/prot.1081. [DOI] [PubMed] [Google Scholar]
- 26.Suhre K., Sanejouand Y. H. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004;32:W610–W614. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wells S., Menor S., Hespenheide B., Thorpe M. F. Constrained geometric simulation of diffusive motion in proteins. Phys. Biol. 2005;2:S127–S136. doi: 10.1088/1478-3975/2/4/S07. [DOI] [PubMed] [Google Scholar]
- 28.Jimenez-Roldan J. E., Freedman R. B., Römer R. A., Wells S. A. Rapid simulation of protein motion: merging flexibility, rigidity and normal mode analyses. Phys. Biol. 2012;9:016008. doi: 10.1088/1478-3975/9/1/016008. [DOI] [PubMed] [Google Scholar]
- 29.Wells S. A., Sartbaeva A. Template-based geometric simulation of flexible frameworks. Materials. 2012;5:415–431. doi: 10.3390/ma5030415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Wells S. A., Jimenez-Roldan J. E., Römer R. A., Zhao Y., Sadler P. J., O’Connor P. B. Protein flexibility is key to cisplatin crosslinking in calmodulin. Protein Sci. 2012;21:1269–1279. doi: 10.1002/pro.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rowe M. L., Ruddock L. W., Kelly G., Schmidt J. M., Williamson R. A., Howard M. J. Solution structure and dynamics of ERp18, a small endoplasmic reticulum resident oxidoreductase. Biochemistry. 2009;48:4596–4606. doi: 10.1021/bi9003342. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 33.Laskowski R. A., Rullmann J. A. C., MacArthur M. W., Kaptein R., Thornton J. M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 34.Holm L., Kaariainen S., Rosenstrom P., Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozlov G., Maattanen P., Schrag J. D., Pollock S., Cygler M., Nagar B., Thomas D. Y., Gehring K. Crystal structure of the bb’ domains of the protein disulfide isomerase ERp57. Structure. 2006;14:1331–1339. doi: 10.1016/j.str.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 36.Kemmink J., Dijkstra K., Mariani M., Scheek R. M., Penka E., Nilges M., Darby N. J. The structure in solution of the b domain of protein disulfide isomerase. J. Biomol. NMR. 1999;13:357–368. doi: 10.1023/a:1008341820489. [DOI] [PubMed] [Google Scholar]
- 37.Wang S., Trumble W. R., Liao H., Wesson C. R., Dunker A. K., Kang C. Crystal structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum. Nat. Struct. Biol. 1998;5:476–483. doi: 10.1038/nsb0698-476. [DOI] [PubMed] [Google Scholar]
- 38.Kozlov G., Määttänen P., Schrag J. D., Hura G. L., Gabrielli L., Cygler M., Thomas D. Y., Gehring K. Structure of the noncatalytic domains and global fold of the protein disulfide isomerase ERp72. Structure. 2009;17:651–659. doi: 10.1016/j.str.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 39.Barak N. N., Neumann P., Sevvana M., Schutkowski M., Naumann K., Malesevic M., Reichardt H., Fischer G., Stubbs M. T., Ferrari D. M. Crystal structure and functional analysis of the protein disulfide isomerase-related protein ERp29. J. Mol. Biol. 2009;385:1630–1642. doi: 10.1016/j.jmb.2008.11.052. [DOI] [PubMed] [Google Scholar]
- 40.Wang L., Wang L., Vavassori S., Li S., Ke H., Anelli T., Degano M., Ronzoni R., Sitia R., Sun F., Wang C.-c. Crystal structure of human ERp44 shows a dynamic functional modulation by its carboxy-terminal tail. EMBO Rep. 2008;9:642–647. doi: 10.1038/embor.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen V. D., Wallis K., Howard M. J., Haapalainen A. M., Salo K. E., Saaranen M. J., Sidhu A., Wierenga R. K., Freedman R. B., Ruddock L. W., Williamson R. A. Alternative conformations of the x region of human protein disulphide-isomerase modulate exposure of the substrate binding b’ domain. J. Mol. Biol. 2008;383:1144–1155. doi: 10.1016/j.jmb.2008.08.085. [DOI] [PubMed] [Google Scholar]
- 42.Wallis A. K., Sidhu A., Byrne L. J., Howard M. J., Ruddock L. W., Williamson R. A., Freedman R. B. The ligand-binding b’ domain of human protein disulphide-isomerase mediates homodimerization. Protein Sci. 2009;18:2569–2577. doi: 10.1002/pro.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kober F. -X., Koelmel W., Kuper J., Drechsler J., Mais C., Hermanns H. H., Schindelin H. The crystal structure of the protein disulfide isomerase family members ERp27 provide insights into its substrate-binding capabilities. J. Mol. Chem. 2012 doi: 10.1074/jbc.M112.410522. doi:10.1074/jbc.M112.410522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lappi A. K., Lensink M. F., Alanen H. I., Salo K. E., Lobell M., Juffer A. H., Ruddock L. W. A conserved arginine plays a role in the catalytic cycle of the protein disulphide isomerases. J. Mol. Biol. 2004;335:283–295. doi: 10.1016/j.jmb.2003.10.051. [DOI] [PubMed] [Google Scholar]
- 45.Jansen G., Määttänen P., Denisov A. Y., Scarffe L., Schade B., Balghi H., Dejgaard K., Chen L. Y., Muller W. J., Gehring K., Thomas D. Y. An interaction map of ER chaperones and foldases. Mol. Cell. Proteomics. 2012;11:710–723. doi: 10.1074/mcp.M111.016550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koradi R., Billeter M., Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.