Abstract
High-throughput screening (HTS) assays that measure the in vitro toxicity of environmental compounds have been widely applied as an alternative to in vivo animal tests of chemical toxicity. Current HTS studies provide the community with rich toxicology information that has the potential to be integrated into toxicity research. The available in vitro toxicity data is updated daily in structured formats (e.g., deposited into PubChem and other data-sharing web portals) or in an unstructured way (papers, laboratory reports, toxicity Web site updates, etc.). The information derived from the current toxicity data is so large and complex that it becomes difficult to process using available database management tools or traditional data processing applications. For this reason, it is necessary to develop a big data approach when conducting modern chemical toxicity research. In vitro data for a compound, obtained from meaningful bioassays, can be viewed as a response profile that gives detailed information about the compound’s ability to affect relevant biological proteins/receptors. This information is critical for the evaluation of complex bioactivities (e.g., animal toxicities) and grows rapidly as big data in toxicology communities. This review focuses mainly on the existing structured in vitro data (e.g., PubChem data sets) as response profiles for compounds of environmental interest (e.g., potential human/animal toxicants). Potential modeling and mining tools to use the current big data pool in chemical toxicity research are also described.
Introduction
With the great progress of combinatorial chemistry since the 1990s, large chemical libraries became the major source of modern drug discovery procedures.1,2 Over the past 10 years, this effort also stimulated the development of high-throughput screening (HTS) techniques.3,4 Traditional toxicity testing protocols using animal models are expensive and time-consuming. Because of the urgent need to use alternative methods in toxicity studies, the U.S. National Research Council (NRC) outlined a new vision and strategies for the increased use of in vitro technologies for chemical risk assessment.5 With its low cost and short testing time, HTS has been viewed as the potential alternative to animal models. In contrast with virtual screening techniques (e.g., QSAR or docking), HTS does not require prior knowledge about potential hits or 3D structures of involved molecular targets.
HTS is a process that screens thousands to millions of compounds using a rapid and standardized protocol. Current HTS techniques are usually combined with robotic methods. Parallel data processing and biological assay miniaturization have become more and more popular in toxicology studies, as they greatly reduce the cost of experimental testing.3,6 It is understandable that some popular compounds, especially those of toxicity interest (e.g., known human toxicants), have been tested multiple times and in many different bioassays. For this reason, the assay response data from multiple resources and/or multiple testing protocols could be viewed as the response profile of the compounds being tested. Figure 1 shows the current data construction of compounds in toxicity testing. Compared to the limited amount of historical animal toxicity data, the chemical–response data space obtained from HTS is much more complex and keeps growing daily.
Figure 1.
Construction of big data for chemical toxicity research.
The term big data describes a collection of data sets that are so large and complex that they are too difficult to process by traditional data analysis tools. Originally, the big data focus was on advanced data storage and handling techniques, such as cloud-based computing or high-speed heterogeneous computational environments.7 Currently, the problem of big data is gaining increasing recognition in clinical studies and other research areas driven by biological data.8,9 Clearly, the progress of HTS and relevant data sharing projects has moved modern chemical toxicity research into the big data era. The need of novel techniques, including data mining/generation, curation, storage, and management, brings new challenges and opportunities to the current toxicology community.
High-Throughput Screening in Chemical Toxicology
There were several important movements by regulatory agencies for the development of HTS assays that are potential alternatives to animal testing. The NIH Roadmap for medical research was launched in 2004.10 Fueled by this initiative, several molecular library screening centers were funded by the NIH Molecular Libraries Common Fund Program. The National Institutes of Health (NIH) Chemical Genomics Center (NCGC), which is now part of the National Center for Advancing Translational Sciences (NCATS), was one of them. In 2005, the National Toxicology Program (NTP) and NCGC started a collaboration to (1) develop a chemical library suitable for quantitative HTS (qHTS), (2) develop HTS assays potentially informative for in vivo toxicity effects, and (3) experimentally test the chemical library by these qHTS assays.4 This was one of the early efforts to systemically use the HTS technique within toxicology studies. During the same period, there were many other HTS projects that were performed by other research groups.11−15 Although these studies were not specifically designed for chemical toxicity, but for drug discovery and other areas, these HTS efforts also generated numerous bioassay data for large chemical libraries. For the early days of HTS development, several reviews are available.3,16−20
In 2006, the U.S. Environmental Protection Agency (EPA) initiated a research program named toxicity forecaster (ToxCast). The goal of this program was to develop methods for utilizing in vitro toxicity tests and various toxicogenomics technologies to quickly evaluate the toxic potential of chemicals and to prioritize candidates for future animal testing.16 Phase I of ToxCast employed a chemical library of 300 unique compounds, most of which were chemicals for agricultural use, such as pesticides, and had relevant animal toxicity testing results available.21 Around 500 cell-free or cell-based assays were used to screen this chemical library. From these, over 600 in vitro end points were measured for each chemical, generating over 200 000 concentration–response data points. In ToxCast phase II, another 767 compounds, including some failed pharmaceuticals, were screened using around 700 HTS assays.22
In 2008, another big collaborative program, called Toxicity Testing in the 21st century (Tox21), was launched by NTP, NCGC, and EPA,23−25 joined later by the U.S. Food and Drug Administration (FDA). The Tox21 collaboration brought together its partners’ expertise in the areas of experimental toxicology, in vitro assays, and informatics.25 The target chemical library of Tox21 screening contains over 8000 unique compounds, including commercial compounds, pesticides, and marketed pharmaceuticals.22 Screening of this extensive chemical library commenced in 2011 at NCGC, with a throughput capacity of approximately 25 assays per year.
Current Toxicity Data Sharing Projects
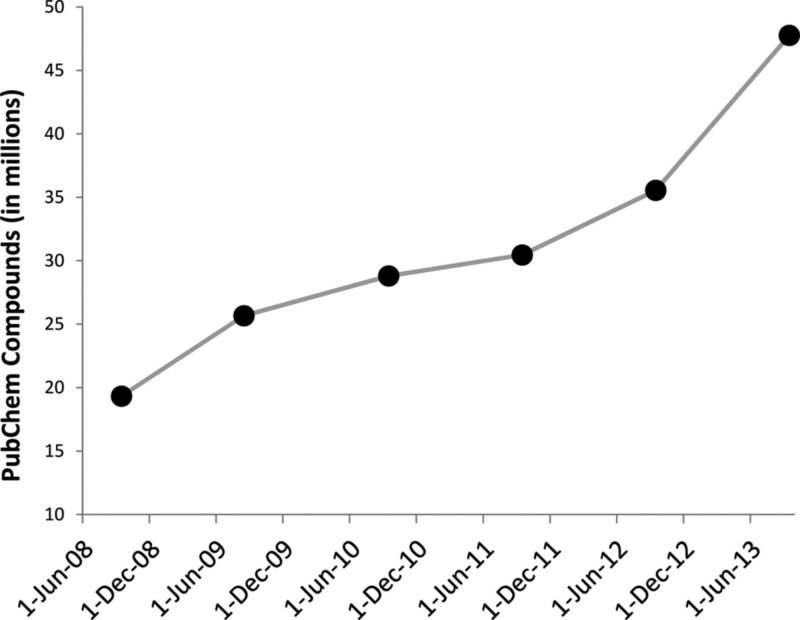
Table 1 summarizes the above data sharing projects and other relevant toxicity sources. Facilitated by the combined efforts of HTS and combinatorial chemical synthesis, modern screening programs produced enormous amounts of biological data, especially the chemical responses on specific targets.26 As a result, several data sharing projects, in parallel to the generation of HTS toxicity data, were also initiated during the past 10 years. For example, PubChem is a public repository for chemical structures and their biological properties.27,28 Most of the HTS data (e.g., those generated from the above toxicology programs) were shared through PubChem. Figure 2 shows the yearly increase of PubChem compounds.29−34 Over the past 5 years, the number of PubChem compounds increased from 19 million in September 200829 to 48 million in September 2013.33 During the same period, the number of bioassays that were deposited into PubChem increased from 1197 in September 200829 to over 700 000, resulting in over five terabytes of data, in September 2013.33
Table 1. Available Public Toxicity Data Resources.
| name | general information | data description |
|---|---|---|
| PubChem27,28 | Around 47 million compounds, over 700 000 bioassays, over 13 billion data points | Toxicity, pharmaceutical, genomics, and literature data |
| ChEMBL88 | Over 600 000 compounds, 3.3 million bioassay readout data | Literature data |
| ACToR89,90 | The toxicity results from 100 various data resources | Both in vitro and in vivo toxicity data |
| ToxNET91 | Over 50 000 environmental compounds from 16 different resources | Both in vitro and in vivo toxicity data |
| SEURAT92 | Over 5500 cosmetic-type compounds in the current COSMOS database web portal | Animal toxicity data |
| CTD37−40 | Over 13 000 compounds, over 32 000 genes, over 6000 diseases | Compound, gene, and disease relationships |
| CEBS35 | About 10 000 toxicity bioassays from various sources | Gene expression data |
| DrugMatrix93 | About 600 drug molecules and 10 000 genes | Gene expression data |
| Cmap94 | About 1300 compounds and 7000 genes | Gene expression data |
Figure 2.
Increase of compounds recorded in PubChem within 5 years (from September 2008 to September 2013).
The Chemical Effects in Biological Systems (CEBS) database developed by the National Institute of Environmental Health Sciences (NIEHS) is now the public repository for all NTP conventional toxicology and carcinogenicity data as well as NCGC HTS data.35,36 Along with the Comparative Toxicogenomics Database (CTD) at Mount Desert Island Biological Laboratory, CEBS aims to promote comparative studies of genes and proteins across species.37−40
All of these data together can be viewed as the current toxicity big data pool, which contains over 70 million compounds, over 1 million bioassays, and around 50 billion data points. The information within this pool is being updated daily and increases rapidly (e.g., the progress of PubChem, as shown in Figure 2).
Characterizing Toxicants by Multiple Bioassay Data
The direct consequence of the HTS testing efforts over the past 10 years is the massive amount of available biological data for organic compounds, especially those of environmental interest. A significant number of those compounds have been tested multiple times. For example, Table 2 shows 20 toxicants obtained from the Integrated Risk Information System (IRIS) database.41 On the basis of the search result on PubChem,42 these toxicants were reported to be tested in hundreds of PubChem bioassays. For example, chlordecone (CAS 143-50-0), an insecticide banned from the market, showed active responses in 328 bioassays (Table 2). Other toxicants have similarly rich response information in PubChem (Table 2).
Table 2. Twenty Human Toxicants with Their Relevant PubChem Bioassay Responses.
| chemicals | CAS | no. of active responses | no. of inactive responses |
|---|---|---|---|
| Chlordecone | 143-50-0 | 328 | 539 |
| Toxaphene | 8001-35-2 | 294 | 112 |
| Hexachlorocyclopentadiene | 77-47-4 | 208 | 262 |
| Dichlorvos | 62-73-7 | 181 | 633 |
| Pentachlorophenol | 87-86-5 | 95 | 690 |
| Heptachlor | 76-44-8 | 85 | 624 |
| DDT, p,p′- | 50-29-3 | 76 | 386 |
| DDD, p,p′- | 72-54-8 | 70 | 186 |
| Endosulfan | 115-29-7 | 65 | 259 |
| Naphthalene | 91-20-3 | 61 | 890 |
| DDD, o,p′- | 53-19-0 | 61 | 964 |
| 1,4-Dichlorobenzene | 106-46-7 | 57 | 362 |
| 4,6-Dinitro-o-cresol | 534-52-1 | 57 | 213 |
| Phenol | 108-95-2 | 53 | 518 |
| Chlorpyrifos | 2921-88-2 | 48 | 739 |
| Methoxychlor | 72-43-5 | 47 | 710 |
| 2,4-Dinitrophenol | 51-28-5 | 46 | 672 |
| Tetrachlorophenol | 25167-83-3 | 45 | 515 |
| Benzo(a)pyrene | 50-32-8 | 39 | 358 |
| 4,4′-Methylenebis(2-chloroaniline) | 101-14-4 | 32 | 431 |
Although there are notable cases when individual in vitro assays are predictive of in vivo outcomes (e.g., assays for skin sensitization43 and endocrine disruption44), for many complex toxicity end points, single-assay data is not sufficient. The multiple bioassay data of a single compound can be viewed as its biological profile, reflecting its interactions. Profiling compounds, especially the toxicants, to study their toxicity potential is the most straightforward way to use the available bioassay data. ToxCast phase I screened over 300 unique compounds, mostly food pesticides, in 467 bioassays. The resulting data was used to profile screened compounds for their potential to induce carcinogenicity,45 developmental toxicity,46,47 reproductive toxicity,48 and endocrine disruption.44,49 In these studies, good correlations could be found between some bioassays and animal toxicity. For example, a model was developed by using peroxisome proliferator-activated receptor signaling assays to predict rodent carcinogenicity of 33 compounds.45 However, although ToxCast is the most comprehensive and biggest toxicity screening project so far, using all of the available ToxCast assay data to develop a global predictive model for chemical toxicants is still questionable.50
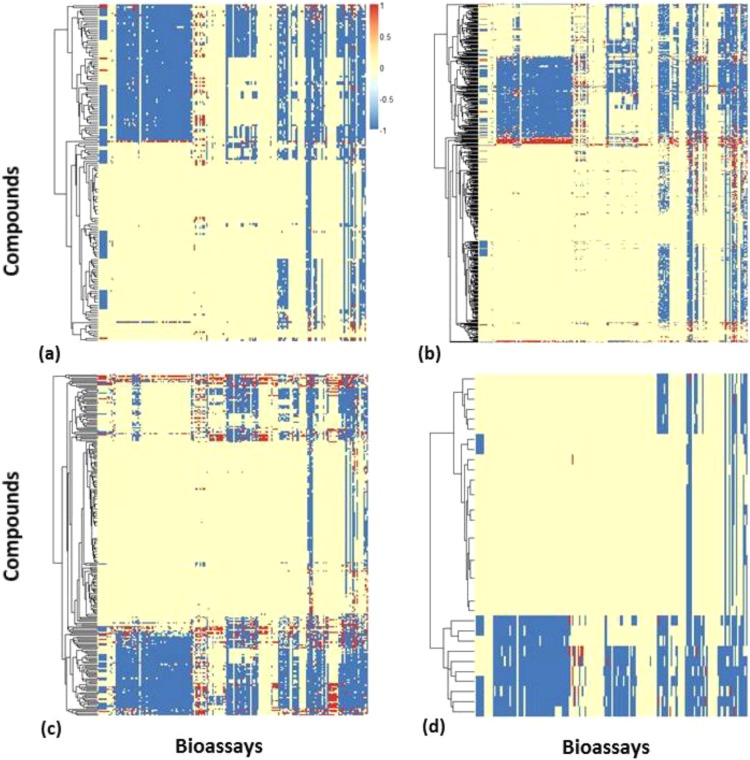
Besides the bioassay data generated by the ToxCast program, the Tox21 compounds have been tested in other screening projects (e.g., NCI60).51 In the current big data era, the bioassay response profile can be very large for some compounds (e.g., those well-known toxicants shown in Table 1). The initial response space can be large, complex, and unorganized. For example, if we searched PubChem bioassay data for the 962 ToxCast compounds, we could identify 193 PubChem assays that have at least five actives among these compounds (accessed in December of 2013). By classifying the ToxCast compounds into four major categories,52 we could compare the response profiles, consisting of the 193 PubChem assays, of different types of compounds (Figure 3). Compared to phthalate plasticizers, the pharmaceutical compounds and pesticides have been studied in most bioassays, and the active response ratios are relatively high.
Figure 3.
Response spaces of different ToxCast compound categories represented by the data obtained from 193 PubChem bioassays: (a) 171 consumer use chemicals (not including pharmaceuticals or pesticides), (b) 470 pesticides, (c) 245 pharmaceuticals, and (d) 34 phthalate plasticizers and alternatives.
It is understandable that most areas within the initial response map are either “no testing” or “inconclusive” (indicating that no conclusion could be made about the relevant compounds based on the testing data) because many bioassays have been applied only to a small portion of this large chemical set. Furthermore, the nature of HTS assays, many of which represent very specific interactions, results in a biased distribution of responses for the target chemicals (many more “inactive” than “active” data entries). Because not all of the bioassay data are relevant or useful for a particular type of toxicity, additional rational selection steps are needed to select useful information from the bulk of available big data.
The Use of Bioassay Data To Prioritize Animal Toxicants
There have been some studies that use current bioassay data to identify likely animal toxicants and/or prioritize them for future experimental animal testing. For example, the currently available ToxCast bioassays have been organized into a global scoring system, called Toxicological Priority Index (ToxPi), to identify potential toxicants by their responses in these assays.44,47,49,52,53 ToxPi, which is a dimensionless index score, was calculated as a weighted combination of all data sources that represents a formalized, rational integration of information from different types of ToxCast bioassays results.49 Furthermore, toxicity pathways could also be generated, linking relevant bioassays together by analyzing their biological targets.54,55 ToxCast phase I is the first time there has been a big data effort to generate and systemically use large scale bioassay data in chemical toxicity studies. In ToxCast phase II, similar efforts continued with the new 767 target chemicals, including 111 failed pharmaceutical drug molecules.52 In the recent Tox21 program, the results obtained from ToxCast were used to select the most useful bioassays as the testing battery for a much larger database.24,56,57
There are other research groups and agencies that use bioassays to study various in vivo toxicities, such as acute toxicity,58−61 developmental toxicity,62 and drug–drug interactions.63 One example is the AcuteTox collaborative project initiated within the European Union. Its purpose is to develop alternative testing strategies that could replace animal testing for predicting human acute oral systemic toxicity.61,64−68 Similar to ToxCast, AcuteTox generated large scale in vitro toxicity data from multiple bioassays.65 All of these efforts contributed to the initial pool of big data for chemical toxicants.
The authors of this review have also utilized bioassay data to predict animal toxicity of organic compounds. In the first two of our studies, multiple qHTS data from NCGC bioassays were used as biological descriptors to develop predictive models for various animal toxicity end points.69,70 The models with hybrid (combination of chemical and biological) descriptors showed better predictivity than the traditional quantitative structure–activity relationship (QSAR) models using only chemical descriptors. In another study, the biological descriptors obtained from toxicogenomics data were used to model animal hepatotoxicity.71
Progress of Toxicology in the Big Data Era
The clear limitation of extrapolating results from in vitro assays to a whole organism is that each in vitro assay generally considers only one or several target sites rather than a comprehensive organism consisting of hundreds of potential targets.72,73 The practical solution is to form a large battery of diverse in vitro assays for a specific animal toxicity, such as the ToxCast strategy.16,22 In the toxicant profiling studies described above, each project was limited to the use of the data generated by its own HTS assays. This lack of data integration across multiple related toxicity databases is clearly a big and open issue. How to integrate large scale data sets from various sources is a key question that needs to be addressed in the current big data scenario. To realize this goal, novel data storage and management methods need to be developed. For example, it is clear that all HTS assays contain certain noise associated with experimental data. Experimental noise varies from assay to assay, but it is rarely less than 15% as is seen, for example, for the Ames mutagenicity test.74 Even in the most recent HTS project, e.g., ToxCast, random errors still exists in the original dose–response data.75 To solve this problem, quality control (QC) review is a necessary step to remove experimental errors. Furthermore, automatic data processing methods have been developed to identify hits (i.e., actives or toxic compounds) from quality-assured HTS data and to increase the data dissemination and reproducibility.76 Future data management tools, which are specifically useful for big data analysis, should be able to normalize the HTS data from different sources into benchmark end points. Currently, some preliminary studies (e.g., CurveP70) have been reported.
In 2007, the NRC envisioned a new paradigm in which biologically important perturbations in key toxicity pathways would be evaluated with new approaches in modern toxicology studies.5 In this book, the NRC defined the toxicity pathway as a cellular response pathway that would result in an adverse health effect when sufficiently perturbed. This was the first time the concept of toxicity pathways, and emphasis on the use of pathways to explain the complex toxicity mechanisms, was introduced. In the above section, some preliminary studies of toxicity pathway studies (e.g., ToxCast project) were introduced.16 Driven by the urgent requirements of the mode of action (MOA) analysis in toxicity studies, the Organization for Economic Development and Cooperation (OECD) has funded the recent development of adverse outcome pathway (AOP) development, which greatly advanced the area of ecotoxicology.77 The AOP models have been successfully applied to skin sensitization evaluations.78
To incorporate more toxicity data, especially the daily updated big data pool as shown in Table 1, into toxicity models (e.g., toxicity pathways), novel data mining tools need to be developed to extract useful data from different resources. Wild and his co-workers developed a framework called Chem2Bio2RDF to link several data resources, such as DrugBank, PubChem, ChEMBL, and others.79 This framework, including other similar data mining tools developed in the same group, was used to create complex systems biology models (e.g., for drug adverse effects).79−82 Recently, Fourches et al. reported a newly developed software, named HTS Navigator, to extract, visualize, and analyze HTS data from various resources.83 Among emerging approaches specific to big data analysis, two key developments are semantic text-mining,84 which helps bridge the language barrier across data sources with different ontologies, and large scale network analysis, to identify groups of related entities for further processing.85
In the current big data scenario, the most critical issue is to identify useful in vitro data. In principle, this could be done by a human expert using the knowledge of the design and quality of each particular bioassay (e.g., confidence score assigned during manual curation to each assay in ChEMBL). We, however, believe that data-driven approaches would provide more efficient ways to accomplish this. One possible strategy is to select assays based on their in vitro–in vivo relationships. Because there are multiple mechanisms behind each toxicity phenotype, each bioassay is likely to show only partial correlation with in vivo effect. For example, if a bioassay represents a receptor that belongs to a toxicity pathway relevant to the target animal toxicity, then this bioassay should provide useful information, such as receptor/pathway perturbation. However, if compounds show inactive results in a particular bioassay, then they can still be toxic because they may bind to other target sites (Figure 4). Indeed, our previous study showed that the bioassay results have a low false-positive rate to predict the relevant animal toxicity.86 The false-negative rate, on the contrary, is high. On the basis of this study, we recently developed an automatic bioassay system to evaluate and extract the relevant bioassay data based on the in vitro–in vivo relationship. In our most recent publications, we developed an approach that could identify the most relevant bioassays as alternatives for animal toxicity models.87 In this study, we initially extracted over 12 000 bioassays and around half million data points for 2000 compounds with animal toxicity results. By using the in vitro–in vivo relationship, which was described in Figure 4, as the criteria, we developed a novel approach to select the most relevant bioassays to acute rat toxicity. The top 47 bioassays could be used to prioritize animal toxicants for future experimental testing in animals. Interestingly, the top potential animal toxicants, which have similar active responses in these bioassays, have dissimilar chemical structures.
Figure 4.
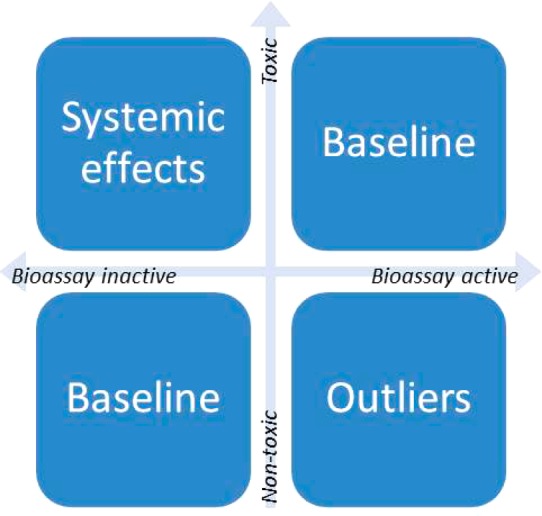
A potential in vitro–in vivo relationship in toxicology studies.
Conclusions
Current innovative technologies enable rapid synthesis and high-throughput screening of large libraries of compounds. Daily updated toxicity bioassay data have transformed current toxicology studies into big data analysis. Fueled by the recent input from the U.S. and European governments, there are many ongoing data-generation and data-sharing programs, accompanied by the development of data curation and automated data management (e.g., “EMBL-EBI” KNIME workflow nodes for ChEMBL, “rpubchem” R package to PubChem) approaches that could be used to sample HTS data in meaningful formats to facilitate chemical toxicity studies. New scoring and modeling methods are also under way to take advantage of the massive amount of bioassay data. Although the use of bioassay data in most current toxicological research projects is still limited to a small portion of well-sampled HTS data, several novel approaches have been reported to be able to access and integrate multiple bioassay data resources to profile toxicants. Under the current big data scenario, it is expected that modern toxicology research will be able to better estimate the systemic effects of compounds on whole organisms and to translate this into better informed regulation of toxicants for animals and humans.
Glossary
Abbreviations
- ACToR
aggregated computational toxicology resource
- AOP
adverse outcome pathway
- CEBS
chemical effects in biological systems
- cmap
connectivity map
- CTD
comparative toxicogenomics database
- EBI
European Bioinformatics Institute
- EPA
Environmental Protection Agency
- FDA
Food and Drug Administration
- HTS
high-throughput screening
- IRIS
integrated risk information system
- MOA
mode of action
- NCATS
National Center for Advancing Translational Sciences
- NCCT
National Center for Computational Toxicology
- NCGC
NIH Chemical Genomics Center
- NIEHS
National Institute of Environmental Health Sciences
- NIH
National Institutes of Health
- NLM
National Library of Medicines
- NRC
National Research Council
- NTP
National Toxicology Program
- OECD
Organization for Economic Development and Cooperation
- QC
quality control
- qHTS
quantitative high-throughput screening
- QSAR
quantitative structure–activity relationship
- SEURAT
safety evaluation ultimately replacing animal testing
- SIS
specialized information services
- Tox21
toxicity testing in the 21st century
- ToxCast
toxicity forecaster
- ToxPi
toxicological priority index
This work was supported, in part, by the National Institute of Environmental Health Sciences of the National Institutes of Health under award no. R15ES023148 and the Colgate–Palmolive Grant for Alternative Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Patel D. V.; Gordon E. M. (1996) Applications of small-molecule combinatorial chemistry to drug discovery. Drug Discovery Today 1, 134–144. [Google Scholar]
- Schreiber S. L. (2000) Target-oriented and diversity-oriented organic synthesis in drug discovery. Science 287, 1964–1969. [DOI] [PubMed] [Google Scholar]
- Malo N.; Hanley J. A.; Cerquozzi S.; Pelletier J.; Nadon R. (2006) Statistical practice in high-throughput screening data analysis. Nat. Biotechnol. 24, 167–175. [DOI] [PubMed] [Google Scholar]
- Inglese J.; Auld D. S.; Jadhav A.; Johnson R. L.; Simeonov A.; Yasgar A.; Zheng W.; Austin C. P. (2006) Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc. Natl. Acad. Sci. U.S.A. 103, 11473–11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee on Toxicity Testing and Assessment of Environmental Agents, N.R.C. (2007) Toxicity testing in the 21st century: a vision and a strategy, The National Academies Press, Washington, DC. [Google Scholar]
- Szymański P.; Markowicz M.; Mikiciuk-Olasik E. (2012) Adaptation of high-throughput screening in drug discovery—toxicological screening tests. Int. J. Mol. Sci. 13, 427–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E. E.; Linderman M. D.; Sorenson J.; Lee L.; Nolan G. P. (2011) Cloud and heterogeneous computing solutions exist today for the emerging big data problems in biology. Nat. Rev. Genet. 12, 224. [DOI] [PubMed] [Google Scholar]
- Marx V. (2013) Biology: the big challenges of big data. Nature 498, 255–260. [DOI] [PubMed] [Google Scholar]
- Swarup V.; Geschwind D. H. (2013) Alzheimer’s disease: from big data to mechanism. Nature 500, 34–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin C. P.; Brady L. S.; Insel T. R.; Collins F. S. (2004) NIH molecular libraries initiative. Science 306, 1138–1139. [DOI] [PubMed] [Google Scholar]
- Fliri A. F.; Loging W. T.; Thadeio P. F.; Volkmann R. A. (2005) Analysis of drug-induced effect patterns to link structure and side effects of medicines. Nat. Chem. Biol. 1, 389–397. [DOI] [PubMed] [Google Scholar]
- Fliri A. F.; Loging W. T.; Thadeio P. F.; Volkmann R. A. (2005) Biospectra analysis: model proteome characterizations for linking molecular structure and biological response. J. Med. Chem. 48, 6918–6925. [DOI] [PubMed] [Google Scholar]
- Fliri A. F.; Loging W. T.; Thadeio P. F.; Volkmann R. A. (2005) Biological spectra analysis: linking biological activity profiles to molecular structure. Proc. Natl. Acad. Sci. U.S.A. 102, 261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. C.; Delaney J. S.; Robinson M. P.; Rice M. J. (2005) Targeting chemical inputs and optimizing hts for agrochemical discovery. Comb. Chem. High Throughput Screening 8, 577–587. [DOI] [PubMed] [Google Scholar]
- Janzen W. P.; Hodge C. N. (2006) A chemogenomic approach to discovering target-selective drugs. Chem. Biol. Drug Des. 67, 85–86. [DOI] [PubMed] [Google Scholar]
- Dix D. J.; Houck K. A.; Martin M. T.; Richard A. M.; Setzer R. W.; Kavlock R. J. (2007) The toxcast program for prioritizing toxicity testing of environmental chemicals. Toxicol. Sci. 95, 5–12. [DOI] [PubMed] [Google Scholar]
- Macarron R.; Banks M. N.; Bojanic D.; Burns D. J.; Cirovic D. A.; Garyantes T.; Green D. V. S.; Hertzberg R. P.; Janzen W. P.; Paslay J. W.; Schopfer U.; Sittampalam G. S. (2011) Impact of high-throughput screening in biomedical research. Nat. Rev. Drug Discovery 10, 188–195. [DOI] [PubMed] [Google Scholar]
- Macarrón R.; Hertzberg R. P. (2011) Design and implementation of high throughput screening assays. Mol. Biotechnol. 47, 270–285. [DOI] [PubMed] [Google Scholar]
- Stein R. L. (2003) High-throughput screening in academia: the harvard experience. J. Biomol. Screen. 8, 615–619. [DOI] [PubMed] [Google Scholar]
- Macarron R. (2006) Critical review of the role of hts in drug discovery. Drug Discovery Today 11, 277–279. [DOI] [PubMed] [Google Scholar]
- Judson R. S.; Houck K. A.; Kavlock R. J.; Knudsen T. B.; Martin M. T.; Mortensen H. M.; Reif D. M.; Rotroff D. M.; Shah I.; Richard A. M.; Dix D. J. (2010) In vitro screening of environmental chemicals for targeted testing prioritization: the toxcast project. Environ. Health Perspect. 118, 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavlock R.; Chandler K.; Houck K.; Hunter S.; Judson R.; Kleinstreuer N.; Knudsen T.; Martin M.; Padilla S.; Reif D.; Richard A.; Rotroff D.; Sipes N.; Dix D. (2012) Update on EPA’s toxcast program: providing high throughput decision support tools for chemical risk management. Chem. Res. Toxicol. 25, 1287–1302. [DOI] [PubMed] [Google Scholar]
- Collins F. S.; Gray G. M.; Bucher J. R. (2008) Toxicology. transforming environmental health protection. Science 319, 906–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher J. R. (2013) Regulatory forum opinion piece: Tox21 and toxicologic pathology. Toxicol. Pathol. 41, 125–127. [DOI] [PubMed] [Google Scholar]
- Shukla S. J.; Huang R.; Austin C. P.; Xia M. (2010) The future of toxicity testing: a focus on in vitro methods using a quantitative high-throughput screening platform. Drug Discovery Today 15, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klekota J.; Brauner E.; Roth F. P.; Schreiber S. L. (2006) Using high-throughput screening data to discriminate compounds with single-target effects from those with side effects. J. Chem. Inf. Model. 46, 1549–1562. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Xiao J.; Suzek T. O.; Zhang J.; Wang J.; Bryant S. H. (2009) PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 37, W623–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Bolton E.; Dracheva S.; Karapetyan K.; Shoemaker B. A.; Suzek T. O.; Wang J.; Xiao J.; Zhang J.; Bryant S. H. (2010) An overview of the PubChem BioAssay resource. Nucleic Acids Res. 38, D255–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E. W.; Barrett T.; Benson D. A.; Bryant S. H.; Canese K.; Chetvernin V.; Church D. M.; Dicuccio M.; Edgar R.; Federhen S.; Feolo M.; Geer L. Y.; Helmberg W.; Kapustin Y.; Landsman D.; Lipman D. J.; Madden T. L.; Maglott D. R.; Miller V.; Mizrachi I.; Ostell J.; Pruitt K. D.; Schuler G. D.; Sequeira E.; Sherry S. T.; Shumway M.; Sirotkin K.; Souvorov A.; Starchenko G.; Tatusova T. A.; Wagner L.; Yaschenko E.; Ye J. (2009) Database resources of the national center for biotechnology information. Nucleic Acids Res. 37, D5–D15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E. W.; Barrett T.; Benson D. A.; Bolton E.; Bryant S. H.; Canese K.; Chetvernin V.; Church D. M.; Dicuccio M.; Federhen S.; Helmberg W.; Kapustin Y.; Landsman D.; Lipman D. J.; Lu Z. Y.; Madden T. L.; Madej T.; Maglott D. R.; Marchler-Bauer A.; Miller V.; Mizrachi I.; Ostell J.; Panchenko A.; Phan L.; Pruitt K. D.; Schuler G. D.; Sequeira E.; Sherry S. T.; Shumway M.; Sirotkin K.; Slotta D.; Souvorov A.; Starchenko G.; Tatusova T. A.; Wagner L.; Wang Y. L.; Wilbur W. J.; Yaschenko E.; Ye J. A. (2011) Database resources of the national center for biotechnology information. Nucleic Acids Res. 39, D38–D51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D. L.; Barrett T.; Benson D. A.; Bryant S. H.; Canese K.; Chetvernin V.; Church D. M.; DiCuccio M.; Edgar R.; Federhen S.; Geer L. Y.; Kapustin Y.; Khovayko O.; Landsman D.; Lipman D. J.; Madden T. L.; Maglott D. R.; Ostell J.; Miller V.; Pruitt K. D.; Schuler G. D.; Sequeira E.; Sherry S. T.; Sirotkin K.; Souvorov A.; Starchenko G.; Tatusov R. L.; Tatusova T. A.; Wagner L.; Yaschenko E. (2007) Database resources of the national center for biotechnology information. Nucleic Acids Res. 35, D5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2013) Database resources of the national center for biotechnology information. Nucleic Acids Res. 41, D8–D20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2014) Database resources of the national center for biotechnology information. Nucleic Acids Res. 42, D7–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E. W.; Barrett T.; Benson D. A.; Bolton E.; Bryant S. H.; Canese K.; Chetvernin V.; Church D. M.; Dicuccio M.; Federhen S.; Helmberg W.; Kapustin Y.; Landsman D.; Lipman D. J.; Lu Z.; Madden T. L.; Madej T.; Maglott D. R.; Marchler-Bauer A.; Miller V.; Mizrachi I.; Ostell J.; Panchenko A.; Pruitt K. D.; Schuler G. D.; Sequeira E.; Sherry S. T.; Shumway M.; Sirotkin K.; Slotta D.; Souvorov A.; Starchenko G.; Tatusova T. A.; Wagner L.; Wang Y.; John W. W.; Yaschenko E.; Ye J. (2010) Database resources of the national center for biotechnology information. Nucleic Acids Res. 38, D5–D16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M.; Stasiewicz S.; Merrick B. A.; Tomer K.; Bushel P.; Paules R.; Stegman N.; Nehls G.; Yost K. J.; Johnson C. H.; Gustafson S. F.; Xirasagar S.; Xiao N.; Huang C.-C.; Boyer P.; Chan D. D.; Pan Q.; Gong H.; Taylor J.; Choi D.; Rashid A.; Ahmed A.; Howle R.; Selkirk J.; Tennant R.; Fostel J. (2008) CEBS-chemical effects in biological systems: a public data repository integrating study design and toxicity data with microarray and proteomics data. Nucleic Acids Res. 36, D892–D900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute of Environmental Health Sciences. Chemical Effects in Biological Systems (CEBS). http://www.niehs.nih.gov/research/resources/databases/cebs/index.cfm. [PubMed]
- Mattingly C. J.; Colby G. T.; Forrest J. N.; Boyer J. L. (2003) The comparative toxicogenomics database (CTD). Environ. Health Perspect. 111, 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly C. J.; Rosenstein M. C.; Davis A. P.; Colby G. T.; Forrest J. N.; Boyer J. L. (2006) The comparative toxicogenomics database: a cross-species resource for building chemical-gene interaction networks. Toxicol. Sci. 92, 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly C. J.; Colby G. T.; Rosenstein M. C.; Forrest J. N.; Boyer J. L. (2004) Promoting comparative molecular studies in environmental health research: an overview of the comparative toxicogenomics database (CTD). Pharmacogenomics J. 4, 5–8. [DOI] [PubMed] [Google Scholar]
- Mattingly C. J.; Rosenstein M. C.; Colby G. T.; Forrest J. N.; Boyer J. L. (2006) The comparative toxicogenomics database (CTD): a resource for comparative toxicological studies. J. Exp. Zool., Part A 305, 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Environmental Protection Agency. Integrated Risk Information System (IRIS). http://www.epa.gov/IRIS/ (accessed Jul 1, 2014).
- PubChem BioAssay, NCBI. http://www.ncbi.nlm.nih.gov/pcassay/ (accessed Dec 1, 2013).
- Natsch A.; Ryan C. A.; Foertsch L.; Emter R.; Jaworska J.; Gerberick F.; Kern P. (2013) A dataset on 145 chemicals tested in alternative assays for skin sensitization undergoing prevalidation. J. Appl. Toxicol. 33, 1337–1352. [DOI] [PubMed] [Google Scholar]
- Rotroff D. M.; Dix D. J.; Houck K. A.; Knudsen T. B.; Martin M. T.; McLaurin K. W.; Reif D. M.; Crofton K. M.; Singh A. V.; Xia M.; Huang R.; Judson R. S. (2013) Using in vitro high throughput screening assays to identify potential endocrine-disrupting chemicals. Environ. Health Perspect. 121, 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstreuer N. C.; Dix D. J.; Houck K. A.; Kavlock R. J.; Knudsen T. B.; Martin M. T.; Paul K. B.; Reif D. M.; Crofton K. M.; Hamilton K.; Hunter R.; Shah I.; Judson R. S. (2013) In vitro perturbations of targets in cancer hallmark processes predict rodent chemical carcinogenesis. Toxicol. Sci. 131, 40–55. [DOI] [PubMed] [Google Scholar]
- Kleinstreuer N. C.; Judson R. S.; Reif D. M.; Sipes N. S.; Singh A. V.; Chandler K. J.; Dewoskin R.; Dix D. J.; Kavlock R. J.; Knudsen T. B. (2011) Environmental impact on vascular development predicted by high-throughput screening. Environ. Health Perspect. 119, 1596–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipes N. S.; Martin M. T.; Reif D. M.; Kleinstreuer N. C.; Judson R. S.; Singh A. V.; Chandler K. J.; Dix D. J.; Kavlock R. J.; Knudsen T. B. (2011) Predictive models of prenatal developmental toxicity from toxcast high-throughput screening data. Toxicol. Sci. 124, 109–127. [DOI] [PubMed] [Google Scholar]
- Martin M. T.; Knudsen T. B.; Reif D. M.; Houck K. A.; Judson R. S.; Kavlock R. J.; Dix D. J. (2011) Predictive model of rat reproductive toxicity from toxcast high throughput screening. Biol. Reprod. 85, 327–339. [DOI] [PubMed] [Google Scholar]
- Reif D. M.; Martin M. T.; Tan S. W.; Houck K. A.; Judson R. S.; Richard A. M.; Knudsen T. B.; Dix D. J.; Kavlock R. J. (2010) Endocrine profiling and prioritization of environmental chemicals using toxcast data. Environ. Health Perspect. 118, 1714–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R. S.; Black M. B.; Li L.; Healy E.; Chu T.-M.; Bao W.; Andersen M. E.; Wolfinger R. D. (2012) A comprehensive statistical analysis of predicting in vivo hazard using high-throughput in vitro screening. Toxicol. Sci. 128, 398–417. [DOI] [PubMed] [Google Scholar]
- Shoemaker R. H. (2006) The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 6, 813–823. [DOI] [PubMed] [Google Scholar]
- Sipes N. S.; Martin M. T.; Kothiya P.; Reif D. M.; Judson R. S.; Richard A. M.; Houck K. A.; Dix D. J.; Kavlock R. J.; Knudsen T. B. (2013) Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem. Res. Toxicol. 26, 878–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif D. M.; Sypa M.; Lock E. F.; Wright F. A.; Wilson A.; Cathey T.; Judson R. R.; Rusyn I. (2013) ToxPi GUI: an interactive visualization tool for transparent integration of data from diverse sources of evidence. Bioinformatics 29, 402–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R. S.; Kavlock R. J.; Setzer R. W.; Hubal E. A. C.; Martin M. T.; Knudsen T. B.; Houck K. A.; Thomas R. S.; Wetmore B. A.; Dix D. J. (2011) Estimating toxicity-related biological pathway altering doses for high-throughput chemical risk assessment. Chem. Res. Toxicol. 24, 451–462. [DOI] [PubMed] [Google Scholar]
- Judson R. S.; Mortensen H. M.; Shah I.; Knudsen T. B.; Elloumi F. (2012) Using pathway modules as targets for assay development in xenobiotic screening. Mol. BioSyst. 8, 531–542. [DOI] [PubMed] [Google Scholar]
- Betts K. S. (2013) Tox21 to date: steps toward modernizing human hazard characterization. Environ. Health Perspect. 121, A228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attene-Ramos M. S.; Miller N.; Huang R.; Michael S.; Itkin M.; Kavlock R. J.; Austin C. P.; Shinn P.; Simeonov A.; Tice R. R.; Xia M. (2013) The Tox21 robotic platform for the assessment of environmental chemicals—from vision to reality. Drug Discovery Today 18, 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. V.; Jones P. A. (2003) In-house assessment of a modified in vitro cytotoxicity assay for higher throughput estimation of acute toxicity. Toxicol. In Vitro 17, 717–722. [DOI] [PubMed] [Google Scholar]
- Jones P. A.; King A. V. (2003) High throughput screening (HTS) for phototoxicity hazard using the in vitro 3T3 neutral red uptake assay. Toxicol. In Vitro 17, 703–708. [DOI] [PubMed] [Google Scholar]
- Schirmer K.; Tanneberger K.; Kramer N. I.; Völker D.; Scholz S.; Hafner C.; Lee L. E. J.; Bols N. C.; Hermens J. L. M. (2008) Developing a list of reference chemicals for testing alternatives to whole fish toxicity tests. Aquat. Toxicol. 90, 128–137. [DOI] [PubMed] [Google Scholar]
- Sjöström M.; Kolman A.; Clemedson C.; Clothier R. (2008) Estimation of human blood LC50 values for use in modeling of in vitro–in vivo data of the acutetox project. Toxicol. In Vitro 22, 1405–1411. [DOI] [PubMed] [Google Scholar]
- Piersma A. H.; Janer G.; Wolterink G.; Bessems J. G. M.; Hakkert B. C.; Slob W. (2008) Quantitative extrapolation of in vitro whole embryo culture embryotoxicity data to developmental toxicity in vivo using the benchmark dose approach. Toxicol. Sci. 101, 91–100. [DOI] [PubMed] [Google Scholar]
- Bjornsson T. D.; Callaghan J. T.; Einolf H. J.; Fischer V.; Gan L.; Grimm S.; Kao J.; King S. P.; Miwa G.; Ni L.; Kumar G.; McLeod J.; Obach R. S.; Roberts S.; Roe A.; Shah A.; Snikeris F.; Sullivan J. T.; Tweedie D.; Vega J. M.; Walsh J.; Wrighton S. A. (2003) The conduct of in vitro and in vivo drug–drug interaction studies: a Pharmaceutical Research and Manufacturers of America (PHRMA) perspective. Drug Metab. Dispos. 31, 815–832. [DOI] [PubMed] [Google Scholar]
- Kolman A.; Clemedson C. (2013) Human in vivo database now on acutetox home page. Toxicol. In Vitro 27, 2350–2351. [DOI] [PubMed] [Google Scholar]
- Kinsner-Ovaskainen A.; Prieto P.; Stanzel S.; Kopp-Schneider A. (2013) Selection of test methods to be included in a testing strategy to predict acute oral toxicity: an approach based on statistical analysis of data collected in phase 1 of the ACuteTox project. Toxicol. In Vitro 27, 1377–1394. [DOI] [PubMed] [Google Scholar]
- Clothier R.; Gómez-Lechón M. J.; Kinsner-Ovaskainen A.; Kopp-Schneider A.; O’Connor J. E.; Prieto P.; Stanzel S. (2013) Comparative analysis of eight cytotoxicity assays evaluated within the ACuteTox project. Toxicol. In Vitro 27, 1347–1356. [DOI] [PubMed] [Google Scholar]
- Kopp-Schneider A.; Prieto P.; Kinsner-Ovaskainen A.; Stanzel S. (2013) Design of a testing strategy using non-animal based test methods: lessons learnt from the ACuteTox project. Toxicol. In Vitro 27, 1395–1401. [DOI] [PubMed] [Google Scholar]
- Prieto P.; Kinsner-Ovaskainen A.; Stanzel S.; Albella B.; Artursson P.; Campillo N.; Cecchelli R.; Cerrato L.; Díaz L.; Di Consiglio E.; Guerra A.; Gombau L.; Herrera G.; Honegger P.; Landry C.; O’Connor J. E.; Páez J. A.; Quintas G.; Svensson R.; Turco L.; Zurich M. G.; Zurbano M. J.; Kopp-Schneider A. (2013) The value of selected in vitro and in silico methods to predict acute oral toxicity in a regulatory context: results from the European project ACuteTox. Toxicol. In Vitro 27, 1357–1376. [DOI] [PubMed] [Google Scholar]
- Zhu H.; Rusyn I.; Richard A.; Tropsha A. (2008) Use of cell viability assay data improves the prediction accuracy of conventional quantitative structure–activity relationship models of animal carcinogenicity. Environ. Health Perspect. 116, 506–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedykh A.; Zhu H.; Tang H.; Zhang L.; Richard A.; Rusyn I.; Tropsha A. (2011) Use of in vitro HTS-derived concentration–response data as biological descriptors improves the accuracy of qsar models of in vivo toxicity. Environ. Health Perspect. 119, 364–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low Y.; Uehara T.; Minowa Y.; Yamada H.; Ohno Y.; Urushidani T.; Sedykh A.; Muratov E.; Kuz’min V.; Fourches D.; Zhu H.; Rusyn I.; Tropsha A. (2011) Predicting drug-induced hepatotoxicity using qsar and toxicogenomics approaches. Chem. Res. Toxicol. 24, 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murk A. J.; Rijntjes E.; Blaauboer B. J.; Clewell R.; Crofton K. M.; Dingemans M. M. L.; Furlow J. D.; Kavlock R.; Köhrle J.; Opitz R.; Traas T.; Visser T. J.; Xia M.; Gutleb A. C. (2013) Mechanism-based testing strategy using in vitro approaches for identification of thyroid hormone disrupting chemicals. Toxicol. In Vitro 27, 1320–1346. [DOI] [PubMed] [Google Scholar]
- Jomaa B.; Aarts J. M. M. J.; de Haan L. H. J.; Peijnenburg A. A. C. M.; Bovee T. F. H.; Murk A. J.; Rietjens I. M. C. M. (2013) In vitro pituitary and thyroid cell proliferation assays and their relevance as alternatives to animal testing. Altex-Alternatives to Anim. Exp. 30, 293–307. [DOI] [PubMed] [Google Scholar]
- Piegorsch W., and Zeiger E. (1991) Measuring intra-assay agreement for the Ames Salmonella assay, in Statistical Methods in Toxicology (Hothorn L., Ed.) Vol. 43, pp 35–41, Springer, Berlin. [Google Scholar]
- Zhu H., Kim M., Zhang L., and Sedykh A. (2014) Computers instead of cells: computational modeling of chemical toxicity, in Reducing, Refining and Replacing the Use of Animals in Toxicity Testing (Allen D., and Waters M. D., Eds.) Chapter 5, pp 163–182, The Royal Society of Chemistry, Cambridge. [Google Scholar]
- Sneddon T. P.; Zhe X. S.; Edmunds S. C.; Li P.; Goodman L.; Hunter C. I. (2014) GigaDB: promoting data dissemination and reproducibility. Database 2014, bau018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankley G. T.; Bennett R. S.; Erickson R. J.; Hoff D. J.; Hornung M. W.; Johnson R. D.; Mount D. R.; Nichols J. W.; Russom C. L.; Schmieder P. K.; Serrrano J. A.; Tietge J. E.; Villeneuve D. L. (2010) Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem. 29, 730–741. [DOI] [PubMed] [Google Scholar]
- MacKay C.; Davies M.; Summerfield V.; Maxwell G. (2013) From pathways to people: applying the adverse outcome pathway (AOP) for skin sensitization to risk assessment. ALTEX 30, 473–486. [DOI] [PubMed] [Google Scholar]
- Chen B.; Dong X.; Jiao D.; Wang H.; Zhu Q.; Ding Y.; Wild D. J. (2010) Chem2Bio2RDF: a semantic framework for linking and data mining chemogenomic and systems chemical biology data. BMC Bioinf. 11, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild D. J.; Ding Y.; Sheth A. P.; Harland L.; Gifford E. M.; Lajiness M. S. (2012) Systems chemical biology and the semantic web: what they mean for the future of drug discovery research. Drug Discovery Today 17, 469–474. [DOI] [PubMed] [Google Scholar]
- Chen B.; Ding Y.; Wild D. J. (2012) Improving integrative searching of systems chemical biology data using semantic annotation. J. Cheminf. 4, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourches D.; Sassano M. F.; Roth B. L.; Tropsha A. (2014) HTS navigator: freely accessible cheminformatics software for analyzing high-throughput screening data. Bioinformatics 30, 588–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N. C.; Hemminger B. M. (2010) Mining connections between chemicals, proteins, and diseases extracted from medline annotations. J. Biomed. Inform. 43, 510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Terfve C.; Rose J. C.; Markowetz F. (2011) HTSanalyzeR: an R/Bioconductor package for integrated network analysis of high-throughput screens. Bioinformatics 27, 879–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H.; Ye L.; Richard A.; Golbraikh A.; Wright F. A.; Rusyn I.; Tropsha A. (2009) A novel two-step hierarchical quantitative structure–activity relationship modeling work flow for predicting acute toxicity of chemicals in rodents. Environ. Health Perspect. 117, 1257–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Hsieh J.-H.; Zhu H. (2014) Profiling animal toxicants by automatically mining public bioassay data: a big data approach for computational toxicology. PLoS One 9, e99863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton A.; Bellis L. J.; Bento A. P.; Chambers J.; Davies M.; Hersey A.; Light Y.; McGlinchey S.; Michalovich D.; Al-Lazikani B.; Overington J. P. (2012) ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 40, D1100–D1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R. S.; Martin M. T.; Egeghy P.; Gangwal S.; Reif D. M.; Kothiya P.; Wolf M.; Cathey T.; Transue T.; Smith D.; Vail J.; Frame A.; Mosher S.; Cohen Hubal E. A.; Richard A. M. (2012) Aggregating data for computational toxicology applications: the U.S. Environmental Protection Agency (EPA) aggregated computational toxicology resource (ACToR) system. Int. J. Mol. Sci. 13, 1805–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R.; Richard A.; Dix D.; Houck K.; Elloumi F.; Martin M.; Cathey T.; Transue T. R.; Spencer R.; Wolf M. (2008) ACToR—aggregated computational toxicology resource. Toxicol. Appl. Pharmacol. 233, 7–13. [DOI] [PubMed] [Google Scholar]
- Fonger G. C.; Stroup D.; Thomas P. L.; Wexler P. (2000) TOXNET: a computerized collection of toxicological and environmental health information. Toxicol. Ind. Health 16, 4–6. [DOI] [PubMed] [Google Scholar]
- Kohonen P.; Benfenati E.; Bower D.; Ceder R.; Crump M.; Cross K.; Grafstrom R. C.; Healy L.; Helma C.; Jeliazkova N.; Jeliazkov V.; Maggioni S.; Miller S.; Myatt G.; Rautenberg M.; Stacey G.; Willighagen E.; Wiseman J.; Hardy B. (2013) The ToxBank data warehouse: supporting the replacement of in vivo repeated dose systemic toxicity testing. Mol. Inform. 32, 47–63. [DOI] [PubMed] [Google Scholar]
- Deparatment of Health and Human Services: National Toxicology Program. DrugMatrix. https://ntp.niehs.nih.gov/drugmatrix/index.html.
- Lamb J.; Crawford E. D.; Peck D.; Modell J. W.; Blat I. C.; Wrobel M. J.; Lerner J.; Brunet J.-P.; Subramanian A.; Ross K. N.; Reich M.; Hieronymus H.; Wei G.; Armstrong S. A.; Haggarty S. J.; Clemons P. A.; Wei R.; Carr S. A.; Lander E. S.; Golub T. R. (2006) The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935. [DOI] [PubMed] [Google Scholar]